Abstract
Plants, as sessile organisms, need to sense and adapt to heterogeneous environments and have developed sophisticated responses by changing their cellular physiology, gene regulation, and genome stability. Recent work demonstrated heritable stress effects on the control of genome stability in plants—a phenomenon that was suggested to be of epigenetic nature. Here, we show that temperature and UV-B stress cause immediate and heritable changes in the epigenetic control of a silent reporter gene in Arabidopsis. This stress-mediated release of gene silencing correlated with pronounced alterations in histone occupancy and in histone H3 acetylation but did not involve adjustments in DNA methylation. We observed transmission of stress effects on reporter gene silencing to non-stressed progeny, but this effect was restricted to areas consisting of a small number of cells and limited to a few non-stressed progeny generations. Furthermore, stress-induced release of gene silencing was antagonized and reset during seed aging. The transient nature of this phenomenon highlights the ability of plants to restrict stress-induced relaxation of epigenetic control mechanisms, which likely contributes to safeguarding genome integrity.
Keywords: Abiotic stress, epigenome stability, histone acetylation, gene silencing
INTRODUCTION
Previous findings from animals suggest that adaptation at the genome level involves variations in the control of epigenetic imprints in response to environmental cues, which can be short-lived or persistent, resulting in their transmission into subsequent generations (Weaver et al., 2004; Anway et al., 2005; Rando and Verstrepen, 2007). It is well documented in plants that adverse environmental conditions cause long-term effects on genome stability, leading to heritable alterations in genome structure (Cullis, 1986; Ries et al., 2000; Kovalchuk et al., 2000; Takeda et al., 2001; Lucht et al., 2002; Kashkush et al., 2003). In Arabidopsis, for example, stress-induced genetic instability is transmitted to non-stressed daughter generations, which was suggested to arise as a consequence of an altered epigenetic status affecting the somatic recombination machinery (Molinier et al., 2006; Boyko et al., 2007; Lukens and Zhan, 2007). More recently, however, this view has been challenged by another report, suggesting that transgenerational stress effects are not a general response in Arabidopsis and may occur in a rather stochastic manner (Pecinka et al., 2009). Moreover, direct evidence for a heritable impact of stress on the epigenetic status in plants has not been provided so far. We therefore determined activity and epigenetic status of a transcriptionally silenced GUS transgene (TS–GUS) together with that of silent endogenous loci in stressed Arabidopsis populations (Morel et al., 2000; see Methods).
We demonstrate that temperature and UV-B stress resulted in an immediate release of gene silencing of the transgene as well as of endogenous loci. Transcriptional reactivation correlated with changes in chromatin conformation and histone acetylation, but was not associated with pronounced alterations in cytosine methylation. Stress effects on the control of transgene silencing were heritable, but limited to areas of a small number of cells, restricted to only two progeny generations and could further be antagonized and reset by seed aging. Our findings demonstrate a transient transgenerational impact of stress on epigenetic regulatory mechanisms, which highlights the ability of plants to balance the tolerance of epigenetic relaxation to induce diversity and to safeguard genome integrity.
RESULTS
Abiotic Stress Alleviates Silencing of TS–GUS
To determine the impact of adverse environments on the control of transgene silencing, we analyzed effects of abiotic stress on the activity of TS–GUS (Supplemental Table 1). Both UV-B radiation and extreme temperatures resulted in a strong reactivation of TS–GUS activity (Figure 1), reflected in large GUS-positive areas specifically after 1 week of recovery from stress treatments. By contrast, only small GUS-positive areas could be observed in non-stressed control plants (Figure 1).
Figure 1.
Phenotypes and TS–GUS Activity of Transgenic Arabidopsis thaliana Plants Immediately and 1 Week after UV-B (A), Heat (B), and Freezing (C) Stress.
Blue areas indicate alleviation of TS–GUS silencing, which is very pronounced in response to heat and UV-B stress ctr.: non-stressed individuals in the same developmental stage. Arrow indicates GUS-positive organs that developed after stress application. Bar = 2 mm.
The elevated levels of TS–GUS activity correlated with a pronounced transcriptional induction of the transgene as determined by qRT–PCR analysis (Figure 2A). In line with the observation that various stresses induce the TS–GUS transgene, we found that several silent endogenous transposable elements including LINEs and TSI (for TRANSCRIPTIONALLY SILENT INFORMATION; Steimer et al., 2000) exhibited a pronounced alleviation of gene silencing upon stress treatment (Figure 2A–2C). These findings indicate that environmental cues can modify the epigenetic status of a silent transgene as well as of endogenous loci. Persistent alterations in the expression status of silenced loci have been correlated with adjustments in their DNA methylation pattern (Vaillant and Paszkowski, 2007). However, a systematic analysis of the methylation status of the TS–GUS locus and of LINEs did not reveal any pronounced changes in symmetric (CG and CHG) and asymmetric (CHH) cytosine methylation, under conditions when transcription of these loci was drastically induced (Figure 3A and Supplemental Figure 1). Similarly, analysis of global cytosine methylation of the Arabidopsis genome did not reveal significant alterations in response to our temperature stress conditions (Supplemental Figure 2). Taken together, these findings demonstrate that transcriptional reactivation of silent loci upon stress treatment does not necessarily correlate with prominent variations in DNA methylation.
