Abstract
The immunoreactivity of EchiTAb-Plus-ICP, an antivenom developed for the treatment of snakebite envenoming in sub-Saharan Africa, to venoms of seven Echis and Bitis species, was assessed by “antivenomics.” This proteomic approach is based on the ability of an antivenom to immunodeplete homologous or heterologous venom proteins. Our results show an extensive cross-reactivity of this antivenom against all Echis and Bitis venoms studied, as revealed by the complete immunodepletion of the majority of venom components, including metalloproteinases, serine proteinases, C-type lectin-like proteins, some phospholipases A2 and L-amino acid oxidase. However, some phospholipases A2, disintegrins and proteinase inhibitors were immunodepleted to only a partial extent. These results support the hypothesis that immunizing horses with a mixture of the venoms of Echis ocellatus, Bitis arietans, and Naja nigricollis generates antibodies capable of recognizing the majority of components of medically-relevant homologous and heterologous viperid venoms of the genera Bitis and Echis from sub-Saharan Africa.
Introduction
Snake envenoming represents a highly neglected public health problem in sub-Saharan Africa, where the number of snakebite cases have been estimated to be as high as 1,000,0001 and annual fatalities may range between 3,500 and 32,000.2 In addition, unknown numbers of patients, as many as 36% in one community-based study,3 are left with permanent physical or psychological sequelae, an undocumented and largely forgotten aspect of this pathology.4–6
Animal-derived antivenom constitutes the only validated therapy for snakebite envenoming.6–9 However, there is a current crisis in antivenom supply to sub-Saharan Africa, because of multiple causes that include lack of commercial incentives for manufacturers, deficient purchasing systems, ignorance of true antivenom requirements, high costs of some available products, loss of confidence of antivenom therapeutic efficacy, and safety caused by the marketing of ineffective products and inadequate regulatory systems.5,6,10–13
The seriousness of this problem has prompted a number of initiatives, fostered by the World Health Organization (WHO), to confront this serious health issue.6,7,11,14,15 Several manufacturers have responded developing antivenoms for sub-Saharan Africa. Thus, in addition to laboratories traditionally producing antivenoms for Africa, such as EgyVac (Egypt), Sanofi-Pasteur (France), and South African Vaccine Producers (South Africa),16 other manufacturers have recently developed new antivenoms for this region, e.g., MicroPharm (UK),17 Instituto Bioclon (Mexico),11,18 Instituto Clodomiro Picado (Costa Rica),19,20 and Instituto Butantan (Brazil) (Dias-da-Silva W, personal communication). However, there was a large heterogeneity in the design and composition of the venoms used in the immunization mixtures to prepare the above antivenoms, an issue complicated by the complexity of sub-Saharan herpetofauna and by the diversity of African snake venom proteomes (“venoms”), including intraspecies venom variability in those species with a wide geographical distribution.21,22 Thus, the selection of venom mixtures appropriate for raising an immune response with wide cross-reactivity against many snake venoms in sub-Saharan Africa is an important task that should be approached initially through a rigorous analysis of the cross-reactivity of antivenoms against the medically most important snake venoms from this region. In the end, however, antivenom safety and efficacy have to be demonstrated in clinical trials.
The study of cross-neutralization of venoms by antivenoms is classically performed, at the preclinical level, by assessing the ability of a particular antivenom to neutralize the most important and clinically relevant toxicological activities of snake venoms using standard laboratory tests in experimental animals.7,23–26 In the case of viperid snake venoms, which inflict the highest toll of envenoming in sub-Saharan Africa,4 preclinical analysis of the neutralizing efficacy of antivenoms should include the neutralization of lethal, hemorrhagic, coagulant, defibrinogenating, and necrotising effects. In the case of EchiTAb-Plus-ICP antivenom, produced by immunizing horses with a mixture of the venoms of Echis ocellatus, Bitis arietans, and Naja nigricollis from Nigeria,19,20 preclinical analyses have already showed its effectiveness in the neutralization not only of these three venoms,19 but also of the venoms of other Echis saw-scaled viper species (Echis leucogaster, Echis pyramidum leakeyi) and other Bitis viper species (Bitis gabonica, Bitis rhinoceros, and Bitis nasicornis).20 Moreover, this antivenom proved clinically effective and safe in a powerful trial performed in Nigeria in patients systemically envenomed by E. ocellatus.27
In addition to preclinical neutralization tests, a complementary approach to assess the immunoreactivity of antivenoms against individual venom components has been recently developed. It is known as “antivenomics” and is based on the application of proteomic analytical tools for studying venoms and antivenoms.21,28,29 Venom and antivenom are incubated together and antivenom-venom protein complexes are then immunoprecipitated by the addition of anti-equine IgG. The supernatant, which contains the non-immunoprecipitated venom components, is analyzed by proteomic techniques and its composition is compared with the chromatographic pattern of the native venom. In this way, venom components escaping immunodepletion by the antivenom can be identified easily and unambiguously. In this work, we have applied an antivenomic approach to assess the cross-reactivity of EchiTAb-Plus-ICP toward the venoms of seven species of sub-Saharan snakes of the family Viperidae. Results provide evidence of a high degree of cross-reactivity of this antivenom against all venoms tested, but also identified a small number of venom proteins that were only partially immunodepleted by the antivenom.
Materials and Methods
Venoms and antivenom.
The venoms of the following viperid snakes were used: Echis ocellatus (Nigeria), E. leucogaster (Mali), E. pyramidum leakeyi (Kenya), Bitis arietans arietans (from Ghana and Nigeria), B. gabonica gabonica, B. rhinoceros, and B. nasicornis. The venom of B. rhinoceros was a gift from César Olmos Jiménez (Entomo Zoo Fauna Arcana, S.L., Cullera, Valencia, Spain), and the venoms of B. gabonica gabonica and B. nasicornis were obtained from Latoxan (Valence, France). The other venoms were from specimens kept at the herpetarium of the Liverpool School of Tropical Medicine, and correspond to venoms pooled from several adult specimens. All venoms were lyophilized and stored at −20°C until used.
