Abstract
The paramyxovirus F protein is a class I viral membrane fusion protein which undergoes a significant refolding transition during virus entry. Previous studies of the Newcastle disease virus, human parainfluenza virus 3 and parainfluenza virus 5 F proteins revealed differences in the pre- and post-fusion structures. The NDV Queensland (Q) F structure lacked structural elements observed in the other two structures, which are key to the refolding and fusogenic activity of F. Here we present the NDV Australia-Victoria (AV) F protein post-fusion structure and provide EM evidence for its folding to a pre-fusion form. The NDV AV F structure contains heptad repeat elements missing in the previous NDV Q F structure, forming a post-fusion six helix bundle (6HB) similar to the post-fusion hPIV3 F structure. Electrostatic and temperature factor analysis of the F structures point to regions of these proteins that may be functionally important in their membrane fusion activity.
Keywords: Paramyxovirus, membrane fusion, virus entry, class I viral fusion proteins, NDV
Introduction
The Paramyxoviridae is a large family of enveloped negative strand RNA viruses with the genome typically encoding 8 to 9 proteins. Members of this family of viruses infect humans and animals causing a variety of respiratory and sometimes systemic diseases (Lamb and Parks, 2007). Paramyxoviruses that infect humans include, among others, measles virus, mumps virus, parainfluenza viruses 1-4, respiratory syncytial virus (RSV) and Nipah and Hendra viruses, while those that predominantly infected animals include Newcastle disease virus (NDV), canine distemper virus, Sendai virus and other recently identified mammalian, avian and fish viruses. The viral lipid envelope is derived from the host cell plasma membrane and contains the viral spike glycoproteins. These glycoproteins consist of the viral attachment protein (variably called HN, H or G) and the fusion (F) glycoprotein (Lamb and Parks, 2007). For most paramyxoviruses, both surface glycoproteins are required for efficient infection of cells, with the fusion glycoprotein mediating the penetration of the cellular membrane during viral entry into cells (Lamb and Jardetzky, 2007; Lamb and Parks, 2007).
The F glycoprotein belongs to the class I fusion protein group (White et al., 2008) and F undergoes a large, irreversible conformational change/refolding event, which promotes the merger of the viral and cellular bilayers, opening a pore to deliver the viral genome into the cytoplasm (Lamb and Jardetzky, 2007). We have previously described the crystal structures of two conformations of the F glycoprotein -- the metastable, pre-fusion conformation of the PIV5 F protein, stabilized by appending a C-terminal trimerization domain (Harbury et al., 1994) to the F ectodomain (F-GCNt), and a post-fusion conformation of a secreted hPIV3 F ectodomain (solF) (Lamb and Jardetzky, 2007; Yin et al., 2005; Yin et al., 2006). The post-fusion hPIV3 F contains a six helix bundle (6HB), formed by two internal heptad repeat sequences (HRA and HRB) (Baker et al., 1999; Dutch et al., 1999; Joshi et al., 1998; Yin et al., 2005), which have been shown to form a highly-stable structure at the time of bilayer merger (Russell et al., 2001). In the pre-fusion PIV5 F structure these two segments are spatially separated from each other. HRB forms a three helix bundle stalk and HRA is collapsed into a globular arrangement around a core beta-sheet structure in domain III (DIII) (Lamb and Jardetzky, 2007; Yin et al., 2006).
Previous crystallographic studies of a secreted NDV F protein provided a partial structure of the ectodomain, initially interpreted to represent a pre-fusion form (Chen et al., 2001; Colman and Lawrence, 2003). The partial NDV F structure corresponds to the head and neck elements also observed in the hPIV3 F structure (Yin et al., 2005), but did not include the 6HB at the end of the helical stalk region, thereby lacking a key structural element identifying this conformation definitively as the post-fusion form.
We have produced the NDV F protein in both pre- and post-fusion conformations, using analogous constructs that yielded the pre-fusion PIV5 F structure and a post-fusion hPIV3 F structure. We demonstrate that the two protein constructs exhibit the pre- and post-fusion forms through EM analysis and we present the crystal structure of the post-fusion form of the NDV F protein. In contrast to the previously determined NDV F structure, our new crystal structure contains the 6HB at the base of the stalk region, consistent with the EM observations and our previously determined hPIV3 F structure. Global superposition of the NDV and hPIV3 structures demonstrates maximum correspondence between distal portions of the structures, with orientation adjustments made in linking domains and the extended helical HRA stalk. Electrostatic profiles of the NDV, hPIV3, and PIV5 F structures show elements of conserved charge distributions despite significant sequence differences in these glycoproteins, which may be important for their common functionality.
