Abstract
Here, we determined the Toxoplasma gondii genotype in amniotic fluid, placenta, and cerebrospinal fluid samples from 14 congenital toxoplasmosis cases in Tunisia, North Africa. Direct genotypic characterization of T. gondii strains was performed by polymerase chain reaction (PCR) amplification of six genetic markers (3′SAG2, 5′ SAG2, SAG3, BTUB, GRA6, and APICO) and thereafter, was analyzed by restriction fragment-length polymorphism (RFLP). Samples were sequenced to resolve strain type whenever there were unclear enzyme digestion results. Multilocus analysis revealed that only one specimen harbored the type I allele in all studied loci, whereas the 13 others gave mixed genotype results with different alleles at different markers. Seven specimens produced RFLP profile of the recombinant strains I/III, and three produced a profile of I/II recombinant strains. The last three specimens produced complex digestion patterns. In these cases, sequence analysis revealed double peaks at known polymorphic sites, indicating the presence of multiple alleles.
Introduction
Toxoplasmosis is a common parasitic disease caused by the protozoan parasite Toxoplasma gondii. Its prevalence in humans varies from region to region depending on ecological and cultural factors.1 In Tunisia, North Africa, an average seroprevalence rate of 58% with a progressive rise from 24% at 10 years of age to 70% at 30 years of age was reported.2 This epidemiological feature suggests that even if infection is a frequent event in childhood, many women at childbearing age stay susceptible to toxoplasmosis.
The possible relationship between congenital toxoplasmosis and T. gondii genotype has been approached in several studies.3 In France, where a systematic diagnosis of congenital toxoplasmosis was performed, more than 80% of infections were caused by the archetypal genotype II.4 The main factor determining the severity of congenital infection with this genotype remained the stage of pregnancy at which it was acquired.5 In South America, where non-archetypal strains predominate, the few reports about isolates from congenital cases indicate the role of type I, atypical, or recombinant I/III strains in severe cases.6 Little is known about genetics of Toxoplasma strains from Africa. The first reports about T. gondii genotypes in fowl from Egypt and several other African countries found a predominance of type III genotypes, but the analysis was restricted to the single SAG2 marker.7,8 Current African isolation and multilocus genotyping of T. gondii from naturally exposed chickens revealed the predominance of archetypal lineages, notably type III genotypes.9 Some strains isolated from African patients possess a combination of type I and type III alleles.10,11 There is only one reported study of strains from congenital toxoplasmosis cases in Africa. This single study about Egyptian female patients with abortion and intrauterine fetal death indicated that strains possessing the type II allele at the SAG2 locus were found in 87% of infections.12
Among the various markers used for Toxoplasma genotyping, the SAG2 marker provides accurate genotyping for most strains within the clonal lineages. However, it cannot detect recombinant strains or those with unusual genotypes.13 This problem can be alleviated by multilocus analysis of T. gondii strains with multiple markers. Furthermore, multilocus nested polymerase chain reaction (nPCR) analysis can detect as few as five parasite genomes and thus, is applicable to low-volume samples containing few parasites, which is typical of clinical specimens.14
The aim of this present study is to determine the genotypes of T. gondii associated with congenital toxoplasmosis in Tunisia using a multilocus genotyping consisting of nPCR amplification of six different restriction fragment-length polymorphism (RFLP) markers: 3′SAG2, 5′SAG2, GRA6, SAG3, BTUB, and APICO.
Material and methods
Clinical samples.
Fourteen cases of congenital toxoplasmosis diagnosed at the Department of Parasitology of Pasteur Institute of Tunis between 2004 and 2008 were included in the study. Congenital infections were identified through maternal pre-natal screening on the basis of pre-natal diagnosis (real-time quantitative PCR; genes B1 and AF487550) and/or neonatal screening (PCR on placenta; serology in newborns) as previously described.15,16 Parasite DNA used for genotyping was obtained from amniotic fluid (AF) in 11 cases, placentas (PL) in 2 cases, and cerebrospinal fluid (CSF) in 1 case. No clinical data were available for newborns in 11 cases. Two newborns had no toxoplasmic clinical signs, and one newborn developed chorioretinitis (Table 1).
Table 1.
