Abstract
The merozoite surface protein 1 (MSP1) is the principal surface antigen of the blood stage form of the Plasmodium parasite. Antibodies recognizing MSP1 are frequently detected following Plasmodium infection, making this protein a significant component of malaria vaccines and diagnostic tests. Although the MSP1 gene sequence has been reported for Plasmodium falciparum and Plasmodium vivax, this gene has not been identified for the other two major human-infectious species, Plasmodium malariae and Plasmodium ovale. MSP1 genes from these two species were isolated from Cameroon blood donor samples. The genes are similar in size to known MSP1 genes and encode proteins with interspecies conserved domains homologous to those identified in other Plasmodium species. Sequence and phylogenetic analysis of all available Plasmodium MSP1 amino acid sequences clearly shows that the Po and Pm MSP1 sequences are truly unique within the Plasmodium genus and not simply Pf or Pv variants.
Introduction
Most cases of human malaria are caused by infection with the parasitic Plasmodium species Plasmodium falciparum and Plasmodium vivax, and to a lesser extent with Plasmodium malariae and Plasmodium ovale.1 Recently, a growing number of human infections with Plasmodium knowlesi have been reported,2 a species typically infecting Old World monkeys. With the exception of P. vivax, which is largely absent from west and central Africa, P. falciparum, P. vivax, and P. malariae infections are globally distributed throughout tropical and subtropical regions. Plasmodium ovale is present primarily in sub-Saharan Africa, India, Southeast Asia, and the western Pacific.3,4 Plasmodium knowlesi is also found in Southeast Asia,5 and though currently thought to cause mostly zoonotic infections in humans,2 it could emerge as a true fifth human malaria parasite.6
The predominant antigen on the parasite surface during the erythrocytic phase of infection is merozoite surface protein 1 (MSP1),7 which is present in all examined Plasmodium species. Genes encoding the P. falciparum and P. vivax MSP1 (Pf MSP1 and Pv MSP1) have been characterized. However, no gene sequence information is available for P. malariae or P. ovale MSP1 (Pm MSP1 and Po MSP1), with the exception of a short coding segment near the 5′ end of the Pm MSP1 gene.8 In general, MSP1 is produced as a large protein (~200 kDa) covalently attached by the carboxyl terminus to the cell membrane by a glycophosphatidylinositol (GPI) anchor.9 Around the time of merozoite release from the red blood cell (RBC), MSP1 undergoes proteolytic maturation, generating three or four fragments, which remain associated as a complex on the cell surface by interaction with the anchored C-terminal fragment (p42).10 The free merozoites rapidly attach to, and invade, new RBCs. This process involves secondary cleavage of the p42 peptide to generate p33, which is shed along with the rest of the MSP1 complex, and p19, which remains anchored to the parasite membrane as it invades the cell.11,12 The apparent molecular masses of homologs to the p42, p33 and p19 peptides from P. falciparum vary somewhat between Plasmodium species.12,13 However, for the sake of simplicity they will be referred to as p42, p33, and p19, regardless of species origin.
Probably because of its high abundance and prominent display on the cell surface, MSP1 is a primary target of the host immune system, and antibodies recognizing various regions of the protein are detected in a large percentage of individuals from endemic areas.14 Epitopes within the p19 peptide are primarily conformational,15 and likely are dependent upon proper folding of two universally conserved epidermal growth factor (EGF)-like domains maintained by multiple disulfide bonds. Portions of MSP1 have been studied as vaccine candidates,16,17 and the p19 polypeptide, which exhibits relatively high intra-species conservation, has been incorporated into malaria antibody immunoassays.18,19
Because of the divergence of MSP1 amino acid sequence across Plasmodium species, a pan-specific Plasmodium antibody test to detect human exposure to the Plasmodium parasite would likely require recombinant MSP1s, not only from P. falciparum and P. vivax, but also from P. malariae and P. ovale. Likewise, a broad-based vaccine would potentially incorporate portions of MSP1 from all four species. Thus, isolation of the Pm and Po MSP1 genes is critical. Cameroon is a malaria-endemic region in sub-Saharan Africa, and a potential source of specimens from individuals infected with P. malariae and P. ovale. The incidence of P. falciparum, P. malariae, and P. ovale infection, as determined by polymerase chain reaction (PCR), in asymptomatic pregnant women from Cameroon was reported to be 91.8%, 7.6%, and 2.5%, respectively.20 In the current study, whole blood samples from Cameroon blood donors infected with P. malariae or P. ovale were identified. The MSP1 genes from these two species were isolated and characterized, and their deduced protein sequences compared with those of other Plasmodium species.
Materials and Methods
Identification of blood donors infected with P. malariae and P. ovale.
DNA was extracted from the whole blood of 82 human immunodeficiency virus (HIV)-negative normal donor samples from Cameroon using the QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA) according to the package insert. Subsequently, PCR targeting the 18S ribosomal RNA (rRNA) gene was used to screen the samples for the presence of Plasmodium DNA, and when detected, to make a species assignment based on melting curve analysis essentially as described.21 For comparison, four control PCRs containing individual plasmid clones of the 18S rRNA genes from P. falciparum, P. vivax, P. malariae, or P. ovale were used as comparators for the donor samples. The plasmid clones were obtained through the ATCC/Malaria Research and Reference Reagent Resource Center (MRA catalog nos. MRA-177, MRA-178, MRA-179, and MRA-180, respectively, deposited by P. A. Zimmerman). For specimens identified as potentially containing P. ovale DNA, the 18S rRNA amplicons were purified using the Gene Clean Spin Kit (MP Biochemcials, Solon, OH) according to the package insert and sequenced. Analysis of the sequences was performed to confirm species identity by BLAST.22 All sequences described here were obtained directly from amplicons, using an ABI Prism Big Dye Terminator Cycle Sequencing Reaction Kit and the ABI Prism 3130xl Genetic Analyzer (Applied Biosystems, Foster City, CA).