Figure 2.
Expression of TS–GUS and Silenced Transposable Elements in Response to Abiotic Stress.
(A) qRT–PCR performed on TS–GUS plants in the stressed S0 generation and in non-stressed S1 progeny. Transcript levels of TS–GUS and of a non-LTR retrotransposon (LINE039) show a significant increase in the S0 but not in the S1 generation. Expression levels were normalized to non-stressed controls (ctr). Standard deviations are indicated as bars. Similarly, transcript levels of TSI (B) and of additional non-LTR retrotransposons ((C); LINE018, LINE118, LINE315) show a significant increase in the S0 but not in the S1 generation.
Figure 3.
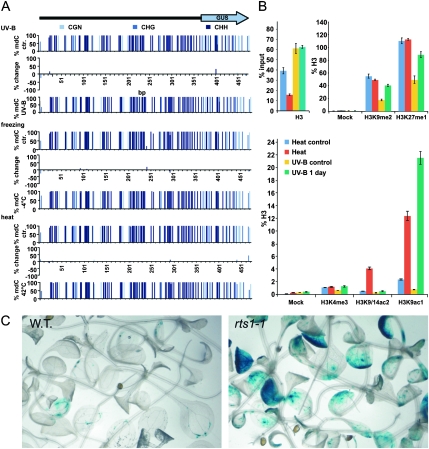
Analysis of DNA Methylation and of the Chromatin Status at the TS–GUS Locus.
(A) Graphic representation of the DNA methylation of TS–GUS in response to UV-B and temperature stress. Ordinates correspond to percent methylation in the entire set of samples analyzed by bisulfite sequencing (n = 20). Differences between samples derived from control (top) and stressed (bottom) plants are indicated as bars (middle). Nucleotide positions are indicated at the x-axes.
(B) Chromatin immunoprecipitation performed with chromatin derived from UV-B, heat-stressed, and non-stressed control plants. Top left: histone H3 occupancy at the TS–GUS locus under ambient (control) and stressed conditions. Shown are values after normalization to the corresponding input fraction. Top right; bottom: histone H3 modifications at the TS–GUS locus under ambient (control) and stressed conditions. Values are normalized to control IPs performed with non-discriminating antibodies against histone H3. Standard deviations are indicated as bars.
(C) Comparison of TS–GUS activity in 14-day-old wild-type (WT, left) and rts1-1 (right) plantlets.
In further experiments, we asked whether alterations in histone modifications might correlate with stress-mediated release of TS–GUS silencing and with activation of silent LINEs. No stress effects on global histone acetylation could be detected in Western blots performed with chromatin derived from stressed and control plants (Supplemental Figure 3). We therefore analyzed chromatin directly at specific loci. To this end, chromatin was precipitated from UV-B-, heat-stressed, and control TS–GUS plants by using antibodies specific for histone H3 and some of its post-translational modifications. Normalization of histone H3 precipitated from stressed and control samples to the corresponding input fraction revealed a reduction of histone H3 at TS–GUS and LINE039 specifically in heat-stressed samples, suggestive of a less condensed chromatin conformation (Figure 3B and Supplemental Figure 4). Assessment of histone H3 acetylation revealed a dramatic increase in the numbers of these chromatin modifications that are typically associated with actively transcribed loci (Figure 3B and Supplemental Figure 4; Berger, 2007). Remarkably, heat and UV-B stress triggered distinct, locus-specific effects on histone acetylation, with heat stress causing an increase in the amounts of histone H3 acetylated at lysine 9 (H3K9ac1) or at lysine 9 and 14 (H3K9/14ac2), whilst UV-B predominantly affected H3K9ac1 levels (Figure 3B and Supplemental Figure 4). In comparison to the dramatic changes of histone acetylation, levels of activating and repressive forms of histone H3 methylation at TS–GUS and LINE039 showed only minor changes or remained unchanged in response to stress (Figure 3B and Supplemental Figure 4).
Our findings indicate that stress-induced variations in histone acetylation correlate with a release of TS–GUS silencing. To identify genetic determinants involved in the control of TS–GUS activity, we introduced the transgene into plants deficient for the histone deacetylase HDA6, which exhibits defects in transcriptional gene silencing and in environmentally controlled variations of chromatin conformation (Aufsatz et al., 2002; Probst et al., 2004; Tessadori et al., 2009). We observed a pronounced increase in reporter activity in rts1-1 (a likely hda6 null allele; Aufsatz et al., 2002), providing a genetic link between the control of histone acetylation and the transcriptional status of TS–GUS (Figure 3C).