The polyspecific EchisTAb-Plus-ICP antivenom was manufactured by caprylic acid fractionation of the plasma of four horses that had been immunized with a mixture (at a weight ratio of 1:1:1.33) of the venoms of Echis ocellatus, Bitis arietans, and Naja nigricollis collected from Nigeria.19 The particular antivenom batch used in this preclinical study (Batch 4260308PALQ) was formulated to have the following composition: protein concentration 69.6 g/L, sodium chloride 7.6 g/L, phenol 1.86 g/L, and pH 6.78. The antivenom batch passed all the quality control requirements at the Quality Control Laboratory of Instituto Clodomiro Picado.
Antivenomics: Immunodepletion of venom proteins by the polyspecific EchiTAb-Plus-ICP antivenom.
We have coined the term “antivenomics” to describe our proteomic protocol for identifying venom proteins bearing epitopes reactive with an antivenom.21,28–32 Briefly, two milligrams of whole venom were dissolved in 70 μL of 20 mM phosphate buffer, pH 7.0, mixed with 4 mg of purified polyvalent antivenom IgGs, and incubated with gentle stirring for 1 hr at 37°C. The IgG concentration was determined spectrophotometrically using an extinction coefficient (ɛ) of 1.4 for a 1 mg/mL IgG concentration at 280 nm using a 1 cm light pathlength cuvette.33 Thereafter, 6 mg of rabbit anti-horse IgG antiserum (Sigma, Tres Cantos, Madrid, Spain) in 350 μL of 20 mM phosphate buffer, pH 7.0, were added, and the mixture was incubated for another 1 hr at 37°C. Immunocomplexes were precipitated by centrifugation at 13,000 rpm for 30 min in an Eppendorf centrifuge (Eppendorf Ibérica, Madrid, Spain) and the supernatant was submitted to reverse-phase separation.
Isolation and characterization of venom proteins.
For reverse-phase high-performance liquid chromatography (HPLC) separations, 2 mg of crude, lyophilized venom or its immunodepleted fraction were separated using an ETTAN LC HPLC system (GE Healthcare, Uppsala, Sweden) and a Lichrosphere RP100 C18 column (250 mm × 4 mm, 5 μm particle size) eluted at 1 mL/min with a linear gradient of 0.1% TFA in water (solution A) and acetonitrile (solution B) (5%B for 10 min, followed by 5–15%B over 20 min, 15–45%B over 120 min, and 45–70%B over 20 min). Protein detection was at 215 nm and peaks were collected manually and dried in a Speed-Vac (Savant, Thermo Electron, Madrid, Spain). Isolated protein fractions were subjected to N-terminal sequence analysis (using a Procise instrument, Applied Biosystems, Foster City, CA) following the manufacturer's instructions. Amino acid sequence similarity searches were performed against the available databanks using the BLAST program34 implemented in the WU-BLAST2 search engine at http://www.bork.embl-heidelberg.de. The molecular masses of the purified proteins were determined by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (on 12% or 15% polyacrylamide gels), and by electrospray ionization (ESI) mass spectrometry using an Applied Biosystems QTrap 2000 mass spectrometer35 operated in Enhanced Multiple Charge mode in the range m/z 600–1,700.
Proteins exhibiting a blocked N-terminus were excised from Coomassie Brilliant Blue-stained SDS-PAGE gels and subjected to automated reduction with DTT and alkylation with iodoacetamide, and in-gel digestion with sequencing grade porcine pancreas trypsin (Promega, Madrid, Spain) using a ProGest digestor (Genomic Solutions Inc., Cambridgeshire, United Kingdom) following the manufacturer's instructions. The 0.65 μL of the tryptic peptide mixtures (from a total volume of ~20 μL) were spotted onto a MALDI-TOF sample holder, mixed with an equal volume of a saturated solution of α-cyano-4-hydroxycinnamic acid (Sigma) in 70% acetonitrile containing 0.1% TFA, air-dried, and analyzed with an Applied Biosystems Voyager-DE Pro MALDI-TOF mass spectrometer, operated in delayed extraction and reflector modes.
For peptide sequencing, the protein digest mixture was loaded onto a nanospray capillary column and subjected to ESI mass spectrometric analysis using a QTrap 2000 mass spectrometer (Applied Biosystems)35 equipped with a nanospray source (Protana, Denmark). Doubly- or triply-charged ions of selected peptides from the MALDI-TOF mass fingerprint spectra were analyzed in Enhanced Resolution MS mode and the monoisotopic ions were fragmented using the Enhanced Product Ion tool with Q0 trapping. Enhanced Resolution was performed at 250 amu/s across the entire mass range. Settings for MS/MS experiments were as follows: Q1- unit resolution; Q1-to-Q2 collision energy −30–40 eV; Q3 entry barrier −8 V; LIT (linear ion trap) Q3 fill time −250 ms; and Q3 scan rate –1,000 amu/s. Collision-induced dissociation (CID) spectra were interpreted manually or using a licensed version of the MASCOT program (http://www.matrixscience.com) against a private database containing 1,083 viperid protein sequences deposited in the SwissProt/TrEMBL database (UniProtKB/Swiss-Prot Release 56.7 of 20-Jan-2009; http://us.expasy.org/sprot/) plus the previously assigned peptide ion sequences from snake venomics projects carried out in our laboratory.21,28–30,31,36–43 MS/MS mass tolerance was set to ±0.6 Da. Carbamidomethyl cysteine and oxidation of methionine were fixed and variable modifications, respectively.
Results
Figures 1 and 2 display reverse-phase HPLC separation patterns of the venoms sampled, along with HPLC chromatograms of the supernatants obtained after subjecting each venom to immunoprecipitation by the EchiTAb-Plus-ICP antivenom. Detailed proteomic characterization of the venoms of E. ocellatus, B. arietans, B. gabonica, B. rhinoceros, and B. nasicornis has been reported previously.21,31,43 According to their antivenomic immunoreactivity profile, venom proteins were classified as C-toxins (completely immunodepleted toxins), P-toxins (partly immunodepleted toxins), and N-toxins (non-immunodepleted toxins).39 Our results show that the vast majority of venom components, in all venoms tested, are C-toxins (Figures 1 and 2). Based on data accrued from previous proteomic analyses of these venoms, these C-toxins comprise mainly P-I and P-III snake venom metalloproteinases (SVMPs), serine proteinases, C-type lectin-like proteins, and some phospholipases A2 (PLA2s), as well as minor components such as L-amino acid oxidase (LAAO) and cysteine-ruch secretory proteins (CRISPs).21,31,43 Only a small number of Echis and Bitis venom components corresponded to P-toxins (Figures 1 and 2). These proteins were identified by proteomic analysis to be primarily PLA2s, disintegrins and proteinase inhibitors. There were no toxins identified as N-toxins, i.e., not being recognized by the antivenom at all (Figures 1 and 2).
