Biological context
Ubiquitination is a post-translational process in eukaryotes that covalently modifies substrate proteins with ubiquitin. Ubiquitin can act as a tag that signals the protein-transport machinery to shuttle the protein to the proteasome for degradation. Within the ubiquitin receptor family of proteins is a group that can directly connect ubiquitinated proteins to the proteasome for degradation. Of these receptors, the best-studied are the ubiquitin-like (UBL)-ubiquitin-associated (UBA) proteins, Rad23A, PLIC2, and Ddi1. These proteins contain an UBL domain that interacts with the proteasome and the UBA domain that recognizes mono and polyubiquitinated proteins [see review (Su and Lau 2009)].
Recently, a potentially new member of this UBL-UBA domain-containing protein family, named connexin43-interacting protein of approximately 75 kDa (CIP75), was identified in a yeast-two hybrid screen (mouse embryonic c-DNA library) to interact with the gap junction protein connexin43 (Cx43) (Li et al. 2008). CIP75 contains an UBL domain at its N-terminus and an UBA domain at its C-terminus; as well as a heat shock chaperonin-binding domain and a PEST sequence. PEST sequences have been shown to direct the ubiquitination and subsequent degradation of proteins undergoing rapid turnover (Roth et al. 1998). CIP75 has 596 amino acids and exhibits high sequence homology (75%) with the human A1Up protein (Davidson et al. 2000). The general domain organization of CIP75 also shows high similarity with the UBL-UBA domain-containing protein family members Ubiquilin-1, PLIC2, Rad23A, Rad23B, and the yeast protein Dsk2 (Li et al. 2008). CIP75 functions in a similar manner as these family members in that the UBL domain of CIP75 is essential for the interaction with the S2/RPN1 and S5a/RPN10 subunits of the 19S subunit from the 26S proteasome complex. However, the UBA domain may associate with non-ubiquitinated Cx43, and this interaction appears to still regulate the turnover of Cx43 through the proteasomal pathway (Li et al. 2008).
UBA domains typically consist of approximately 45 amino acids and are characterized by relatively poor sequence conservation (Hofmann and Bucher 1996). UBA domains adopt a common, compact fold comprising a bundle of three helices and the hydrophobic surface formed by the C-terminus of α-helix 1, loop 1, and α-helix 3, is the principal interface with ubiquitin [see review (Hurley et al. 2006)]. Studies using the two UBA domains from HHR23A established that there are functional differences (i.e. differential protein partners) between UBA domains (Withers-Ward et al. 2000), suggesting the hydrophobic surface may be a more general protein–protein interaction module. Support for this is provided from a survey of the ubiquitin interaction properties of 30 UBA domains, which identified that ~30% did not bind ubiquitin (Raasi et al. 2005). Because primary sequence conservation in the UBA domain is low, different family members might present distinctive binding epitopes on a conserved three-helical scaffold. The conserved residues contribute to the common structure of UBA domains, while divergent residues on the surface permit selective interactions with different target proteins. Here, we solved the solution structure of the UBA domain of CIP75 to better understand how a UBA domain can recognize a non-ubiquitinated protein partner that is fated for proteasomal degradation.
