Abstract
Pemphigus foliaceus is a life threatening skin disease that is associated with autoimmunity to desmoglein, a skin protein involved in the adhesion of keratinocytes. This disease is endemic in certain areas of South America, suggesting the mediation of environmental factors triggering autoimmunity. Among the possible environmental factors, exposure to bites of black flies, in particular Simulium nigrimanum has been suggested. In this work, we describe the sialotranscriptome of adult female S. nigrimanum flies. It reveals the complexity of the salivary potion of this insect, comprised by over 70 distinct genes within over 30 protein families, including several novel families, even when compared with the previously described sialotranscriptome of the autogenous black fly, S. vittatum. The uncovering of this sialotranscriptome provides a platform for testing pemphigus patient sera against recombinant salivary proteins from S. nigrimanum and for the discovery of novel pharmacologically active compounds.
Introduction
Pemphigus foliaceus, also known by the Portuguese name “fogo selvagem,” is a life threatening autoimmune disease leading to keratinocyte detachment and skin blistering.1 Patients have increased antibody against desmoglein, a structural protein important in intercellular adhesion.2,3 This is a rare disease in general, but endemic in some Amerindian reservations in Brazil. The patchy spatial distribution of this condition in genetically related groups indicated an environmental component causing the disease.4 Among the environmental variables, exposure to black fly bites, in particular to the anthropophilic species Simulium nigrimanum, was shown to correlate with anti-desmoglein titers in an epidemiological study.5
Saliva of blood-sucking arthropods contain a vast array of pharmacologically active compounds that disarms their hosts hemostasis, the physiological reaction preventing blood loss after tissue injury, and inflammation, which can lead to enhanced hemostasis and host defense reactions.6,7 The advent of transcriptome methods has led to characterization of the complexity of the salivary potion of blood feeding arthropods, which number several dozen different proteins for sand flies, near 100 for mosquitoes and black flies, and hundreds in the case of hard ticks.8 This salivary potion contains enzymes such as apyrase, which degrades adenosine diphosphate (ADP) and adenosine triphosphate (ATP) (powerful agonists of platelet and neutrophil aggregation), hyaluronidase (which facilitates the spreading of the salivary pharmaceuticals into the feeding pool), vasodilators in various forms, different platelet aggregation antagonists, complement activation inhibitors, and agonist chelators, or kratagonists, proteins that bind different hemostasis and inflammation agonists such as biogenic amines (serotonin, norepinephrine, and histamine), inflammatory leukotrienes, and thromboxane A2, a potent platelet agonist.8
Comparative transcriptome analyses of different mosquito species and genera indicate that the evolution of salivary proteins has occurred at a very fast pace, possibly caused by the immune pressure of their hosts. Indeed, comparisons between Anopheles, Culex, and Aedes showed that each contained genus-specific protein families, and even subgenus-specific families.9–14 Regarding black flies, a single sialotranscriptome (from the Greek sialo = saliva) has been analyzed so far, from the North American species Simulium vittatum.15 It is the goal of this work to describe the sialotranscriptome of S. nigrimanum, to compare it with that of S. vittatum, and to provide candidate proteins that might be the trigger of anti-desmoglein antibodies in fogo selvagem.
Materials and Methods
Chemicals.
Standard laboratory chemicals were purchased from Sigma Chemicals (St. Louis, MO) if not specified otherwise.
Black flies.
Adult S. nigrimanum (113 flies) were collected near streams and homes in the Terena Ameridian community of Aldeia Limão Verde, municipality of Aquidauana, Mato Grosso do Sul state, Brazil. From October 6 to 8, 2007, flies were captured with aspirators from the exposed limbs of coauthor, DPE. The salivary glands were dissected in phosphate-buffered saline (PBS), immediately transferred to 50 μL of RNAlater (Ambio, Inc., Austin, TX) and kept refrigerated for 7 days. After transport to the United States, the glands were frozen and shipped to the Laboratory of Malaria and Vector Research, Rockville, MD.
Library construction.
Salivary gland RNA, extracted from 193 intact glands, was isolated using the Micro-FastTrack mRNA isolation kit (Invitrogen, San Diego, CA). The polymerase chain reaction (PCR)-based cDNA library was made following the instructions for the SMART cDNA library construction kit (Clontech, Palo Alto, CA). This system uses oligoribonucleotide (SMART IV) to attach an identical sequence at the 5′ end of each reverse-transcribed cDNA strand. This sequence is then used in subsequent PCR reactions and restriction digests.
First-strand synthesis was carried out using PowerScript reverse transcriptase at 42°C for 1 hour in the presence of the SMART IV and CDS III (3′) primers. Second-strand synthesis was performed using a long distance (LD) PCR-based protocol, using Advantage Taq polymerase (Clontech) mix in the presence of the 5′ PCR primer and the CDS III (3′) primer. The cDNA synthesis procedure resulted in creation of SfiI A and B restriction enzyme sites at the ends of the PCR products that are used for cloning into the phage vector (lambda TriplEx2 vector, Clontech). The PCR conditions were as follows: 95°C for 1 min; 24 cycles of 95°C for 10 sec, 68°C for 6 min. A small portion of the cDNA obtained by PCR was analyzed on a 1.1% agarose gel to check quality and range of cDNA synthesized. Double-stranded cDNA was immediately treated with proteinase K (0.8 μg/mL) at 45°C for 20 min, and the enzyme was removed by ultrafiltration though a Microcon (Amicon Inc., Beverly, CA) YM-100 centrifugal filter device. The cleaned, double-stranded cDNA was then digested with SfiI at 50°C for 2 hours, followed by size fractionation on a ChromaSpin-400 column (Clontech) into small (S), medium (M), and large (L) transcripts based on their electrophoresis profile on a 1.1% agarose gel. Selected fractions were pooled and concentrated using a Microcon YM-100.
The concentrated cDNA mixture was ligated into the λ TriplEx2 vector (Clontech), and the resulting ligation mixture was packaged using the GigaPack III Plus packaging extract (Stratagene, La Jolla, CA) according to the manufacturer's instructions. The packaged library was plated by infecting log-phase XL1- Blue Escherichia coli cells (Clontech). The percentage of recombinant clones was determined by blue-white selection screening on LB/MgSO4 plates containing X-gal/IPTG. Recombinants were also determined by PCR, using vector primers, PT2F1 (AAG TAC TCT AGC AAT TGT GAG C) and PT2R1 (CTC TTC GCT ATT ACG CCA GCT G flanking the inserted cDNA, with subsequent visualization of the products on a 1.1% agarose/EtBr gel.
cDNA sequencing.
Twenty-four 96-well plates were prepared for cyclo sequencing, each containing 94 clones and two DNA controls, as follows: The cDNA library was plated on LB/MgSO4 plates containing X gal/IPTG to an average of 250 plaques per 150 mm Petri plate. Recombinant (white) plaques were randomly selected and transferred to 96-well microtiter plate (Nunc, Rochester, NY) containing 75 μL of ultrapure water (KD Medical, Columbia, MD) per well. The plates were covered and placed on a gyrating shaker for 30 min at room temperature. The phage suspension was either immediately used for PCR or stored at 4°C for future use.
To amplify the cDNA using a PCR reaction, 4 μL of the phage sample was used as a template. The primers were sequences from the λ TriplEx2 vector and named PT2F1 (AAG TAC TCT AGC AAT TGT GAG C) and PT2R1 (CTC TTC GCT ATT ACG CCA GCT G), positioned at the 5′ end and the 3′ end of the cDNA insert, respectively. The reaction was carried out in a 96-well PCR microtiter plate (Applied Biosystems, Inc., Foster City, CA) using FastStart Taq polymerase (Roche Diagnostics) on a GeneAmp PCR system 9700 (Perkin Elmer Corp., Foster City, CA). The PCR conditions were 1 hold of 75°C for 3 min; 1 hold 94°C for 4 min, 33 cycles of 94°C for 1 min, 49°C for 1 min; 72°C for 2 min. The amplified products were analyzed on a 1.5% agarose/EtBr gel. Clones were PCR amplified, and the ones showing single band were selected for sequencing. Approximately 200–250 ng of each PCR product was transferred to a 96-well PCR microtiter plate (Applied Biosystems) and frozen at −20°C. Samples were shipped on dry ice to the Rocky Mountain Laboratories Genomics Unit with primer (PT2F3, TCT CGG GAA GCG CGC CAT TGT) and template combined together in an ABI 96-well Optical Reaction Plate (P/N 4306737) following the manufacturer's recommended concentrations. Sequencing reactions were set up as recommended by Applied Biosystems BigDye Terminator v3.1 Cycle Sequencing Kit by adding 1 μL ABI BigDye Terminator Ready Reaction Mix v3.1 (P/N 4336921), 1.5 μL 5× ABI Sequencing Buffer (P/N 4336699), and 3.5 μL of water for a final volume of 10 μL. Cycle sequencing was performed at 96°C for 10 sec, 50°C for 5 sec, 60°C for 4 min for 27 cycles on either a Bio-Rad Tetrad 2 (Bio-Rad Laboratories, Hercules, CA) or ABI 9700 (Applied Biosystems) thermal cycler. Fluorescently-labeled extension products were purified following Applied Biosystems BigDye XTerminator Purification protocol and subsequently processed on an ABI 3730xL DNA Analyzer (Applied Biosystems). The AB1 file generated for each sample from the 3730xL DNA Analyzer was provided to researchers in Rockville, MD through a secure network drive for all subsequent downstream sequencing analysis. In addition to the sequencing of the cDNA clones, primer extension experiments were performed in selected clones to further extend sequence coverage.
Bioinformatic tools and procedures.
Expressed sequence tags (EST) were trimmed of primer and vector sequences. The BLAST tool,16 CAP3 assembler,17 and ClustalW18 software were used to compare, assemble, and align sequences, respectively. Phylogenetic analysis and statistical neighbor-joining (NJ) bootstrap tests of the phylogenies were done with the Mega package.19 For functional annotation of the transcripts we used the tool BlastX16 to compare the nucleotide sequences to the non-redundant (NR) protein database of the National Center for Biotechnology Information (NCBI, National Library of Medicine, NIH,) and to the Gene Ontology (GO) database.20 The tool, reverse position specific Blast (RPS-BLAST)16 was used to search for conserved protein domains in the Pfam,21 SMART,22 Kog,23 and conserved domains databases (CDD).24 We have also compared the transcripts with other subsets of mitochondrial and rRNA nucleotide sequences downloaded from NCBI. Segments of the three-frame translations of the EST (because the libraries were unidirectional, 6-frame translations were not used), starting with a methionine found in the first 300 predicted amino acids (AAs), or the predicted protein translation in the case of complete coding sequences, were submitted to the SignalP server25 to help identify translation products that could be secreted. O-glycosylation sites on the proteins were predicted with the program NetOGlyc.26 Functional annotation of the transcripts was based on all the comparisons above. Following inspection of all these results, transcripts were classified as either Secretory (S), Housekeeping (H), or of Unknown (U) function, with further subdivisions based on function and/or protein families. Codon volatility was calculated as previously described.27
Results and Discussion
cDNA library characteristics.