Amplification and sequencing of the MSP1 genes.
The initial goal was to establish islands of known sequence near the 5′ and 3′ ends of the P. malariae and P. ovale MSP1 genes. Such an island existed only for the 5′ end of the P. malariae gene.8 Because no other MSP1 sequences were available for P. malariae or P. ovale, degenerate primers (described later) were used to amplify sequences near the remaining unknown termini. Detailed protocols for all methods mentioned herein will be made available upon request. Plasmodium-specific degenerate primers As1 and As28 were used to amplify a short region near the 5′ end of the MSP1 gene from a P. ovale-infected donor, resulting in an amplicon of about 420 bp. This is within the predicted size range, based on spacing of the As primer pair within MSP1 gene sequences from other Plasmodium species. The amplicon was gel purified, sequenced to confirm the presence of an MSP1 coding region, and P. ovale-specific primers PoMSP1-F1 and PoMSP1-F2 were designed. Table 1 contains the sequences of all oligonucleotides used herein.
Table 1.
Oligonucleotide sequences
| Designation | Sequence (5′ to 3′) |
|---|---|
| Degenerate | |
| As1 | TTTTTYRTTATCAAWTGWCAATGTGWAAC* |
| As2 | GCYCTYAAYAAATCATARTRRAAGTT |
| MSP1-R1 | GAAGCTGGARGARCTRCAGAA |
| MSP1-R2 | GAAGAACTRCAGAAAAYWCCMTC |
| Plasmodium malariae | |
| PmMSP1-F1 | AATGAAGATTATGAACAACTYATTCAA* |
| PmMSP1-F2 | AAGTTGGGAAAACTGGAGGAAC† |
| PmMSP1-R2 | GAAGGCTCTTTATCTTCAACAC |
| PmMSP1-R3 | CAACACATCCTCCATCAMCTAG |
| Plasmodium ovale | |
| PoMSP1-F1 | CAAGTGGTTAGCAAGTTAACAG |
| PoMSP1-F2 | CGGACATAAGATTCCTTCTATC |
| PoMSP1-R1 | TTGGTACACATGTTTCTCCAAC |
| PoMSP1-R2 | ATCTCCATTCTTCTCGTCCATC |
As1 and PmMSP1-F1 (aka Mmsp1) sequences are slightly different than the published primer sequences.8
PmMSP1-F2 was designed based on GenBank sequence AF138881.1.
To obtain Plasmodium-specific primers from the 3′ end of the MSP1 gene, nucleotide sequences near the 3′ end of the MSP1 open reading frame for 14 Plasmodium species (Table 2, excluding P. malariae and P. ovale, which were unknown) were collected from GenBank. The sequences were aligned and degenerate reverse primers MSP1-R1 and MSP1-R2 derived from the most conserved region were designed. Amplifications using combinations of these degenerate reverse and forward primers specific for P. malariae (PmMSP1-F18 and PmMSP1-F2) or P. ovale (PoMSP1-F1 and PoMSP1-F2) were performed on DNA extracted from P. malariae or P. ovale parasitemic specimens, respectively. When necessary, a second round of PCR was performed on the first round products, using nested or hemi-nested primer combinations. First or second round products, consistent with the predicted size of a MSP1 gene amplicon (~5 kb), were gel purified and sequences near the 3′ end of the genes were obtained using the MSP1-R1 or MSP1-R2 primer. Based on these sequences, species-specific reverse primers near the MSP1 3′ end were designed for P. malariae (PmMSP1-R2 and PmMSP1-R3), and P. ovale (PoMSP1-R1 and PoMSP1-R2).
Table 2.
Merozoite surface protein 1 (MSP1) sequences of Plasmodium species from GenBank
| Accession no. | |||
|---|---|---|---|
| Plasmodium species | Host | Amino acid | Nucleotide |
| Plasmodium yoelii | Rodent (Thamnonys rutilans) | EAA17822.1 | AABL01001865.1 |
| Plasmodium berghei | Rodent (Grammomys surdaster) | AAF13063.1 | AF187232.1 |
| Plasmodium chabaudi | Rodent (Thamnonys rutilans) | AAA29499.1 | L22982.1 |
| Plasmodium coatneyi | Primate (Macaca fascicularis) | BAF74048.1 | AB266180.1 |
| Plasmodium knowlesi | Primate (Macaca fascicularis) | BAF74052.1 | AB266184.1 |
| Plasmodium fragile | Primate (Macaca sinica) | BAF74049.1 | AB266181.1 |
| Plasmodium cynomolgi | Primate (Macaca sinica) | BAF74063.1 | AB266195.1 |
| Plasmodium simiovale | Primate (Macaca sinica) | BAF74053.1 | AB266185.1 |
| Plasmodium vivax (Belem) | Human (Homo sapiens) | AAN86208.1 | AF435594.1 |
| Plasmodium vivax (Sal1) | Human (Homo sapies) | EDL45115.1 | AAKM01000007.1 |
| Plasmodium hylobati | Primate (Hylobati moloch) | BAF74050.1 | AB266182.1 |
| Plasmodium inui | Primate (Cynopithecus niger) | BAF74051.1 | AB266183.1 |
| Plasmodium malariae* | Human (Homo sapiens) | AAD42067.1 | AF138881.1 |
| Plasmodium malariae (MM1A) | Human (Homo sapiens) | TBD | FJ824669 |
| Plasmodium ovale (OM1A) | Human (Homo sapiens) | TBD | FJ824670 |
| Plasmodium ovale (OM1B) | Human (Homo sapiens) | TBD | FJ824671 |
| Plasmodium falciparum (K1) | Human (Homo sapiens) | CAA27070.1 | X03371.1 |
| Plasmodium falciparum (MAD20) | Human (Homo sapiens) | A26868 | X05624.2 |
| Plasmodium reichenowi | Chimpanzee (Pan troglodytes) | CAH10285.1 | AJ786604.1 |
| Plasmodium gallinaceum | Bird (Gallus gallus) | CAH10838.1 | AJ809338.1 |
Only partial sequence available.