Methods and results
The UBA domain of CIP75 (residues M549-S596) was subcloned into the bacterial expression vector pGEX-6P-2 (GST-tagged; Amersham Biosciences, Piscataway, NJ) by standard PCR methods. BL21(DE3) bacterial cells (Novagen, Madison, WI) transformed with the pGEX-6P-2 vector containing the CIP75 UBA domain gene were grown in 1 L of minimum M63 media in the presence of 15N-ammonium chloride and 13C-glucose and induced with 120 mg of IPTG at a cell density of 0.6 OD600 for 4 h. The bacterial cells were pelleted and resuspended in 1× phosphate buffered saline (PBS) containing the Complete Protease Inhibitor (Roche Molecular Biochemicals, Mannheim, Germany), 1 mM PMSF, and 1% NP-40. The cells were lysed by a French Press and the lysate was collected by centrifugation (12,000g, 60 min). The GST-tagged CIP75 UBA domain was incubated with High-affinity GST resin (GenScript, Piscataway, NJ) and the GST-tag was cleaved by incubation with 160 units of PreScission Protease (Amersham Biosciences, Piscataway, NJ) in PBS buffer overnight. The CIP75 UBA domain was then concentrated using an Amicon Ultra 3 K filter and the pH was adjusted to 5.8. Nuclear magnetic resonance (NMR) data were acquired at 7°C using a Varian INOVA 600 spectrometer fitted with a cold probe. Backbone sequential assignments were obtained using the following 3D experiments: HNCACB, CBCA(CO)NH, HNCO, HN(CA)CO, and HNHA spectra. All backbone signals were assigned except for the amide protons of E551 and V552 due to intermediate exchange. The side chain chemical shifts were obtained from the 3D 13C-HCCH-TOCSY, 15N-TOCSY-HSQC, C(CO)NH, and HBHA(CO)NH experiments. Aromatic side chains were assigned using a 2D TOCSY and 3D 13C-edited NOESY (aromatic). The RCSB PDB accession code for the coordinates of the UBA structure is 2KNZ. The chemical shift assignments were deposited in the BioMagResBank (accession code: 16484). All data were processed with the NMRPipe program (Delaglio et al. 1995) and analyzed with NMRView (Johnson 1994).
Distance constraints were derived from NOEs observed in the 15N-NOESY-HSQC and 13C-NOESY-HSQC spectra, each with mixing times of 150 ms. Model structures were calculated by using the program ARIA 1.2 (Linge et al. 2001), as implemented in the Crystallography & NMR System (CNS) 1.1 software (Brunger et al. 1998; Linge et al. 2001). The input data consisted of the chemical shifts obtained from the resonance assignments, cross-peak positions and volumes from the 15N-NOESY-HSQC and 13C-NOESY-HSQC spectra without assignment, 3JHN-Hα coupling constants from the HNHA spectrum, and hydrogen bonds based on temperature coefficient experiments. During the first ARIA run, an automated calibration was performed, unassigned NOE peaks were assigned, distance restraints were generated, and the 3D protein structures were calculated. At the end of each run, rejected restraints and residual NOE violations were analyzed. The new assignments were checked and introduced or omitted in the subsequent run. This procedure of assignment/refinement was repeated iteratively until the completion of the NO-ESY spectra assignments. In the final iteration, the 50 structures with the lowest restraint energy values were further refined by molecular dynamics simulation in water to remove artifacts (Linge et al. 2003). The final restraint file containing both ambiguous and unambiguous restraints was used as a reference for the next calculation.
For the NMR titration studies, unlabeled ubiquitin (from bovine erythrocytes, Sigma–Aldrich, St. Louis, MO) or the carboxyl terminal domain of rat Cx43 (Cx43CT; residues S255-I382) were diluted in a PBS buffer solution (pH 5.8) containing 15N-UBA to a final 1:1 and 1:20 M ratio (final UBA concentration, 50 μM), respectively, and 15N-HSQC spectra were acquired. The 15N-HSQC is a 2D NMR experiment where each amino acid (except proline) gives one signal (or chemical shift) that corresponds to the N–H amide group. These chemical shifts are sensitive to the chemical environment and even small changes in structure and/or dynamics can change the chemical shift of an amino acid.