A total of 2,039 clones out of 2,350 that were sequenced yielded good quality sequences and were used to assemble a database (Supplemental Table S1) that yielded 827 clusters of related sequences, 651 of which contained only one EST. The consensus sequence of each cluster is named either a contig (deriving from two or more sequences) or a singleton (deriving from a single sequence). For sake of simplicity, this work uses “cluster” or “contig” to denote sequences derived from both consensus sequences and singletons. The 827 clusters were compared using the program BlastX, BlastN, or RPS-BLAST16 to the non-redundant protein database of the NCBI (NR), a gene ontology database (GO),20 the conserved domains database of the NCBI (CDD),24 and a custom-prepared subset of the NCBI nucleotide database containing either mitochondrial or rRNA sequences.
Because the libraries used are unidirectional, three-frame translations of the dataset were also derived, and open reading frames (ORFs) starting with a methionine and longer than 40 AAs residues were submitted to SignalP server25 to help identify putative-secreted proteins. The EST assembly, BLAST, and signal peptide results were loaded into an Excel (Microsoft, Redman, OR) spreadsheet for manual annotation and are provided in Supplemental Table S1.
Four categories of expressed genes derived from the manual annotation of the contigs were created (Table 1 and Figure 1). The putatively secreted (S) category contained 29.6% of the clusters and 61.5% of the sequences, with an average number of 5.1 sequences per cluster. The housekeeping (H) category had 27.9% and 20.2% of the clusters and sequences, respectively, and an average of 1.8 sequences per cluster. Four singletons were classified as transposable element/Viral (TEV) products, constituting less than 0.5% of the ESTs or contigs. Transposable elements (TE) have been a common finding in sialotranscriptomes and most probably reflect regulatory transcripts repressing transposition rather than active transposition.28 For example, the transcript sn_contig_826 matches a Gypsy transposon polymerase, but has a stop codon. More specific to Simulium is the transcript sn-contig_406 that matches polyproteins of picorna-like virus specific to insects.29 The picornaviridae is a ancient family affecting prokaryotes and eukaryotes, both animal and plants30 and are responsible for varied diseases such as hepatitis (Hepatitis A) and poliomyelitis31 in humans and paralytic diseases in insects.29,32 Finally, 42% of the clusters, containing 18% of all sequences, were classified as unknown (U), because no functional assignment could be made. This category had an average of 1.1 sequences per cluster. A good proportion of these transcripts could derive from 3′ or 5′ untranslated regions of genes of the previous two categories, as was recently indicated for a sialotranscriptome of Anopholes gambiae.12
Table 1.
Functional classification of transcripts originating from the salivary glands of Simulium nigrimanum
| Class | Number of contigs | Number of ESTs | EST's/contig |
|---|---|---|---|
| Salivary | 245 | 1255 | 5.1 |
| Housekeeping | 231 | 411 | 1.8 |
| Transposable element/virus | 4 | 4 | 1.0 |
| Unknown | 347 | 369 | 1.1 |
Figure 1.
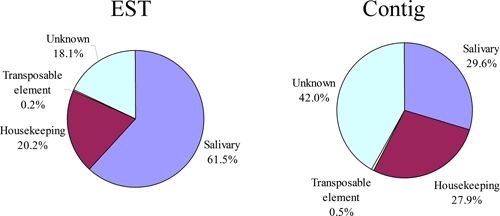
Functional class distribution of expressed sequence tags (EST) or assembled contigs (Contigs) deriving from a salivary gland cDNA library from adult Simulium nigrimanum black flies. This figure appears in color at www.ajtmh.org.
Housekeeping (H) genes.
The 231 clusters (comprising 411 ESTs) attributed to H genes expressed in the salivary glands of S. nigrimanum were further divided into 18 subgroups according to function (Table 2). Not surprisingly for an organ specialized for the secretion of polypeptides, the two larger sets were associated with protein synthesis machinery (49 clusters containing 117 ESTs) and energy metabolism (43 clusters containing 80 ESTs), a pattern also observed in other sialotranscriptomes.13,33,34 We have arbitrarily included a group of 111 ESTs (41clusters) in the H category that represent highly conserved proteins of unknown function, presumably associated with cellular function. They are named conserved proteins of unknown function in Supplemental Table S1, immediately preceding the clusters of the TEV class. These sets may help functional identification of the “conserved hypothetical” proteins as previously reviewed by Galperin and Koonin.35 The complete list of all 231 gene clusters, along with further information about each, is given in Supplemental Table S1.
Table 2.
Functional classification of Housekeeping transcripts originating from the salivary glands of Simulium nigrimanum
| Class | Number of contigs | Number of ESTs | EST's/ contig |
|---|---|---|---|
| Protein synthesis machinery | 49 | 117 | 2.4 |
| Unknown, conserved | 41 | 111 | 2.7 |
| Metabolism, energy | 43 | 80 | 1.9 |
| Signal transduction | 23 | 25 | 1.1 |
| Protein modification machinery | 14 | 14 | 1.0 |
| Protein export machinery | 12 | 12 | 1.0 |
| Transcription factor | 9 | 11 | 1.2 |
| Nuclear regulation | 6 | 6 | 1.0 |
| Proteasome machinery | 5 | 6 | 1.2 |
| Transporters/storage | 6 | 6 | 1.0 |
| Cytoskeletal | 6 | 6 | 1.0 |
| Transcription machinery | 4 | 4 | 1.0 |
| Metabolism, carbohydrate | 3 | 3 | 1.0 |
| Metabolism, nucleotide | 3 | 3 | 1.0 |
| Oxidant metabolism/detoxification | 2 | 2 | 1.0 |
| Metabolism, amino acid | 2 | 2 | 1.0 |
| Metabolism, lipid | 2 | 2 | 1.0 |
| Metabolism, intermediate | 1 | 1 | 1.0 |
| Total | 231 | 411 |
Possibly secreted (S) class of expressed genes.
Inspection of Supplemental Table S1 indicates the expression of several expanded gene families, including those coding for Kunitz-domain containing polypeptides, antigen-5 family members, odorant-binding/D7 protein families, vasodilatory proteins of the Simulium vittatum erythema protein (SVEP) family, low complexity protein families, and several protein families previously found only in S. vittatum15 (Table 3).
Table 3.
Functional classification of putative secreted transcripts originating from the salivary glands of Simulium nigrimanum
| Class | Number of ESTs |
|---|---|
| Subclass | |
| Protease inhibitor domains | |
| Kunitz domains | 91 |
| Enzymes | |
| Serine proteases | 82 |
| Endonuclease | 1 |
| Destabilase | 5 |
| 5′-nucleotidase/apyrase | 19 |
| Amylase | 52 |
| Antigen 5 family | 31 |
| Lipocalin | 1 |
| Immunity related | |
| Pattern recognition | 2 |
| Antimicrobial | 36 |
| Families unique to blood sucking Nematocera | |
| D7/OBP | 317 |
| Aegyptin family | 43 |
| Families unique to black flies | |
| SVEP* | 86 |
| Other Simulium-specific proteins | 171 |
| Families with low complexity repeats | |
| PolyQ family | 17 |
| Glycine histidine-rich | 38 |
| Collagen-like peptide | 38 |
| Other putative secreted peptides | 224 |
| Phenoloxidase inhibitor | 1 |
SVEP = Simulium vittatum erythema protein.
Detailed analysis of the S. nigrimanum sialome.
Several clusters of sequences coding for housekeeping and putative secreted polypeptides indicated in Supplemental Table S1 are abundant and complete enough to extract novel consensus sequences. Additionally, we have performed primer extension studies in several clones to obtain full- or near full-length sequences of products of interest. A total of 117 novel sequences, 72 of which code for putative secreted proteins, are grouped together in Supplemental Table S2.
Ubiquitous protein families of characterized function(s).
Enzymes.
Enzymes have been found in the salivary secretion of blood-sucking insects and ticks. In blood-sucking Nematocera, which also feed on sugar solutions, glycosidases such as maltase and amylases are commonly found. Enzymes can also function in antimicrobial functions, such as lysozyme and serine proteases that are possibly related to activation of the propheloloxidase cascade.36,37 Endonucleases, together with hyaluronidases may also be present, possibly decreasing the skin viscosity and helping the diffusion of salivary pharmaceuticals and formation of the feeding cavity in pool-feeding insects such as sand flies and black flies.13,38–43 Apyrase is also commonly found; it hydrolyzes host ATP and ADP to adenosine monophosphate (AMP), thus depleting these agonists of neutrophil and platelet aggregation.6,7,44 The sialotranscriptome of S. nigrimanum reveals the presence of at least two glycosidases, possibly polymorphic as demonstrated by assembly of related coding sequences from multiple ESTs (Supplemental Table S1). Mosquitoes and S. vittatum sialotranscriptomes also reveal two glycosidases in their transcriptomes, consistently.
The salivary apyrase of blood-sucking arthropods is a typical case of convergent evolution. The salivary activity derives from at least three different genes. In mosquitoes and triatomine bugs of the genus Triatoma, members of the 5′ nucleotidase family have been co-opted for this function.45–48 In sand flies and bed bugs a new gene product was characterized, in the form of the Cimex-type apyrase family.49,50 In fleas it has been proposed that the activity derives from the CD39 protein family.51 Previous sialotranscriptome analysis of S. vittatum found three ESTs coding for members of the 5′-nucleotidase family, and it was shown that the carboxy terminus of these putative enzymes lacked the inositol-phosphatide anchoring domain46,52,53 typical of membrane-bound enzymes of this family, a situation previously shown in the salivary apyrase of the mosquito Aedes aegypti. The sialotranscriptome of S. nigrimanum reveals 19 ESTs associated with at least two different genes, allowing deduction of two 3′ truncated CDS, Sim-187, which is 76% identical to its S. vittatum homolog, and Sim-214, which is only 37% identical to the same protein in S. vittatum. Interestingly, while Sim-187 produces matches to mosquito and tabanid salivary apyrases, Sim-214 produces matches to S. vittatum and to bacterial proteins. Alignment of the simulid, mosquito, and bacterial proteins producing these best matches (Figure 2A) allows production of a bootstrapped phylogram showing strong support for 4 clades (Figure 2B); clade I contains the mosquito proteins; clade II has Sim-187 and the S. vittatum protein, joining the mosquito clade with 55% bootstrap support; clade III has four of the bacterial proteins; and strangely enough, clade IV contains Sim-214 together with the Desulvovibrio piger enzyme, with 81% bootstrap support, suggesting that Sim-187 may have arisen in S. nigrimanum by horizontal transfer (HT), as it has been proposed for some salivary gland expressed genes from mosquitoes.10,12 However, the fact that Sim-214 produces a best match to the S. vittatum protein, which in turn produces best matches to mosquito salivary apyrases suggests more conservatively that Sim-214 arose by gene duplication from a common fly ancestral gene that evolved divergently from its parent to now resemble a bacterial protein.
Figure 2.