Limited Inheritance of Stress-Induced Release of TS–GUS Silencing
To investigate whether stress-induced release of TS–GUS silencing can be transmitted to non-stressed daughter generations, three successive generations were analyzed for transgene reactivation. When we compared the progeny of stressed and of non-stressed parental plants, we observed an increased number of small areas of cells showing GUS activity, indicating a less stringent control of the silenced status of TS–GUS in the progeny of stressed plants (Figure 4A, 4C, and Supplemental Tables 1 and 2). The F1 progeny of reciprocal crosses, in which gametes of stressed wild-type parents were combined with those of non-stressed TS–GUS plants, also exhibited a significant increase in TS–GUS reactivation frequency (Figure 4B and Supplemental Figure 5), demonstrating that changed TS–GUS activity is controlled by inherited, stress-modulated trans-acting factors.
Figure 4.
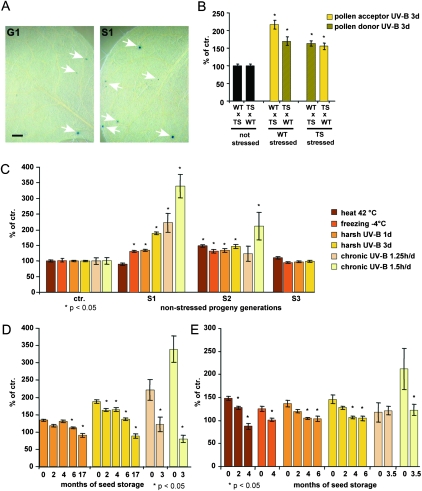
Heritability and Resetting of Stress Effects on TS–GUS Activity.
(A) TS–GUS activity (arrows) on rosette leaves of non-stressed progeny (G1, left; S1, right) derived from UV-B stressed (S0) and non-stressed (G0) parental plants. Bar = 0.5 mm.
(B) TS–GUS reactivation in F1 plants of reciprocal crosses, for which one parent has been exposed to UV-B stress. Asterisks indicate a significant difference (p < 0.05) between the progeny of these crosses and the F1 progeny derived from crosses performed with non-stressed parents (= 100%).
(C) Quantitative comparison of TS–GUS reactivation in the progeny of stressed plants (S1–S3). All values are normalized to GUS-positive areas of same-generation, non-stressed controls grown and scored in parallel (six). UV-B and freezing stress caused a significant increase in GUS reactivation in the S1 and S2 progeny, which was no longer detectable in the S3 generation. In heat-stressed plants, a significant increase in TS–GUS reactivation was observed in the S2 generation, which was no longer detected in the S3 generation. Asterisks indicate a significant difference (p < 0.05) between the progeny of stressed plants and their respective non-stressed controls (= 100%).
(D, E) Seed age correlates with resetting of TS–GUS reactivation in the S1 (D) and the S2 (E) generation. Asterisks indicate a significant difference (p < 0.05) when comparing TS–GUS activity in seedlings scored immediately after seed harvesting with TS–GUS activity in seedlings after seed storage for the time periods indicated. All values are normalized to GUS-positive areas of non-stressed controls (G1 and G2) grown in parallel (= 100%).
The increased number of GUS-positive areas remained detectable for only up to two progeny generations not exposed to stress. In the third generation, the number of GUS staining areas was comparable to control plants (Figure 4C). Moreover, transcript levels of either TS–GUS or endogenous LINEs were similar in the progeny of stressed and non-stressed parental plants (Figure 2), further suggesting that on the whole-plant level, heritable stress effects are limited and that most somatic cells do not exhibit a compromised control of gene silencing.
Heritable Release of Gene Silencing Is Antagonized by Seed Aging
Our results provide evidence for environmental parameters heritably affecting the status of an epigenetic read-out in higher plants. However, in contrast to the pronounced responses that we observed in plants directly after stress exposure, heritability of these effects was limited. We therefore searched for regulatory switches that might restrict the inheritance of stress effects on TS–GUS activity. The lifecycle of Arabidopsis involves a stage of seed dormancy, characterized by a shutdown of most metabolic activities, to endure environments unfavorable for germination (Finkelstein et al., 2008). To test for a potential influence of this resting stage on transgenerational stress effects, we scored TS–GUS activity in S1 and S2 plants germinated either immediately or several months after seed harvest (see Methods and Supplemental Tables 1 and 2). These experiments revealed that heritable effects of UV-B and temperature stress on TS–GUS reactivation reproducibly declined in dependence on the duration of seed storage (Figure 4D and 4E). Collectively, these findings further demonstrate the ability of plants to restrict stress-induced relaxation of epigenetic control mechanisms, which likely contributes to safeguarding genome integrity.