We have characterized the structure of the UBA domain of CIP75 (M549-S596). The backbone stereoview of the final 10 structures of the CIP75 UBA domain for residues M549-S593 is displayed in Fig. 1A (residues Q594-S596 were unfolded and therefore not included). The structure was determined from a total of 1,824 NOE-derived distance restraints and 40 backbone hydrogen bonds (Table 1). The ensemble of the final 10 structures is well defined with a backbone and heavy atoms root-mean-square deviations (RMSDs) of 0.36 ± 0.08 Å and 0.89 ± 0.10 Å, respectively. The CIP75 UBA domain adopts a compact three-helix bundle (Fig. 1B). The helices are packed against each other through a well-defined hydrophobic core formed by residues F554, L558, L561, F566, A571, A575, L576, A586, I587, and L590. In addition to these core-forming residues, hydrogen bonds help stabilize the structure. For e.g. loop 1 adopts a characteristic turn that is stabilized by a hydrogen bond between the amide group of G565 and the side chain carbonyl group of N562 in α-helix 1. Additionally, the backbone carbonyl group of L558 in α-helix 1 forms a hydrogen bond with the side chain amide group of N572 in α-helix 2. Helices 1 and 2 are arranged at an angle of 125.7° (equivalent to −54.3°) and helices 2 and 3 exhibit an angle of 121.7° (or −58.3°). These values are close to the optimum of 52° reported for a “knob in hole” packing of the side chains. In contrast, the interhelical angle between helices 1 and 3 is only about 53.6° (or −126.4°). Finally, the surface of the CIP75 UBA domain contains two hydrophobic patches (epitopes 1 & 2) (Fig. 2B), which comprise ~ 1,325 Å 2 of the total ~3,052 Å 2 (43%) solvent accessible surface area.
Fig. 1.
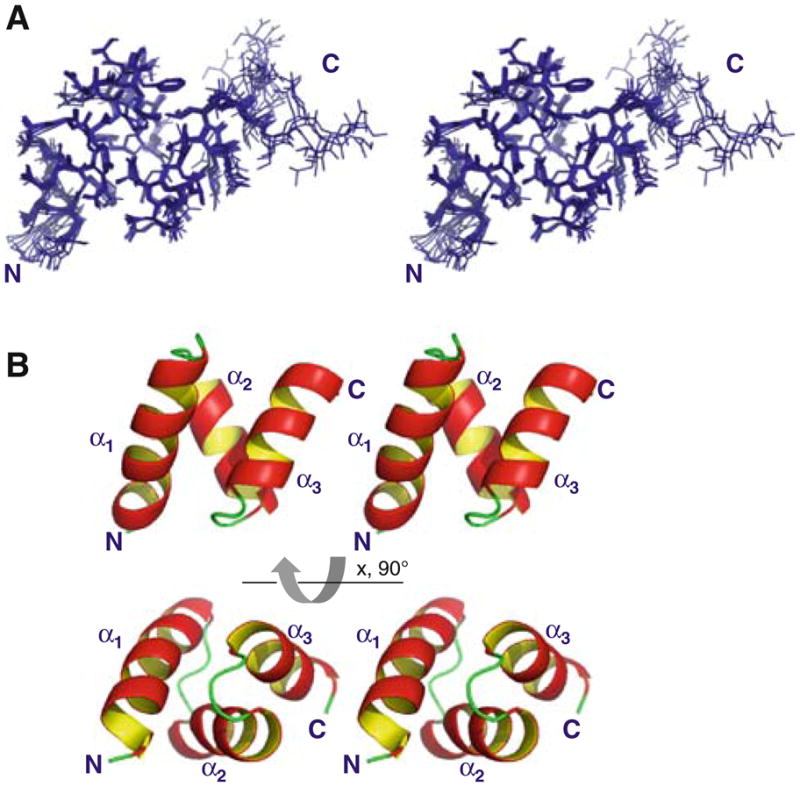
Solution structure of the UBA domain of CIP75. A. Stereo-view of the 10 lowest energy structures which have been superposed according to the backbone atoms (region M549-S593). B. Stereoview of the ribbon diagram of the lowest energy CIP75 UBA domain structure. The three α-helices (α1, α2, and α3) and the amino (N) and carboxyl (C) terminal domains have been labeled
Table 1.