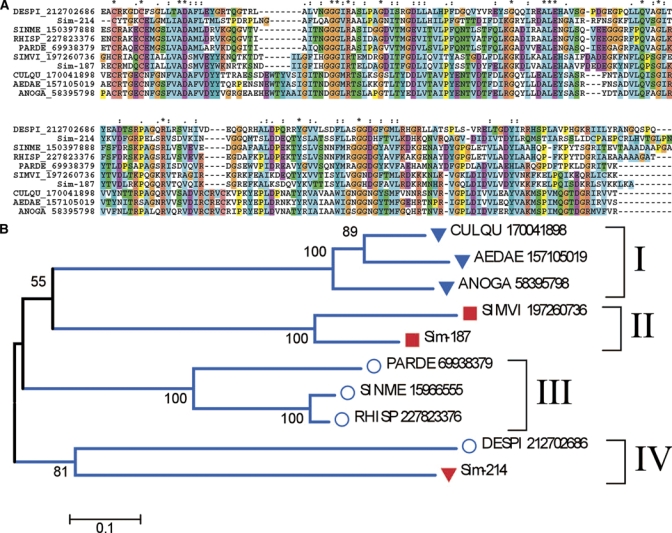
The 5′-nucleotidase/apyrase family of black flies, mosquitoes, and prokaryotes. (A) Clustal alignment. The Simulium nigrimanum proteins are Sim-214 and Sim187. The remaining sequences are named with the first three letters from the genus name followed by two letters from the species name and by their National Center for Biotechnology Information (NCBI) protein accession number. The symbols above the alignment indicate: (*) identical sites; (:) conserved sites; (.) less conserved sites. (B) Neighbor joining bootstrapped phylogram of the alignment in A, showing 4 clades. The numbers on the branches represent the percent bootstrap support. The bar in the bottom represents 10% amino acid divergence. Mosquito proteins are indicated by a triangle, Simulium proteins by a square, and bacterial proteins by a circle. For more details, see text. This figure appears in color at www.ajtmh.org.
The carboxy terminal regions of Simulium salivary proteins of the 5′-nucleotidase family were aligned with mosquito and tabanid (Chrysoptin) salivary apyrases, and with 5′-nucleotidases of human, mouse, cow, and Drosophila (Figure 3), showing that all salivary apyrases of blood-sucking Diptera lack the hydrophobic terminal region where the inositol anchor is located, indicating these salivary enzymes to be secreted and not membrane bound.
Figure 3.

The 5′-nucleotidase/apyrase family of black flies, mosquitoes, tabanids compared with Drosophila and mammalian enzymes. The Simulium nigrimanum proteins are Sim-214 and Sim187. The remaining sequences are named with the first three letters from the genus name followed by two letters from the species name and by their National Center for Biotechnology Information (NCBI) protein accession number. The box shows the deletion of the membrane anchor region in the salivary apyrases of Diptera. The symbols above the alignment indicate: (*) identical sites; (:) conserved sites; (.) less conserved sites. This figure appears in color at www.ajtmh.org.
Comparison of the number of ESTs coding for members of the 5′-nucleotidase in the S. nigrimanum and S. vittatum sialotranscriptomes is informative. The previously described sialome of S. vittatum, a set of 1,483 ESTs, had only three sequences coding for this type of enzyme,15 whereas in S. nigrimanum 13 ESTs were found out of 1,204 ESTs. The χ2 derived from these numbers is 8.5917 with a P = 0.00337, indicating a highly significant difference, in accordance with enzyme measurements of salivary apyrase of New World black flies that indicated higher enzyme activity in vectors of human onchocerciasis.54
Destabilase is an endo-ɛ-(γ-Glu)-Lys isopeptidase, which cleaves isopeptide bonds formed by transglutaminase (Factor XIIIa) between Gln γ-carboxamide and the ɛ-amino group of lysine. This enzyme activity leads to dissolution of stabilized fibrin. Destabilase was first described in the salivary glands of the leech Hirudo medicinalis,55 later shown to be the product of a multigene family that is related to the lysozyme superfamily.56,57 Five ESTs in the S. nigrimanum sialotranscriptome have the destabilase CDD motif, possibly coding for two alleles. Whether these proteins function as the leech proteins do or as classical lysozymes remains to be determined.
Eight serine proteases sequences, four of which are full length, are presented in supplemental file S2, possibly deriving from 6 different genes. Sim-192 and Sim-193 appear to be alleles, whereas Sim-166 and Sim-167 appear to be differential transcripts of the same gene, or the product of recent gene duplications. These enzymes may function in immunity-related protease cascades or in blood feeding, such as in a fibrinolytic function.
Immunity-related gene products.
Within this group, supplemental file S2 presents the full-length sequences of an antimicrobial peptide of the cecropin family, two lysozyme sequences (possibly alleles), and two members of the Gram-negative bacteria-binding protein, which function as bacterial recognition molecules that initiate immunity reactions. We also found a single EST matching a mosquito protein annotated as phenoloxidase inhibitor, and 54% identical to the Musca tyrosinase inhibitor deposited at SwissProt. Sn-307 has the sequence VGD flanked by cysteines (CVRVGDWC), which is similar to snake disintegrins that block the adhesion of alpha4beta1 integrins.58,59 Accordingly, Sn-307 may function as an inhibitor of vascular adhesion, as their snake venom counterparts.
Kunitz-domain containing peptides.
The ubiquitous Kunitz domain is associated with proteins having serine protease inhibitor activity,60,61 and ion-channel inhibitory activity.62–65 Many tick anticlotting peptides from this family were described.66–68 No transcripts coding for Kunitz domains have been found in transcriptomes of mosquitoes or sand flies, where anti-clotting activity was associated with serpins69 or the anopheles-specific proteins of the anophelin family.70,71 Kunitz peptides, however, were found in the sialotranscriptomes of the biting midge Culicoides sonorensis72 and S. vittatum, but none of these proteins have been functionally characterized. Figure 4 shows the full-length sequences of four Kunitz domain-containing peptides from S. nigrimanum, deducted from the assembly of 7-41 ESTs, indicating these transcripts are relatively abundant. Alignment of the four S. nigrimanum proteins with the two Kunitz proteins described from S. vittatum clearly shows two polypeptide families (Figure 4A), one of them containing an extended carboxy terminal domain populated by basic peptides, reminiscent of the basic tail family of tick salivary proteins. This basic tail may lead the peptides to associate to negatively charged phospholipids such as those on the surface of activated platelets that serve as a matrix for assembly of blood coagulation proteolytic complexes such as the Xase and prothrombinase complexes.73–75 The phylogram of the alignment (Figure 4B) shows strong support for 2 clades of Simulium proteins (marked I and II), indicating that the two black flies shared a common ancestor already containing the two different genes. The phylogram also indicates that S. nigrimanum has two genes in each clade, or more probably each has at least two common alleles.
Figure 4.
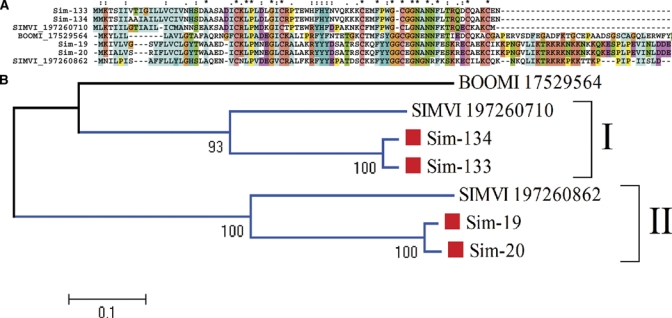
The Simulium Kunitz salivary protein family. (A) Clustal alignment of 4 S. nigrimanum proteins (starting with Sim-) with 2 Simulium vittatum proteins, indicated by SIMVI_X where X is their National Center for Biotechnology Information (NCBI) accession numbers. The tick anti-thrombin peptide named boophilin (BOOMI_17529564) is included as an out-group. The symbols above the alignment indicate identity (*), high similarity (:), and similarity (.) of residues in the indicated alignment position. (B) Phylogram derived from the alignment in A. Simulium nigrimanum sequences are marked with a square. The numbers on the tree bifurcations indicate the percentage bootstrap support above 50%. The bar at the bottom represents 10% amino acid substitution. Protein sequences were aligned by the Clustal program39 and the dendrogram was done with the Mega package41 after 10,000 bootstraps with the neighbor joining (NJ) algorithm. For more details, see text. This figure appears in color at www.ajtmh.org.
Antigen 5 protein family.
AG5-related salivary products are ubiquitous in sialotranscriptomes of blood-feeding arthropods, being members of a group of secreted proteins that belong to the CAP family (Cys-rich secretory proteins; AG5 proteins of insects; pathogenesis-related protein 1 of plants).76 The majority of these animal proteins have no known function. The notable exceptions include proteolytic activity in Conus,77 smooth muscle-relaxing activity in snake venoms,78,79 and salivary neurotoxin activity in the venomous lizard Heloderma horridum.80 Recently, an antigen-5 protein from the saliva of a tabanid fly81 was shown to inhibit platelet aggregation by the unusual acquisition of a typical RGD domain that is known to prevent fibrinogen binding to platelets and its ensuing aggregation,82 and a stable fly salivary protein of this family was shown to bind host immunoglobulins and may function as an inhibitor of the classical pathway of complement activation.83 These diversity of functions prevent generalizations regarding this protein family. The sialotranscriptome of S. nigrimanum produced 24 ESTs matching members of this family, which produce best matches to salivary proteins of Culicoides, sand flies, and mosquitoes.
Protein families exclusive to blood-feeding Diptera.
D7/odorant-binding protein (OBP) family.
The D7 salivary family is typical of blood-feeding Nematocera.84 It is related to the OBP family,85 but containing two additional helices per domain.86 Members of this family have been found in all mosquito, Culicoides, sand fly, and black fly transcriptomes so far studied. Single- or double-domain proteins exist, constituting the short and long D7 subfamilies.87 Anopheles gambiae has. Eight genes encoding these proteins, with 3 genes encoding the long forms and 5 encoding the short forms.12 Mosquito proteins have recently been shown to bind biogenic amines88 and inflammatory lipids89 thus helping blood feeding by sequestering vasoconstrictory, inflammatory and platelet aggregation agonists. In S. vittatum both short and long D7 proteins were also found,15 one of which was demonstrated in a patent to be an anti-clotting agent.90 The S. nigrimanum sialotranscriptome reveals both long and short forms of the D7 family, with 34 deducted protein sequences being available in supplemental file S2, many of which appear to be alleles. This family is also the most represented in the sialotranscriptome, with a total of 317 EST of the total of 1,255 attributed to secreted products (Table 3).
Phylogenetic analysis of the D7 sequences of S. nigrimanum combined with those from S. vittatum reveals 9 clades with strong bootstrap support (supplemental file S2 and Figure 5). Some of these clades, such as I, IV, V, VII, and IX have closely related sequences that could be alleles, whereas others such as II, III, VII, and VIII contain sequences with over 15% amino acid divergence indicative of two or more genes. Accordingly, at least 13 genes must exist in S. nigrimanum coding for D7 proteins, possibly 14 if we count 3 genes in Clade II. Notice also the multispecies clades III, VI, and VIII, indicating the ancestor of S. vittatum and S. nigrimanum already had these genes. The phylogram also indicates that the majority of the short sequences coalesce with strong bootstrap support under one super clade, named Short in Figure 5, containing clades I–V. Long D7 sequences cluster both with other long sequences, but also with short S. nigrimanum sequences. It is also to be noticed that the two related short sequences in clade VII, Sim-188 and Sim-189 have a cluster of basic AAs in their carboxy terminal region that might lead these proteins to associate with negatively charged lipids that are important for assembly of blood clotting enzymes.73,91 Finally, the S. vittatum sequence gi|197260866, the salivary anti-thrombin of S. vittatum,90 localizes in Clade III, suggesting the homologous proteins of S. nigrimanum, which are 67% identical in AAs sequence, may also display anti-thrombin activity.