Origin of samples and information about biological diagnosis of congenital toxoplasmosis
| Code | Sample | Maternal infection | Pre-natal diagnosis | Neonatal screening | Post-natal follow-up | ||||
|---|---|---|---|---|---|---|---|---|---|
| Ultrasound | AF puncture (PCR) | Placenta (PCR) | Serology | Serology | Clinics | ||||
| 1 | AF06/06 | AF | 1st T | N | + | ND | |||
| 2 | AF44/05 | AF | 1st T | N | + | ND | + | + | N |
| 3 | CSF01/08 | CSF | 3rd T | N | ND | ND | + | + | N |
| 4 | AF08/08 | AF | 2nd T | N | + | ND | |||
| 5 | AF26/04 | AF | 1st T | N | + | ND | |||
| 6 | AF08/06 | AF | 1st T | N | + | ND | |||
| 7 | AF33/05 | AF | 3rd T | N | + | ND | |||
| 8 | AF07/06 | AF | 1st T | N | + | ND | |||
| 9 | PL05/06 | PL | 3rd T | N | + | + | |||
| 10 | AF07/02 | AF | 2nd T | N | + | ND | |||
| 11 | AF19/04 | AF | 2nd T | N | + | ND | + | + | chorioretinitis |
| 12 | PL04/06 | PL | 2nd T | N | + | + | |||
| 13 | AF10/08 | AF | 1st T | N | + | ND | |||
| 14 | AF16/07 | AF | 1st T | N | + | ND | |||
AF = amniotic fluid; CSF = cerebrospinal fluid; PL = placenta; T = trimester; N = normal; ND = not done.
Experimental samples.
Reference strains used for each of the clonal lineages (provided by Pr. Marie Laure Dardé, Biologic Resources Center (BRC) Toxoplasma; http://www.toxobrc.com/) were: RH (type I), Prugniaud (type II), and NED (type III). Parasites were grown and maintained in Swiss mice by intraperitoneal inoculations. Tachyzoites were harvested from the peritoneal cavities (RH strain) or brains (NED and Prugniaud strains) in phosphate-buffered saline (PBS); then, they were pooled, centrifuged, and washed two times at 2,000 g for 10 minutes. The parasite pellets were processed for DNA extraction using Qiamp DNA Mini Kit (Qiagen, GmbH, Germany).
PCR–RFLP analysis.
Strain typing was performed by PCR amplification of six genetic markers (3′SAG2, 5′ SAG2, SAG3, BTUB, GRA6, and APICO) and thereafter, was analyzed by RFLP. First, the lineage type was determined using two nPCRs amplifying separately the 5′ and 3′ ends of the SAG2 gene as previously described (Table 2).17 Thereafter, a multiplex nPCR with a set of three-way markers (SAG3, BTUB, GRA6, and APICO) was used (Table 2).14,18 The initial round of amplification with external primers for the four different markers was carried out in 25 μL of mixture containing 1× PCR buffer, 3 mM MgCl2, 400 µM each of the deoxyribonucleoside triphosphates (dNTPs), 1 µg bovine serum albumin (BSA), 0.2 µM each of the forward and reverse primers, 1.5 units of Hot Start DNA polymerase (Qiagen), and 4 µL of DNA sample. Amplification was conducted at 94°C for 15 minutes followed by 40 cycles of 94°C for 45 seconds, 55°C for 1 minute, and 72°C for 1 minute. The last extension step was at 60°C for 10 minutes. PCR products were diluted and used for second-round amplification of each marker separately in a 25-μL volume mixture containing 1× PCR buffer, 2.5 mM MgCl2, 200 µM each of the dNTPs, 0.2 µM of each primer, and 0.75 units of Hot Start DNA polymerase. The amplification protocol was 94°C for 15 minutes followed by 40 cycles of 94°C for 45 seconds, annealing temperature for 45 seconds, and 72°C for 1 minute. The overextension step was at 72°C for 5 minutes. The annealing temperature was 50°C for Apico, 55°C for GRA6, and 62°C for BTUB and SAG3. PCR products were examined by electrophoresis on 2% agarose gel stained with ethidium bromide and visualized under ultraviolet (UV) light. The amplified fragments of nPCRs were digested with appropriate restriction enzymes for different markers (Table 2). Controls were carried out from RH, Prugniaud, and NED strains. The polymorphism within each locus was analyzed by RFLP patterns used to distinguish each type strain. The digested PCR products were resolved by 3% agarose gel stained with ethidium bromide.