Results
EM analysis of pre- and post-fusion NDV F
Two constructs for the expression of the NDV Australia-Victoria (AV) F ectodomain were generated (Figure 1), following the approaches used to determine the post-fusion hPIV3 and pre-fusion PIV5 F structures (Yin et al., 2005; Yin et al., 2006). For the post-fusion form, the NDV F ectodomain was truncated at the C-terminal end of the HRB segment and a C-terminal protease cleavage site and 6His tag appended. The potential pre-fusion construct was generated by appending a helical trimerization tag derived from a reengineered GCN4 sequence (GCNt) in proper heptad repeat alignment with the F HRB helix. Proteins were expressed and purified using the baculovirus expression system.
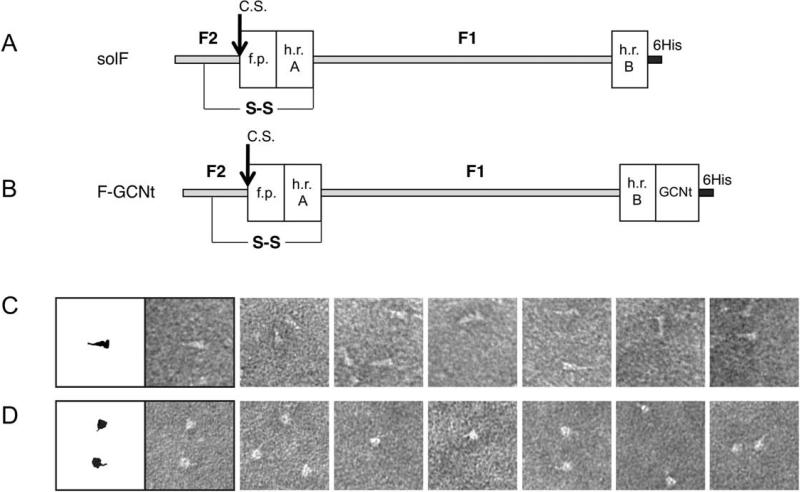
Figure 1. Protein construct design for NDV F constructs.
(A, B). Construct design of the NDV fusion proteins, solF and F-GCNt, which are expressed in the post-fusion and pre-fusion conformations, respectively. The solF construct is truncated before the F transmembrane and cytoplasmic domains, contains a mutated cleavage activation site of a single R residue, indicated with an arrow labeled C.S., and contains an appended protease cleavage site and 6His tag for purification indicated in dark grey and labeled in the figure. The F1 and F2 fragments generated after proteolytic activation of F are indicated, along with the key structural elements for fusion HRA, HRB and the fusion peptide. The NDV F-GCNt construct contains a C-terminal GCN-trimerization domain in frame with the native HRB heptad repeat sequence, upstream of the 6His tag. The inserted GCNt trimerization domain is added to stabilize the protein in the pre-fusion state. (C, D). Negative stained electron microscopy of the NDV pre-fusion and post-fusion F proteins. (C). The phenotype of the NDV post-fusion F construct is that of an elongated triangle or golf tee as seen previously for the PIV5 and hPIV3 post-fusion F proteins. (D). The phenotype of the NDV pre-fusion F protein is that of a spherical head with a thin tail observed only in select orientations similar to the lollipop phenotype observed for the PIV5 pre-fusion construct studied previously.
Samples of the NDV solF and F-GCNt were analyzed by negative stain EM (Figures 1C, D). Samples of the solF (Figure 1C) were homogeneous and revealed the golf-tee like shape previously observed for the hPIV3 protein and for the heat-treated PIV5 protein, consistent with the solF protein adopting a post-fusion configuration. The samples of the F-GCNt protein (Figure 1D) revealed a non-homogeneous mixture of apparently pre- and post-fusion forms, suggesting that the NDV F remains incompletely stabilized by the GCNt domain or that the folding pathway for the NDV F protein bifurcates and produces two populations of F in the pre-and post-fusion conformations.
