Abstract
Profiles of Burkholderia pseudomallei persistence and antibodies in blood, spleen, liver, and lungs of infected BALB/c mice were investigated. Animals were infected intraperitoneally with low (6 colony-forming units) or high (230 colony-forming units) doses of B. pseudomallei. In the high-dose infected group, bacteria were found by culture in 100% of blood, liver, spleen and lung samples at 24 and 48 hours after infections; blood samples were 100% positive by polymerase chain reaction after 60 hours. Antibody responses in the high-dose infected group were low. These responses were detected in the low-dose infected group after 5 days, peaked at 7–14 days, and showed persistence until 28 days post-infection. Bacterial loads and antibody profiles varied according to the level of bacterial infections. This kinetic study, although in animals, provides crucial knowledge that might be useful for the development of a sensitive and specific diagnostic assay for patients with melioidosis.
Burkholderia pseudomallei is a facultative, intracellular, gram-negative bacteria. It causes melioidosis, which is found in humans and animals. The disease is most common in tropical regions, especially in Southeast Asia and northern Australia,1 where it is a major cause of community acquired septicemia and acute pneumonia. In northeastern Thailand, B. pseudomallei is a major cause of morbidity and mortality and causes one-fifth of all community-acquired septicemias.2 Infection is acquired by contact with contaminated soil or water when exposed to a skin abrasion or inhalation of B. pseudomallei from environmental sources.3 The range of clinical presentations and the potential for asymptomatic infection depend on the differences in the route of inoculation, inoculum size, virulence of the infecting strains, and immune competence and genetic pre-disposition of the host. Moreover, clinical symptoms of the disease resemble several diseases such as tuberculosis and leptospirosis. Therefore, rapid and specific diagnosis is crucial for this disease.
Antibiotic therapy is complicated by antibiotic resistance of some clinical isolates, frequently resulting in relapse or reactivation decades later.3 One of the major limitations of antibiotic therapy for B. pseudomallei infection is the lack of knowledge on the persistence of the bacterial cells and antibodies in blood samples that could be used to monitor the effectiveness of treatment in eradication of the organism. This knowledge would be of great importance in evaluating the progression of melioidosis. Furthermore, current laboratory diagnosis of melioidosis still depends upon culture as the gold standard. Although serologic and molecular biological methods have been developed, the sensitivity and specificity of methods varied from one laboratory to another, and none of the methods developed gave satisfactory results when compared with culture.4,5 Moreover, these methods are not rapid enough for diagnosis of septic melioidosis, which has a high mortality rate.
One factor that makes all tests unsatisfactory is the unknown time and quantities of B. pseudomallei cells and antibodies present in the blood of patients. Several questions concerning the immune responses and the antigens in the circulation after exposure to the organisms are still controversial. These include 1) how long B. pseudomallei persists in blood, 2) when the antibody response occurs, and 3) how long it persists. For antigen and antibody detections to be useful in the diagnosis of melioidosis, the kinetics of antibodies and antigens should be investigated.
In this study, BALB/c mice were injected intraperitoneally with either low (0.3 50% lethal dose [LD50]) or high (12 LD50) infective doses of B. pseudomallei strain A2 isolated from a septic patient. The protocol was reviewed and approved by the animal ethics committee of the Faculty of Medicine at Khon Kaen University. Kinetic growth of B. pseudomallei strain A2 and its DNA in the blood of BALB/c mice was determined by conventional culture and then identified by biochemical tests, immunoreactivity with a monoclonal antibody,6 and a polymerase chain reaction (PCR). The specific antibody responses were measured in plasma of BALB/c mice by an enzyme-linked immunosorbent assay (ELISA) using culture-filtered antigens.7
The PCR amplification of B. pseudomallei DNA was performed using a DNA thermal cycler machine (2400; Perkin-Elmer, Norwalk, CT). The primers used were wcbG-for (5′-AACGAGTCGGTCATTTCCCTGA-3′) and wcbG-rev (5′-CCGATATTGCCGACTTCCACTGTGAT-3′), which amplified a 323-basepair fragment of B. pseudomallei capsule gene.8 The reaction was carried out in a total volume of 50 μL containing 5 μL of 10× PCR buffer (20 mM Tris-HCl, pH 8.4, 50 mM KCl), 2.5 μL of deoxynucleoside triphosphates (1 mM each), 2.5 μL of each primer (10 μM each), 5 μL of sample, and 5 units of Taq DNA polymerase. The template DNA was initially denatured at 94°C for 5 minutes. The amplification procedure was 40 cycles at 94°C for 30 seconds (denaturation), 60°C for 30 seconds (annealing), and 72°C for 45 seconds (extension).