Figure 5.
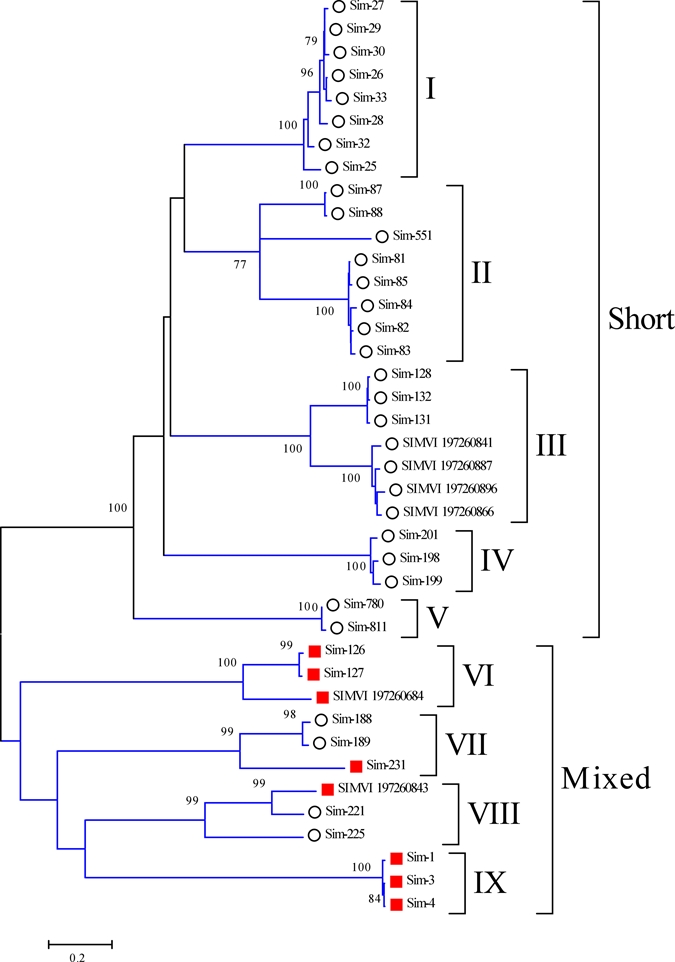
The Simulium D7 salivary protein family. Phylogram deriving from the alignment of S. nigrimanum proteins (starting with Sim-) with those of Simulium vittatum, indicated by SIMVI_X where X is their National Center for Biotechnology Information (NCBI) accession numbers. The numbers on the tree bifurcations indicate the percentage bootstrap support above 75%. The bar at the bottom represents 20% amino acid substitution. Members of the short D7 family are marked with a circle, those from the long family with a square. Protein sequences were aligned by the Clustal program39 and the dendrogram was done with the Mega package41 after 10,000 bootstraps with the neighbor joining (NJ) algorithm. For more details, see text. This figure appears in color at www.ajtmh.org.
30. kDa antigen/Aegyptin family.
A salivary immunogenic protein from Ae. aegypti named 30 kDa antigen92 is similar to anopheline proteins named GE-rich protein for its abundance in glycine and glutamate residues. Recent functional analysis of members of this protein family from Aedes and Anopheles revealed they are antagonists of collagen-induced platelet aggregation.93,94 Alignment of black fly and mosquito proteins (Figure 6) shows the previously described distinct domains,93 including the signal peptide region, a glycine/aspartate/glutamate-rich region, and a relatively more conserved carboxyterminus, where the conserved pattern T-Y-x(6)-L-x(19,22)-Q-x(18,19)-I-x(2)-C-F-x(20)-C-x(3,10)-C-x(20,21)-C is found. This unique family supports a common origin of hematophagy between black flies and mosquitoes, as proposed by Grimaldi and Engels.95
Figure 6.
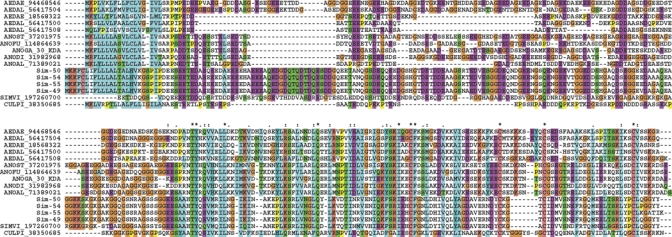
The 30 kDa antigen/Aegyptin family of black flies and mosquitoes. The Simulium nigrimanum proteins are indicated by Sim-XX where XX is the number indicated in supplemental file S2. The remaining sequences are named with the first three letters from the genus name followed by two letters from the species name and by their National Center for Biotechnology Information (NCBI) protein accession number. The symbols above the alignment indicate: (*) identical sites; (:) conserved sites; (.) less conserved sites. For more details, see text. This figure appears in color at www.ajtmh.org.
Hyp16 family.
The first member of this family was identified in the sialotranscriptome of Anopheles. stephensi,14 followed by identification in the sialome of Aedes albopictus,11 and more recently, in the sialome of the stable fly, Stomoxys calcitrans where it was found abundantly expressed.41 Sim-692 is 29% identical and 48% similar in its sequence to the An. stephensi protein. Homologues in An. gambiae and Ae. aegypti are found in the deducted proteomes of these mosquitoes. Alignment of these protein sequences reveal the conserved pattern G-x(12,13)-C-D-x(3)-C-P-x(5)-C-x(3)-K-x(12,15)-C-x(4)-G-x(4)-K-x(17,19)-E that might help to identify members of this family, none of which have known function (Figure 7).
Figure 7.
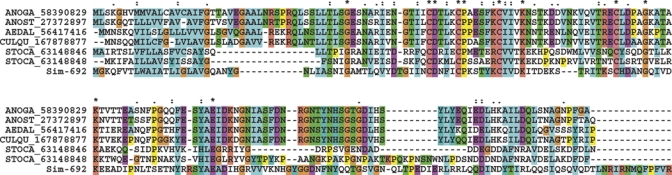
The Hyp16 family of blood-sucking Diptera. The Simulium nigrimanum protein is indicated by Sim-692. The remaining sequences are named with the first three letters from the genus name followed by two letters from the species name and by their National Center for Biotechnology Information (NCBI) protein accession number. The symbols above the alignment indicate: (*) identical sites; (:) conserved sites; (.) less conserved sites. For more details, see text. This figure appears in color at www.ajtmh.org.
Protein families exclusive to black flies.
SVEP.
The salivary vasodilator of S. vittatum, named SVEP for S. vittatum erythema protein, has been previously identified as a novel protein96 and the recombinant protein expressed and functionally characterized as a vasodilator possibly activating ATP-dependent K+ channels.97 The sialotranscriptome of S. vittatum identified SVEP to belong to a diverse multigene family containing at least 5 genes.15 As expected, S. nigrimanum has homologues to SVEP, varying from 45–66% identity. Alignment of the Simulium SVEPs shows similar sized sequences with relatively few conserved AAs spread over the length of the sequence (Figure 8A). The phylogram also indicates that each species has at least 5 genes coding for members of the family, and that their products group within their own species, even though they can be very distant from each other, showing a S. vittatum and a S. nigrimanum clade, and an intermediate S. nigrimanum clade that might derive from a gene closer to the ancestral gene (Figure 8B). This scenario indicates either that the gene duplications in each species occurred after the split from a common ancestor, or that gene conversion events within each species occurred to keep some degree of homogeneity among the genes.
Figure 8.
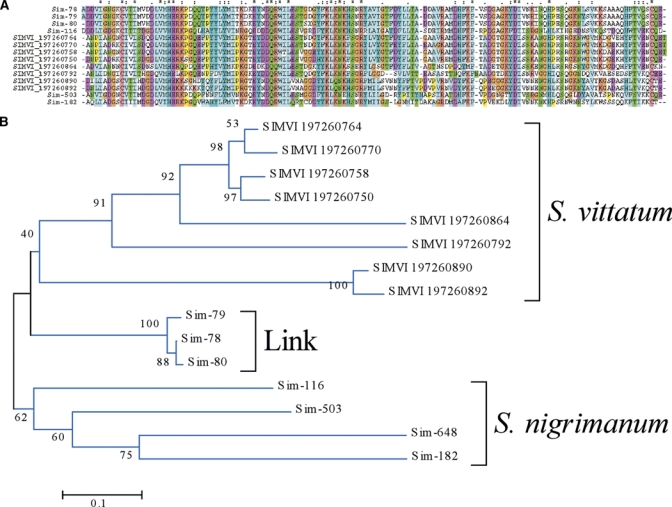
The Simulium vittatum erythema protein (SVEP) superfamily of Simulium. (A) Clustal aligment. The symbols above the alignment indicate: (*) identical sites; (:) conserved sites; (.) less conserved sites. (B) Bootstrapped phylogram. The Simulium nigrimanum proteins are indicated by a square. The S. vittatum sequences are named SIMVI followed by their National Center for Biotechnology Information (NCBI) protein accession number. The vasodilator SVEP is marked with a triangle. The numbers on the tree bifurcations indicate the percentage bootstrap support. The bar at the bottom represents 10% amino acid substitution. Protein sequences were aligned by the Clustal program39 and the dendrogram was done with the Mega package41 after 10,000 bootstraps with the neighbor joining (NJ) algorithm. For more details, see text. This figure appears in color at www.ajtmh.org.
To attempt an insight into the origins of the SVEP family, members of the S. nigrimanum proteins were submitted to PSI-BLAST querying the NR protein database from NCBI. As expected, the protein Sim-116, which belongs to the canonical SVEP family, retrieved members of the S. vittatum SVEP family, but in the second round it retrieved a bacterial insect toxin with low e-value. Further iterations led to retrieving many additional bacterial proteins annotated as glycosidases and ricin, suggesting the SVEP family originated from a carbohydrate domain-containing ancestor. Although C-type lectins are a common finding in sialotranscriptomes of mosquitoes and sand flies, a ricin member in a sialotranscriptome is a novelty.
Collagen-like family.
This protein family was previously identified in the S. vittatum sialome. It consists of basic proteins (pI = 10) rich in glycine, lysine, and proline, with over 20% of the protein consisting of Pro+Gly. No cysteine residues are found in the mature proteins. The amino terminal region has a relatively conserved region (Figure 9A), but the carboxy terminus region determines at least three subfamilies, as follows: 1) there are two long S. vittatum proteins, responsible for the insert/gap regions seen in Figure 8A, and marked as clade Long in Figure 9B. 2) Two shorter S. vittatum proteins possess a basic tail of lysine-rich residues that might drive these proteins to binding anionic phospholipids73; these are marked as Basic tail clade in Figure 9B. The S. nigrimanum sequences form a single uniform clade (Figure 9B), and their sequences are more homogeneous (Figure 9A). The S. vittatum sequence gi|197260678 does not belong to any of the two strong clades, representing a possible link between the two groups of sequences. Functional analysis of recombinant proteins from this family should take into consideration that they may be modified into hydroxyl proline and lysine, as collagen is modified.98
Figure 9.