Figure 1.
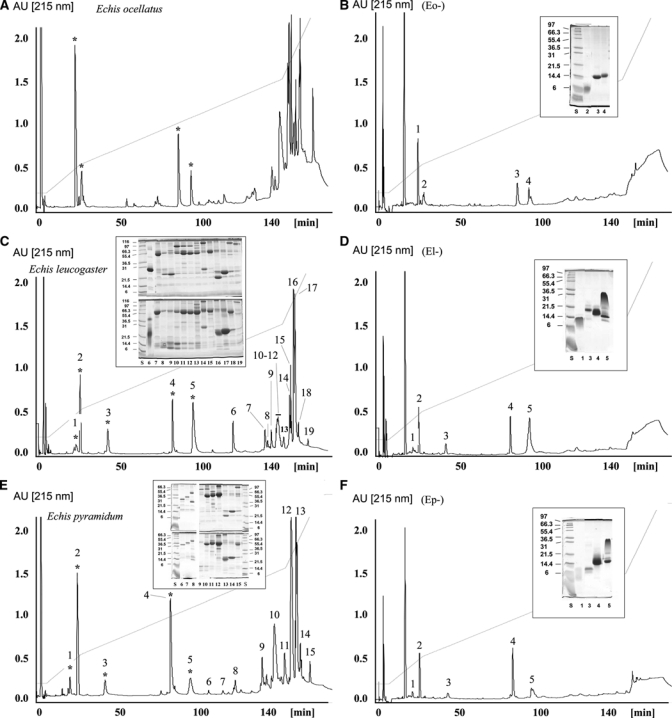
Immunodepletion of venom proteins from Echis taxa by the polyspecific EchiTAb-Plus-ICP antivenom. Panels B, D, and F show, respectively, reverse-phase separations of the proteins recovered after incubation of the crude venoms of (A) E. ocellatus, (C) E. leucogaster, and (E) E. pyramidum leakeyi with the polyspecific EchiTAb-Plus-ICP antivenom, followed by rabbit anti-horse IgG antiserum and immunoprecipitation. The inset shows SDS-PAGE analyses of β-mercaptoethanol-reduced fractions labeled as in the respective chromatograms. Molecular mass markers (in kDa) are indicated at the side of each gel. Protein fraction numbering is as in Table 1.
Figure 2.
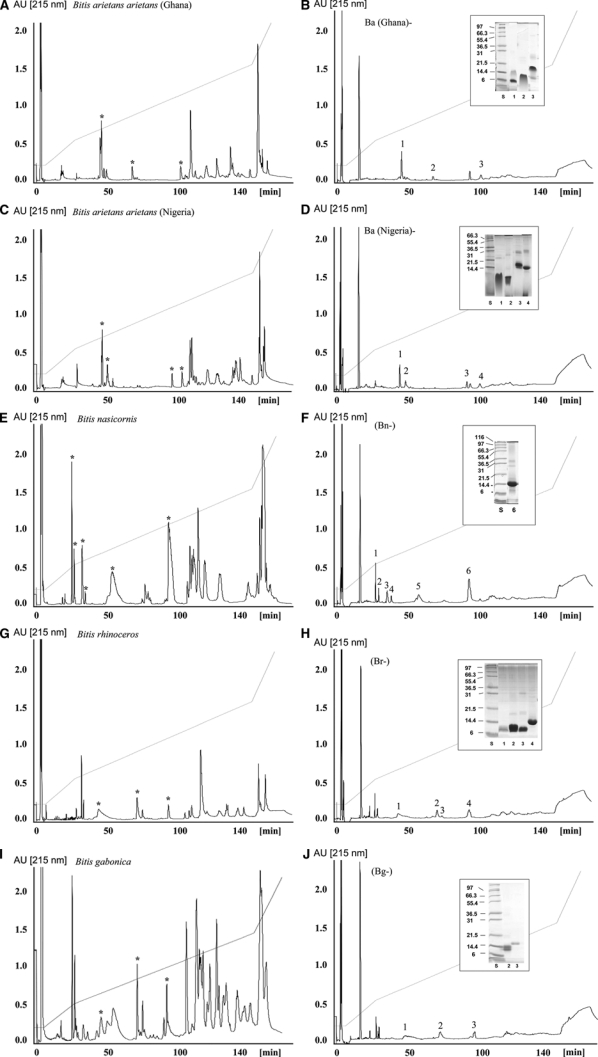
Immunodepletion of venom proteins from Bitis taxa by the polyspecific EchiTAb-Plus-ICP antivenom. Panels B, D, F, H, and J show, respectively, reverse-phase separations of the proteins recovered after incubation of the crude venoms of (A) B. arietans (from Ghana), (C) B. arietans (Nigeria), (E) B. nasicornis, (G) B. rhinoceros, and (I) B. gabonica gabonica with the polyspecific EchiTAb-Plus-ICP antivenom, followed by rabbit anti-horse IgG antiserum and immunoprecipitation. The inset shows SDS-PAGE analyses of β-mercaptoethanol-reduced fractions labeled as in the respective chromatograms. Protein fraction numbering is as in Table 1.
Discussion
Proteomic techniques such as “venomics” and “antivenomics” are valuable tools for analyzing the complex composition of snake venoms and the immunological reactivity of antivenom antibodies against specific venom components.29,37,38,44 These proteomic approaches represent potential tools for designing suitable venom mixtures to be used in immunization protocols for generating monospecific and polyspecific antivenoms exhibiting a wide profile of cross-reactivity. In the case of sub-Saharan Africa, this is of particular significance because of the diversity of snake species and the large complexity of venom proteomes. Thus, the question of which is the simplest combination of venoms that would generate an antivenom capable of neutralizing the largest possible number of viperid snake venoms in sub-Saharan Africa is highly relevant in the context of current efforts being performed by manufacturers to produce antivenoms for this region.