Blood samples were processed as described.9 Briefly, 100-µL blood samples used for PCR were centrifuged (12,000 × g for 5 minutes). After the serum was removed, erythrocytes were lysed by addition of 200 µL of sterile distilled water, vortexed and centrifuged at 12,000 × g for 5 minutes and repeated three times to completely remove hemoglobin. Pellets were washed twice with TE buffer (10 mM Tris-HCl, 10 mM EDTA, pH 8.0), re-suspended in 10 μL of TE buffer, and boiled for 10 minutes; 1 μL was then used in the PCR.
An overnight culture of B. pseudomallei K96243 in Luria Bertani broth was used for genomic DNA extraction by digestion with proteinase K and extraction with phenol-chloroform;10 2 pg/μL was used as a positive control. To study the growth kinetics of B. pseudomallei and specific antibody responses at the high infective dose, 40 BALB/c mice were infected with 230 colony-forming units (CFU) (12 LD50) of B. pseudomallei. The mice were divided into five groups of eight mice/group. Groups 1–5 were infected with B. pseudomallei, and their blood and organs were collected after 12, 24, 48, 60, and 72 hours of infection. Phosphate-buffered saline (PBS) was used for injection in a control group of six mice. In the low-dose experiment, 15 BALB/c mice were infected with 6 CFU (0.3 LD50) of B. pseudomallei. Fifteen other mice were injected with PBS and used as controls. Heparinized blood was aseptically collected from the tail veins of infected mice on days 1, 3, 5, 7, 14, 21, and 28 post-infection. All samples were used to determine the kinetics of specific antibodies by ELISA and the presence of B. pseudomallei in blood by a plate count technique and PCR.
Results for control mice that received PBS showed no detectable bacteremia. Therefore, data are not shown. In the high infection dose group, bacteria were detected, even in a low number (mean ± SE = 8 ± 8 CFU/mL), in blood and organs by culture starting at 12 hours after infection (Figure 1A). Bacteremia increased and peaked at 48–60 hours post-infection; high bacterial counts were found (mean ± SE = 1.1 ± 0.85 × 104 CFU/mL) (Figure 1A). Bacteria were found in all organs tested, but were present at a higher level and for a longer time in spleens. Within 24 hours after infection, 100% of mice were positive for the bacteria in blood, spleen, and liver (Figure 1B). The PCR showed that 20% of blood samples were positive for B. pseudomallei as early as 12 hours after infection. However, bacteria were not detected in any organs (Figure 1C). At 24 hours after infection, 40% of blood samples and 80% of spleen samples were positive, and 100% were positive at 60 hours and 48 hours post-infection, respectively (Figure 1C). Therefore, for 100% detection, the culture method was found to be more sensitive because it detected bacteria 36 hours earlier than the PCR (Figure 1).
Figure 1.