Crystallization and structure determination of the secreted solF
Attempts to crystallize the F-GCNt protein were unsuccessful, likely due to the heterogeneity of the F conformations observed by EM. However, the solF protein was crystallized and the crystals diffracted X-rays to a resolution of 3.5 Å. The solF crystals belong to space group R3, with cell dimensions a=b=83.3 Å and c=461.4 Å. The structure was solved by molecular replacement using the partial structure of the NDV F trimer determined previously (Chen et al., 2001). The model for the F structure was refined to a final Rfree of 29.4% with overall good geometry. The statistics for the data collection and final model refinement are collected in Table 1.
Table 1.
Crystallographic data and refinement statistics
| Data collection | |
|---|---|
| Source | APS-5IDB |
| Wavelength (Å) | 0.99998 |
| Space group | R3 |
| Unit-cell parameters | |
| a (Å) | 83.3 |
| b (Å) | 83.3 |
| c (Å) | 461.4 |
| α(°) | 90 |
| β(°) | 90 |
| γ(°) | 120 |
| Resolution range (Å) | 50.0-3.5 |
| Rmerge (%) | 9.5 (47.5)† |
| I/σ(I) | 15.3 (3.5)† |
| Completeness (%) | 99.9 |
| Redundancy | 6.2 (6.0)† |
| Refinement | |
| Resolution (Å) | 3.5 |
| No. unique reflections (free) | 14966 (1004) |
| R factor/Rfree | 0.261 / 0.294 |
| No. of residues, atoms | 418, 3022 |
| Average B-factors | |
| Main-chain atoms (Å2) | 142 |
| Side-chain atoms (Å2) | 139 |
| R.m.s. deviations in bond lengths (Å) | 0.002 |
| R.m.s. deviations in bond angles (°) | 0.406 |
| Ramachandran plot statistics (%) | |
| Residues in preferred regions | 90.5 |
| Residues in allowed regions | 9.5 |
last shell (3.63-3.5 Å) statistics in parentheses.
Structure of the NDV solF
The NDV F crystals contain two independent F subunits per asymmetric unit, with the F trimers lying along crystallographic threefold axes (Figure 2A). Due to the moderate resolution of the diffraction data, the two subunits were refined with non-crystallographic symmetry restraints and exhibit minimal structural differences. The NDV solF trimer (Figure 2B) consists of a head, neck and stalk region, in a golf-tee like configuration, similar to that observed in the EM analysis (Figure 1C). At the base of the stalk, the structure contains the characteristic 6HB formed by HRA and HRB segments, which assemble as the F protein adopts the post-fusion structure during membrane fusion (Lamb and Jardetzky, 2007; Lamb and Parks, 2007; Russell and Jardetzky and Lamb, 2001). Examples of electron density observed for the final NDV F model are shown in Figure 2.
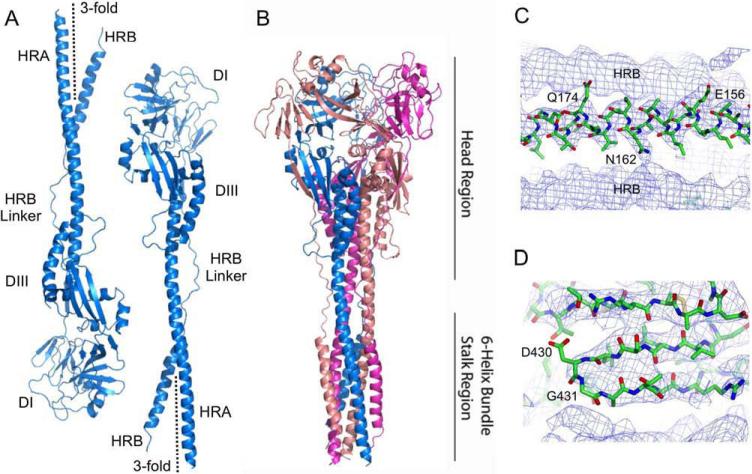
Figure 2. Crystal structure of the NDV AV F protein.