Extensive sequence could not be obtained from the 5 kb Pm and Po MSP1 amplicons produced using degenerate primers. Therefore, amplification of the near full-length genes was repeated using species-specific forward and reverse primer pairs. The resulting amplicons (~5 kb) from each species were gel purified and served as templates for sequencing. Additional species-specific primers were designed from the newly acquired sequence for use in the subsequent reactions (primers not shown). This process was continued until sequence was obtained across the entirety of each gene except for the extreme termini. Specific primers generated during the sequencing of Po MSP1 gene from the initial P. ovale isolate enabled rapid determination of a second Po MSP1 gene sequence derived from a separate blood donor.
Adaptor-mediated or inverse PCR was used to generate amplicons encompassing the extreme ends of the MSP1 open reading frames.23 Sequence from these amplicons enabled final assembly of the complete coding regions for the Pm and Po MSP1 genes using Sequencher (version 4.5; Gene Codes, Ann Arbor, MI).
Sequence analysis of MSP1.
GenBank was searched for protein sequences related to the deduced Pm and Po MSP1 amino acid sequences by protein BLAST.22 Sixteen MSP1 sequences, representing 14 Plasmodium species, were included in the dataset used for subsequent protein sequence comparisons (Table 2). Pairwise alignments for determination of percent amino acid sequence identity were generated using the GAP program. Alignment of full-length MSP1 sequences from 16 Plasmodium species (Pm and Po included) was performed using the PILEUP program. The alignment was then manually refined for sequences bordering regions of low similarity (gaps) to maximize overall amino acid similarity within those regions. An average amino acid similarity score within a 10 residue sliding window was calculated for each position across the manually refined alignment of full-length MSP1 proteins using PLOTSIMILARITY and the blosum62.cmp scoring matrix, and the resulting values plotted versus alignment position. A shorter de-gapped alignment (1,280 positions) was generated by deletion of any position of the manually refined full-length MSP1 alignment in which a gap occurred. Both full-length and de-gapped alignments are available upon request. GAP, PILEUP, PRETTY, and PLOTSIMILARITY software programs are all members of the Wisconsin GCG package.24 MEGA4 software25 was used to align the two Po MSP1 DNA sequences using CLUSTALW. Alignment positions containing gaps were deleted before performing a codon-based Z-test on the de-gapped DNA alignment (5,124 nucleotides after gap deletion encoding 1,708 amino acids) using the bootstrap method (2,000 replicates, Nei-Gojobori p-distance model)26 to determine the degree of selection operating on this gene. The deduced protein sequences from Pm and Po MSP1 were analyzed for likely GPI anchor sites and signal peptide sequences using Big PI Predictor27 and SignalP,28 respectively.
Phylogenetic analysis of the manually refined de-gapped MSP1 amino acid sequence alignment was performed using MEGA4 software.25 Tree inference was made using neighbor-joining (NJ) methods. Distances were calculated by using the Jones-Taylor-Thornton29 amino acid substitution matrix and assuming uniform rates of substitution among sites. Assessment of tree robustness was determined by using the bootstrap test (1000 replicates). A bootstrap score of at least 70% was considered strong support for relatedness.30
Results
Identification of parasitemic donors.
Whole blood from Cameroon normal blood donors was screened by PCR for the presence of the Plasmodium 18S rRNA gene. Of the 82 specimens tested 30 (37%) were PCR positive, indicating the donors were parasitemic. Based on melting curve analysis of the 18S rRNA amplicons (data not shown), three potential P. malariae-infected donors, and four potential P. ovale-infected donors were identified. The three P. malariae-positive donors and two of the P. ovale-positive donors appeared to be co-infected with P. falciparum, as indicated by the presence of bimodal Tm profiles. The other two P. ovale-positive donors (OM1A and OM1B) were apparent single infections. The remaining 23 donors appeared to be infected only with P. falciparum.
Sequences of the 18S ribosomal DNA (rDNA) gene amplicons were determined for P. ovale specimens OM1A and OM1B to confirm the presence of P. ovale in these two donors, and to determine if they were derived from the classic- or variant-type P. ovale stains.31 BLAST analysis showed 100% homology of the amplicon sequences with “classic-type” P. ovale 18S rRNA gene sequences in GenBank (data not shown). For specimens identified as co-infected with P. falciparum and P. malariae, the 18S rRNA gene sequence chromatograms suggested the presence of multiple sequences. However, sequence obtained near the 5′ end of the MSP1 gene from one of these samples (MM1A) (see Materials and Methods) was greater than 96% identical to the 387 nucleotide P. malariae MSP1 sequence in GenBank (AF138881.1), confirming the presence of P. malariae in this sample.
Characteristics of the deduced Pm MSP1 and Po MSP1 proteins.
The complete Pm MSP1 (isolate MM1A) and Po MSP1 (isolate OM1A) genes encode proteins of 1,751 and 1,730 amino acids in length, respectively, each beginning with a predicted 19 amino acid signal peptide sequence. A nearly complete MSP1 gene sequence from a second P. ovale-infected specimen (OM1B) was also determined. Only the extreme 5′ end sequence was not obtained, which based on comparison to the OM1A Po MSP1 gene, corresponds to the first 8 amino acids of the signal peptide. The OM1B Po MSP1 gene encodes a 1,726 amino acid protein, assuming the additional 8 amino acids at the N-terminus. The two Po MSP1 sequences are 98.9% and 97.4% identical at the DNA and protein levels, respectively.
Sequence comparison.