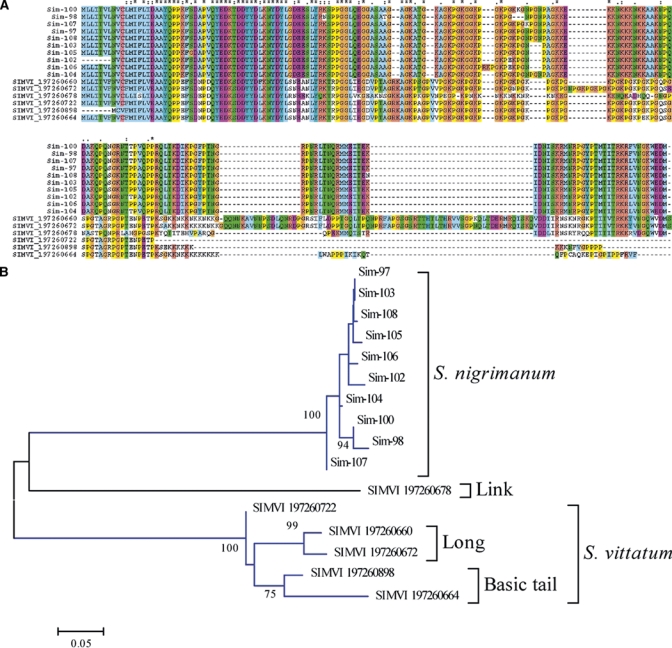
The collagen-like superfamily of Simulium. (A) Clustal aligment. The symbols above the alignment indicate: (*) identical sites; (:) conserved sites; (.) less conserved sites. (B) Bootstrapped phylogram. The Simulium nigrimanum proteins are indicated by Sim- and their identification number. The Simulium vittatum sequences are named SIMVI followed by their National Center for Biotechnology Information (NCBI) protein accession number. The numbers on the tree bifurcations indicate the percentage bootstrap support above 75%. The bar at the bottom represents 5% amino acid substitution. Protein sequences were aligned by the Clustal program39 and the dendrogram was done with the Mega package41 after 10,000 bootstraps with the neighbor joining (NJ) algorithm. For more details, see text. This figure appears in color at www.ajtmh.org.
Acidic H P Q E-rich proteins of low complexity.
This protein family was also identified in the S. vittatum sialotranscriptome, where it comprised its most abundant cluster of ESTs. These have low complexity and abound in histidine, proline and glutamic acid residues, with regions of Gly-His or Pro-His repeats. They are possibly related to mosquito and Culicoides proteins that also show Pro-His and Gly-His repeats. The S. nigrimanum proteins are tilde;65% identical in primary sequence to their S. vittatum homologues. It is possible that the His repeats may function as antimicrobials by chelating Zn or other trace element ions.99–101
Simulium mucins.
Mucins are low complexity proteins rich in serine and threonine residues that can accept N-acetyl-galactosamine residues.26 The S. nigrimanum sialotranscriptome reveals the presence of such proteins, over one-third of their residues consisting of Ser+Thr, and displaying 33–136 putative galactosylation sites.
Simulium basic 7–13 kDa family.
The S. nigrimanum sialotranscriptome revealed seven protein sequences with two or more ESTs each coding for basic (pI 8.1–10.6) proteins ranging from 7 to 13 kDa in predicted mature MW. This group of proteins clusters when they are compared at 30% similarity over 50% of the smaller length, indicating they are possibly related. These seven sequences belong to two more closely related groups, one with four sequences, the other with three sequences. The second group contains sequences that are tilde;65% identical in primary sequence to a previously described orphan S. vittatum protein.
Sv 7.8 kDa family.
A second group of six proteins also coding for basic proteins of size varying from 7 to 11 kDa can also be identified by their 30% similarity level. Members of this family match previously described S. vittatum proteins varying in identity from 40% to 57%, which were named Sv 7.8 kDa family previously. Other members of this group of S. nigrimanum proteins match S. vittatum proteins classified as orphans, indicating this group may derive from a very divergent multifamily gene from Simulium. The 7.8 kDa family clusters at the borderline 25% similarity level with the above described 7–13 kDa family, but no conserved AAs can be identified in the alignment, indicating if these two families share a common ancestor, the family has diverged beyond recognition.
Basic 13 kDa family.
Five deducted protein sequences from the S. nigrimanum sialotranscriptome match a previously described orphan protein from S. vittatum, constituting a new protein family here named the basic 13 kDa family. They do not match significantly any other known protein.
Sv 7 kDa family.
Five sequences from the S. nigrimanum sialotranscriptome match two smaller sequences from the S. vittatum sialome previously named the Sv 7 kDa family. The S. nigrimanum sequences have a glycine-rich insert when compared with the S. vittatum relatives (Figure 10). This family is Ser+Thr-rich, and have eight potential N-acetyl-galactosylation sites.
Figure 10.

The Sv 7kDa family of black flies. The Simulium nigrimanum proteins are indicated by Sim-XX where XX is the number indicated in supplemental file S2. The remaining sequences are from Simulium vittatum, with indicated National Center for Biotechnology Information (NCBI) protein accession number. The symbols above the alignment indicate: (*) identical sites; (:) conserved sites; (.) less conserved sites. For more details, see text. This figure appears in color at www.ajtmh.org.
Sv 4.8 kDa family.
This Simulium family produces shorter peptides in S. nigrimanum then in S. vittatum (Figure 11). All mature peptides are devoid of cysteines.
Figure 11.

The Sv 4.8kDa family of black flies. The Simulium nigrimanum proteins are indicated by Sim-XX where XX is the number indicated in supplemental file S2. The remaining sequences are from Simulium vittatum, with indicated National Center for Biotechnology Information (NCBI) protein accession number. The symbols above the alignment indicate: (*) identical sites; (:) conserved sites; (.) less conserved sites. For more details, see text. This figure appears in color at www.ajtmh.org.
Other deorphanized Simulium proteins.
Three additional protein families were found in common between S. vittatum and S. nigrimanum, and no other known protein. These constitute the 5 Cys, the basic 13 kDa and the basic 7 kDa family. These proteins do not produce significant similarities to other proteins in the NR database.
Families exclusive to S. nigrimanum.
Five distinct protein families, all with two or more ESTs, were found in the S. nigrimanum sialotranscriptome, plus seven additional orphan proteins, as follows:
Sn actinohivin-like.
Sim-177 and Sim-178 have 32% and 31% identities to an actinomycete protein named Actinohivin, and lesser identities to other bacterial proteins annotated as xylanases. Actinohivin is a lectin with anti-human immunodeficiency virus (HIV) properties.102 This suggests the fly proteins to be possible sugar ligands, which is further suggested by their CDD match to the ricin motif. Sim-177 and actinohivin blast alignments were used to build a search model for Psiblast, which produced matches to proteins annotated as glycosydases or lectins, including ricin, with small e-values on the second round, further suggesting these black fly proteins belong to the ricin lectin family of proteins. These results suggest that the Sn actinohivin-like family may be related to the SVEP family, having evolved beyond recognition at the primary structure level.
The similarity of this family to actinomycete proteins suggests it derives from a microbial contaminant, or horizontal transfer, or convergent evolution. Because a total of nine ESTs were obtained for this family, it would be expected that many more microbial contaminants would be found in the assembled database. Confirmation of these sequences at the fly's genome level may help to sort out these possibilities.
8–10 CysW family.
Eight deducted proteins from the S. nigrimanum sialotranscriptome have similar sizes and have a common framework of tryptophans cysteines and other aliphatic residues (Figure 12). Two subfamilies are clearly distinguished, one containing 10 conserved cysteines and five conserved tryptophans, the other containing eight and six conserved C and W residues, respectively. They produce only low score matches to proteins in the NR or Swissprot databases. To obtain further insight on this family, the two best matches from the NR database were used to build a Psiblast search model with each member of the 8–10 Cys family. Interestingly, the 8 Cys member Sim-159 retrieves, after one iteration of Psiblast, proteins annotated as nicotinic acetylcholine receptor, with small e-values (5e−43), whereas Sim-145 processed similarly retrieves proteins annotated as junctional adhesion molecules, with similar small e-values. Interestingly, whereas autoimmunity to desmoglein is proposed as the cause of pemphigus, other targets of autoimmunity have been considered, including the nicotinic acetylcholine receptor,103,104 while junctional molecules are profoundly involved in skin and vascular epithelium adhesion.105 Although these comparisons may not be robust, they provide for candidate proteins that could be implicated in the etiology of pemphigus foliaceus.
Figure 12.
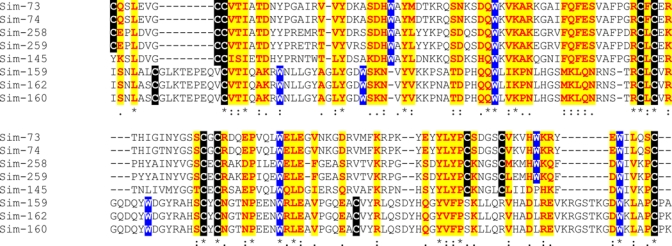
The 8-10CysW family of Simulium nigrimanum. The signal peptide region is not shown. The symbols below the alignment indicate: (*) identical sites; (:) conserved sites; (.) less conserved sites. Cysteines, tryptophanes, and conserved residues are marked with different backgrounds Protein sequences were aligned by the Clustal program39. For more details, see text. This figure appears in color at www.ajtmh.org.
Additional orphan S. nigrimanum proteins.
We additionally identified seven coding sequences in the S. nigrimanum sialotranscriptome that code for putative secreted proteins, based on 1–4 EST each. These are annotated as Orphan S. nigrimanum proteins in supplemental file S2. We additionally report four additional similarly orphan proteins, which are show alleles, based on multiple ESTs. These are named the acidic 28 kDa, the basic 28 kDa, the Sn basic 4.4 kDa, and the Sn basic 17 kDa families. They have no significant matches to any database queried in Supplemental Table S2.
Comparison of the S. nigrimanum sialome to desmogleins.
To help identify possible candidate proteins that could induce anti-desmoglein immunity, we obtained a subset of the NR database that contained the word desmoglein in its fasta definition line and used the tool BLASTP to search the sialome against this database (using the switches –W2 and –FF, meaning word size for search = 2 and filter of low complexity turned off). The results are shown in column AV of Supplemental Table S2. The protein family named Collagen-like produced matches with e-values lower than 0.0001, mainly caused by the Gly repeats in the protein. The Simulium mucin family also produced small e-values in the comparisons, again caused by the glycine repeats. To the extent this family is glycosylated (observe the serine and threonines in between the glycines), these modifications should be taken into account. The Sv7.0 family also shows similarities to desmoglein, caused by glycine and also aspartyl repeats. It has been pointed out previously the possible similarities of the 8 Cys family to the nicotinic acetylcholine receptor and to junctional adhesion molecule families, which may be implicated in pemphigus pathology.