DISCUSSION
Environmental cues induce changes in metabolism, energy allocation, and growth (Beana-Gonzalez, 2010) that are partially caused through epigenetic reprogramming and result in short- or long-lived changes of transcriptional states (Takeda et al., 2001; Liu et al., 2009; Alexandre et al., 2009). Stress induces the reactivation of silent transposable elements, which, in the case of subsequent transposition, results in heritable effects on genome stability and on endogenous gene expression (Takeda et al., 2001; Kashkush et al., 2003). In addition, more recent results show that environmental stress promotes genetic instability by heritable epigenetic effects on the plant somatic recombination machinery (Molinier et al., 2006; Boyko et al., 2007) and on the activity of epialleles (Lukens and Zhan, 2007). Thus, transgenerational effects of stress on epigenetic regulation can affect the control of genome stability in multiple ways and consequently play a relevant role in adaptive evolution by creating genetic diversity (McClintock, 1984; Rando and Verstrepen, 2007).
This study addresses immediate and heritable effects of abiotic stresses, such as high and low-temperature stress and UV-B irradiation, on the epigenetic regulation of the silent TS–GUS transgene locus (Morel et al., 2000). Our results reveal strong immediate but limited heritable stress effects on the control of plant gene silencing: in the stressed generation, the silent TS–GUS transgene as well as endogenous silent transposons were transcriptionally reactivated. Although strong with respect to its initial amplitude, the reactivated state persisted only transiently and was eventually reversed in tissue formed after stress treatments in the recovery phase (Supplemental Figure 6). Alleviation of TS–GUS silencing was associated with prominent changes in histone acetylation but not paralleled by changes in DNA methylation. Such persisting cytosine methylation, which is inherited over mitotic cell divisions, could serve as a platform for the recruitment of histone-modifying factors that mediate the re-establishment of silent chromatin resulting in re-silencing in recovered tissue after stress conditions cease (Bird and Wolffe, 1999; Fuks et al., 2003; Li et al., 2006). However, our findings indicate that a stress-mediated increase in histone acetylation is sufficient to transiently overrule the silenced status of even hypermethylated loci, such as TS–GUS or LINEs. We cannot rule out that transient changes in DNA methylation occur at some point during the stress treatments and are important for the establishment of transcriptional reactivation. For heat-stressed plants, however, methylation and expression data were obtained from plants without a recovery phase directly after stress (see Methods), which would leave only a very narrow time frame for transient demethylation and subsequent reestablishment of methylation.
Our results with regard to stress-mediated increase in histone acetylation levels are consistent with a previous report demonstrating an increase of histone H3 lysine 9 acetylation at stress-responsive promoters under drought stress conditions (Kim et al., 2008). Similar observations have been made with regard to UV-B effects on histone acetylation and chromatin remodeling in maize and Arabidopsis (Casati et al., 2008; Cloix and Jenkins, 2008). Moreover, the importance of histone acetylation states for plant acclimation and stress tolerance has been highlighted by a recent report demonstrating that mutants in HOS15, a likely constituent of repressive histone deacetylase complexes, are hypersensitive to freezing temperatures (Zhu et al., 2008).
In non-stressed progeny, heritable stress effects resulted in a significant increase in small areas showing GUS activity. These areas are restricted to a few neighboring cells, which might reflect a single reactivation event that is clonally inherited or the coordinated response of adjacent cells to positional cues. The small area size suggests that this failure of TS–GUS silencing occurs late in development and argues against the inheritance of an active TS–GUS epiallele from previous stressed generations, which would be expected to result in much larger areas (Stam, 2009). Consistently, reciprocal crosses of non-stressed TS–GUS plants and stressed wild-type plants also exhibit an elevated number of GUS-positive areas in the F1 generation. This suggests that the observed heritable stress effects on TS–GUS are mediated by one or more trans-acting factors that are modulated upon stress exposure to affect TS–GUS silencing in non-stressed progeny. Our data show that the histone deacetylase HDA6 could represent one of those trans-acting determinants that might impinge on the epigenetic status of TS–GUS and of endogenous loci as well. In this context, it is remarkable that besides its role in the control of histone acetylation, HDA6 also acts as a transducer of environmental signals on overall chromatin organization (Tessadori et al., 2009). Similar to the role of histone deacetylases in promoting flowering (vernalization; Bond et al., 2009) and in mediating salt and drought tolerance (Sridha and Wu, 2006), it seems possible that a stressful environment modulates expression or selectivity of HDA6 and additional constituents of epigenetic integrity. This could account for a less stringent control of the epigenetic status, which then is reset in remote progeny generations of stress-exposed plants. Our results are consistent with findings by Molinier and coworkers, who demonstrated heritable effects of both abiotic and biotic stresses on somatic homologous recombination (SHR; Molinier et al., 2006). Yet, we observed that heritable stress effects on epigenetic markers are highly restricted, which is in line with a recent report demonstrating a limited heritable impact of abiotic stress on SHR (Pecinka et al., 2009). Similar to TS–GUS, silencing of several tested LINE elements was relaxed upon stress treatment. Thus, it seems possible that heritable stress effects on LINE expression are also manifested in progeny plants.