To identify regions of interspecies conservation and variability across the entire MSP1 protein, an alignment of complete MSP1 amino acid sequences from 16 Plasmodium species was performed. This alignment was further aligned manually (available upon request). A plot of the average amino acid sequence similarity at each position within the final alignment is presented in Figure 1, revealing the presence of multiple interspecies conserved regions. These are interspersed with regions of greater divergence that encompass imperfect amino acid repeats varying in length and sequence. Relative positions of previously reported interspecies conserved MSP1 sequence blocks32 are shown, along with the positions of differences observed between the two Po MSP1 protein sequences (see Discussion).
Figure 1.
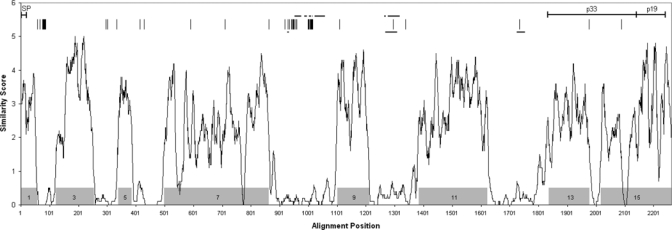
Interspecies comparison of MSP1 protein sequences. Full-length deduced MSP1 amino acid sequences from 16 Plasmodium species (Table 2) were aligned and an average amino acid similarity score was determined for successive 10 amino acid segments (alignment available upon request). Plasmodium falciparum, P. vivax, and P. ovale were represented by strains K1, Belem, and OM1A, respectively. Shaded boxes indicate locations of reported conserved sequence blocks.32 Along the top, positions of differences between deduced MSP1 amino acid sequences from P. ovale strains OM1A and OM1B are denoted by vertical lines. Locations of imperfect repeats (horizontal lines) are indicated for P. malariae (upper lines) and P. ovale (OM1A isolate, lower lines). Signal peptide (SP), p33 and p19 regions are also shown.
Pairwise alignments of complete MSP1 protein sequences representing all four major human-infectious Plasmodium species and P. knowlesi were performed, and the percent identity for each pair was determined (Table 3). The P. falciparum protein (K1 strain) shows the highest divergence with percent identities ranging from 44.1% to 45.7% versus the other four species. Percent identities using Pf MSP1 from the MAD20 strain were slightly lower (data not shown). Interspecies comparisons of all the other MSP1s exhibit percent identities in the 48.8% to 54.2% range, except for P. vivax and P. knowlesi, which are more closely related. Similar relative homologies were obtained when comparisons were limited to the more highly conserved p19 region.
Table 3.
Merozoite surface protein 1 (MSP1) amino acid sequence comparison
| (% identity) | |||||
|---|---|---|---|---|---|
| P. falciparum | P. vivax | P. knowlesi | P. malariae | P. ovale | |
| Plasmodium falciparum* | 51.7† | 51.1 | 48.3 | 52.3 | |
| Plasmodium vivax* | 45.4† | 82.0 | 55.6 | 58.4 | |
| Plasmodium knowlesi | 45.7 | 71.1 | 56.2 | 56.8 | |
| Plasmodium malariae | 44.1 | 51.4 | 48.8 | 52.8 | |
| Plasmodium ovale* | 44.7 | 54.2 | 53.5 | 51.9 | |
P. falciparum strain (K1), P. vivax strain (Belem), and P. ovale strain (OM1A).
Values below the diagonal = complete MSP1 open reading frame; values above the diagonal = MSP1-p19 only (ending at glycophosphatidylinositol [GPI] attachment residue).
The N-terminal sequences of MSP1 p42 and p19 have been experimentally determined for P. falciparum (strainT9/94) and the rodent parasite Plasmodium chabaudi (strain AS).12,33 Alignments of the regions encompassing the p42 and p19 cleavage sites for these two strains indicate that the cleavage positions are conserved within the context of the limited surrounding homology.10 Based on comparisons with amino acid sequences encompassing the P. falciparum and P. chabaudi cleavage sites, probable p42 and p19 cleavage sites for the Pm and Po MSP1 proteins were assigned (Figure 2, see Discussion).
Figure 2.
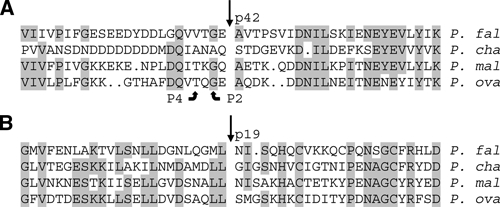
Predicted sites of primary and secondary merozoite surface protein 1 (MSP1) cleavage. Sequence alignments encompassing the known N-terminal cleavage sites (straight arrows) for (A) MSP1-p42 and (B) MSP1-p19 from Plasmodium falciparum strain T9/94 (P. fal) and Plasmodium chabaudi strain AS (P. cha), and the predicted cleavage sites for the homologous regions from Plasmodium malariae strain MM1A (P. mal) and Plasmodium ovale strain OM1A (P. ova) are shown. Aligned residues conserved in at least 3 of the 4 species are shaded, and the P2 and P4 positions are indicated for MSP1-p42.
The Pm and Po MSP1s contain regions near the C-termini that are homologous to the p33 and p19 segments of other MSP1s. Within the p19 region, the number and relative positions of cysteine residues is conserved compared with the same region of most other Plasmodium species (Figure 3). Likewise, the highly conserved sequence surrounding the C-terminal GPI anchor site of Plasmodium MSP1s is present in the P. malariae and P. ovale proteins.
Figure 3.

Alignment of C-terminal p19 regions of merozoite surface protein 1 (MSP1). The deduced amino acid sequences encompassing the predicted MSP1-p19 C-terminal regions (underlined) of the indicated Plasmodium species (denoted by P. and the first 3 letters of the species name) were aligned. P. falA and P. falB refer to Plasmodium falciparum strains K1 and MAD20, respectively, and P. viv refers to the Plasmodium vivax Belem strain. Amino acid differences (lower case letters) vs. the Consensus sequence (upper case letters) are shown. Gaps introduced to maximize alignment (dashes), positions identical to the Consensus sequence (dots), and positions within the Consensus lacking a plurality (X) are indicated. Positions of conserved cysteine residues are shaded. The predicted GPI anchor site (arrow) and the conserved residues (box) surrounding it are indicated.