Table 2.
Summary of markers used for genotyping T. gondii
| Marker (location) | External primers | Internal primers | Restriction enzymes |
|---|---|---|---|
| SAG2 (VIII)5′3′15 | F4: GCTACCTCGAACAGGAACAC | F: GAAATGTTTCAGGTTGCTGC | Sau 3AI |
| R4: GCATCAACAGTCTTCGTTGC | R2: GCAAGAGCGAACTTGAACAC | ||
| F3: TCTGTTCTCCGAAGTGACTCC | F2: ATTCTCATGCCTCCGCTTC | HhaI | |
| R3: TCAAAGCGTGCATTATCGC | R: AACGTTTCACGAAGGCACAC | ||
| SAG3 (XII)12,16 | P43S1: AACTCTCACCATTCCACCC | P43AS2: TCTTGTCGGGTGTTCACTCA | Nci I |
| P43AS1: GCGCGTTGTTAGACAAGACA | P43S2: CACAAGGAGACCGAGAAGGA | ||
| GRA6 (X) | GRA6-F1x: ATTTGTGTTTCCGAGCAGGT | GRA6-F1: TTTCCGAGCAGGTGACCT | Mse I |
| GRA6-R1: GCACCTTCGCTTGTGGTT | GRA6-R1x: TCGCCGAAGAGTTGACATAG | ||
| BTUB (IX) | Btb (ext) F: TCCAAAATGAGAGAAATCGT | Btb-F: GAGGTCATCTCGGACGAACA | Taq I BsiE I |
| Btb (ext) R: AAATTGAAATGACCGAAGAA | Btb-R: TTGTAGGAACACCCCGACGC | ||
| APICO (plastid) | Apico-oF: TGGTTTTAACCCTAGATTGTGG* | Apico-F: TGCAAATTCTTGAATTCTCAGTT | Afl II Dde I |
| Apico-oR: AAACGGAATTAATGAGATTTGAACT* | Apico-R: GGGATTCGAACCCTTGATA |
This study.
To avoid possible contamination, several measures were taken—separate spaces were used to set up PCRs, filter tips, and UV radiations; also, tests were run with few samples in the same time. Different negative controls (no DNA, uninfected sample, and extracted no DNA) were also used. Positive controls from different strains of T. gondii were never used in the same run with clinical samples.
DNA sequencing.
Samples were sequenced to resolve strain type whenever there were unclear enzyme digestion results, and sequencing was also performed on selected samples to screen for possible new polymorphisms. nPCR products from T. gondii strains as well as clinical samples were used for DNA sequencing. The fragments were sequenced with both internal primers SAG3 and GRA6 to obtain forward and reverse sequences. Sequencing was done three times with different PCR products from the same isolate to avoid any contamination. The kit ABI Prism BigDye Terminator Cycle Sequencing Reaction (Applied Biosystems, Branchburg, NJ) was used, and electrophoreses were run on a polyacrylamide gel POP7 in a four-capillary Applied Biosystems 3130 Genetic Analyzer. Nucleotide sequences were aligned for comparison using “Clustal W” from Bio-Edit Sequence Alignment Editor. The sequences were deposited in GenBank, and the accession numbers of the nucleotide sequences are cited later.
Results
PCR analysis.
Small amount or no amplification products were observed after the first step of amplification because of the low amount of parasite DNA present in clinical samples (data not shown). After the second round of amplification, positive amplicons were obtained for all samples with SAG2 (3′ and 5′), GRA6, and APICO PCRs. Only 13 and 8 samples were amplified using SAG3 and BTUB markers, respectively (Table 3). Negative controls remained free of amplified products.
Table 3.