EchiTAb-Plus-ICP antivenom is generated by immunizing horses with a mixture of the venoms of Echis ocellatus, Bitis arietans, and Naja nigricollis from Nigeria. Our present antivenomics results show that EchiTAb-Plus-ICP completely immunodepleted the vast majority of components from the sampled venoms, and partially immunodepleted some additional components. The results also show that all components in all Echis and Bitis venoms investigated were either completely or partially immunodepleted, and there were no components that were not reactive with the antivenom antibodies. However, keep in mind that immunodepletion does not necessarily imply therapeutic neutralization of these venoms and, therefore, antivenomic analysis has to be complemented by preclinical neutralization studies, followed by appropriate clinical trials. In this respect, preclinical studies with the venoms analyzed in this work performed with EchiTAb-Plus-ICP,20 showed that this antivenom neutralizes the most clinically relevant toxic (lethal, hemorrhagic, coagulant, and necrotizing) activities of these venoms,19,20 and also of the venoms of other medically important sub-Saharan African species, i.e., E. leucogaster, E. pyramidum leakeyi, B. gabonica, B. rhinoceros, and B. nasicornis.20 We conclude, therefore, that partially immunodepleted toxins may not compromise the in vivo neutralizing efficacy of EchiTAb-Plus-ICP antivenom.
The complete immunodepletion of SVMPs and serine proteinases is particularly relevant, because these proteinases are abundant in the venomes of African Echis and Bitis species31,43 and are responsible for some of the most serious pathophysiological manifestations of envenoming by species of these genera. The SVMPs comprise 30–66% of the venom proteins of Echis and Bitis species, whereas serine proteinases represent 20–25% of the venom proteins of Bitis species, and only 2% of the venom proteins of E. ocellatus.31,43 The SVMPs, particularly P-III SVMPs, induce local and systemic hemorrhage, and severe hemostatic disturbances resulting from prothrombin activation.45,46 A number of hemorrhagic and procoagulant SVMPs have been isolated and characterized from the venoms of Echis sp.47–50 Serine proteinases, in turn, exert a number of activities, such as defibrinogenation caused by the action of thrombin-like enzymes.51 Bleeding and coagulopathy are the most important systemic pathophysiological manifestations of these envenomings, creating the risk of provoking hemorrhagic strokes, haemodynamic disturbances, cardiovascular collapse, and disseminated intravascular coagulation.4 Thus, the effective immunodepletion of SVMPs and serine proteinases by the EchiTAb-Plus-ICP antivenom correlates well with our earlier reports of the preclinical venom-neutralizing effectiveness of this antivenom against these important African vipers (using assays of venom-induced lethality, hemorrhage, and coagulation19,20 and its clinical efficacy in treating human victims of E. ocellatus envenoming.27
The partial immunological reactivity to some PLA2s, disintegrins, and proteinase inhibitors revealed the presence of low affinity or low abundance of antibodies against these components in the antivenom. Previous studies on antivenoms that target a number of venoms of Central and South American viper species showed evidence that PLA2s and other low-molecular-mass components were poorly immunogenic, thus resulting in the generation of low-affinity antibodies.28,30,32,39 To properly assess the therapeutic consequences of this observation, it is necessary to study the potential relevance of these proteins as toxins, a subject that deserves further study. The percentage of PLA2s in the characterized venom proteomes of Bitis and Echis venoms ranges from 4.3%, in the case of B. arietans, to 20.1% for B. nasicornis.31,43 Some PLA2s in snake venoms are toxic and exert various pathophysiological effects, such as neurotoxic, myotoxic, hypotensive, and hemostasis-perturbing activities.52 Other PLA2s, particularly the acidic ones, are devoid of toxicity and are thought to be more likely to play a predominantly digestive function. In the case of Echis sp. and Bitis sp. venoms, a myotoxic Ser49 PLA2 homologue was characterized from the venom of E. carinatus sochureki,53 and PLA2s devoid of toxicity were purified from the venom of B. nasicornis.54 EchiTAb-Plus-ICP antivenom fully neutralized the myotoxic and lethal activity of E. ocellatus and B. arietans venoms,19 thus suggesting that the partially immunodepleted PLA2s may not play a major pathogenic role in envenoming by these vipers. Despite this, it is necessary to search for novel strategies aimed at enhancing the immune response against venom PLA2s. These might be based on chemical modification of antigens, the use of novel adjuvants and immunization schemes, or the introduction of recombinant toxins or DNA immunization strategies.55,56 The challenge will be to identify strategies that are immuno-potentiating without necessarily increasing the final cost of these antivenoms, which are destined for use in rural, subsistence farming communities in Africa.
Disintegrins, which were also only partially immunodepleted, comprise between 3.4% and 8.5% of the venoms of some Echis and Bitis species.31,43 Some disintegrins inhibit platelet aggregation,57 although their role in the pathophysiology of envenoming remains unclear. The observation that some disintegrins were not completely immunodepleted from Echis and Bitis venoms is probably attributable to their low molecular mass and structural compactness, making them poorly immunogenic. The other venom components that were only partially immunodepleted, i.e., proteinase inhibitors, are also low molecular mass peptides whose main role is to control proteinase activity in the venom gland58 and are, consequently, not likely to play a significant role in envenoming.
In conclusion, the antivenomics approach used in this study might become a useful tool to assess the profile of immune reactivity of antivenoms against individual venom components, thus complementing preclinical studies on the neutralizing capability of antivenoms. Our immunochemical observations showed evidence that antibodies raised in horses against the venoms of E. ocellatus and B. arietans are capable of recognizing all venom components in some of the viperid snake venoms of greatest medical importance in sub-Saharan Africa, further supporting the hypothesis that a pool of venoms of these two species constitutes a good immunizing mixture for the generation of antivenoms aimed at treating viperid envenomings in this region. The preclinical assessment of EchiTAb-Plus-ICP, both in its neutralization of toxic activities and its antivenomic profile, strongly suggests that this antivenom may be effective for the treatment of envenomings by a number of Bitis and Echis species in sub-Saharan Africa, a hypothesis that must be addressed by clinical trials.
Table 1.