Phylogenetic analysis.
To further examine the relatedness of the new MSP1 protein sequences to those from other Plasmodium species, we performed phylogenetic analysis on the manually refined de-gapped alignment described previously. As shown in Figure 4, the taxa from the reference dataset of MSP1 sequences (not including P. malariae or P. ovale MSP1) are segregated into four distinct clades encompassing rodents, Old World monkeys, birds, and Great Apes. The human parasite P. vivax is subsumed by the sequences derived from the Old World monkey clade. Bootstrap support for these clusters is 100%. This MSP1 sequence-based topology is consistent with that described by others.32 Plasmodium ovale and P. malariae occupy a bifurcating branch with a node deep in the tree but with low bootstrap support, and so do not show relatedness to a specific clade. The reason for this is unclear but may reflect unique sequence features/substitution patterns among MSP1 sequences from Po and Pm not observed among members of the other clades.
Figure 4.
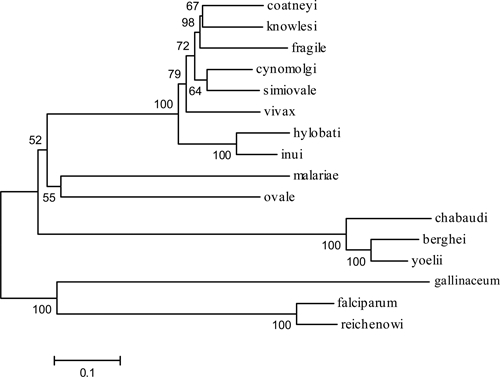
MSP1 phylogenetic tree. The evolutionary relatedness of MSP1 sequences was inferred using the neighbor-joining method. Only the amino acid sequences comprising the interspecies conserved blocks of MSP1 were examined, all regions containing gaps were not considered in the analysis. The evolutionary distances were computed using the JTT matrix-based method and are in the units of the number of amino acid substitutions per site. There were a total of 1,280 positions in the final dataset. The optimal tree is shown with the percentage of replicate trees in which the associated taxa cluster together based on the bootstrap test (1,000 replicates) are shown next to the branches.
Discussion
The purpose of this study was to obtain the MSP1 sequences from the remaining two human infectious Plasmodium species responsible for causing malaria in humans, namely, P. ovale and P. malariae. MSP1 genes from Plasmodium species infecting mammals exhibit sizes in the range of about 4,900 to 5,800 bp, encoding proteins of 1,630 to 1,929 amino acids (187 kD to 213 kD). Both Pm and Po MSP1 genes described in this study fall within these ranges. Furthermore, based on the presence of single long open reading frames and the lack of any extended gaps (outside of the variable regions) in the deduced amino acid sequences when aligned with other MSP1 proteins, the Pm and Po MSP1 genes are unlikely to contain introns, consistent with the absence of introns from all Plasmodium MSP1 genes in GenBank.
Previously, interspecies conserved blocks (ICBs) were identified on comparison of MSP1 amino acid sequences from all of the Plasmodium species listed in Table 2,32 with the exception of P. malariae and P. ovale. As would be expected, all these ICBs reside within regions of high interspecies amino acid sequence similarity described here (Figure 1). However, even within the ICBs (7 and 15 in particular), regions of very low similarity exist. A similarity profile based on alignment of full-length MSP1 protein sequences from the four human-infectious Plasmodium species plus P. knowlesi was very similar to the one in Figure 1 (data not shown), indicating that the interspecies pattern of conserved and divergent regions between MSP1s is maintained for P. malariae and P. ovale, and is representative of Plasmodium MSP1 as a whole.
A common feature of Plasmodium MSP1s is the presence of imperfect tandem amino acid repeats of varying sizes, sequences, and copy numbers, located within regions of high interspecies divergence.34,35 The Pm and Po MSP1s also harbor imperfect tandem peptide repeats, ranging in size from 2 to 19 amino acids, which are present in two regions of low interspecies similarity flanked between ICBs 7, 9, and 11. An additional repeat domain is observed further downstream for the Po MSP1 in a third non-conserved region between ICBs 11 and 13 (Figure 1).
Of the three unique amino acids immediately flanking the predicted Pm and Po MSP1 p42 cleavage sites, all are present at analogous positions flanking the P. falciparum or P. chabaudi sites (Figure 2). Furthermore, glycine and threonine residues are present in the P2 and P4 positions, respectively. This is consistent with the substrate specificity of the protease responsible for primary cleavage of Pf MSP1 to produce p42 (PfSUB1),36 which has orthologs in all Plasmodium species examined.37 Glycine or alanine in the P2 position is required for PfSUB1 activity, whereas valine, isoleucine, leucine, or threonine is present in the P4 position of all demonstrated PfSUB1 cleavage sites. Little is known about the substrate specificity of the PfSUB2 protease, which cleaves Pf MSP1 p42 to form p33 and p19.38 However, in P. falciparum and P. chabaudi this cleavage occurs immediately after a leucine residue that, with the exception of Plasmodium gallinaceum, is conserved among MSP1 alignments of all examined Plasmodium species,10 including P. malariae and P. ovale (Figure 3).
Upon invasion of the RBC, the MSP1 complex is shed, except for the p19 fragment, which remains covalently attached to the parasite membrane through a GPI anchor.10 A short peptide sequence “FCSSS” near the C terminus of MSP1 appears to be involved in the transamidase-mediated cleavage of the protein, generating a new carboxyl terminus to which the GPI anchor is attached. To date, this MSP1 sequence has been conserved across all Plasmodium species and isolates (Figure 3), with the exception of Plasmodium inui in which the middle serine residue is a tyrosine. Interestingly, both the Pm and Po MSP1s are predicted to have glycine substituted at the first serine residue. This serine is likely the site of GPI attachment for Pf MSP1,39 and presumably for all other Plasmodium species. Although glycine at this position of MSP1 is unique to the P. malariae and P. ovale proteins, GPI-mediated anchorage for other proteins through carboxyl glycine residues is not uncommon.40 Furthermore, software for predicting GPI attachment sites (Big-PI Predictor)27 identifies the glycine at this position as the most likely candidate for both the Pm and Po MSP1s.