(A). Structures of the two antiparallel NDV F monomers from two different trimers present in the asymmetric unit. The two trimers lie along crystallographic 3-fold axes, which are indicated in the Figure. (B) Structure of the biological NDV F trimer observed when the crystallographic symmetry mates of one of the monomers are generated. Chains are colored in blue, pink and magenta. The head and 6-helix bundle domain stalk regions are indicated to the right. (C) Example sigmaA weighted electron density maps calculated using the final refined structure. The electron density map is contoured at 0.9σ and shows a segment of the 6HB with a model for the central HRA helix (residues E156, N162 and Q174 are indicated) flanked by two HRB helices from neighboring crystallographically-related symmetry mates. (D) An example of poor electron density observed in the 430 loop is shown. The electron density map represented is identical to that in panel (C). Electron density for residues G431 and D430 is partial and may be due to the flexibility of G431 and the low resolution (3.5 Å) of the diffraction data obtained.
Dissolved crystals of NDV solF migrate as a single band on SDS/PAGE with an electrophoretic mobility consistent with that expected for uncleaved Fo. However, although electron density was observed for residues 34 to 102 and 145 to 493 in both chains within the asymmetric unit, no electron density was observed for residues 32 to 33, 103 to 144 and also for the C-terminal amino acids including the purification tag that was retained during crystallization. The internal missing residues (103 to 144) begin 13 residues N-terminal to the cleavage/activation site at residue 116 and extend through the fusion peptide (residues 117 to 141) and the beginning 3 residues of HRA. Electron density for similar residues were also missing in the electron density map for the hPIV3 post-fusion F (Yin et al., 2005), and it was considered likely that these residues did not form a defined structure in solution but were draped flexibly along the exterior of the hPIV3 stalk. Evidence in support of this notion is provided by the finding that the hPIV3 solF can be cleaved at the cleavage/activation site by trypsin. The F protein molecules formed on cleavage associate with liposomes and in solution form rosettes due to aggregation of the hydrophobic fusion peptides (Connolly et al., 2006). Liposome insertion and rosette formation occur spontaneously, without any requirement for prior heat treatment, due to release of the covalent restraints on the fusion peptide. In contrast, the prefusion PIV5 F structure does not exhibit similar liposome insertion or rosette formation unless heated to 50° C, causing its conversion to a post-fusion form (Connolly et al., 2006). It is also important to note that the heat driven conversion of PIV5 pre-fusion F to post-fusion F is an intrinsic property of the purified metastable pre-fusion F protein and it does not require participation of cellular proteins such as folding chaperones or disulfide isomerases (Connolly et al., 2006).
Comparison of the NDV AV and NDV Q F structures
The NDV AV F structure was superimposed on the previously determined NDV Queensland V4 (Q) F structure (Chen et al., 2001). The NDV F protein from the AV and Q strains share approximately 96% sequence identity over approximately 469 ectodomain residues. Overall, the correspondence between the two structures is very good with an overall RMSD on Cα atoms of 0.74 Å over 340 residues. Structural differences in loops located in D1, D2, and D3 are evident, including residues near amino acids 72, 310, 404-414, and 421 to 425.
The major difference between the two NDV F structures is located at the base of the stalk region. The NDV AV F structure extends further to reveal an intact 6HB formed by the HRA and HRB regions that were not present in the original NDV Q model (Figure 3). The new NDV F structure includes the previously not modeled HRA residues 145-170 and HRB residues 455-493. The presence of the NDV AV F 6HB demonstrates conclusively that the secreted F protein adopts a stable post-fusion configuration similar to that previously described for the hPIV3 F protein, resolving ambiguities in the original interpretation of the NDV Q F protein structure.