Structural statistics of the 10 lowest energy structures of the UBA domain
| Conformational restraints | ||
| NOE distance restraints | ||
| Total | 1,824 | |
| Intra-residue (|i–j| = 0) | 565 | |
| Sequential (|i–j| = 1) | 410 | |
| Medium range (2 ≤ |i–j| < 5) | 410 | |
| Long range (|i–j| ≥ 5) | 435 | |
| Unambiguous restraints | 1,820 | |
| Ambiguous restraints | 4 | |
| Backbone hydrogen bonds | 40 | |
| Residual violations | Average number per residue | |
| Distance restraints > 0.3 Å | 0 | |
| Distance restraints > 0.5 Å | 0 | |
| RMSD from standard geometry | ||
| Bond lengths (Å) | 0.0052 ± 0.0002 | |
| Bond angles (degrees) | 0.66 ± 0.02 | |
| Impropers (degrees) | 1.90 ± 0.10 | |
| Energies | ||
| NOE | 63 kcal/mol ± 6 | |
| Van der Waals | −455 kcal/mol ± 5 | |
| Electrostatic | −2,029 kcal/mol ± 30 | |
| Ramachandran Maps | ||
| Residues in most favored regions (M549-S593) | 93.5% | |
| Residues in additional allowed regions (M549-S593) | 5.7% | |
| Average RMSD | Backbone (Å) | All non-hydrogens (Å) |
| UBA domain (M549-S593) | 0.36 ± 0.08 | 0.89 ± 0.10 |
RMSD root mean square deviation
Fig. 2.
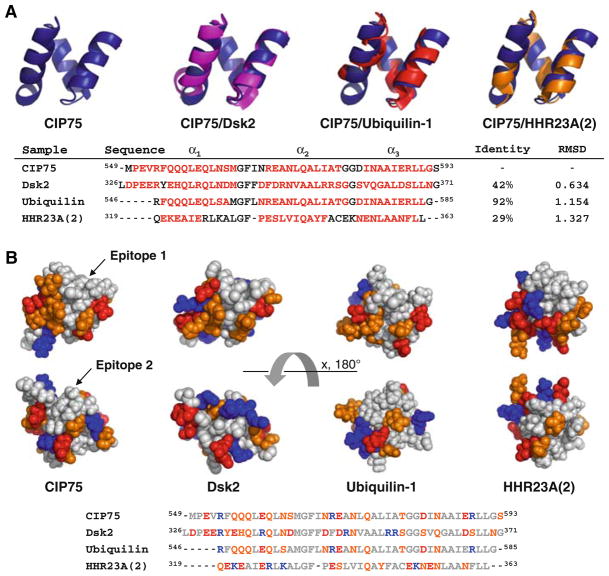
Comparison of the CIP75 UBA domain structure with previously solved UBA domains. A. Presented is the ribbon diagram structure of the CIP75 UBA domain alone and aligned with the UBA domains from Dsk2 (PDB: 2BWB) (purple), Ubiquilin-1 (PDB: 2JY5) (red), and HHR23A(2) (PDB: 1DV0) (orange). Below the structures is an alignment of their sequences, with the percent identity and RMSD compared with the CIP75 UBA domain. The RMSD is for alignment of the backbone atoms. Residues comprising the three α-helices are colored in red. B. Hydrophobic surface view of the CIP75, Dsk2, Ubiquilin-1, and HHR23A(2) UBA domains showing the hydrophobic epitopes 1 and 2. Hydrophobic residues (A, G, F, I, L, M, P, V) are colored grey, negatively charged residues (D, E) in red, positively charged (K, R) in blue, and polar residues (N, Q, S, T, Y, H) in orange. Below the structures is an alignment of their sequences, colored according to the hydrophobic surface view
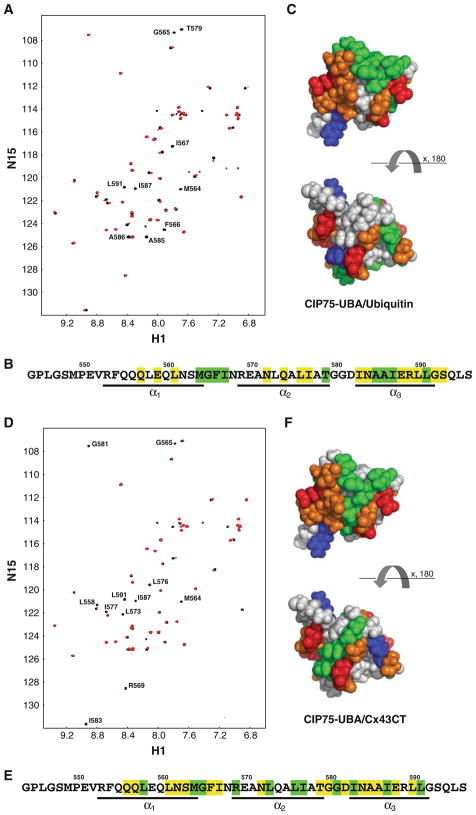
Next, we examined if the CIP75 UBA domain could associate with ubiquitin, and defined the UBA binding interface with the Cx43CT domain. Based on our NMR titration data, the binding surface of CIP75 UBA with ubiquitin comprises mainly residues M564-I567 in loop 1 and A585-L590 of α-helix 3 (epitope 1), which broaden beyond detection upon addition of ubiquitin (Fig. 3A–C). As well, T579 also shows a signal disappearance. Some residues in α-helices 1 and 2 exhibit some changes in chemical shift or signal attenuation, however noticeably weaker, thus indicating that these elements of the CIP75 UBA construct are probably not directly involved in the ubiquitin binding. NMR titration studies also identified that the Cx43CT binds the CIP75 UBA domain in the same location as ubiquitin. After determining the strongly affected residues (Fig. 3D–F), like what was observed for ubiquitin, they mapped to the main hydrophobic patch (epitope 1) of CIP75 UBA. However, Cx43CT also interacts with the hydrophobic patch localized on the opposite side of the main one (epitope 2). Unlike ubiquitin, Cx43CT strongly affected residues in α-helix 2 of the CIP75-UBA domain (residues R569, L576, L576, and I577).