Assignment of reverse-phase isolated proteins from the non-immunodepleted HPLC fractions of the venoms of Echis ocellatus (Figure 1B), E. leucogaster (Figure 1D), E. pyramidum leakeyi (Figure 1F), B. arietans arietans (Ghana) (Figure 2B), B. arietans arietans (Nigeria) (Figure 2D), Bitis nasicornis (Figure 2F), B. rhinoceros (Figure 2H), and B. gabonica gabonica (Figure 2I), to protein families by N-terminal Edman sequencing, mass spectrometry and collision-induced fragmentation by nESI-MS/MS of selected peptide ions from in-gel digested protein bands
| HPLC fraction | N-terminal sequence | Molecular mass | Peptide ion | MS/MS-derived sequence | Protein family | % Of immunodepletion | |
|---|---|---|---|---|---|---|---|
| m/z | z | ||||||
| Echis ocellatus | |||||||
| Eo-1 | Blocked | 444.1 | 444.1 | 1 | ZKW | Inhibitor of SVMP | 42 |
| Eo-2 | DCESGPCCDNCKFLK | 5494, 5592 | Disintegrin ocellatusin [Q3BER] | 62 | |||
| Eo-3 | SVVELGKMIIQETGKS | 13825 | PLA2 [CAQ72890] | 67 | |||
| Eo-4 | SVIEFGTMIIEETGRSPF | 13866 | PLA2 [CAQ72891] | 45 | |||
| Echis leucogaster | |||||||
| El-1 | DCESGPCCRDCKFLK | 5458 | Disintegrin leucogastin B [P0C7A8] | 60 | |||
| El-2 | Blocked | 444.1 | 444.1 | 1 | ZKW | Inhibitor of SVMP | 38 |
| El-3 | DCASGPCCRDCKFLE | 5426 | Disintegrin leucogastin A [P0C7A7] | 65 | |||
| El-4 | NLYQFGKMIKNKTGK | 14066 | PLA2 | 40 | |||
| El-5 | SVIELGKMIIQLTNK | 13696 | PLA2 | 25 | |||
| Echis pyramidum leakeyi | |||||||
| Ep-1 | DCASGPCCRDCKFLKEGT | 5555 | Disintegrin pyramidin A [P0C6R7] | 66 | |||
| Ep-2 | Blocked | 444.1 | 444.1 | 1 | ZKW | Inhibitor of SVMP | 48 |
| Ep-3 | DCASGPCCRDCKFLEE | 5434 | Disintegrin pyramidin B [P0C6R8] | 60 | |||
| Ep-4 | NLYQFGKMIKNKTGK | 14103 | PLA2 [P59172] | 40 | |||
| Ep-5 | SVIELGKMIIQLTNK | 13696 | PLA2 | 30 | |||
| Bitis arietans arietans (Ghana) | |||||||
| BaG-1 | SPPVCGNKILEQGED | 8991 | Disintegrin bitistatin D1 [P17497] | 61 | |||
| BaG-2 | Blocked | 6942 | 396.2 | 2 | TPEECR | Kunitz-type inhibitor | 72 |
| BaG-3 | SLVEFGQMIQEETER | 13905 | PLA2 | 66 | |||
| Bitis arietans arietans (Nigeria) | |||||||
| BaN-1 | SPPVCGNKILEQGED | 8991 | Disintegrin bitistatin D1 [P17497] | 65 | |||
| BaN-2 | SPPVCGNEELEEGEE | 8950 | Disintegrin bitistatin D3 [Q4JCS0] | 79 | |||
| BaN-3 | HLNQFMEMIQ | 14038 | PLA2 | 63 | |||
| BaN-4 | SLVEFGQMIQEETER | 13905 | PLA2 | 72 | |||
| Bitis nasicornis | |||||||
| Bn-1-5 | n.p. | ||||||
| Bn-6 | SLLEFAKMIKEETGF | 13828 | PLA2 | 68 | |||
| Bitis rhinoceros | |||||||
| Br-1 | KKRPNFCYLPADPG | 7 kDa▼ | Kunitz-type inhibitor | 66 | |||
| Br-2 | NSAHPCCDPVTCK | 15184 | ~ Disintegrin Bitisgabonin-1 [Q6T6T3] | 62 | |||
| Br-3 | NSAHPCCDPVTCK | 15111 | ~ Disintegrin Bitisgabonin-1 [Q6T6T2] | 80 | |||
| Br-4 | SLEEFAKMIKEETG | 13891 | PLA2 | 45 | |||
| Bitis gabonica gabonica | |||||||
| Bg-1 | KKRPDFCYLPADTGP | 7 kDa▼ | Kunitz-type inhibitor [Q6T6T5] | 85 | |||
| Bg-2 | NSAHPCCDPVTCKPK | 15184 | Disintegrin Bitisgabonin-1 [Q6T6T3] | 95 | |||
| Bg-3 | HLEQFGNMIDHVSGR | 13967 | PLA2 [Q6T7B8] | ||||
X, Ile or Leu; Z, pyrrolidone carboxylic acid. Unless otherwise stated, for MS/MS analyses, cysteine residues were carbamidomethylated; molecular masses were determined by electrospray-ionization mass spectrometry or SDS-PAGE of reduced (▼) samples; n.p. = non-peptidic material found.
Footnotes
Financial support: This study was supported by Vicerrectoría de Investigación, Universidad de Costa Rica (project 741-A9-003), CRUSA-CSIC (project 2007CR0004), Ministerio de Innovación y Ciencia (Madrid, Spain) (grant BFU2007-61563), and the EchiTAb Study Group in partnership with the Federal Ministry of Health, Republic of Nigeria.
Authors' addresses: Juan J. Calvete, Pedro Cid, and Libia Sanz, Instituto de Biomedicina de Valencia, C.S.I.C., Valencia, Spain, E-mails: jcalvete@ibv.csic.es, pcid@ibv.csic.es, and libia.sanz@ibv.csic.es. Álvaro Segura, Mauren Villalta, María Herrera, Guillermo León, and José María Gutiérrez, Instituto Clodomiro Picado, Facultad de Microbiología, Universidad de Costa Rica, San José, Costa Rica, E-mails: asegura@icp.ucr.ac.cr, mvillalta@icp.ucr.ac.cr, mariah@icp.ucr.ac.cr, guillermo.leon@ucr.ac.cr, and jose.gutierrez@ucr.ac.cr. Robert Harrison and R. David G. Theakston, Alistair Reid Venom Research Unit, Liverpool School of Tropical Medicine, Liverpool, UK, E-mails: r.harrison@liverpool.ac.uk and r.d.g.theakston@liverpool.ac.uk. Nandul Durfa and Abdusalami Nasidi, Federal Ministry of Health, Abuja, Nigeria, E-mails: ndurfa2004@yahoo.com.uk and nasidia@hotmail.com. David A. Warrell, Nuffield Department of Clinical Medicine, University of Oxford, Oxford, UK, E-mail: david.warrell@ndm.ox.ac.uk.