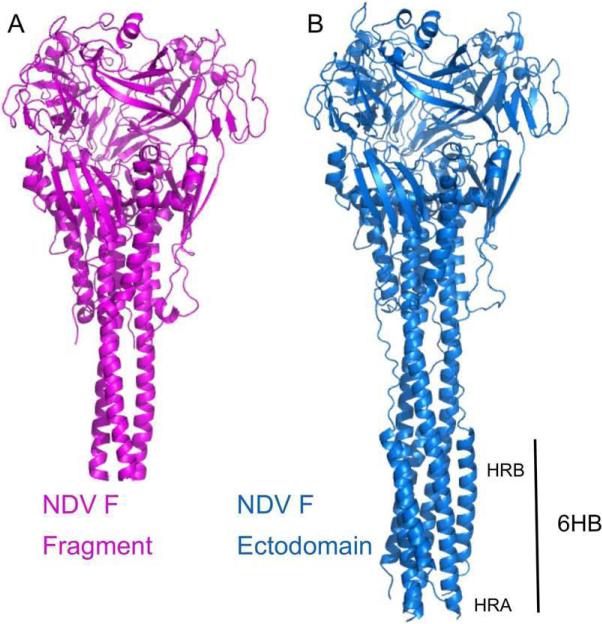
Figure 3. Comparison of the NDV AV and partial NDV Q F ectodomain structures.
(A). Structure of the previously determined NDV Q F protein. (B). Structure of the NDV AV F ectodomain. Note the absence of the N-terminal region of the HRA three-helix bundle and the entire HRB helix in the NDV Q F structure, leading to a shortening of the stalk region. The NDV AV F structure shows overall very close correspondence with the NDV Q structure in the head and neck regions, while containing the post-fusion 6HB.
Comparison to the hPIV3 post-fusion F structure
We have compared previously structures of the hPIV3 and NDV Q F proteins, but this comparison did not include the full NDV HRA and HRB regions. The global superposition of the NDV AV and hPIV3 F structures is shown in Figure 4. The best superposition generated by the program TOPP (CCP4, 1994) corresponds to an overall RMSD of 2.02 Å on 240 Cα atoms of the ectodomain residues. The structural diversity provides another measure of the overall differences between the two structures, taking into account the number of residues matched in the superposition by a minimum distance. It is defined as the root mean square deviation times the square of the ratio of the number of residues in the first molecule (Nmol1) divided by the number of residues matched (Nfit). For the top match, the structural diversity is 6.14. The second alignment generated by TOPP corresponds to a better topological match with 14 matched secondary structure elements, but with a higher structural diversity score of 32.4, with fewer residues matched in the alignment (174 versus 240). The secondary alignment corresponds to a better superposition of the F head domains, but leads to large differences in the HRA and HRB positions.
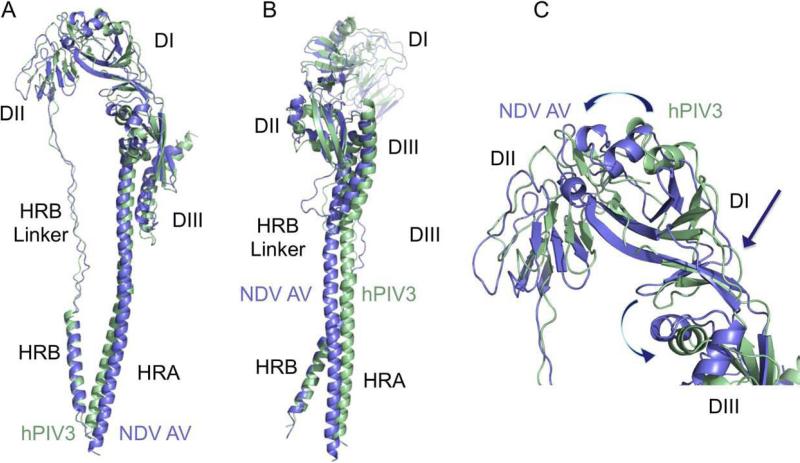
Figure 4. Comparison of the NDV F and hPIV3 post-fusion structures.
(A, B) Global superposition of the NDV AV F (blue) and hPIV3 F post-fusion structures (green). Best correspondence is observed between the combination of DIII and HRB segments, leading to displacement of HRA, DII, and portions of DI. (C). Close-up of the overlay of DI and DII domains. Positional displacements, relative to the superposed DIII domains, can be viewed as twisting motions (indicated by arrows) at the junctions of DI/DIII and DI/DII domains.
Overall, the largest differences in the best global alignment are located in DII, and deviations in the core three helix HRA bundle and the adjacent HRB linker region are also evident (Figure 4). Otherwise there is generally very good correspondence between the hPIV3 and NDV F structures.