Fig. 3.
Interaction of the CIP75 UBA domain with ubiquitin and the Cx43CT domain. A. The CIP75 UBA domain was titrated with ubiquitin to a 1:1 M ratio. The control spectrum, UBA only (black), has been overlapped with a spectrum obtained when both molecules were present at a 1:1 M ratio (red). The resonance peaks that completely disappeared have been labeled. B. A summary of all the UBA residues affected by ubiquitin. Residues which completely disappeared or slightly disappeared/shifted are colored in green and yellow, respectively. Residues comprising the three α-helices are underlined. C. Hydrophobic surface view of the CIP75 UBA domain with the UBA residues which completely disappeared in the presence of ubiquitin colored in green. Hydrophobic residues (A, G, F, I, L, M, P, V) are colored grey, negatively charged residues (D, E) in red, positively charged (K, R) in blue, and polar residues (N, Q, S, T, Y, H) in orange. D. The CIP75 UBA domain was titrated with Cx43CT to a 1:20 M ratio. The control spectrum, UBA only (black), has been overlapped with a spectrum obtained when both molecules were present at a 1:20 M ratio (red). The resonance peaks that completely disappeared have been labeled. E. A summary of all the UBA residues affected by the Cx43CT domain. F. Hydrophobic surface view of the CIP75 UBA domain with the UBA residues which completely disappeared in the presence of Cx43CT colored in green
Discussion and conclusions
The CIP75 UBA domain exhibits the same three-helix bundle as previously solved UBA domains (see review (Hurley et al. 2006)). For example, the RMSDs between the CIP75 UBA domain and the UBA domains from Dsk2, Ubiquilin-1, and HHR23A(2; second UBA domain) are 0.63, 1.15, and 1.33 Å, respectively (Fig. 2A). This structural conservation is in despite of their relatively low sequence conservation (e.g. 42% sequence identity between CIP75 UBA and Dsk2 UBA). Interestingly, while the overall fold is the same, the hydrophobic surfaces that have been previously identified as being important for the recognition of molecular partners are different. As displayed in Fig. 2B, from one point-of-view (top), the structures have a wide hydrophobic patch (epitope 1) at the top that narrows in the middle, but in the case of HHR23A(2), does not reach the bottom surface. This top portion contains the conserved MGF motif (loop 1) and a di-leucine motif (α-helix 3), all involved in binding ubiquitin [see review (Hurley et al. 2006)]. UBA domains also have a hydrophobic patch (epitope 2) on the opposite side of the one described above (Fig. 2B, bottom); however, it is not as compact and so far has not been reported as being involved in protein–protein interactions. For the UBAs described here, the total surface area of hydrophobic patches is rather large, comprising between 33% for Dsk2 and up to 45% of the total protein surface for Ubiquilin-1. CIP75 UBA is in the upper part of the range as 43% of its surface is formed by hydrophobic residues. Large hydrophobic patches are not that common and when present are often binding sites for other proteins. Accordingly, all the binding surfaces described for the UBA/ubiquitin interaction are on the hydrophobic surface of epitope 1 (Fig. 2B, top view).