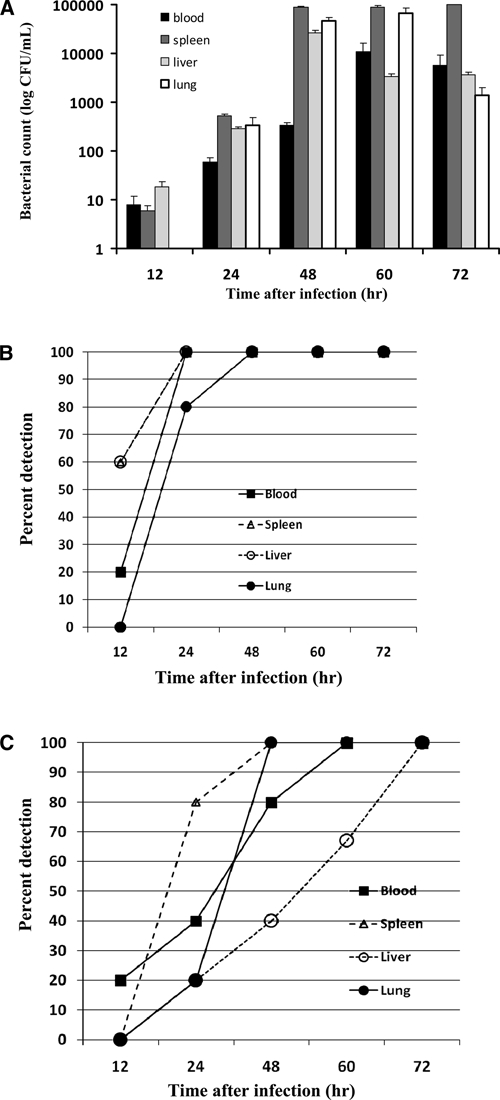
Kinetics of bacterial loads in blood and organs of high-dose Burkholderia pseudomallei-infected mice. Forty BALB/c mice were infected intraperitoneally with 12 50% lethal doses (230 colony-forming units) of B. pseudomallei A2. Mice were divided into 5 groups (8 mice/group). Groups 1–5 were infected with B. pseudomallei and their blood (black box), spleen (dark gray box), liver (light gray box), and lungs (white box) were collected after 12, 24, 48, 60, and 72 hours after infection. Mice were monitored for bacterial load by culture on Ashdown's selective medium and identified by using monoclonal antibodies (A). Each bar represents the logarithm of the mean ± SE colony-forming units. Control mice are not shown because no bacteria were detected in their blood samples. Percentage of positive results in blood and organs as detected by culture (B) or polymerase chain reaction (C) at various times after infections were plotted against time. Results were obtained from two independent experiments.
The sensitivity of wcbG-for/wcbG-rev primers specific for a capsule gene of B. pseudomallei was evaluated by a 10-fold serial dilution of the B. pseudomallei genomic DNA ranging from 320 ng to 3.2 fg. Sensitivity was found to be at 32 fg. When this PCR was evaluated in blood spiked with live bacteria or bacterial DNA, the lower limit of detection was 5 CFU or 1 pg. This capsular gene (wcbG; BPSL2801) coded for 313 amino acids and was found by BLAST and comparative genomics to be unique for B. pseudomallei and B. mallei, a closely related obligate parasite that is not found in Thailand. It is present in a single copy on B. pseudomallei chromosome I of 668, K96243, MHSR346, 1710b, and 1106a sequenced strains.
The specificity of the primers was evaluated by PCRs with 12 other bacteria (Escherichia coli, Klebsiella pneumonia, Enterobacter sp., Salmonella typhi, Shigella dysenteriae, Pseudomonas aeruginosa, Staphyloccocus aureus, Group A and B β-hemolytic Streptococcus, Proteus mirabilis, B. mallei, and a closely related non-pathogenic, B. thailandensis). The PCR results indicated that the primers were specific for B. pseudomallei because they did not show amplifications with other bacteria except B. mallei, as predicted. The wcbG putative capsular polysaccharide biosynthesis gene was demonstrated to be related to the virulence of the bacteria in infected hamsters8 and is found only in B. pseudomallei and B. mallei but not in B. thailandensis.11 Therefore, detection of this gene can reflect active melioidosis.
For ELISA detection, the level of specific antibodies detected in mice within 72 hours of infection was low and not different from the control because there was no antibody production (Figure 2A). Therefore, in acute melioidosis infection, the suitable methods for diagnosis are culture or PCR. Antibody detection was an inferior method, especially for primary infection. Because bacteria were found in the blood for a short period (7–10 days),12 culture and PCR results might be false-negative results when samples were taken after 10 days of infection. Therefore, sample collection times are important in making decisions as to what kind of tests should be used. This finding is the major reason why laboratory diagnosis of melioidosis is still not satisfactory.
Figure 2.