Sequencing of a second Po MSP1 gene (OM1B isolate) allowed comparison of the two deduced protein sequences, revealing 97% sequence identity. However, interesting differences do exist, consisting of amino acid substitutions from one to five residues in length, in addition to deletions/insertions (Figure 1). These latter differences occur most often within the three imperfect repeat regions, and reflect variability in the number of repeats within each block. Most differences between the two Po MSP1s occur in the first three quarters of the protein, with only two differences (single amino acid substitutions) occurring within p33, and none within p19. With one or two exceptions, all of the sequence differences lie within regions of low interspecies similarity. There were a total of 6 synonymous and 51 non-synonymous mutations resulting in values for dS (the number of synonymous substitutions per synonymous site) and dN (the number of non-synonymous substitutions per non-synonymous site) of 0.00539 (std. error 0.00215) and 0.01197 (std. error 0.00164), respectively. A codon-based Z-test performed on a de-gapped alignment of the two Po MSP1 DNA sequences indicates that the probability of rejecting the null hypothesis of strict neutrality (dN = dS) is high (P ≤ 0.01). This, along with the fact that dN > dS, supports that the observed non-synonymous differences are caused by positive selection. Positive selection has also been reported to be acting upon both Pf and Pv MSP1 genes.32,41
On the basis of analysis of sequences from three different P. ovale genes, a recent report segregates P. ovale into two clades, classic and variant, that are as divergent as separate species.31 Analysis of partial 18S rRNA gene sequences from the two P. ovale isolates described here indicates they are both members of the classic clade (data not shown). It would be interesting to obtain the MSP1 sequence for a member of the P. ovale variant clade, and compare it to the sequences presented here to see if it supports the contention that the two clades represent separate species.
Phylogenetic analysis of Po and Pm MSP1 amino acid sequences against a reference data set of previously known sequences placed Pm and Po MSP1s on a separate deeply rooted bifurcated branch (Figure 4). However, because of the low bootstrap values and the long branch lengths, relatedness of these sequences with each other or with the reference clades could not be determined. These observations are consistent with Plasmodium phylogenies based on sequences of other genes, in which no common consensus is evident for the placement of P. malariae or P. ovale within the trees.42–45 The topology of the tree (Figure 4) with regards to the reference sequence branches leading to the Old World monkey MSP1 sequences (which include P. vivax), the rodent sequences, Great Ape/human, and bird sequences is supported by high bootstrap values (i.e., 100%), and is in agreement with that shown for MSP1 by Tanabe and others,32 and is in agreement with trees produced from other Plasmodium genes including cytochrome b,44,45 18S rDNA,42 and mitochondrial DNA.43 The reasons for the inability to clearly establish the placement of Pm and Po MSP1s in the tree is unclear, but may be caused by sampling bias as described for inclusion of long branch lengths in a dataset46 because both Po and Pm sequences provide relatively long branch lengths as compared with those taxa within a clade defined by high bootstrap support. However, these analyses show that the P. ovale and P. malariae MSP1 sequences are clearly unique and are not variants of Pf, Pv, or any other known Plasmodium isolates.
In conclusion, we have provided convincing evidence that the MSP1 genes from P. malariae and P. ovale have now been isolated, thus completing the inventory of MSP1 genes for the human-infectious Plasmodium species. Both deduced proteins exhibit sequence patterns typical of the MSP1 family, yet neither is so closely related to previously known family members, or to each other, that they would be considered species variants. Availability of these two new MSP1 proteins will be of considerable benefit for vaccine development and in assembly of an immunoassay directed to detect antibodies against the four major human-infectious Plasmodium species. To this end, studies evaluating and comparing recombinant MSP1 proteins from all four human-infectious Plasmodium species for antibody detection are underway.
Acknowledgments
We are grateful to Gerald Schochetman and Catherine Brennan of Abbott Laboratories, USA; and Dora Mbanya, University of Yaoundé I, Yaoundé, Cameroon; Lazare Kaptué, Université des Montagnes, Bangangté, Cameroon; and Lutz G. Gürtler, Goethe University, Frankfurt, Germany for providing the clinical samples used in this investigation.
Footnotes
Authors' addresses: Larry Birkenmeyer, A. Scott Muerhoff, George Dawson, and Suresh M. Desai, Abbott Diagnostics, Infectious Diseases R&D, Abbott Park, IL, E-mails: larry.birkenmeyer@abbott.com, scott.muerhoff@abbott.com, george.dawson@abbott.com, and suresh.desai@abbott.com.
References
- 1.Korenromp E, Miller J, Nahlen B, Wardlaw T, Young M. World Malaria Report. 2005. http://rbm.who.int/wmr2005/html/toc.htm Available at.