Reprint requests: Juan J. Calvete, Instituto de Biomedicina de Valencia, C.S.I.C., Jaume Roig 11, 46010 Valencia, Spain, Tel: 34-96-339-1778, Fax: 34-96-369-0800, E-mail: jcalvete@ibv.csic.es.
References
- 1.Chippaux JP. Snake-bites: appraisal of the global situation. Bull World Health Organ. 1998;76:515–524. [PMC free article] [PubMed] [Google Scholar]
- 2.Kasturiratne A, Wickremasinghe AR, de Silva N, Gunawardena NK, Pathmeswaran A, Premaratna R, Savioli L, Lalloo DG, da Silva HJ. The global burden of snakebite: a literature analysis and modeling based on regional estimates of envenoming and deaths. PLoS Med. 2008;5:e218. doi: 10.1371/journal.pmed.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Snow RW, Bronzan R, Roques T, Nyamawi C, Murphy S, Marsh K. The prevalence and morbidity of snake bite and treatment-seeking behaviour among a rural Kenyan population. Ann Trop Med Parasitol. 1994;88:665–671. doi: 10.1080/00034983.1994.11812919. [DOI] [PubMed] [Google Scholar]
- 4.Warrell DA. In: Handbook of Clinical Toxicology of Animal Venoms and Poisons. Meier J, White J, editors. Boca Raton, FL: CRC Press; 1995. pp. 433–492. (Clinical toxicology of snakebite in Africa and the Middle East/Arabian peninsula). [Google Scholar]
- 5.Chippaux JP. In: Perspectives in Molecular Toxinology. Ménez A, editor. England: John Wiley; 2002. pp. 457–472. (The treatment of snake bites: analysis of requirements and assessment of therapeutic efficacy in tropical Africa). [Google Scholar]
- 6.World Health Organization . Rabies and Envenomings: A Neglected Public Health Issue. Geneva: World Health Organization; 2007. [Google Scholar]
- 7.Theakston RD, Warrell DA, Griffiths E. Report of a WHO workshop on the standardization and control of antivenoms. Toxicon. 2003;41:541–557. doi: 10.1016/s0041-0101(02)00393-8. [DOI] [PubMed] [Google Scholar]
- 8.Lalloo DG, Theakston RD. Snake antivenoms. J Toxicol Clin Toxicol. 2003;41:317–327. doi: 10.1081/clt-120021113. [DOI] [PubMed] [Google Scholar]
- 9.Gutiérrez JM, León G. In: Animal Toxins: State of the Art Perspectives in Health and Biotechnology. de Lima ME, Pimenta AM, Martin-Euclairte MF, Zingali RB, Rochat H, editors. Belo Horizonte: Editora UFMG; 2009. pp. 393–421. (Snake antivenoms. Technological, clinical and public health issues). [Google Scholar]
- 10.Theakston RD, Warrell DA. Crisis in antivenom supply for Africa. Lancet. 2000;356:2104. doi: 10.1016/s0140-6736(05)74319-1. [DOI] [PubMed] [Google Scholar]
- 11.Stock RP, Massougbodji A, Alagón A, Chippaux JP. Bringing antivenoms to sub-Saharan Africa. Nat Biotechnol. 2007;25:173–177. doi: 10.1038/nbt0207-173. [DOI] [PubMed] [Google Scholar]
- 12.Visser LE, Kyei-Faried S, Belcher DW, Geelhoed DW, van Leeuwen JS, van Roosmalen J. Failure of a new antivenom to treat Echis ocellatus snake bite in rural Ghana: the importance of quality surveillance. Trans R Soc Trop Med Hyg. 2008;102:445–450. doi: 10.1016/j.trstmh.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 13.Warrell DA. Unscrupulous marketing of snake bite antivenoms in Africa and Papua New Guinea: choosing the right product–‘what's in a name?'. Trans R Soc Trop Med Hyg. 2008;102:397–399. doi: 10.1016/j.trstmh.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 14.Gutiérrez JM, Theakston RD, Warrell DA. Confronting the neglected problem of snake bite envenoming: the need for a global partnership. PLoS Med. 2006;3:e412. doi: 10.1371/journal.pmed.0030150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams D, Gutiérrez JM, Harrison RA, Warrell DA, White J, Winkel KD, Gopalakrishnakone P. An antidote for snake bite: The Global Snake Bite Initiative. Lancet. 2010;375:89–91. doi: 10.1016/S0140-6736(09)61159-4. [DOI] [PubMed] [Google Scholar]
- 16.Meier J. In: Handbook of Clinical Toxicology of Animal Venoms and Poisons. Meier J, White J, editors. Boca Raton, FL: CRC Press; 1995. pp. 689–721. (Commercially available antivenoms (“hyperimmune sera,” “antivenins,” “antisera”) for antivenom therapy). [Google Scholar]
- 17.Meyer WP, Habib AG, Onayade AA, Yakubu A, Smith DC, Nasidi A, Daudu IJ, Warrell DA, Theakston RD. First clinical experiences with a new ovine Fab Echis ocellatus snake bite antivenom in Nigeria: randomized comparative trial with Institute Pasteur serum (Ipser) Africa antivenom. Am J Trop Med Hyg. 1997;56:291–300. doi: 10.4269/ajtmh.1997.56.291. [DOI] [PubMed] [Google Scholar]
- 18.Ramos-Cerrillo B, de Roodt AR, Chippaux JP, Olguín L, Casasola A, Guzmán G, Paniagua-Solís J, Alagón A, Stock RP. Characterization of a new polyvalent antivenom (Antivipmyn Africa) against African vipers and elapids. Toxicon. 2008;52:881–888. doi: 10.1016/j.toxicon.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 19.Gutiérrez JM, Rojas E, Quesada L, León G, Núñez J, Laing GD, Sasa M, Renjifo JM, Nasidi A, Warrell DA, Theakston RD, Rojas G. Pan-African polyspecific antivenom produced by caprylic acid purification of horse IgG: an alternative to the antivenom crisis in Africa. Trans R Soc Trop Med Hyg. 2005;99:468–475. doi: 10.1016/j.trstmh.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 20.Segura Á, Villalta M, Herrera M, León G, Harrison R, Durfa N, Nasidi A, Calvete JJ, Theakston RD, Warrell DA, Gutiérrez JM. Preclinical assessment of the efficacy of a new antivenom (EchiTAb-Plus-ICP) for the treatment of viper envenoming in sub-Saharan Africa. Toxicon. 2010;55:369–374. doi: 10.1016/j.toxicon.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 21.Calvete JJ, Escolano J, Sanz L. Snake venomics of Bitis species reveals large intragenus venom toxin composition variation. Application to taxonomy of congeneric taxa. J Proteome Res. 2007;6:2732–2745. doi: 10.1021/pr0701714. [DOI] [PubMed] [Google Scholar]