Multilocus genotypic characterization of T. gondii strains associated with congenital toxoplasmosis
| Sample | Alleles | Final genotype | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 3′SAG2 | 5′SAG2 | SAG3 | Accession No. | GRA6 | Accession No. | BTUB | APICO | |||
| 1 | AF 06/06 | 1 | 1 | 1 | 139468 | 1 | 1 | 1 | I | |
| 2 | AF 44/05 | 1 | 1 | 3 | 139467 | 1 | 1 | 1 | I/III | |
| 3 | CSF 01/08 | 1 | 1 | 3 | 139476 | 1 | ND | 1 | I/III | |
| 4 | AF 08/08 | 1 | 1 | 3 | 139477 | 1 | ND | 1 | I/III | |
| 5 | AF 26/04 | 1 | 1 | 3 | 139464 | 1 | 3 | 1 | I/III | |
| 6 | AF 08/06 | 1 | 1 | 3 | 139473 | 1 | 3 | 3 | I/III | |
| 7 | AF 33/05 | 1 | 1 | 1 + 3 | 139466 139465 | 1 | 3 | 1 | I/III | |
| 8 | AF 07/06 | 1 | 1 | 3 | 139467 | 1 + 3 | 139482 139483 | 1 | 1 | I/III |
| 9 | PL 05/06 | 1 | 1 | 2 | 139472 | 1 + 2 | 139484 139485 | ND | 1 | I/II |
| 10 | AF 07/02 | 1 | 1 | ND | 1 + 2 | 139486 139487 | 1 + 2 | 1 | I/II | |
| 11 | AF 19/04 | 1 | 1 | 1 + 2 | 139462 139463 | 1 + 2 | 139480 139481 | 1 + 2 | 1 | I/II |
| 12 | PL 04/06 | 1 | 1 | 2 + 3 | 139471 139470 | 1 | ND | 1 | Mix of I/II and I/III | |
| 13 | AF 10/08 | 1 | 1 | 2 + 3 | 139478 139479 | 1 | ND | 1 | Mix of I/II and I/III | |
| 14 | AF 16/07 | 1 | 1 | 1 + 2 + 3 | 139474 139475 | 1 | ND | 1 | Mix of I/II and I/III | |
RFLP analysis.
Genotyping of samples by SAG2 PCR–RFLP revealed that the 14 samples were infected with SAG2 Type I parasites (Table 3). SAG3 marker showed that only one specimen harbored the type I allele, whereas six harbored the type III, one harbored the type II, and five harbored a mixture of alleles (Figure 1 and Table 3). GRA6 marker showed that 10 specimens harbored the type I allele, whereas four samples harbored a mixture of alleles (Figure 1 and Table 3). BTUB marker showed that three specimens harbored the type I allele, whereas three samples harbored the type III and two samples harbored a mixture of alleles (Figure 1 and Table 3). APICO marker showed that 13 specimens harbored the type I allele, whereas one sample harbored type III (Table 3). When results from each locus were combined, only one specimen harbored the type I allele in all studied loci. All the remaining samples had genotypes consisting of different combinations of alleles seen in the clonal types. Seven specimens were shown to possess a chimerical combination of allele types I–III, and three possessed a combination of allele types I–II. The last three specimens produced complex RFLP patterns with a mixture of two to three genotypes in the same sample (Figure 1 and Table 3).
Figure 1.
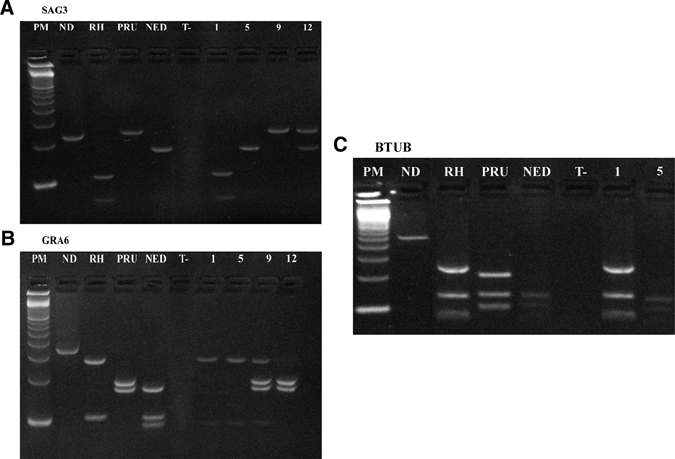
Restriction analysis of PCR products directly amplified from clinical samples: PCR amplifications of SAG3 marker (A), GRA6 marker (B), and BTUB marker (C). Lane PM = 100-basepair (bp) DNA ladder (Amersham, Buckinghamshire, UK); lane ND = undigested PCR product; lanes RH, PRU, and NED = digested PCR products representative of T. gondii strain types I, II, and III, respectively; lane T = negative control; lanes 1, 5, 9, and 12 = digested PCR products of positive samples AF 06/06, AF 26/04, PL 05/06, and PL 04/06 genotyped as type I, type I/III, type I/II, and mix of strains, respectively. Note that the last two specimens produce a complex digestion pattern with GRA6 and SAG3, respectively.