Epigenetic memory—namely the inheritance of epigenetic effects into subsequent generations—has been demonstrated in flies (Maurange and Paro, 2002) as well as in mammalian cells (Feng et al., 2006) and seemingly arises as a result of an incomplete erasure of epigenetic marks during gametogenesis and/or embryogenesis. Likewise, our results suggest that epigenetic modulation of the TS–GUS silencing machinery resists reversion in both the plant male and female germline to a certain extent. However, we also found that the observed heritability of TS–GUS reactivation is highly sensitive to resetting at another stage of plant development during seed aging. Epigenetic regulation plays an essential role during seed development (Zhang and Ogas, 2009). In this context, resetting mechanisms could act via pathways establishing seed dormancy (Finkelstein et al., 2008) or via the germination-coupled repair of double-strand DNA breaks that accumulate in aged seeds (Costa et al., 2001). Such resetting of stress-mediated effects resembles transgenerational reprogramming in gametes of non-plant organisms, which is essential for implementing developmental programs during embryogenesis (Youngson and Whitelaw, 2008) and, recently, similar effects have been described for plants (Jahnke and Scholten, 2009). By analogy, a mechanism, acting in the erasure of stress-induced epigenetic memory allows modulation of its inheritance to progeny, which could be essential for safeguarding plant genome integrity in subsequent generations. Nevertheless, the nature of mechanisms by which stress could reversibly affect inheritance of epigenetic states remains to be determined.
METHODS
Plant Material and Growth Conditions
The homozygous Arabidopsis thaliana line L5 (ecotype Columbia) harbors multiple copies of a transcriptionally silenced 35Spro::GUS marker gene (TS–GUS; Elmayan et al., 1998; Morel et al., 2000; Probst et al., 2004). Wild-type Arabidopsis plants (Columbia accession) were used to determine the effects of stress treatments on plant phenotypes. S1 plants are defined as the non-stressed progeny of stress-exposed S0 plants. S2 and S3 plants correspond to the subsequent non-stressed generations. G1, G2, and G3 correspond to non-stressed control generations that were analyzed in parallel to the stressed populations.
Eight to 10-day-old in vitro grown plants were transferred to soil and cultivated further until stress treatment. Stress treatments were administered before bolting on 3-week-old plants. For heat stress, plants grown under long-day conditions (16/8 h light/dark, 40 μmol m−2 s−1 white light) were exposed to 42°C for 48 h. For freezing stress, plants grown under short-day conditions (8/16 h light/dark, 70 μmol m−2 s−1 white light) were acclimated at 4°C for 1 week (8/16 h light/dark, 50 μmol m−2 s−1 white light) and were then subjected to freezing stress at –4°C for 24 h in the dark. For harsh short-term UV-B stress, plants grown under long-day conditions (16/8 h light/dark, 100 μmol m−2 s−1 white light) were exposed for 1 or 3 d to 4 μmol m−2 s−1 UV-B (Philips TL 20W/12RS lamps) for 6 h/day. For chronic UV-B stress, 4 μmol m−2 s−1 UV-B for either 1.25 or 1.5 h/day was applied to plants during their entire vegetative growth phase. After stress application, plants were returned to pre-stress growth conditions for seed production. For each stress condition, a minimum of 50 S0 plants was generated. Dry seeds were harvested and stored either at 4°C or at room temperature.
Semi-Quantitative Scoring of Somatic Reactivation of Transcriptional Gene Silencing
Somatic reactivation of TS–GUS in stressed S0 plants was determined by GUS staining (Jefferson et al., 1987) of whole rosettes, either directly after stress or after 1 week of recovery. For UV-B and freezing stress, this was done after 1 week of recovery, for heat stress directly after stress. Non-stressed controls (G0) were stained in parallel. Reactivation frequencies in the progeny of stressed plant populations (S1, S2, S3) and non-stressed controls (G1, G2, G3) were quantified by counting GUS-positive (blue) areas under a dissecting microscope. For UV-B stress, 11–15-day-old in vitro cultivated seedlings were scored, whereas for temperature stress, we used leaves of 3-week-old soil-grown plants. Since TS–GUS reactivation frequencies can only be compared within individual experiments, and to account for variations of the histochemical GUS assays, the number of GUS-positive areas was normalized (±SE) to those in non-stressed controls and their progeny (G0, G1, G2, G3) that were grown and scored in parallel. Two or three biological repetitions were performed for each stress experiment. On average, 40–60 plants were scored for each dataset in the individual experiments (Supplemental Table 1). Statistical analysis was performed with a Student's t-test, and p < 0.05 was used as the threshold for significance.
Analysis of GUS and LINE Expression by qRT–PCR
Expression analysis was done with plant material harvested after 1 week of recovery (UV-B, freezing stress) or directly after stress (heat stress) along with material from non-stressed control plants. Total RNA from Arabidopsis leaves was isolated with the NucleoSpin RNA plant kit (Macherey-Nagel). 900 ng of total RNA were reverse transcribed with RevertAid M-MuLV reverse transcriptase (Fermentas). qPCR was performed on a Biorad iQ5 cycler with 15-μl reactions that were set up using the SensiMix Plus SYBR Kit and Fluorescein (Peqlab). The primers used for the amplification of portions of GUS and of different transposons are denoted in Supplemental Table 3. ROC3 (At2g16600) was used as a reference gene for temperature-stressed material, UBC21 and UBC28 (At5g25760, At1g64230) for UV-B stressed material. Primer sequences are given in Supplemental Table 3. All reactions were done in quadruplicate. Normalized fold expression of GUS and transposon ORFs was calculated with the iQ5 optical system software Version 2.0 according to the method of Pfaffl (2001). PCR efficiencies were calculated from serial dilutions of the cloned amplicons that were run in parallel with the samples. Normalization to multiple reference genes (UV-B stressed material) followed the algorithm of Vandesompele et al. (2002).