Electrostatics of the pre- and post-fusion F structures
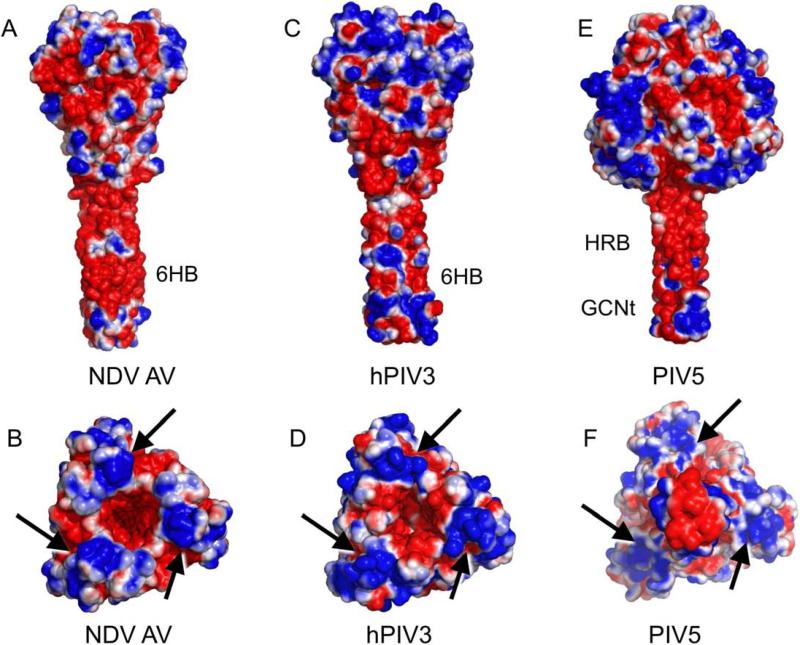
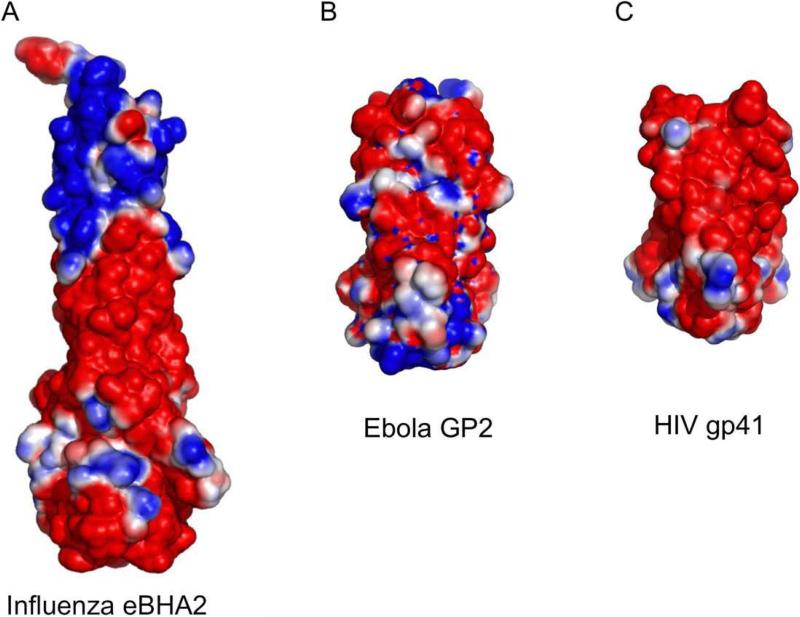
The electrostatic profiles of the NDV, hPIV3, and PIV5 F structures calculated using the program APBS (Baker et al., 2001) are shown in Figure 5. All three structures reveal a common negatively charged surface in their stalk regions despite the fact that there is low sequence identity and that the conformations represent both pre-fusion and post-fusion forms. The NDV (AV) F sequence identity with PIV5 and HPIV3 F proteins is 34% and 29%, respectively, and the corresponding sequence similarity is 52% and 47%. The stalks of the NDV and hPIV3 F proteins display the HRB segment on the exterior of the post-fusion 6HB, while the PIV5 pre-fusion stalk consists of an isolated HRB three helix bundle. Since the heptad repeat residues of HRB are used to form packing interactions in both the pre-fusion three helix bundle and the post-fusion 6HB, the two conformational states present a similar external surface of HRB. Therefore, the negative charge of the stalk regions can be ascribed to the nature of the residues in HRB. Similar negatively charged profiles are observed for the post-fusion structures of other class I viral fusion proteins including the influenza hemagglutinin (Bullough et al., 1994; Chen et al., 1999), gP41 of HIV (Malashkevich et al., 1998; Weissenhorn et al., 1997), and Gp2 of Ebola virus (Malashkevich et al., 1999; Weissenhorn et al., 1998) (Figure 6).