- 22.Currier RB, Harrison RA, Rowley RD, Laing GD, Wagstaff SC. Intra-species variation in venom composition, immunoreactivity and enzyme function of the African Puff Adder (Bitis arietans) Toxicon. 2010;55:864–873. doi: 10.1016/j.toxicon.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Theakston RD, Reid HA. Development of simple standard assay procedures for the characterization of snake venoms. Bull World Health Organ. 1983;61:949–956. [PMC free article] [PubMed] [Google Scholar]
- 24.Theakston RD. In: Natural Toxins–Animal, Plant and Microbial. Harris JB, editor. Oxford: Clarendon Press; 1986. pp. 287–303. (Characterization of venoms and standardization of antivenoms). [Google Scholar]
- 25.Gutiérrez JM, Rojas G, Lomonte B, Gené JA, Chaves F, Alvarado J, Rojas E. Standardization of assays for testing the neutralizing ability of antivenoms. Toxicon. 1990;28:1127–1129. doi: 10.1016/0041-0101(90)90110-s. [DOI] [PubMed] [Google Scholar]
- 26.Gutiérrez JM, Rojas G, Bogarín G, Lomonte B. In: Envenomings and Their Treatments. Bon C, Goyffon M, editors. Lyon: Fondation Marcel Mérieux; 1996. pp. 223–231. (Evaluation of the neutralizing ability of antivenoms for the treatment of snakebite envenoming in Central America). [Google Scholar]
- 27.Abubakar SB, Abubakar IS, Habib AG, Nasidi A, Durfa N, Yusuf PO, Larnyang S, Garnvwa J, Sokomba E, Salako L, Laing GD, Theakston RD, Juszczak E, Alder N, Warrell DA. Nigeria-UK EchiTab Study Group, 2010. Pre-clinical and preliminary dose-finding and safety studies to identify candidate antivenoms for treatment of envenoming by saw-scaled or carpet vipers (Echis ocellatus) in northern Nigeria. Toxicon. 55:719–723.. doi: 10.1016/j.toxicon.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 28.Lomonte B, Escolano J, Fernández J, Sanz L, Angulo Y, Gutiérrez JM, Calvete JJ. Snake venomics and antivenomics of the arboreal neotopical pitvipers Bothriechis lateralis and Bothriehis schlegelii. J Proteome Res. 2008;7:2445–2457. doi: 10.1021/pr8000139. [DOI] [PubMed] [Google Scholar]
- 29.Gutiérrez JM, Lomonte B, León G, Alape-Girón A, Flores-Díaz M, Sanz L, Angulo Y, Calvete JJ. Snake venomics and antivenomics: proteomic tools in the design and control of antivenoms for the treatment of snakebite envenoming. J Proteomics. 2009;72:165–182. doi: 10.1016/j.jprot.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 30.Gutiérrez JM, Sanz L, Escolano J, Fernández J, Lomonte B, Angulo Y, Rucavado A, Warrell DA, Calvete JJ. Snake venomics of the Lesser Antillean pit vipers Bothrops caribbaeus and Bothrops lanceolatus: correlation with toxicological activities and immunoreactivity of a heterologous antivenom. J Proteome Res. 2008;7:4396–4408. doi: 10.1021/pr8003826. [DOI] [PubMed] [Google Scholar]
- 31.Calvete JJ, Marcinkiewicz C, Sanz L. Snake venomics of Bitis gabonica gabonica. Protein family composition, subunit organization of venom toxins, and characterization of dimeric disintegrins bitisgabonin-1 and bitisgabonin-2. J Proteome Res. 2007;6:326–336. doi: 10.1021/pr060494k. [DOI] [PubMed] [Google Scholar]
- 32.Núñez V, Cid P, Sanz L, De La Torre P, Angulo Y, Lomonte B, Gutiérrez JM, Calvete JJ. Snake venomics and antivenomics of Bothrops atrox venoms from Colombia and the Amazon regions of Brazil, Perú and Ecuador suggest the occurrence of geographic variation of venom phenotype by a trend towards paedomorphism. J Proteomics. 2009;73:57–78. doi: 10.1016/j.jprot.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 33.Fasman DG. Practical Handbook of Biochemistry and Molecular Biology. Boston, MA: CRC Press; 1992. [Google Scholar]
- 34.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le Blanc JC, Hager JW, Ilisiu AM, Hunter C, Zhong F, Chu I. Unique scanning capabilities of a new hybrid linear ion trap mass spectrometer (QTRAP) used for high sensitivity proteomics applications. Proteomics. 2003;3:859–869. doi: 10.1002/pmic.200300415. [DOI] [PubMed] [Google Scholar]
- 36.Juárez P, Wagstaff SC, Oliver J, Sanz L, Harrison RA, Calvete JJ. Molecular cloning of disintegrin-like transcript BA-5A from Bitis arietans venom gland cDNA library: a putative intermediate in the evolution of the long chain disintegrin bitistatin. J Mol Evol. 2006;63:142–152. doi: 10.1007/s00239-005-0268-z. [DOI] [PubMed] [Google Scholar]
- 37.Calvete JJ, Juárez P, Sanz L. Snake venomics. Strategy and applications. J Mass Spectrom. 2007;42:1405–1414. doi: 10.1002/jms.1242. [DOI] [PubMed] [Google Scholar]
- 38.Calvete JJ, Sanz L, Angulo Y, Lomonte B, Gutiérrez JM. Venoms, venomics, antivenomics. FEBS Lett. 2009;583:1736–1743. doi: 10.1016/j.febslet.2009.03.029. [DOI] [PubMed] [Google Scholar]