- 2.Collins WE, Barnwell JW. Plasmodium knowlesi: finally being recognized. J Infect Dis. 2009;199:1107–1108. doi: 10.1086/597415. [DOI] [PubMed] [Google Scholar]
- 3.Lysenko AJ, Beljaev AE. An analysis of the geographical distribution of Plasmodium ovale. Bull World Health Organ. 1969;40:383–394. [PMC free article] [PubMed] [Google Scholar]
- 4.Mueller I, Zimmerman PA, Reeder JC. Plasmodium malariae and Plasmodium ovale–the “bashful” malaria parasites. Trends Parasitol. 2007;23:278–283. doi: 10.1016/j.pt.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox-Singh J, Davis TM, Lee KS, Shamsul SS, Matusop A, Ratnam S, Rahman HA, Conway DJ, Singh B. Plasmodium knowlesi malaria in humans is widely distributed and potentially life threatening. Clin Infect Dis. 2008;46:165–171. doi: 10.1086/524888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White NJ. Plasmodium knowlesi: the fifth human malaria parasite. Clin Infect Dis. 2008;46:172–173. doi: 10.1086/524889. [DOI] [PubMed] [Google Scholar]
- 7.Florens L, Washburn MP, Raine JD, Anthony RM, Grainger M, Haynes JD, Moch JK, Muster N, Sacci JB, Tabb DL, Witney AA, Wolters D, Wu Y, Gardner MJ, Holder AA, Sinden RE, Yates JR, Carucci DJ. A proteomic view of the Plasmodium falciparum life cycle. Nature. 2002;419:520–526. doi: 10.1038/nature01107. [DOI] [PubMed] [Google Scholar]
- 8.Fandeur T, Volney B, Peneau C, de Thoisy B. Monkeys of the rainforest in French Guiana are natural reservoirs for P. brasilianum/P. malariae malaria. Parasitology. 2000;120:11–21. doi: 10.1017/s0031182099005168. [DOI] [PubMed] [Google Scholar]
- 9.Gerold P, Schofield L, Blackman MJ, Holder AA, Schwarz RT. Structural analysis of the glycosyl-phosphatidylinositol membrane anchor of the merozoite surface proteins-1 and -2 of Plasmodium falciparum. Mol Biochem Parasitol. 1996;75:131–143. doi: 10.1016/0166-6851(95)02518-9. [DOI] [PubMed] [Google Scholar]
- 10.Blackman MJ. Proteases involved in erythrocyte invasion by the malaria parasite: function and potential as chemotherapeutic targets. Curr Drug Targets. 2000;1:59–83. doi: 10.2174/1389450003349461. [DOI] [PubMed] [Google Scholar]
- 11.Blackman MJ, Whittle H, Holder AA. Processing of the Plasmodium falciparum major merozoite surface protein-1: identification of a 33-kilodalton secondary processing product which is shed prior to erythrocyte invasion. Mol Biochem Parasitol. 1991;49:35–44. doi: 10.1016/0166-6851(91)90128-s. [DOI] [PubMed] [Google Scholar]
- 12.O'Dea KP, McKean PG, Harris A, Brown KN. Processing of the Plasmodium chabaudi chabaudi AS merozoite surface protein 1 in vivo and in vitro. Mol Biochem Parasitol. 1995;72:111–119. doi: 10.1016/0166-6851(95)00090-n. [DOI] [PubMed] [Google Scholar]
- 13.Blackman MJ, Dennis ED, Hirst EM, Kocken CH, Scott-Finnigan TJ, Thomas AW. Plasmodium knowlesi: secondary processing of the malaria merozoite surface protein-1. Exp Parasitol. 1996;83:229–239. doi: 10.1006/expr.1996.0069. [DOI] [PubMed] [Google Scholar]
- 14.Tolle R, Fruh K, Doumbo O, Koita O, N'Diaye M, Fischer A, Dietz K, Bujard H. A prospective study of the association between the human humoral immune response to Plasmodium falciparum blood stage antigen gp190 and control of malarial infections. Infect Immun. 1993;61:40–47. doi: 10.1128/iai.61.1.40-47.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egan AF, Chappel JA, Burghaus PA, Morris JS, McBride JS, Holder AA, Kaslow DC, Riley EM. Serum antibodies from malaria-exposed people recognize conserved epitopes formed by the two epidermal growth factor motifs of MSP1(19), the carboxy-terminal fragment of the major merozoite surface protein of Plasmodium falciparum. Infect Immun. 1995;63:456–466. doi: 10.1128/iai.63.2.456-466.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faber BW, Remarque EJ, Morgan WD, Kocken CH, Holder AA, Thomas AW. Malaria vaccine-related benefits of a single protein comprising Plasmodium falciparum apical membrane antigen 1 domains I and II fused to a modified form of the 19-kilodalton C-terminal fragment of merozoite surface protein 1. Infect Immun. 2007;75:5947–5955. doi: 10.1128/IAI.01804-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stowers AW, Cioce V, Shimp RL, Lawson M, Hui G, Muratova O, Kaslow DC, Robinson R, Long CA, Miller LH. Efficacy of two alternate vaccines based on Plasmodium falciparum merozoite surface protein 1 in an Aotus challenge trial. Infect Immun. 2001;69:1536–1546. doi: 10.1128/IAI.69.3.1536-1546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim S, Ahn HJ, Kim TS, Nam HW. ELISA detection of vivax malaria with recombinant multiple stage-specific antigens and its application to survey of residents in endemic areas. Korean J Parasitol. 2003;41:203–207. doi: 10.3347/kjp.2003.41.4.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitchen AD, Lowe PH, Lalloo K, Chiodini PL. Evaluation of a malarial antibody assay for use in the screening of blood and tissue products for clinical use. Vox Sang. 2004;87:150–155. doi: 10.1111/j.1423-0410.2004.00561.x. [DOI] [PubMed] [Google Scholar]
- 20.Walker-Abbey A, Djokam RR, Eno A, Leke RF, Titanji VP, Fogako J, Sama G, Thuita LH, Beardslee E, Snounou G, Zhou A, Taylor DW. Malaria in pregnant Cameroonian women: the effect of age and gravidity on submicroscopic and mixed-species infections and multiple parasite genotypes. Am J Trop Med Hyg. 2005;72:229–235. [PubMed] [Google Scholar]