Figure 5. Electrostatic profiles of the NDV, hPIV3 and PIV5 protein structures.
(A,B) NDV F. (C,D). hPIV3 F. (E,F). PIV5 F. “Side” and “end-on” views of the proteins are shown in paired panels A&B, C&D and E&F. Arrows point to conserved regions of positive charge on the apices of the pre-and post-fusion F timers. Note that the PIV5 F trimer in panel (F) is oriented with the pre-fusion stalk pointed toward the reader out of the plane of view, in order to maintain a consistent orientation of the DI-DIII head domains. Electrostatic charge is contoured at -1 to +1 kT/e.
Figure 6. Electrostatic charge on the surfaces of the post-fusion influenza HA2, Ebola GP2 and HIV gp41.
(A) The bacterially expressed influenza HA2 fragment structure. (2) The Ebola virus GP2 protein. (C) The HIV gp41 six-helix bundle. All structures are contoured at -1 to +1 kT/e.
Figures 5B and D shows top views of the NDV and hPIV3 F head regions, revealing a central cavity that is negatively charged with three positively charged (blue regions indicated with arrows) lobes projecting from the trimer axis. The charge distribution between NDV and hPIV3 structures appears remarkably similar despite their low sequence identity of approximately 29%.
The PIV5 F structure in Figure 5F shows a view from the bottom of the pre-fusion stalk, which projects outward from the plane of view. This provides an analogous orientation of the head domains shown for the NDV and hPIV3 structures in Figures 5B and 5D, because of the inversion of the orientation of the stalk region during the F conformational change. Interestingly, the PIV5 F structure shows a similar distribution of positive charge on the three lobes of the trimer as observed for the two post-fusion structures.
Overall, all of the F protein structures exhibit similar negatively charged stalk regions and positively charged lobe regions. Whether these electrostatic profiles influence the progression of membrane fusion remains to be established, but the conservation of the distribution of these charged features across distantly related F protein family members suggests that there may be an underlying functional role.
Analysis of the temperature factor profiles in the pre-and post-fusion F structures
Because of the large structural differences in the F protein in its two conformations, inter-domain and inter-chain packing interactions change substantially and thereby may influence the mobility of specific regions. The atomic temperature factors (or B factors) refined during the structure determination provide insight into the relative flexibility of different protein regions, and these might highlight important functional sites of F in its two conformational states. For example, dynamic or disordered regions in the prefusion F protein stalk, or head, could indicate a site associated with the initiation of the pre- to post-fusion conformational change. The comparison of the distribution of highly flexible, and highly ordered, regions in the pre- and post fusion F structures is therefore of interest in furthering our understanding of the F protein conformational change.