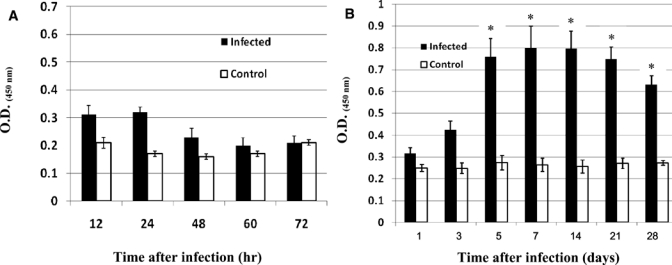
Enzyme-linked immunosorbent assay (ELISA) analysis of specific antibody responses in low-and high-dose Burkholderia pseudomallei-infected mice. BALB/c mice were infected with 12 50% lethal doses (LD50) (230 colony-forming units) (A) or 0.3 LD50 (6 colony-forming units) (B) of B. pseudomallei by intraperitoneal injection. Plasma samples on hours or days indicated were collected and specific antibody to culture-filtered antigens were measured by ELISA. Results are expressed as mean ± SE (optical density at 450 nm). Mice injected with phosphate-buffered saline were used as control. Asterisks represent statistically significant differences (P < 0.05) compared with controls. Results were obtained from two independent experiments.
When animals were infected with a low number of bacteria (0.3 LD50), blood culture was positive only on days 1, 3 and 5 after infection (6.7%, 20%, and 6.7%, respectively) (Table 1), and all PCR results were negative. Five mice positive for bacteremia died (1 on days 3 and 7, and 3 on day 5 after infection) (Table 1). This finding may reflect acute infections. Therefore, results for these animals were excluded from analysis. Chronic infections developed in 10 other mice; these mice survived more than one 1 month. This observation was confirmed by positive cultures of organs from all five mice. In addition, five other mice showed positive blood cultures when injected with antibodies against interferon-γ (Srisurat N, unpublished data). Antibody detection was found to be useful because its results were significantly different from those of controls, especially after 5 days of infection (Figure 2B). However, antibody detection was dependent on the infective dose.
Table 1.
Bacteremia in 15 BALB/c mice infected with low-dose (0.3 LD50) used to produce a chronic infection by Burkholderia pseudomallei*
| Time | Bacteremia (CFU/mL) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | |
| Day 1 | NG | NG | NG | NG | NG | NG | NG | 1.5 × 102 | NG | 10 | NG | NG | NG | NG | NG |
| Day 3 | NG | NG | NG | NG | 4.5 × 103 | NG | NG | Died | NG | 10 | 10 | NG | NG | NG | NG |
| Day 5 | NG | NG | 20 | NG | Died | NG | NG | NG | Died | Died | NG | NG | NG | NG | |
| Day 7 | NG | NG | Died | NG | NG | NG | NG | NG | NG | NG | NG | ||||
| Day 14 | NG | NG | NG | NG | NG | NG | NG | NG | NG | NG | |||||
| Day 21 | NG | NG | NG | NG | NG | NG | NG | NG | NG | NG | |||||
| Day 28 | NG | NG | NG | NG | NG | NG | NG | NG | NG | NG | |||||
LD50 = 50% lethal dose, CFU colony-forming units; NG = no growth.
Our results demonstrated that culture and PCR were useful in detecting acute (usually with a moderate to high dose of infection) infections and providing an early diagnosis, and antibody detection was useful in detecting sub-acute or chronic infections. The time of specimen collections was important for test selection. Results of this study, although from an animal model, might be used as guidelines for developing sensitive and specific laboratory diagnostic protocols to monitor progression of melioidosis, leading to better treatment for patients with this disease.
Acknowledgments
We thank Emeritus Professor James A Will (University of Wisconsin-Madison) for editing the manuscript and Dr. Apichai Tuanyok (Center for Microbial Genetics and Genomics, Northern Arizona University, Flagstaff, AZ) for designing the wcbG primer.
Footnotes
Financial support: This study was supported by the Commission on Higher Education, Thailand (project CHE-RG), and Melioidosis Research Center of Khon Kaen University.