The location of the CIP75 UBA interaction with ubiquitin is similar with those observed for the Dsk2 UBA and Ubiquilin-1 UBA/ubiquitin interaction (Ohno et al. 2005; Zhang et al. 2008). This shared binding interface for ubiquitin supports a common regulatory function for UBA domains in the ubiquitin/proteasome pathway. The UBL domain of the UBL-UBA protein family was shown to be able to interact with the UBA domain (Kang et al. 2007; Lowe et al. 2006). These studies suggest that full length UBL-UBA proteins could adopt a ‘close’ conformation upon binding between the UBA and UBL domains. This interaction however is weaker, so that ubiquitin would disrupt it by binding the UBA domain and therefore allowing UBL to interact with the proteasome subunit. Consequently, only UBL-UBA carrying an ubiquitinated protein would be targeted by the proteasome. However, in the case of CIP75, it was shown that its UBA domain can interact with non-ubiquitinated Cx43 and that this interaction appears to play a role in regulating the turnover of Cx43 through the proteasomal pathway (Kang et al. 2007; unpublished data).
The Cx43CT domain interacts in the same hydrophobic patch (epitope 1) as ubiquitin; however, this interaction also occurs with the other hydrophobic patch (epitope 2). These differences of binding interface between ubiquitin and another protein partner were also described in the case of HHR23A(2) with HIV-1 Vpr (Withers-Ward et al. 2000). This interaction was shown to be similar, but not identical to the binding interface with ubiquitin (Mueller and Feigon 2002). However, as described for HHR23A(2), both interaction areas are in close enough proximity, and as they clearly overlap in the case of CIP75 UBA, suggests that each would interfere with the binding of the other. We could therefore propose that the interaction between ubiquitin or the UBL and UBA domains of CIP75 is competing with Cx43CT (i.e. mimicking ubiquitin), which could explain why Cx43CT can be targeted by the proteasome degradation pathway even without being ubiquitinated.
Acknowledgments
This work was supported by grants from the National Institutes of Health (GM072631, P.L.S.; CA052098, A.F.L.).
Contributor Information
Fabien Kieken, Department of Biochemistry and Molecular Biology and Eppley Cancer Center, University of Nebraska Medical Center, Omaha, NE 68198, USA.
Gaëlle Spagnol, Department of Biochemistry and Molecular Biology and Eppley Cancer Center, University of Nebraska Medical Center, Omaha, NE 68198, USA.
Vivian Su, Natural Products and Cancer Biology Program, Cancer Research Center of Hawaii, University of Hawaii at Manoa, Honolulu, HI 96813, USA.