Sequencing analysis.
To confirm SAG3 type, all restriction fragment products were subjected to direct DNA sequence determination. All sequences were submitted to Genbank (accession numbers = GU139462–GU139479). The sample was identified, because allele 1 was identical to type I TgUgCh62 strain (accession number = EF585690). The sample identified as type 2 was identical to type II TgUgCh68 strain (accession number = EF585684), and the six samples identified as type 3 gave homology of 99–100% with type III TgUgCh64 strain (accession number = EF585687). Samples with restriction profile showing a mixture of two to three archetypal alleles revealed double peaks at known polymorphic sites, indicating the concomitant presence of two alleles in the same sample (Figure 2). It was interesting to note that the last sample with restriction profile showing a mixture of three archetypal alleles also revealed the presence of two strains; the first had 99% homology with the natural recombinant I/III P-Br strain (accession number = AY187280), and the second had 98% homology with type II TgUgCh68 strain (accession number = EF 585684). No new single-nucleotide polymorphisms (SNPs) were found. For the GRA6 marker, any product that did not produce a single type after restriction analysis was sequenced. In four cases, double picks at polymorphic sites were noted, revealing a double infection. Three were caused by type I allele (identical to TgUgCh83 strain; accession number = EF585715) and type II allele (identical to TgUgCh78 strain; accession number = EF585712), and one was caused by type I allele (identical to TgUgCh83 strain) and type III allele (giving 99% homology with TgUgCh64 strain; accession number = EF585705).
Figure 2.
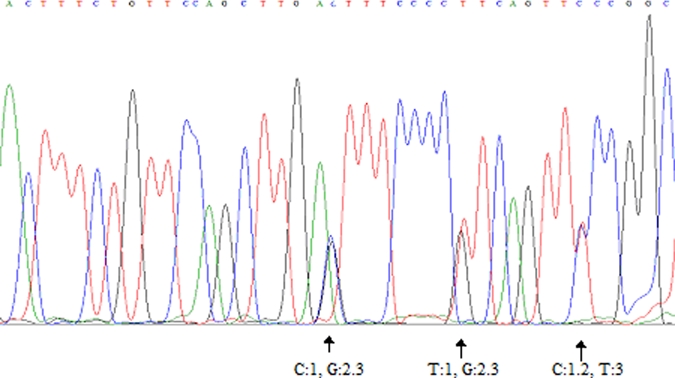
Sequence analysis of the SAG3 marker using reverse primer, which reveals double peaks at known polymorphic sites (sample AF 33/05). This figure appears in color at www.ajtmh.org.
Discussion
Very few studies concerning genotypic characterization of Toxoplasma strains associated with human toxoplasmosis in Africa have been reported. Two studies genotyped the SAG2 locus and were performed on samples from Ugandan human immunodeficiency virus (HIV) patients and Egyptian female patients with abortion and intrauterine fetal death, respectively.12,19 Both reported the predominance of genotype II. Using the same marker, all Tunisian samples were classified as SAG2 type I. Similar results were mentioned in some Mediterranean countries, such as Spain, where strains possessing the type I allele at the SAG2 locus were found in 6 of 13 cases of congenital infection.20 However, this marker is not able to detect recombinant or exotic strains, causing misclassification of them as genotype I.
The advantage of using a combination of different independent markers is that it is much more likely to detect recombinant genotypes. Recently, multiplex PCR of microsatellites for typing was developed.21 However, this method has some limitations; a special sequencer and analysis software are needed, and it has a low level of analytic sensitivity (at least 50 parasites in a sample). In contrast, the multilocus PCR–RFLP genotyping method developed by Khan and others14 offers the advantages that it is simple, can be conducted on small amounts of different types of samples, and has good sensitivity (detecting between 5 and 10 parasites). Because our samples contained small numbers of organisms (data not shown), this technique was more suitable for their typing, and precautions were taken to avoid contamination.