Authors' addresses: Nuttiya Srisurat, Unchalee Tatawasart, and Surasakdi Wongratanacheewin, Department of Microbiology and Melioidosis Research Center, Faculty of Medicine, Khon Kaen University, Khon Kaen, Thailand, E-mails: nsrisurat@yahoo.com, unchalee@kku.ac.th, and sura_wng@kku.ac.th. Rasana W. Sermswan, Department of Biochemistry and Melioidosis Research Center, Faculty of Medicine, Khon Kaen University, Khon Kaen, Thailand, E-mail: rasana@kku.ac.th.
References
- 1.Cheng AC, Currie BJ. Melioidosis: epidemiology, pathophysiology, and management. Clin Microbiol Rev. 2005;18:383–416. doi: 10.1128/CMR.18.2.383-416.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaowagul W, White NJ, Dance DA, Wattanagoon Y, Naigowit P, Davis TM, Looareesuwan S, Pitakwatchara N. Melioidosis: a major cause of community-acquired septicemia in northeastern Thailand. J Infect Dis. 1989;159:890–899. doi: 10.1093/infdis/159.5.890. [DOI] [PubMed] [Google Scholar]
- 3.White NJ. Melioidosis. Lancet. 2003;361:1715–1722. doi: 10.1016/s0140-6736(03)13374-0. [DOI] [PubMed] [Google Scholar]
- 4.Sirisinha S, Anuntagool N, Dharakul T, Ekpo P, Wongratanacheewin S, Naigowit P, Petchclai B, Thamlikitkul V, Suputtamongkol Y. Recent developments in laboratory diagnosis of melioidosis. Acta Trop. 2000;74:235–245. doi: 10.1016/s0001-706x(99)00076-5. [DOI] [PubMed] [Google Scholar]
- 5.Kunakorn M, Raksakait K, Sethaudom C, Sermswan RW, Dharakul T. Comparison of three PCR primer sets for diagnosis of septicemic melioidosis. Acta Trop. 2000;74:247–251. doi: 10.1016/s0001-706x(99)00077-7. [DOI] [PubMed] [Google Scholar]
- 6.Anuntagool N, Naigowit P, Petkanchanapong V, Aramsri P, Panichakul T, Sirisinha S. Monoclonal antibody-based rapid identification of Burkholderia pseudomallei in blood culture fluid from patients with community-acquired septicaemia. J Med Microbiol. 2000;49:1075–1078. doi: 10.1099/0022-1317-49-12-1075. [DOI] [PubMed] [Google Scholar]
- 7.Sermswan RW, Wongratanacheewin S, Anuntagool N, Sirisinha S. Comparison of the polymerase chain reaction and serologic tests for diagnosis of septicemic melioidosis. Am J Trop Med Hyg. 2000;63:146–149. doi: 10.4269/ajtmh.2000.63.146. [DOI] [PubMed] [Google Scholar]
- 8.Reckseidler-Zenteno SL, DeVinney R, Woods DE. The capsular polysaccharide of Burkholderia pseudomallei contributes to survival in serum by reducing complement factor C3b deposition. Infect Immun. 2005;73:1106–1115. doi: 10.1128/IAI.73.2.1106-1115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rattanathongkom A, Sermswan RW, Wongratanacheewin S. Detection of Burkholderia pseudomallei in blood samples using polymerase chain reaction. Mol Cell Probes. 1997;11:25–31. doi: 10.1006/mcpr.1996.0072. [DOI] [PubMed] [Google Scholar]
- 10.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 11.Kim HS, Schell MA, Yu Y, Ulrich RL, Sarria SH, Nierman WC, DeShazer D. Bacterial genome adaptation to niches: divergence of the potential virulence genes in three Burkholderia species of different survival strategies. BMC Genomics. 2005;6:174. doi: 10.1186/1471-2164-6-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wongratanacheewin S, Kespichayawattana W, Intachote P, Pichyangkul S, Sermswan RW, Krieg AM, Sirisinha S. Immunostimulatory CpG oligodeoxynucleotide confers protection in a murine model of infection with Burkholderia pseudomallei. Infect Immun. 2004;72:4494–4502. doi: 10.1128/IAI.72.8.4494-4502.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