- 39.Calvete JJ, Borges A, Segura A, Flores-Díaz M, Alape-Girón A, Gutiérrez JM, Diez N, De Sousa L, Kiriakos D, Sánchez E, Faks JG, Escolano J, Sanz L. Snake venomics and antivenomics of Bothrops colombiensis, a medically important pitviper of the Bothrops atrox-asper complex endemic to Venezuela: contributing to its taxonomy and snakebite management. J Proteomics. 2009;72:227–240. doi: 10.1016/j.jprot.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 40.Angulo Y, Escolano J, Lomonte B, Gutiérrez JM, Sanz L, Calvete JJ. Snake venomics of Central American pitvipers. Clues for rationalizing the distinct envenomation profiles of Atropoides nummifer and Atropoides picadoi. J Proteome Res. 2008;7:708–719. doi: 10.1021/pr700610z. [DOI] [PubMed] [Google Scholar]
- 41.Sanz L, Ayvazyan N, Calvete JJ. Snake venomics of the Armenian mountain vipers Macrovipera lebetina obtusa and Vipera raddei. J Proteomics. 2008;71:198–209. doi: 10.1016/j.jprot.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 42.Alape-Girón A, Sanz L, Escolano J, Flores-Díaz M, Madrigal M, Sasa M, Calvete JJ. Snake venomics of the lancehead pitviper Bothrops asper: geographic, individual, and ontogenetic variations. J Proteome Res. 2008;7:3556–3571. doi: 10.1021/pr800332p. [DOI] [PubMed] [Google Scholar]
- 43.Wagstaff SC, Sanz L, Juárez P, Harrison RA, Calvete JJ. Combined snake venomics and venom gland transcriptomic analysis of the ocellated carpet viper, Echis ocellatus. J Proteomics. 2009;71:609–623. doi: 10.1016/j.jprot.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 44.Fox JW, Serrano SM. Exploring snake venom proteomes: multifaceted analyses for complex toxin mixtures. Proteomics. 2008;8:909–920. doi: 10.1002/pmic.200700777. [DOI] [PubMed] [Google Scholar]
- 45.Gutiérrez JM, Rucavado A, Escalante T, Díaz C. Hemorrhage induced by snake venom metalloproteinases: biochemical and biophysical mechanisms involved in microvessel damage. Toxicon. 2005;45:997–1011. doi: 10.1016/j.toxicon.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 46.Fox JW, Serrano SM. Structural considerations of the snake venom metalloproteinases, key members of the M12 reprolysin family of metalloproteinases. Toxicon. 2005;45:969–985. doi: 10.1016/j.toxicon.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 47.Nishida S, Fujita T, Kohno N, Atoda H, Morita T, Takeya H, Kido I, Paine MJ, Kawabata S, Iwanaga S. cDNA cloning and deduced amino acid sequence of prothrombin activator (ecarin) from Kenyan Echis carinatus venom. Biochemistry. 1995;7:1771–1778. doi: 10.1021/bi00005a034. [DOI] [PubMed] [Google Scholar]
- 48.Yamada D, Sekiya F, Morita T. Isolation and characterization of carinactivase, a novel prothrombin activator in Echis carinatus venom with a unique catalytic mechanism. J Biol Chem. 1996;271:5200–5207. doi: 10.1074/jbc.271.9.5200. [DOI] [PubMed] [Google Scholar]
- 49.Howes JM, Wilkinson MC, Theakston RD, Laing GD. The purification and partial characterization of two novel metalloproteinases from the venom of the West African carpet viper, Echis ocellatus. Toxicon. 2003;42:21–27. doi: 10.1016/s0041-0101(03)00096-5. [DOI] [PubMed] [Google Scholar]
- 50.Howes JM, Kamiguti AS, Theakston RD, Wilkinson MC, Laing GD. Effects of three novel metalloproteinases from the venom of the West African saw-scaled viper, Echis ocellatus on blood coagulation and platelets. Biochim Biophys Acta. 2005;1724:194–202. doi: 10.1016/j.bbagen.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 51.Markland FS. Snake venoms and the hemostatic system. Toxicon. 1998;36:1749–1800. doi: 10.1016/s0041-0101(98)00126-3. [DOI] [PubMed] [Google Scholar]
- 52.Kini RM. Excitement ahead: structure, function and mechanism of snake venom phospholipase A2 enzymes. Toxicon. 2003;42:827–840. doi: 10.1016/j.toxicon.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 53.Polgár J, Magnenat EM, Peitsch MC, Wells TN, Clemetson KJ. Asp-49 is not an absolute prerequisite for the enzymatic activity of low-Mr phospholipases A2: purification, characterization and computer modeling of an enzymatically active Ser-49 phospholipase A2, ecarpholin S, from the venom of Echis carinatus sochureki (saw-scaled viper) Biochem J. 1996;319:961–968. doi: 10.1042/bj3190961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Joubert FJ, Townshend GS, Botes DP. Snake venoms. Purification, some properties of two phospholipases A2 (CM-I and CM-II) and the amino-acid sequence of CM-II of Bitis nasicornis (horned adder) venom. Hoppe Seylers Z Physiol Chem. 1983;364:1717–1726. doi: 10.1515/bchm2.1983.364.2.1717. [DOI] [PubMed] [Google Scholar]
- 55.Harrison RA. Development of venom toxin-specific antibodies by DNA immunisation: rationale and strategies to improve therapy of viper envenoming. Vaccine. 004;22:1648–1655. doi: 10.1016/j.vaccine.2003.09.046. [DOI] [PubMed] [Google Scholar]
- 56.Wagstaff SC, Laing GD, Theakston RD, Papaspyridis C, Harrison RA. Bioinformatics and multiepitope DNA immunization to design rational snake antivenom. PLoS Med. 2006;3:e184. doi: 10.1371/journal.pmed.0030184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Calvete JJ, Marcinkiewicz C, Monleón D, Esteve V, Celda B, Juárez P, Sanz L. Snake venom disintegrins: evolution of structure and function. Toxicon. 2005;45:1063–1074. doi: 10.1016/j.toxicon.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 58.Wagstaff SC, Favreau P, Cheneval O, Laing GD, Wilkinson MC, Miller RL, Stöcklin R, Harrison RA. Molecular characterization of endogenous snake venom metalloproteinase inhibitors. Biochem Biophys Res Commun. 2008;365:650–656. doi: 10.1016/j.bbrc.2007.11.027. [DOI] [PubMed] [Google Scholar]