Using multilocus analysis, all Tunisian isolates examined in the present study, except one, harbored recombinant I/II and/or I/III strain, which is in concordance with the very few results reported in Ugandan HIV patients and other patients of African origin.10,11,21 Sequencing of different alleles revealed a greater homology than those previously reported in African free-ranging chickens.22,23 Although the very limited clinical data in the present study cannot allow hypothesis testing about strain virulence, it is important to note the presence of strains with close homology to the natural recombinant I/III P-Br strain that was incriminated in severe cases in South America.6 This suggests that this genotype could be a new emerging one in this part of the world. In addition, other recombinant or atypical strains may be present in Africa, which has been proven recently by extensive sequence analysis of eight isolates from Uganda.23 This study found mostly type II genotypes, but they contained novel SNPs that suggested regional allelic variants.23
Perhaps, the most significant finding of our present work compared with previously published data is the high frequency of apparent concomitant infection in clinical samples. Mixed infection with T. gondii strains has been previously reported in England and Wales.24 This finding was explained by the ingestion of more than one type of parasite in food products containing meat originating from multiple animals.25 However, the high proportion of mixed infections shown in African isolates from free-ranging chickens underlines the high frequency in this part of the world of sequential infections with parasites of different types acquired as oocysts directly from the environment.22 It also suggests that intermediate hosts are infected with more than one strain. Congenital toxoplasmosis is, in most cases, the result of a primary infection acquired by an immunologically naïve patient during pregnancy.5 It seems that we can exclude sequential infection as an explanation for the presence of different genotypes in a significant number of patients. Similarly, it is hard to explain the observed frequency of mixed infections as a consequence of exposure to mixed oocysts.26 The possible mechanism leading to the observed mixed infection is the ingestion of tissue cysts from infected meat.27 Mutton is the meat most commonly incriminated in the transmission of toxoplasmosis in Tunisia, and it could be seen as a real source of contamination.28 Genotypic characterization of T. gondii in sheep in Tunisia could help to establish correlation between mixed infections in humans in Tunisia and contaminated mutton.
Acknowledgments
The authors thank Pr. Marie Laure Dardé for the T. gondii strains and for constructive discussions and critical comments on the manuscript.
Footnotes
Financial support: This study was supported by the Ministry of Higher Education, Research and Technology in Tunisia and carried out within the framework of the Research Lab “Parasitoses emergentes” LR 05SP03.
Authors' addresses: Sonia Boughattas, Rym Ben-abdallah, Emna Siala, Olfa Souissi, Karim Aoun, and Aïda Bouratbine, LR “Parasitoses emergentes,” Laboratoire de Parasitologie, Institut Pasteur de Tunis, Tunis Belvedère, Tunisia, E-mails: bsonia0@yahoo.fr, olfasouissi75@yahoo.fr, rym.benabdallah@pasteur.rns.tn, emna.siala@pasteur.rns.tn, karim.aoun@pasteur.rns.tn, and aida.bouratbine@pasteur.rns.tn.
Reprint requests: Aïda Bouratbine, Laboratoire de Parasitologie, Institut Pasteur de Tunis, 13 Place Pasteur, BP 74, 1002 Tunis Belvedère, Tunisia, E-mail: aida.bouratbine@pasteur.rns.tn.