- 21.Mangold KA, Manson RU, Koay ES, Stephens L, Regner M, Thomson RB, Jr, Peterson LR, Kaul KL. Real-time PCR for detection and identification of Plasmodium spp. J Clin Microbiol. 2005;43:2435–2440. doi: 10.1128/JCM.43.5.2435-2440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 24.Genetics Computer Group . Wisconsin Package Version 11.0. San Diego, CA: Accelrys Inc; 2005. [Google Scholar]
- 25.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 26.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 27.Eisenhaber B, Bork P, Eisenhaber F. Prediction of potential GPI-modification sites in proprotein sequences. J Mol Biol. 1999;292:741–758. doi: 10.1006/jmbi.1999.3069. [DOI] [PubMed] [Google Scholar]
- 28.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 29.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 30.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 31.Win TT, Jalloh A, Tantular IS, Tsuboi T, Ferreira MU, Kimura M, Kawamoto F. Molecular analysis of Plasmodium ovale variants. Emerg Infect Dis. 2004;10:1235–1240. doi: 10.3201/eid1007.030411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanabe K, Escalante A, Sakihama N, Honda M, Arisue N, Horii T, Culleton R, Hayakawa T, Hashimoto T, Longacre S, Pathirana S, Handunnetti S, Kishino H. Recent independent evolution of msp1 polymorphism in Plasmodium vivax and related simian malaria parasites. Mol Biochem Parasitol. 2007;156:74–79. doi: 10.1016/j.molbiopara.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 33.Blackman MJ, Ling IT, Nicholls SC, Holder AA. Proteolytic processing of the Plasmodium falciparum merozoite surface protein-1 produces a membrane-bound fragment containing two epidermal growth factor-like domains. Mol Biochem Parasitol. 1991;49:29–33. doi: 10.1016/0166-6851(91)90127-r. [DOI] [PubMed] [Google Scholar]
- 34.Deleersnijder W, Hendrix D, Bendahman N, Hanegreefs J, Brijs L, Hamers-Casterman C, Hamers R. Molecular cloning and sequence analysis of the gene encoding the major merozoite surface antigen of Plasmodium chabaudi chabaudi IP-PC1. Mol Biochem Parasitol. 1990;43:231–244. doi: 10.1016/0166-6851(90)90148-f. [DOI] [PubMed] [Google Scholar]
- 35.Zhong H, Fan JY, Yang S, Davidson EA. Cloning and characterization of the merozoite surface antigen 1 gene of Plasmodium berghei. Am J Trop Med Hyg. 1999;60:994–999. doi: 10.4269/ajtmh.1999.60.994. [DOI] [PubMed] [Google Scholar]
- 36.Koussis K, Withers-Martinez C, Yeoh S, Child M, Hackett F, Knuepfer E, Juliano L, Woehlbier U, Bujard H, Blackman MJ. A multifunctional serine protease primes the malaria parasite for red blood cell invasion. EMBO J. 2009;28:725–735. doi: 10.1038/emboj.2009.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Withers-Martinez C, Jean L, Blackman MJ. Subtilisin-like proteases of the malaria parasite. Mol Microbiol. 2004;53:55–63. doi: 10.1111/j.1365-2958.2004.04144.x. [DOI] [PubMed] [Google Scholar]
- 38.Harris PK, Yeoh S, Dluzewski AR, O'Donnell RA, Withers-Martinez C, Hackett F, Bannister LH, Mitchell GH, Blackman MJ. Molecular identification of a malaria merozoite surface sheddase. PLoS Pathog. 2005;1:241–251. doi: 10.1371/journal.ppat.0010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naik RS, Branch OH, Woods AS, Vijaykumar M, Perkins DJ, Nahlen BL, Lal AA, Cotter RJ, Costello CE, Ockenhouse CF, Davidson EA, Gowda DC. Glycosylphosphatidylinositol anchors of Plasmodium falciparum: molecular characterization and naturally elicited antibody response that may provide immunity to malaria pathogenesis. J Exp Med. 2000;192:1563–1576. doi: 10.1084/jem.192.11.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Udenfriend S, Kodukula K. How glycosylphosphatidylinositol-anchored membrane proteins are made. Annu Rev Biochem. 1995;64:563–591. doi: 10.1146/annurev.bi.64.070195.003023. [DOI] [PubMed] [Google Scholar]
- 41.Hughes AL. Positive selection and interallelic recombination at the merozoite surface antigen-1 (MSA-1) locus of Plasmodium falciparum. Mol Biol Evol. 1992;9:381–393. doi: 10.1093/oxfordjournals.molbev.a040730. [DOI] [PubMed] [Google Scholar]
- 42.Leclerc MC, Hugot JP, Durand P, Renaud F. Evolutionary relationships between 15 Plasmodium species from new and old world primates (including humans): an 18S rDNA cladistic analysis. Parasitology. 2004;129:677–684. doi: 10.1017/s0031182004006146. [DOI] [PubMed] [Google Scholar]
- 43.Ollomo B, Durand P, Prugnolle F, Douzery E, Arnathau C, Nkoghe D, Leroy E, Renaud F. A new malaria agent in African hominids. PLoS Pathog. 2009;5:e1000446. doi: 10.1371/journal.ppat.1000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perkins SL, Schall JJ. A molecular phylogeny of malarial parasites recovered from cytochrome b gene sequences. J Parasitol. 2002;88:972–978. doi: 10.1645/0022-3395(2002)088[0972:AMPOMP]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 45.Rich SM, Leendertz FH, Xu G, LeBreton M, Djoko CF, Aminake MN, Takang EE, Diffo JL, Pike BL, Rosenthal BM, Formenty P, Boesch C, Ayala FJ, Wolfe ND. The origin of malignant malaria. Proc Natl Acad Sci USA. 2009;106:14902–14907. doi: 10.1073/pnas.0907740106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van de Peer Y, Baldauf SL, Doolittle WF, Meyer A. An updated and comprehensive rRNA phylogeny of (crown) eukaryotes based on rate-calibrated evolutionary distances. J Mol Evol. 2000;51:565–576. doi: 10.1007/s002390010120. [DOI] [PubMed] [Google Scholar]