Housekeeping proteins.
Supplemental Table S2 presents sequence information on 99 proteins classified as housekeeping.
Conclusion
Analysis of the sialome of S. nigrimanum, the second done for this family of blood-feeding flies, uncovers both the common and divergent evolutionary pathways taken in producing today's salivary “magic potion” of such arthropods. The previously reported S. vittatum sialotranscriptome revealed protein families unique to Simulium, those unique to blood-feeding Diptera, and ubiquitous protein families.15 The S. nigrimanum sialotranscriptome indeed has these three classes of salivary proteins, including 13 protein families unique to Simulium, but additionally uncovered 13 novel protein classes, including six that are relatively well expressed. It is interesting to notice that most of the S. nigrimanum salivary proteins have only 30–60% identity to their S. vittatum best match, whereas the proteins from the housekeeping class have over 95% identity, thus showing the fast divergence of the salivary proteins.
The finding of many new protein families in S. nigrimanum as compared with S. vittatum may be related to the anthropophilic behavior of S. nigrimanum as compared with S. vittatum, as well as the fact that S. vittatum is autogenous. The dependence of a mammalian host for reproduction could have increased the pressure for the best possible salivary potion in S. nigrimanum as opposed to the more relaxed state of an autogenous species.
It is also to be noted that this sialotranscriptome generated many EST clusters coding for very similar proteins, having only a few AA changes, suggesting the existence of alleles or gene duplications creating very closely related genes, as indicated throughout the Results section. This is in contrast to many sialotranscriptomes performed so far, which derived from colonized material that has suffered bottlenecks and should be more genetically homogeneous. This indicates that salivary gland genes may have a tendency to be polymorphic and might be good population markers; it may also relate to the patchy distribution of Fogo Selvagem to the extent that it may be promoted by specific alleles producing cross-reactivity with self antigens.
Disclaimer: The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organization imply endorsement by the government of the United States of America.
Footnotes
Financial support: This work was supported in part by the Intramural Research Program of the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, and NIH grant RO1 AR32599 to LAD.
Disclosure: Because JGV, VMP, and JMCR are government employees and this is a government work, the work is in the public domain in the United States. Notwithstanding any other agreements, the NIH reserves the right to provide the work to PubMedCentral for display and use by the public, and PubMedCentral may tag or modify the work consistent with its customary practices. You can establish rights outside of the U.S. subject to a government use license.
Authors' addresses: José M. C. Ribeiro, Jesus G. Valenzuela, Van M. Pham, and Lindsay Kleeman, National Institute of Allergy and Infectious Diseases, Laboratory of Malaria and Vector Research, Rockville, MD, E-mails: jribeiro@niaid.nih.gov, vpham@niaid.nih.gov, jvalenzuela@niaid.nih.gov, and kleemanli@niaid.nih.gov. Kent D. Barbian and Amanda J. Favreau, Genomics Unit, Research Technologies Section, Rocky Mountain Laboratories, Hamilton, MT, E-mails: KBarbian@niaid.nih.gov and favreaua@niaid.nih.gov. Donald P. Eaton, Wildlife Conservation Society, Campo Grande, MS, Brazil, E-mail: ksadeaton@yahoo.com. Valeria Aoki and Evandro A. Rivitti, Department of Dermatology, Universidade de São Paulo, Brazil, E-mails: valaoki@hotmail.com and elimaria@hcnet.usp.br. Gunter Hans-Filho, Department of Dermatology, Universidade Federal de Mato Grosso do Sul, Campo Grande, Brazil, E-mail: ghansfilho@hotmail.com. Luis A. Diaz, Department of Dermatology, University of North Carolina at Chapel Hill, Chapel Hill, NC, E-mail: ldiaz@med.unc.edu.
References
- 1.Culton DA, Qian Y, Li N, Rubenstein D, Aoki V, Filho GH, Rivitti EA, Diaz LA. Advances in pemphigus and its endemic pemphigus foliaceus (Fogo Selvagem) phenotype: a paradigm of human autoimmunity. J Autoimmun. 2008;31:311–324. doi: 10.1016/j.jaut.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qian Y, Clarke SH, Aoki V, Hans-Filho G, Rivitti EA, Diaz LA. Antigen selection of anti-DSG1 autoantibodies during and before the onset of endemic pemphigus foliaceus. J Invest Dermatol. 2009;129:2823–2834. doi: 10.1038/jid.2009.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rock B, Labib RS, Diaz LA. Monovalent Fab' immunoglobulin fragments from endemic pemphigus foliaceus autoantibodies reproduce the human disease in neonatal Balb/c mice. J Clin Invest. 1990;85:296–299. doi: 10.1172/JCI114426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hilario-Vargas J, Dasher DA, Li N, Aoki V, Hans-Filho G, dos Santos V, Qaqish BF, Rivitti EA, Diaz LA. Prevalence of anti-desmoglein-3 antibodies in endemic regions of Fogo selvagem in Brazil. J Invest Dermatol. 2006;126:2044–2048. doi: 10.1038/sj.jid.5700388. [DOI] [PubMed] [Google Scholar]
- 5.Eaton DP, Diaz LA, Hans-Filho G, Santos VD, Aoki V, Friedman H, Rivitti EA, Sampaio SA, Gottlieb MS, Giudice GJ, Lopez A, Cupp EW. Comparison of black fly species (Diptera: Simuliidae) on an Amerindian reservation with a high prevalence of fogo selvagem to neighboring disease-free sites in the State of Mato Grosso do Sul, Brazil. The Cooperative Group on Fogo Selvagem Research. J Med Entomol. 1998;35:120–131. doi: 10.1093/jmedent/35.2.120. [DOI] [PubMed] [Google Scholar]
- 6.Ribeiro JM, Francischetti IM. Role of arthropod saliva in blood feeding: sialome and post-sialome perspectives. Annu Rev Entomol. 2003;48:73–88. doi: 10.1146/annurev.ento.48.060402.102812. [DOI] [PubMed] [Google Scholar]
- 7.Ribeiro JM. Blood-feeding arthropods: live syringes or invertebrate pharmacologists? Infect Agents Dis. 1995;4:143–152. [PubMed] [Google Scholar]
- 8.Ribeiro JMC, Arcà B. From sialomes to the sialoverse: an insight into the salivary potion of blood feeding insects. Adv Insect Physiol. 2009;37:59–118. [Google Scholar]
- 9.Calvo E, Pham VM, Marinotti O, Andersen JF, Ribeiro JM. The salivary gland transcriptome of the neotropical malaria vector Anopheles darlingi reveals accelerated evolution of genes relevant to hematophagy. BMC Genomics. 2009;10:57. doi: 10.1186/1471-2164-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ribeiro JM, Arca B, Lombardo F, Calvo E, Phan VM, Chandra PK, Wikel SK. An annotated catalogue of salivary gland transcripts in the adult female mosquito, Aedes aegypti. BMC Genomics. 2007;8:6. doi: 10.1186/1471-2164-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arca B, Lombardo F, Francischetti IM, Pham VM, Mestres-Simon M, Andersen JF, Ribeiro JM. An insight into the sialome of the adult female mosquito Aedes albopictus. Insect Biochem Mol Biol. 2007;37:107–127. doi: 10.1016/j.ibmb.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 12.Arca B, Lombardo F, Valenzuela JG, Francischetti IM, Marinotti O, Coluzzi M, Ribeiro JM. An updated catalogue of salivary gland transcripts in the adult female mosquito, Anopheles gambiae. J Exp Biol. 2005;208:3971–3986. doi: 10.1242/jeb.01849. [DOI] [PubMed] [Google Scholar]
- 13.Ribeiro JM, Charlab R, Pham VM, Garfield M, Valenzuela JG. An insight into the salivary transcriptome and proteome of the adult female mosquito Culex pipiens quinquefasciatus. Insect Biochem Mol Biol. 2004;34:543–563. doi: 10.1016/j.ibmb.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 14.Valenzuela JG, Francischetti IM, Pham VM, Garfield MK, Ribeiro JM. Exploring the salivary gland transcriptome and proteome of the Anopheles stephensi mosquito. Insect Biochem Mol Biol. 2003;33:717–732. doi: 10.1016/s0965-1748(03)00067-5. [DOI] [PubMed] [Google Scholar]
- 15.Andersen JF, Pham VM, Meng Z, Champagne DE, Ribeiro JM. Insight into the sialome of the Black Fly, Simulium vittatum. J Proteome Res. 2009;8:1474–1488. doi: 10.1021/pr8008429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar S, Tamura K, Nei M. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 20.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schultz J, Copley RR, Doerks T, Ponting CP, Bork P. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000;28:231–234. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30:281–283. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 26.Julenius K, Molgaard A, Gupta R, Brunak S. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology. 2005;15:153–164. doi: 10.1093/glycob/cwh151. [DOI] [PubMed] [Google Scholar]
- 27.Friedman R, Hughes AL. Codon volatility as an indicator of positive selection: data from eukaryotic genome comparisons. Mol Biol Evol. 2005;22:542–546. doi: 10.1093/molbev/msi038. [DOI] [PubMed] [Google Scholar]
- 28.Silva JC, Loreto EL, Clark JB. Factors that affect the horizontal transfer of transposable elements. Curr Issues Mol Biol. 2004;6:57–71. [PubMed] [Google Scholar]
- 29.Govan VA, Leat N, Allsopp M, Davison S. Analysis of the complete genome sequence of acute bee paralysis virus shows that it belongs to the novel group of insect-infecting RNA viruses. Virology. 2000;277:457–463. doi: 10.1006/viro.2000.0616. [DOI] [PubMed] [Google Scholar]
- 30.Koonin EV, Wolf YI, Nagasaki K, Dolja VV. The Big Bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups. Nat Rev Microbiol. 2008;6:925–939. doi: 10.1038/nrmicro2030. [DOI] [PubMed] [Google Scholar]