Quantitative Analysis of DNA Methylation and Histone Acetylation
Analysis of DNA methylation and histone acetylation was performed with plant material harvested after 1 week of recovery (UV-B, freezing stress) or directly after stress (heat stress) along with material from non-stressed control plants. For bisulfite sequencing, genomic DNA was isolated with the DNeasy Plant Midi Kit (Qiagen), according to the manufacturer's instructions. 2.5 μg of DNA were digested with XbaI and Cfr42I overnight in 100 μl and subsequently purified with the Wizard SV Gel and PCR Clean-Up System (Promega). 500 ng of pre-cut DNA were bisulfite-treated according to the instructions of the EpiTect Bisulfite Kit (Qiagen). As a control for complete conversion, exon 15 from the PHAVOLUTA locus (At1g30490) that lacks methylated cytosines was analyzed (Bao et al., 2004; Reinders et al., 2008). For this, at least five clones were sequenced per pool of bisulfite-treated DNA. In all cases, conversion of the control locus ranged between 99 and 100%. All PCR amplifications from bisulfite-treated DNA were performed with nested primer pairs, which are designated F1/R1 and F2/R2 in Supplemental Table 3. PCR conditions for amplifying the analyzed loci are available upon request. The final PCR products were cloned into the pGEM-Teasy vector (Promega) and subjected to sequencing. For the tested loci, 20 individual clones were analyzed by sequencing and methylation data were assessed using CyMATE (Hetzl et al., 2007). Quantification of global cytosine methylation was performed by using HPLC as described previously (Rozhon et al., 2008).
For global histone acetylation analysis, a previously described histone-enriched protein extraction procedure was employed (Yan et al., 2007). Western blotting was performed with the following antibodies according to the instructions of the provider: unmodified histone H3 ChIP grade (ab1791; Abcam), histone H3 acetyl K9 ChIP grade (ab10812, Abcam), and histone H3 acetyl K9/K14 (06–599, Millipore).
ChIP was performed as described previously (Huettel et al., 2006). Immunoprecipitation was performed with the following antibodies: rabbit serum for mock IP (R9133; Sigma), unmodified histone H3 ChIP grade (ab1791; Abcam), histone H3 trimethyl K4 ChIP grade (04–745; Millipore), histone H3 dimethyl K9 ChIP grade (ab1220; Abcam), histone H3 monomethyl K27 ChIP grade (gift from T. Jenuwein, MPI of Immunobiology, Freiburg, Germany), histone H3 acetyl K9 ChIP grade (ab10812, Abcam), and histone H3 acetyl K9/K14 (06–599, Millipore). To test for enrichment of TS–GUS in the respective chromatin IPs with antibodies specific for unmodified and modified histone H3 compared to mock IP, qPCR was performed on a Biorad iQ5 cycler with 15-μl reactions that were set up using the SensiMix Plus SYBR Kit and Fluorescein (Peqlab). The primers used to amplify a fragment including upstream control regions and part of the GUS open reading frame are depicted in Supplemental Table 3. Data for unmodified histone H3 were normalized to Input (% Input), while those for the different H3 modifications were normalized to unmodified histone H3 (% H3), respectively.
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
Supported by the GEN-AU program from the Austrian Federal Ministry of Science and Research.
Supplementary Material
Acknowledgments
We thank H. Vaucheret and O. Mittelsten Scheid for the TS–GUS line, T. Jenuwein for donating antibodies specific for histone H3 monomethyl K27, B. Hohn, K. Riha, M. Siomos, and G. Fink for critical comments on the manuscript, and O. Mittelsten Scheid for continuous support and encouragement. No conflict of interest declared.