References
- 1.Dupouy-Camet J, Gavinet MF, Paugam A, Tourte Schaffer CI. Mode de contamination, incidence et prevalence de la toxoplasmose. Med Mal Infect. 1993;23:139–147. [Google Scholar]
- 2.Bouratbine A, Siala E, Chahed MK, Aoun K, Ben Ismail R. Sero-epidemiologic profile of toxoplasmosis in northern Tunisia. Parasite. 2001;8:61–66. doi: 10.1051/parasite/2001081061. [DOI] [PubMed] [Google Scholar]
- 3.Costa JM, Dardé ML, Assouline B, Vidaud M, Bretagne S. Microsatellite in the beta-tubulin gene of Toxoplasma gondii as a new genetic marker for use in direct screening of amniotic fluids. J Clin Microbiol. 1997;35:2542–2545. doi: 10.1128/jcm.35.10.2542-2545.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ajzenberg D, Cogné N, Paris L, Bessières MH, Thulliez P, Filisetti D, Pelloux H, Marty P, Dardé ML. Genotype of 86 Toxoplasma gondii isolates associated with human congenital toxoplasmosis, and correlation with clinical findings. J Infect Dis. 2002;186:684–689. doi: 10.1086/342663. [DOI] [PubMed] [Google Scholar]
- 5.Dunn D, Wallon M, Peyron F, Petersen E, Peckham C, Gilbert R. Mother-to-child transmission of toxoplasmosis: risk estimates for clinical counseling. Lancet. 1999;353:1829–1833. doi: 10.1016/S0140-6736(98)08220-8. [DOI] [PubMed] [Google Scholar]
- 6.Gallego C, Saavedra-Matiz C, Gómez-Marín JE. Direct genotyping of animal and human isolates of Toxoplasma gondii from Colombia (South America) Acta Trop. 2006;97:161–167. doi: 10.1016/j.actatropica.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Dubey JP, Graham DH, Dahl E, Hilali M, El-Ghaysh A, Sreekumar C, Kwok OCH, Shen SK, Lehmann T. Isolation and molecular characterization of Toxoplasma gondii from chickens and ducks from Egypt. Vet Parasitol. 2003;114:89–95. doi: 10.1016/s0304-4017(03)00133-x. [DOI] [PubMed] [Google Scholar]
- 8.Dubey JP, Karhemere S, Dahl E, Sreekumar C, Diabate´ A, Dabire´ KR, Vianna MCB, Kwok OCH, Lehmann T. First biologic and genetic characterization of Toxoplasma gondii isolates from chickens from Africa (Democratic Republic of Congo, Mali, Burkina Faso, and Kenya) J Parasitol. 2005;91:69–72. doi: 10.1645/GE-410R. [DOI] [PubMed] [Google Scholar]
- 9.Velmurugan GV, Dubey JP, Su C. Genotyping studies of Toxoplasma gondii isolates from Africa revealed that the archetypal clonal lineages predominate as in North America and Europe. Vet Parasitol. 2008;155:314–318. doi: 10.1016/j.vetpar.2008.04.021. [DOI] [PubMed] [Google Scholar]
- 10.Genot S, Franck J, Forel JM, Rebaudet S, Ajzenberg D, de Paula AM, Dardé ML, Stein A, Ranque S. Severe Toxoplasma gondii I/III recombinant-genotype encephalitis in a human immunodeficiency virus patient. J Clin Microbiol. 2007;45:3138–3140. doi: 10.1128/JCM.00021-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ajzenberg D, Yera H, Marty P, Paris L, Dalle F, Menotti J, Aubert D, Franck J, Bessières MH, Quinio D, Pelloux H, Delhaes L, Desbois N, Thulliez P, Robert-Gangneux F, Kauffmann-Lacroix C, Pujol S, Rabodonirina M, Bougnoux ME, Cuisenier B, Duhamel C, Duong TH, Filisetti D, Flori P, Gay-Andrieu F, Pratlong F, Nevez G, Totet A, Carme B, Bonnabau H, Dardé ML, Villena I. Genotype of 88 Toxoplasma gondii isolates associated with toxoplasmosis in immunocompromised patients and correlation with clinical findings. J Infect Dis. 2009;199:1155–1167. doi: 10.1086/597477. [DOI] [PubMed] [Google Scholar]
- 12.Abdel-Hameed DM, Hassanein OM. Genotyping of Toxoplasma gondii strains from female patients with toxoplasmosis. J Egypt Soc Parasitol. 2008;38:511–520. [PubMed] [Google Scholar]