- 31.Stanway G, Brown F, Christian P, Hovi T, Hyypiä T, King AMQ, Knowles NJ, Lemon SM, Minor PD, Pallansch MA, Palmenberg AC, Skern T. Family Picornaviridae. Virus Taxonomy. Eighth Report of the International Committee on Taxonomy of Viruses. London: Elsevier/Academic Press; 2005. pp. 757–778. [Google Scholar]
- 32.Tate J, Liljas L, Scotti P, Christian P, Lin T, Johnson JE. The crystal structure of cricket paralysis virus: the first view of a new virus family. Nat Struct Biol. 1999;6:765–774. doi: 10.1038/11543. [DOI] [PubMed] [Google Scholar]
- 33.Francischetti IM, Valenzuela JG, Pham VM, Garfield MK, Ribeiro JM. Toward a catalog for the transcripts and proteins (sialome) from the salivary gland of the malaria vector Anopheles gambiae. J Exp Biol. 2002;205:2429–2451. doi: 10.1242/jeb.205.16.2429. [DOI] [PubMed] [Google Scholar]
- 34.Ribeiro JM, Andersen J, Silva-Neto MA, Pham VM, Garfield MK, Valenzuela JG. Exploring the sialome of the blood-sucking bug Rhodnius prolixus. Insect Biochem Mol Biol. 2004;34:61–79. doi: 10.1016/j.ibmb.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 35.Galperin MY, Koonin EV. “Conserved hypothetical” proteins: prioritization of targets for experimental study. Nucleic Acids Res. 2004;32:5452–5463. doi: 10.1093/nar/gkh885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanost MR, Jiang H, Yu XQ. Innate immune responses of a lepidopteran insect, Manduca sexta. Immunol Rev. 2004;198:97–105. doi: 10.1111/j.0105-2896.2004.0121.x. [DOI] [PubMed] [Google Scholar]
- 37.Soderhall K, Cerenius L. Role of the prophenoloxidase-activating system in invertebrate immunity. Curr Opin Immunol. 1998;10:23–28. doi: 10.1016/s0952-7915(98)80026-5. [DOI] [PubMed] [Google Scholar]
- 38.Volfova V, Hostomska J, Cerny M, Votypka J, Volf P. Hyaluronidase of bloodsucking insects and its enhancing effect on leishmania infection in mice. PLoS Negl Trop Dis. 2008;2:e294. doi: 10.1371/journal.pntd.0000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson AD, Heesom KJ, Mawby WJ, Mellor PS, Russell CL. Identification of abundant proteins and potential allergens in Culicoides nubeculosus salivary glands. Vet Immunol Immunopathol. 2008;122:94–103. doi: 10.1016/j.vetimm.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 40.Ribeiro JM, Charlab R, Rowton ED, Cupp EW. Simulium vittatum (Diptera: Simuliidae) and Lutzomyia longipalpis (Diptera: Psychodidae) salivary gland hyaluronidase activity. J Med Entomol. 2000;37:743–747. doi: 10.1603/0022-2585-37.5.743. [DOI] [PubMed] [Google Scholar]
- 41.Wang X, Ribeiro JM, Broce AB, Wilkerson MJ, Kanost MR. An insight into the transcriptome and proteome of the salivary gland of the stable fly, Stomoxys calcitrans. Insect Biochem Mol Biol. 2009;39:607–614. doi: 10.1016/j.ibmb.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Calvo E, Ribeiro JM. A novel secreted endonuclease from Culex quinquefasciatus salivary glands. J Exp Biol. 2006;209:2651–2659. doi: 10.1242/jeb.02267. [DOI] [PubMed] [Google Scholar]
- 43.Valenzuela JG, Garfield M, Rowton ED, Pham VM. Identification of the most abundant secreted proteins from the salivary glands of the sand fly Lutzomyia longipalpis, vector of Leishmania chagasi. J Exp Biol. 2004;207:3717–3729. doi: 10.1242/jeb.01185. [DOI] [PubMed] [Google Scholar]
- 44.Ribeiro JM. Role of arthropod saliva in blood feeding. Annu Rev Entomol. 1987;32:463–478. doi: 10.1146/annurev.en.32.010187.002335. [DOI] [PubMed] [Google Scholar]
- 45.Sun D, McNicol A, James AA, Peng Z. Expression of functional recombinant mosquito salivary apyrase: a potential therapeutic platelet aggregation inhibitor. Platelets. 2006;17:178–184. doi: 10.1080/09537100500460234. [DOI] [PubMed] [Google Scholar]
- 46.Champagne DE, Smartt CT, Ribeiro JM, James AA. The salivary gland-specific apyrase of the mosquito Aedes aegypti is a member of the 5¢-nucleotidase family. Proc Natl Acad Sci USA. 1995;92:694–698. doi: 10.1073/pnas.92.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Faudry E, Santana JM, Ebel C, Vernet T, Teixeira AR. Salivary apyrases of Triatoma infestans are assembled into homo-oligomers. Biochem J. 2006;396:509–515. doi: 10.1042/BJ20052019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Faudry E, Lozzi SP, Santana JM, D'Souza-Ault M, Kieffer S, Felix CR, Ricart CA, Sousa MV, Vernet T, Teixeira AR. Triatoma infestans apyrases belong to the 5¢-nucleotidase family. J Biol Chem. 2004;279:19607–19613. doi: 10.1074/jbc.M401681200. [DOI] [PubMed] [Google Scholar]
- 49.Valenzuela JG, Belkaid Y, Rowton E, Ribeiro JM. The salivary apyrase of the blood-sucking sand fly Phlebotomus papatasi belongs to the novel Cimex family of apyrases. J Exp Biol. 2001;204:229–237. doi: 10.1242/jeb.204.2.229. [DOI] [PubMed] [Google Scholar]
- 50.Valenzuela JG, Charlab R, Galperin MY, Ribeiro JM. Purification, cloning, and expression of an apyrase from the bed bug Cimex lectularius. A new type of nucleotide-binding enzyme. J Biol Chem. 1998;273:30583–30590. doi: 10.1074/jbc.273.46.30583. [DOI] [PubMed] [Google Scholar]
- 51.Andersen JF, Hinnebusch BJ, Lucas DA, Conrads TP, Veenstra TD, Pham VM, Ribeiro JM. An insight into the sialome of the oriental rat flea, Xenopsylla cheopis (Rots) BMC Genomics. 2007;8:102. doi: 10.1186/1471-2164-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ogata S, Hayashi Y, Misumi Y, Ikehara Y. Membrane-anchoring domain of rat liver 5¢-nucleotidase: identification of the COOH-terminal serine-523 covalently attached with a glycolipid. Biochemistry. 1990;29:7923–7927. doi: 10.1021/bi00486a021. [DOI] [PubMed] [Google Scholar]
- 53.Misumi Y, Ogata S, Ohkubo K, Hirose S, Ikehara Y. Primary structure of human placental 5¢-nucleotidase and identification of the glycolipid anchor in the mature form. Eur J Biochem. 1990;191:563–569. doi: 10.1111/j.1432-1033.1990.tb19158.x. [DOI] [PubMed] [Google Scholar]
- 54.Cupp MS, Cupp EW, Ochoa AJ, Moulton JK. Salivary apyrase in New World blackflies (Diptera: Simuliidae) and its relationship to onchocerciasis vector status. Med Vet Entomol. 1995;9:325–330. doi: 10.1111/j.1365-2915.1995.tb00141.x. [DOI] [PubMed] [Google Scholar]
- 55.Baskova IP, Nikonov GI. Destabilase, the novel epsilon-(gamma-Glu)-Lys isopeptidase with thrombolytic activity. Blood Coagul Fibrinolysis. 1991;2:167–172. doi: 10.1097/00001721-199102000-00025. [DOI] [PubMed] [Google Scholar]
- 56.Zavalova LL, Artamonova II, Berezhnoy SN, Tagaev AA, Baskova IP, Andersen J, Roepstorff P, Egorov Ts A. Multiple forms of medicinal leech destabilase-lysozyme. Biochem Biophys Res Commun. 2003;306:318–323. doi: 10.1016/s0006-291x(03)00896-9. [DOI] [PubMed] [Google Scholar]
- 57.Zavalova LL, Baskova IP, Lukyanov SA, Sass AV, Snezhkov EV, Akopov SB, Artamonova II, Archipova VS, Nesmeyanov VA, Kozlov DG, Benevolensky SV, Kiseleva VI, Poverenny AM, Sverdlov ED. Destabilase from the medicinal leech is a representative of a novel family of lysozymes. Biochim Biophys Acta. 2000;1478:69–77. doi: 10.1016/s0167-4838(00)00006-6. [DOI] [PubMed] [Google Scholar]
- 58.Vija H, Samel M, Siigur E, Aaspollu A, Tonismagi K, Trummal K, Subbi J, Siigur J. VGD and MLD-motifs containing heterodimeric disintegrin viplebedin-2 from Vipera lebetina snake venom. Purification and cDNA cloning. Comp Biochem Physiol B Biochem Mol Biol. 2009;153:253–260. doi: 10.1016/j.cbpb.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 59.Calvete JJ, Moreno-Murciano MP, Theakston RD, Kisiel DG, Marcinkiewicz C. Snake venom disintegrins: novel dimeric disintegrins and structural diversification by disulphide bond engineering. Biochem J. 2003;372:725–734. doi: 10.1042/BJ20021739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ascenzi P, Bocedi A, Bolognesi M, Spallarossa A, Coletta M, De Cristofaro R, Menegatti E. The bovine basic pancreatic trypsin inhibitor (Kunitz inhibitor): a milestone protein. Curr Protein Pept Sci. 2003;4:231–251. doi: 10.2174/1389203033487180. [DOI] [PubMed] [Google Scholar]
- 61.Salier JP. Inter-alpha-trypsin inhibitor: emergence of a family within the Kunitz-type protease inhibitor superfamily. Trends Biochem Sci. 1990;15:435–439. doi: 10.1016/0968-0004(90)90282-g. [DOI] [PubMed] [Google Scholar]
- 62.Paesen GC, Siebold C, Dallas ML, Peers C, Harlos K, Nuttall PA, Nunn MA, Stuart DI, Esnouf RM. An ion-channel modulator from the saliva of the brown ear tick has a highly modified Kunitz/BPTI structure. J Mol Biol. 2009;389:734–747. doi: 10.1016/j.jmb.2009.04.045. [DOI] [PubMed] [Google Scholar]
- 63.Castaneda O, Harvey AL. Discovery and characterization of cnidarian peptide toxins that affect neuronal potassium ion channels. Toxicon. 2009;54:1119–1124. doi: 10.1016/j.toxicon.2009.02.032. [DOI] [PubMed] [Google Scholar]
- 64.Dy CY, Buczek P, Imperial JS, Bulaj G, Horvath MP. Structure of conkunitzin-S1, a neurotoxin and Kunitz-fold disulfide variant from cone snail. Acta Crystallogr D Biol Crystallogr. 2006;62:980–990. doi: 10.1107/S0907444906021123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lucchesi KJ, Moczydlowski E. On the interaction of bovine pancreatic trypsin inhibitor with maxi Ca(2+)-activated K+ channels. A model system for analysis of peptide-induced subconductance states. J Gen Physiol. 1991;97:1295–1319. doi: 10.1085/jgp.97.6.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mans BJ, Neitz AW. Adaptation of ticks to a blood-feeding environment: evolution from a functional perspective. Insect Biochem Mol Biol. 2004;34:1–17. doi: 10.1016/j.ibmb.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 67.Steen NA, Barker SC, Alewood PF. Proteins in the saliva of the Ixodida (ticks): pharmacological features and biological significance. Toxicon. 2006;47:1–20. doi: 10.1016/j.toxicon.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 68.Francischetti IM, Valenzuela JG, Andersen JF, Mather TN, Ribeiro JM. Ixolaris, a novel recombinant tissue factor pathway inhibitor (TFPI) from the salivary gland of the tick, Ixodes scapularis: identification of factor X and factor Xa as scaffolds for the inhibition of factor VIIa/tissue factor complex. Blood. 2002;99:3602–3612. doi: 10.1182/blood-2001-12-0237. [DOI] [PubMed] [Google Scholar]