References
- Alexandre C, Möller-Steinbach Y, Schönrock N, Wilhelm Gruissem W, Hennig L. Arabidopsis MSI1 is required for negative regulation of the response to drought stress. Mol. Plant. 2009;2:675–687. doi: 10.1093/mp/ssp012. [DOI] [PubMed] [Google Scholar]
- Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aufsatz W, Mette MF, van der Winden J, Matzke M, Matzke AJ. HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO J. 2002;21:6832–6841. doi: 10.1093/emboj/cdf663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-Gonzalez E. Energy signaling in the regulation of gene expression during stress. Mol. Plant. 2010;3:300–313. doi: 10.1093/mp/ssp113. [DOI] [PubMed] [Google Scholar]
- Bao N, Lye KW, Barton MK. MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev. Cell. 2004;7:653–662. doi: 10.1016/j.devcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Bird AP, Wolffe AP. Methylation-induced repression—belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- Bond DM, Dennis ES, Pogson BJ, Finnegan EJ. Histone acetylation, VERNALIZATION INSENSITIVE 3, FLOWERING LOCUS C, and the vernalization response. Mol. Plant. 2009;2:724–737. doi: 10.1093/mp/ssp021. [DOI] [PubMed] [Google Scholar]
- Boyko A, Kathiria P, Zemp FJ, Yao Y, Pogribny I, Kovalchuk I. Transgenerational changes in the genome stability and methylation in pathogen-infected plants (virus-induced plant genome instability) Nucleic Acids Res. 2007;35:1714–1725. doi: 10.1093/nar/gkm029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P, Campi M, Chu F, Suzuki N, Maltby D, Guan S, Burlingame AL, Walbot V. Histone acetylation and chromatin remodeling are required for UV-B-dependent transcriptional activation of regulated genes in maize. Plant Cell. 2008;20:827–842. doi: 10.1105/tpc.107.056457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloix C, Jenkins GI. Interaction of the Arabidopsis UV-B-specific signaling component UVR8 with chromatin. Mol. Plant. 2008;1:118–128. doi: 10.1093/mp/ssm012. [DOI] [PubMed] [Google Scholar]
- Costa RM, Morgante PG, Berra CM, Nakabashi M, Bruneau D, Bouchez D, Sweder KS, Van Sluys MA, Menck CF. The participation of AtXPB1, the XPB/RAD25 homologue gene from Arabidopsis thaliana, in DNA repair and plant development. Plant J. 2001;28:385–395. doi: 10.1046/j.1365-313x.2001.01162.x. [DOI] [PubMed] [Google Scholar]
- Cullis CA. Unstable genes in plants. Symp. Soc. Exp. Biol. 1986;40:77–84. [PubMed] [Google Scholar]
- Elmayan T, et al. Arabidopsis mutants impaired in cosuppression. Plant Cell. 1998;10:1747–1758. doi: 10.1105/tpc.10.10.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng YQ, Desprat R, Fu H, Olivier E, Lin CM, Lobell A, Gowda SN, Aladjem MI, Bouhassira EE. DNA methylation supports intrinsic epigenetic memory in mammalian cells. PLoS Genet. 2006;2:e65. doi: 10.1371/journal.pgen.0020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R, Reeves W, Ariizumi T, Steber C. Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 2008;59:387–415. doi: 10.1146/annurev.arplant.59.032607.092740. [DOI] [PubMed] [Google Scholar]
- Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. 2003;278:4035–4040. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- Hetzl J, Foerster AM, Raidl G, Mittelsten Scheid O. CyMATE: a new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. Plant J. 2007;51:526–536. doi: 10.1111/j.1365-313X.2007.03152.x. [DOI] [PubMed] [Google Scholar]
- Huettel B, Kanno T, Daxinger L, Aufsatz W, Matzke AJ, Matzke M. Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J. 2006;25:2828–2836. doi: 10.1038/sj.emboj.7601150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahnke S, Scholten S. Epigenetic resetting of a gene imprinted in plant embryos. Curr. Biol. 2009;19:1677–1681. doi: 10.1016/j.cub.2009.08.053. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush K, Feldman M, Levy AA. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat. Genet. 2003;33:102–106. doi: 10.1038/ng1063. [DOI] [PubMed] [Google Scholar]
- Kim JM, To TJ, Ishida J, Morosawa T, Kawashima M, Matsui A, Toyoda T, Kimura H, Shinozaki K, Seki M. Alterations of lysine modifications on the histone H3 N-tail under drought stress conditions in Arabidopsis thaliana. Plant Cell Physio. 2008;49:1580–1588. doi: 10.1093/pcp/pcn133. [DOI] [PubMed] [Google Scholar]
- Kovalchuk I, Kovalchuk O, Hohn B. Genome-wide variation of the somatic mutation frequency in transgenic plants. EMBO J. 2000;19:4431–4438. doi: 10.1093/emboj/19.17.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Langst G, Grummt I. NoRC-dependent nucleosome positioning silences rRNA genes. EMBO J. 2006;25:5735–5741. doi: 10.1038/sj.emboj.7601454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZQ, Gao J, Dong AW, Shen WH. A truncated Arabidopsis NUCLEOSOME ASSEMBLY PROTEIN 1, AtNAP1;3T, alters plant growth responses to abscisic acid and salt in the Atnap1;3–2 mutant. Mol. Plant. 2009;2:688–699. doi: 10.1093/mp/ssp026. [DOI] [PubMed] [Google Scholar]