Alan F. Lau, Natural Products and Cancer Biology Program, Cancer Research Center of Hawaii, University of Hawaii at Manoa, Honolulu, HI 96813, USA
Paul L. Sorgen, Email: psorgen@unmc.edu, Department of Biochemistry and Molecular Biology and Eppley Cancer Center, University of Nebraska Medical Center, Omaha, NE 68198, USA
References
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Davidson JD, Riley B, Burright EN, Duvick LA, Zoghbi HY, Orr HT. Identification and characterization of an ataxin-1-interacting protein: A1Up, a ubiquitin-like nuclear protein. Hum Mol Genet. 2000;9:2305–2312. doi: 10.1093/oxfordjournals.hmg.a018922. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Hofmann K, Bucher P. The UBA domain: a sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem Sci. 1996;21:172–173. [PubMed] [Google Scholar]
- Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem J. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA, Blevins RA. NMR view: a computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- Kang Y, Zhang N, Koepp DM, Walters KJ. Ubiquitin receptor proteins hHR23a and hPLIC2 interact. J Mol Biol. 2007;365:1093–1101. doi: 10.1016/j.jmb.2006.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Su V, Kurata WE, Jin C, Lau AF. A novel connexin43-interacting protein, CIP75, which belongs to the UbL-UBA protein family, regulates the turnover of connexin43. J Biol Chem. 2008;283:5748–5759. doi: 10.1074/jbc.M709288200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linge JP, O’Donoghue SI, Nilges M. Automated assignment of ambiguous nuclear overhauser effects with ARIA. Methods Enzymol. 2001;339:71–90. doi: 10.1016/s0076-6879(01)39310-2. [DOI] [PubMed] [Google Scholar]
- Linge JP, Habeck M, Rieping W, Nilges M. ARIA: automated NOE assignment and NMR structure calculation. Bioinformatics. 2003;19:315–316. doi: 10.1093/bioinformatics/19.2.315. [DOI] [PubMed] [Google Scholar]
- Lowe ED, Hasan N, Trempe JF, Fonso L, Noble ME, Endicott JA, Johnson LN, Brown NR. Structures of the Dsk2 UBL and UBA domains and their complex. Acta Crystallogr D Biol Crystallogr. 2006;62:177–188. doi: 10.1107/S0907444905037777. [DOI] [PubMed] [Google Scholar]
- Mueller TD, Feigon J. Solution structures of UBA domains reveal a conserved hydrophobic surface for protein-protein interactions. J Mol Biol. 2002;319:1243–1255. doi: 10.1016/S0022-2836(02)00302-9. [DOI] [PubMed] [Google Scholar]
- Ohno A, Jee J, Fujiwara K, Tenno T, Goda N, Tochio H, Kobayashi H, Hiroaki H, Shirakawa M. Structure of the UBA domain of Dsk2p in complex with ubiquitin molecular determinants for ubiquitin recognition. Structure. 2005;13:521–532. doi: 10.1016/j.str.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Raasi S, Varadan R, Fushman D, Pickart CM. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat Struct Mol Biol. 2005;12:708–714. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- Roth AF, Sullivan DM, Davis NG. A large PEST-like sequence directs the ubiquitination, endocytosis, and vacuolar degradation of the yeast a-factor receptor. J Cell Biol. 1998;142:949–961. doi: 10.1083/jcb.142.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su V, Lau AF. Ubiquitin-like and ubiquitin-associated domain proteins: significance in proteasomal degradation. Cell Mol Life Sci. 2009;66:2819–2833. doi: 10.1007/s00018-009-0048-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withers-Ward ES, Mueller TD, Chen IS, Feigon J. Biochemical and structural analysis of the interaction between the UBA(2) domain of the DNA repair protein HHR23A and HIV-1 Vpr. Biochemistry. 2000;39:14103–14112. doi: 10.1021/bi0017071. [DOI] [PubMed] [Google Scholar]
- Zhang D, Raasi S, Fushman D. Affinity makes the difference: nonselective interaction of the UBA domain of Ubiquilin-1 with monomeric ubiquitin and polyubiquitin chains. J Mol Biol. 2008;377:162–180. doi: 10.1016/j.jmb.2007.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]