- 13.Fazaeli A, Carter PE, Pennington TH. Intergenic spacer (IGS) polymorphism: a new genetic marker for differentiation of Toxoplasma gondii strains and Neospora caninum. J Parasitol. 2000;86:716–723. doi: 10.1645/0022-3395(2000)086[0716:ISIPAN]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 14.Khan A, Su C, German M, Storch GA, Clifford DB, Sibley LD. Genotyping of Toxoplasma gondii strains from immunocompromised patients reveals high prevalence of type I strains. J Clin Microbiol. 2005;43:5881–5887. doi: 10.1128/JCM.43.12.5881-5887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siala E, Ben Abdallah R, Delabesse E, Aoun K, Paris L, Bouratbine A. Contribution of the real time PCR in antenatal diagnosis of congenital toxoplasmosis. Tunis Med. 2007;85:385–388. [PubMed] [Google Scholar]
- 16.Ben Abdallah R, Aoun K, Siala E, Souissi O, Maatoug R, Hlioui S, Bouratbine A. Congenital toxoplasmosis: clinical and biological analysis of 11 cases in Tunisia. Arch Pediatr. 2009;16:118–121. doi: 10.1016/j.arcped.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 17.Howe DK, Honoré S, Derouin F, Sibley LD. Determination of genotypes of Toxoplasma gondii strains isolated from patients with toxoplasmosis. J Clin Microbiol. 1997;35:1411–1414. doi: 10.1128/jcm.35.6.1411-1414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su C, Zhang X, Dubey JP. Genotyping of Toxoplasma gondii by multilocus PCR-RFLP markers: a high resolution and simple method for identification of parasites. Int J Parasitol. 2006;36:841–848. doi: 10.1016/j.ijpara.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 19.Lindström I, Kaddu-Mulindwa DH, Kironde F, Lindh J. Prevalence of latent and reactivated Toxoplasma gondii parasites in HIV-patients from Uganda. Acta Trop. 2006;100:218–222. doi: 10.1016/j.actatropica.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 20.Fuentes I, Rubio JM, Ramírez C, Alvar J. Genotypic characterization of Toxoplasma gondii strains associated with human toxoplasmosis in Spain: direct analysis from clinical samples. J Clin Microbiol. 2001;39:1566–1570. doi: 10.1128/JCM.39.4.1566-1570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ajzenberg D, Dumetre A, Dardé ML. Multiplex PCR for typing strains of Toxoplasma gondii. J Clin Microbiol. 2005;43:1940–1943. doi: 10.1128/JCM.43.4.1940-1943.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindström I, Sundar N, Lindh J, Kironde F, Kabasa JD, Kwok OC, Dubey JP, Smith JE. Isolation and genotyping of Toxoplasma gondii from Ugandan chickens reveals frequent multiple infections. Parasitology. 2007;135:39–45. doi: 10.1017/S0031182007003654. [DOI] [PubMed] [Google Scholar]
- 23.Bontell I, Hall N, Ashelford EK, Dubey JP, Boyle PJ, Lindh J, Smith EJ. Whole genome sequencing of a natural recombinant Toxoplasma gondii strain reveals chromosome sorting and local allelic variants. Genome Biol. 2009;10:R53. doi: 10.1186/gb-2009-10-5-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aspinall TV, Guy EC, Roberts KE, Joynson DH, Hyde JE, Sims PF. Molecular evidence for multiple Toxoplasma gondii infections in individual patients in England and Wales: public health implications. Int J Parasitol. 2003;33:97–103. doi: 10.1016/s0020-7519(02)00230-8. [DOI] [PubMed] [Google Scholar]
- 25.Aspinall TV, Marlee D, Hyde JE, Sims PF. Prevalence of Toxoplasma gondii in commercial meat products as monitored by polymerase chain reaction–food for thought? Int J Parasitol. 2002;32:1193–1199. doi: 10.1016/s0020-7519(02)00070-x. [DOI] [PubMed] [Google Scholar]
- 26.Cook AJ, Gilbert RE, Buffolano W, Zufferey J, Petersen E, Jenum PA, Foulon W, Semprini AE, Dunn DT. Sources of Toxoplasma infection in pregnant women: European multicentre case-control study. European Research Network on Congenital Toxoplasmosis. BMJ. 2000;321:142–147. doi: 10.1136/bmj.321.7254.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dubey JP, Sundar N, Hill D, Velmurugan GV, Bandini LA, Kwok OCH, Majumdar D, Su C. High prevalence and abundant atypical genotypes of Toxoplasma gondii isolated from lambs destined for human consumption in the USA. Int J Parasitol. 2008;38:999–1006. doi: 10.1016/j.ijpara.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 28.Ben Rachid MS, Blaha R. Human and animal toxoplasmosis in Tunisia. Tunis Med. 1970;48:101–110. [PubMed] [Google Scholar]