- Lucht JM, Mauch-Mani B, Steiner HY, Metraux JP, Ryals J, Hohn B. Pathogen stress increases somatic recombination frequency in Arabidopsis. Nat. Genet. 2002;30:311–314. doi: 10.1038/ng846. [DOI] [PubMed] [Google Scholar]
- Lukens LN, Zhan S. The plant genome's methylation status and response to stress: implications for plant improvement. Curr. Opin. Plant Biol. 2007;10:317–322. doi: 10.1016/j.pbi.2007.04.012. [DOI] [PubMed] [Google Scholar]
- Maurange C, Paro R. A cellular memory module conveys epigenetic inheritance of hedgehog expression during Drosophila wing imaginal disc development. Genes Dev. 2002;16:2672–2683. doi: 10.1101/gad.242702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- Molinier J, Ries G, Zipfel C, Hohn B. Transgeneration memory of stress in plants. Nature. 2006;442:1046–1049. doi: 10.1038/nature05022. [DOI] [PubMed] [Google Scholar]
- Morel JB, Mourrain P, Beclin C, Vaucheret H. DNA methylation and chromatin structure affect transcriptional and post-transcriptional transgene silencing in Arabidopsis. Curr. Biol. 2000;10:1591–1594. doi: 10.1016/s0960-9822(00)00862-9. [DOI] [PubMed] [Google Scholar]
- Pecinka A, Rosa M, Schikora A, Berlinger M, Hirt H, Luschnig C, Mittelsten Scheid O. Transgenerational stress memory is not a general response in Arabidopsis. PLoS One. 2009;4:e5202. doi: 10.1371/journal.pone.0005202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst AV, et al. Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell. 2004;16:1021–1034. doi: 10.1105/tpc.018754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando OJ, Verstrepen KJ. Timescales of genetic and epigenetic inheritance. Cell. 2007;128:655–668. doi: 10.1016/j.cell.2007.01.023. [DOI] [PubMed] [Google Scholar]
- Reinders J, Delucinge Vivier C, Theiler G, Chollet D, Descombes P, Paszkowski J. Genome-wide, high-resolution DNA methylation profiling using bisulfite-mediated cytosine conversion. Genome Res. 2008;18:469–476. doi: 10.1101/gr.7073008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries G, Heller W, Puchta H, Sandermann H, Seidlitz HK, Hohn B. Elevated UV-B radiation reduces genome stability in plants. Nature. 2000;406:98–101. doi: 10.1038/35017595. [DOI] [PubMed] [Google Scholar]
- Rozhon W, Baubec T, Mayerhofer J, Mittelsten Scheid O, Jonak C. Rapid quantification of global DNA methylation by isocratic cation exchange high-performance liquid chromatography. Anal. Biochem. 2008;375:354–360. doi: 10.1016/j.ab.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Sridha S, Wu K. Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J. 2006;46:124–133. doi: 10.1111/j.1365-313X.2006.02678.x. [DOI] [PubMed] [Google Scholar]
- Stam M. Paramutation: a heritable change in gene expression by allelic interactions in trans. Mol. Plant. 2009;2:578–588. doi: 10.1093/mp/ssp020. [DOI] [PubMed] [Google Scholar]
- Steimer A, Amedeo P, Afsar K, Fransz P, Mittelsten Scheid O, Paszkowski J. Endogenous targets of transcriptional gene silencing in Arabidopsis. Plant Cell. 2000;12:1165–1178. doi: 10.1105/tpc.12.7.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Sugimoto K, Kakutani T, Hirochika H. Linear DNA intermediates of the Tto1 retrotransposon in Gag particles accumulated in stressed tobacco and Arabidopsis thaliana. Plant J. 2001;28:307–317. doi: 10.1046/j.1365-313x.2001.01151.x. [DOI] [PubMed] [Google Scholar]
- Tessadori F, et al. Phytochrome B and histone deacetylase 6 control light-induced chromatin compaction in Arabidopsis thaliana. PLoS Genet. 2009;5:e1000638. doi: 10.1371/journal.pgen.1000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillant I, Paszkowski J. Role of histone and DNA methylation in gene regulation. Curr. Opin. Plant Biol. 2007;10:528–533. doi: 10.1016/j.pbi.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT–PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat. Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Yan D, Zhang Y, Niu L, Yuan Y, Cao X. Identification and characterization of two closely related histone H4 arginine 3 methyltransferases in Arabidopsis thaliana. Biochem. J. 2007;408:113–121. doi: 10.1042/BJ20070786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngson NA, Whitelaw E. Transgenerational epigenetic effects. Annu. Rev. Genomics Hum. Genet. 2008;9:233–257. doi: 10.1146/annurev.genom.9.081307.164445. [DOI] [PubMed] [Google Scholar]
- Zhang H, Ogas J. An epigenetic perspective on developmental regulation of seed genes. Mol. Plant. 2009;2:610–627. doi: 10.1093/mp/ssp027. [DOI] [PubMed] [Google Scholar]
- Zhu J, et al. Involvement of Arabidopsis HOS15 in histone deacetylation and cold tolerance. Proc. Natl Acad. Sci. U S A. 2008;105:4945–4950. doi: 10.1073/pnas.0801029105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.