- 69.Stark KR, James AA. Isolation and characterization of the gene encoding a novel factor Xa-directed anticoagulant from the yellow fever mosquito, Aedes aegypti. J Biol Chem. 1998;273:20802–20809. doi: 10.1074/jbc.273.33.20802. [DOI] [PubMed] [Google Scholar]
- 70.Valenzuela JG, Francischetti IM, Ribeiro JM. Purification, cloning, and synthesis of a novel salivary anti-thrombin from the mosquito Anopheles albimanus. Biochemistry. 1999;38:11209–11215. doi: 10.1021/bi990761i. [DOI] [PubMed] [Google Scholar]
- 71.Francischetti IM, Valenzuela JG, Ribeiro JM. Anophelin: kinetics and mechanism of thrombin inhibition. Biochemistry. 1999;38:16678–16685. doi: 10.1021/bi991231p. [DOI] [PubMed] [Google Scholar]
- 72.Campbell CL, Vandyke KA, Letchworth GJ, Drolet BS, Hanekamp T, Wilson WC. Midgut and salivary gland transcriptomes of the arbovirus vector Culicoides sonorensis (Diptera: Ceratopogonidae) Insect Mol Biol. 2005;14:121–136. doi: 10.1111/j.1365-2583.2004.00537.x. [DOI] [PubMed] [Google Scholar]
- 73.Andersen JF, Gudderra NP, Francischetti IM, Valenzuela JG, Ribeiro JM. Recognition of anionic phospholipid membranes by an antihemostatic protein from a blood-feeding insect. Biochemistry. 2004;43:6987–6994. doi: 10.1021/bi049655t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Buser CA, Kim J, McLaughlin S, Peitzsch RM. Does the binding of clusters of basic residues to acidic lipids induce domain formation in membranes? Mol Membr Biol. 1995;12:69–75. doi: 10.3109/09687689509038498. [DOI] [PubMed] [Google Scholar]
- 75.Kirszberg C, Lima LG, Da Silva de Oliveira A, Pickering W, Gray E, Barrowcliffe TW, Rumjanek VM, Monteiro RQ. Simultaneous tissue factor expression and phosphatidylserine exposure account for the highly procoagulant pattern of melanoma cell lines. Melanoma Res. 2009;19:301–308. doi: 10.1097/CMR.0b013e32832e40fe. [DOI] [PubMed] [Google Scholar]
- 76.Megraw T, Kaufman TC, Kovalick GE. Sequence and expression of Drosophila Antigen 5-related 2, a new member of the CAP gene family. Gene. 1998;222:297–304. doi: 10.1016/s0378-1119(98)00489-2. [DOI] [PubMed] [Google Scholar]
- 77.Milne TJ, Abbenante G, Tyndall JD, Halliday J, Lewis RJ. Isolation and characterization of a cone snail protease with homology to CRISP proteins of the pathogenesis-related protein superfamily. J Biol Chem. 2003;278:31105–31110. doi: 10.1074/jbc.M304843200. [DOI] [PubMed] [Google Scholar]
- 78.Yamazaki Y, Hyodo F, Morita T. Wide distribution of cysteine-rich secretory proteins in snake venoms: isolation and cloning of novel snake venom cysteine-rich secretory proteins. Arch Biochem Biophys. 2003;412:133–141. doi: 10.1016/s0003-9861(03)00028-6. [DOI] [PubMed] [Google Scholar]
- 79.Yamazaki Y, Morita T. Structure and function of snake venom cysteine-rich secretory proteins. Toxicon. 2004;44:227–231. doi: 10.1016/j.toxicon.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 80.Mochca-Morales J, Martin BM, Possani LD. Isolation and characterization of helothermine, a novel toxin from Heloderma horridum horridum (Mexican beaded lizard) venom. Toxicon. 1990;28:299–309. doi: 10.1016/0041-0101(90)90065-f. [DOI] [PubMed] [Google Scholar]
- 81.Xu X, Yang H, Ma D, Wu J, Wang Y, Song Y, Wang X, Lu Y, Yang J, Lai R. Toward an understanding of the molecular mechanism for successful blood feeding by coupling proteomics analysis with pharmacological testing of horsefly salivary glands. Mol Cell Proteomics. 2008;7:582–590. doi: 10.1074/mcp.M700497-MCP200. [DOI] [PubMed] [Google Scholar]
- 82.Scarborough RM, Naughton MA, Teng W, Rose JW, Phillips DR, Nannizzi L, Arfsten A, Campbell AM, Charo IF. Design of potent and specific integrin antagonists. Peptide antagonists with high specificity for glycoprotein IIb-IIIa. J Biol Chem. 1993;268:1066–1073. [PubMed] [Google Scholar]
- 83.Ameri M, Wang X, Wilkerson MJ, Kanost MR, Broce AB. An immunoglobulin binding protein (antigen 5) of the stable fly (Diptera: Muscidae) salivary gland stimulates bovine immune responses. J Med Entomol. 2008;45:94–101. doi: 10.1603/0022-2585(2008)45[94:aibpao]2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Valenzuela JG, Charlab R, Gonzalez EC, de Miranda-Santos IK, Marinotti O, Francischetti IM, Ribeiro JM. The D7 family of salivary proteins in blood sucking diptera. Insect Mol Biol. 2002;11:149–155. doi: 10.1046/j.1365-2583.2002.00319.x. [DOI] [PubMed] [Google Scholar]
- 85.Hekmat-Scafe DS, Dorit RL, Carlson JR. Molecular evolution of odorant-binding protein genes OS-E and OS-F in Drosophila. Genetics. 2000;155:117–127. doi: 10.1093/genetics/155.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mans BJ, Calvo E, Ribeiro JM, Andersen JF. The crystal structure of D7r4, a salivary biogenic amine-binding protein from the malaria mosquito Anopheles gambiae. J Biol Chem. 2007;282:36626–36633. doi: 10.1074/jbc.M706410200. [DOI] [PubMed] [Google Scholar]
- 87.Valenzuela JG, Charlab R, Gonzalez EC, Miranda-Santos IKF, Marinotti O, Francischetti IM, Ribeiro JMC. The D7 family of salivary proteins in blood sucking Diptera. Insect Mol Biol. 2002;11:149–155. doi: 10.1046/j.1365-2583.2002.00319.x. [DOI] [PubMed] [Google Scholar]
- 88.Calvo E, Mans BJ, Andersen JF, Ribeiro JM. Function and evolution of a mosquito salivary protein family. J Biol Chem. 2006;281:1935–1942. doi: 10.1074/jbc.M510359200. [DOI] [PubMed] [Google Scholar]
- 89.Calvo E, Mans BJ, Ribeiro JM, Andersen JF. Multifunctionality and mechanism of ligand binding in a mosquito antiinflammatory protein. Proc Natl Acad Sci USA. 2009;106:3728–3733. doi: 10.1073/pnas.0813190106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cupp MS, Cupp EW. Antithrombin Protein and DNA Sequences from Black Fly. USA: 2000. [Google Scholar]
- 91.Stevenson KJ, Poller L. The procoagulant activity of partial thromboplastin extracts: the role of phosphatidyl serine. Thromb Res. 1982;26:341–350. doi: 10.1016/0049-3848(82)90252-3. [DOI] [PubMed] [Google Scholar]
- 92.Simons FE, Peng Z. Mosquito allergy: recombinant mosquito salivary antigens for new diagnostic tests. Int Arch Allergy Immunol. 2001;124:403–405. doi: 10.1159/000053771. [DOI] [PubMed] [Google Scholar]
- 93.Calvo E, Tokumasu F, Marinotti O, Villeval JL, Ribeiro JM, Francischetti IM. Aegyptin, a novel mosquito salivary gland protein, specifically binds to collagen and prevents its interaction with platelet glycoprotein VI, integrin alpha2beta1, and von Willebrand factor. J Biol Chem. 2007;282:26928–26938. doi: 10.1074/jbc.M705669200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yoshida S, Sudo T, Niimi M, Tao L, Sun B, Kambayashi J, Watanabe H, Luo E, Matsuoka H. Inhibition of collagen-induced platelet aggregation by anopheline antiplatelet protein, a saliva protein from a malaria vector mosquito. Blood. 2008;111:2007–2014. doi: 10.1182/blood-2007-06-097824. [DOI] [PubMed] [Google Scholar]
- 95.Grimaldi D, Engel M. Evolution of the Insects. New York: Cambridge University Press; 2005. [Google Scholar]
- 96.Cupp MS, Ribeiro JM, Cupp EW. Vasodilative activity in black fly salivary glands. Am J Trop Med Hyg. 1994;50:241–246. doi: 10.4269/ajtmh.1994.50.241. [DOI] [PubMed] [Google Scholar]
- 97.Cupp MS, Ribeiro JM, Champagne DE, Cupp EW. Analyses of cDNA and recombinant protein for a potent vasoactive protein in saliva of a blood-feeding black fly, Simulium vittatum. J Exp Biol. 1998;201:1553–1561. doi: 10.1242/jeb.201.10.1553. [DOI] [PubMed] [Google Scholar]
- 98.Pinnell SR. Regulation of collagen biosynthesis by ascorbic acid: a review. Yale J Biol Med. 1985;58:553–559. [PMC free article] [PubMed] [Google Scholar]
- 99.Rydengard V, Andersson Nordahl E, Schmidtchen A. Zinc potentiates the antibacterial effects of histidine-rich peptides against Enterococcus faecalis. FEBS J. 2006;273:2399–2406. doi: 10.1111/j.1742-4658.2006.05246.x. [DOI] [PubMed] [Google Scholar]
- 100.Nishikawa M, Ogawa K. Antimicrobial activity of a chelatable poly(arginyl-histidine) produced by the ergot fungus Verticillium kibiense. Antimicrob Agents Chemother. 2004;48:229–235. doi: 10.1128/AAC.48.1.229-235.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Loomans HJ, Hahn BL, Li QQ, Phadnis SH, Sohnle PG. Histidine-based zinc-binding sequences and the antimicrobial activity of calprotectin. J Infect Dis. 1998;177:812–814. doi: 10.1086/517816. [DOI] [PubMed] [Google Scholar]
- 102.Tanaka H, Chiba H, Inokoshi J, Kuno A, Sugai T, Takahashi A, Ito Y, Tsunoda M, Suzuki K, Takenaka A, Sekiguchi T, Umeyama H, Hirabayashi J, Omura S. Mechanism by which the lectin actinohivin blocks HIV infection of target cells. Proc Natl Acad Sci USA. 2009;106:15633–15638. doi: 10.1073/pnas.0907572106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Femiano F. Pemphigus vulgaris: recent advances in our understanding of its pathogenesis. Minerva Stomatol. 2007;56:215–223. [PubMed] [Google Scholar]
- 104.Grando SA. Cholinergic control of epidermal cohesion. Exp Dermatol. 2006;15:265–282. doi: 10.1111/j.0906-6705.2006.00410.x. [DOI] [PubMed] [Google Scholar]
- 105.Bazzoni G. The JAM family of junctional adhesion molecules. Curr Opin Cell Biol. 2003;15:525–530. doi: 10.1016/s0955-0674(03)00104-2. [DOI] [PubMed] [Google Scholar]