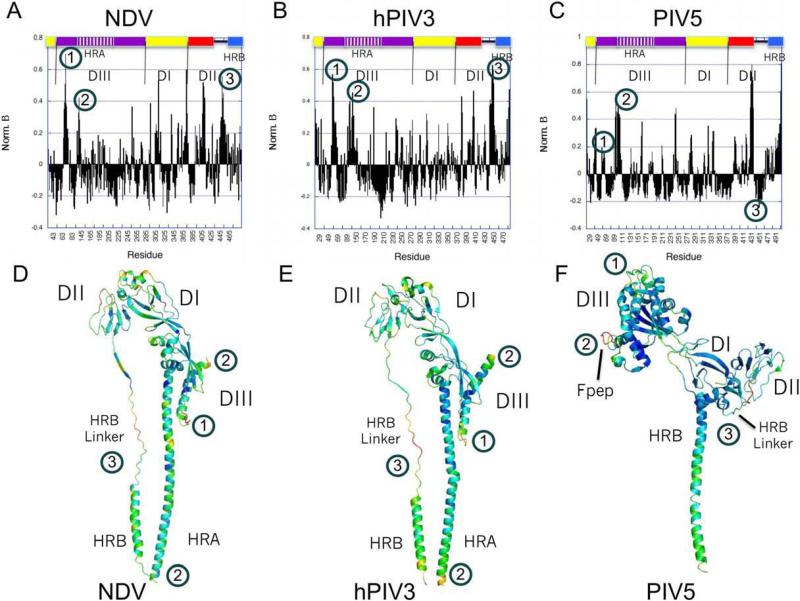
Figure 7 shows an analysis of the temperature factors for each of the three F protein structures. The bar graph plots shown in Figures 7A-C represent the normalized, averaged temperature factors for independently observed chains of each structure. The normalization places the plots on similar scales, while averaging the temperature factors between multiple chains reduces systematic chain differences due to crystal packing interactions. The normalized temperature factors are also shown graphically in color displayed on individual chains of the pre- and post-fusion structures, with blue representing the lowest values ramping to yellow as the highest values (Figure 7 panels D, E and F).
Figure 7. Temperature factor analysis of the pre-and post-fusion F structures.
Normalized temperature factors were calculated from the NDV AV, hPIV3 and PIV5 F structures as described in Materials and Methods. The normalized values are plotted bar graph form in panels A, C and E and also displayed as color ramps on monomers of the three structures in panels B, D and F. Color ramps are from blue (low values) to yellow (high values) of the normalized temperature factors. Note that normalization of the temperature factors is relative to the overall mean and maximum values observed for each structure, yielding values that drop below 0 for regions that are better ordered than average and values above 0 for progressively more disordered regions. (A,B) NDV F. (C,D). hPIV3 F. (E,F). PIV5 F. Regions indicated by numbers 1 to 3 are discussed in the text.
The normalized temperature factor plots show very similar profiles for the NDV and hPIV3 structures, while the PIV5 plot shows some significant differences. Three regions are highlighted in the plots and indicated by numbers. The first region is a loop in DIII immediately preceding a helix (HRC) upstream of the cleavage/activation site adjacent to the fusion peptide. The second region is the cleavage/activation site and the N terminus of the fusion peptide. The third region includes a portion of the HRB Linker segment and the N terminus of HRB.
Region 1 exhibits higher normalized temperature factors in the two post-fusion structures as compared to the PIV5 pre-fusion structure (approximately 0.6 versus 0.2). This is consistent with the local packing interactions of this region, which in the pre-fusion structure involves direct interactions with a loop in the HRA segment that forms a turret-like structure at the top of the head region. In the post-fusion structures, the region 1 loop forms less intimate contacts with the rearranged helical HRA, consistent with the observed increase in average temperature factor.
Region 2 exhibits a similar spike in temperature factor (approximately 0.4), immediately following a region of low temperature factors corresponding to the HRC helix (approximately -0.2). In both conformational states, region 2 corresponds to segments exposed at the F trimer surface and the higher average temperature factor in this region in the post-fusion structure is consistent with its accessibility to cleavage by proteases. The switch between the better-ordered HRC helix and the more disordered cleavage site is conserved in the two structural conformations.
Region 3 corresponds to the linker connecting DII to the helical HRB segment. In the two post-fusion structures, region 3 exhibits generally high temperature factors between 0.3-0.4, while the pre-fusion structure exhibits a more disordered segment of the linker, followed by a segment that is significantly better ordered, with normalized temperature factors approaching -0.2. In the structure of the pre-fusion form, this switch from disordered to ordered segments in region 3 corresponds to the folding of the linker into the base of the head domain and the initial N-terminal residues of HRB. This switch in apparent mobility in this region is somewhat unexpected, as our current models for the refolding of the F protein posit that the first step involves unfolding of the pre-fusion HRB three helix bundle and that the post-fusion six helix bundle forms a more stable structure (Lamb and Jardetzky, 2007; Russell and Jardetzky and Lamb, 2001; Russell et al., 2003; Yin et al., 2006). However, the low normalized temperature factors observed for the C-terminal end of the HRB linker and the N terminus of HRB are consistent with a role for the pre-fusion head domain in stabilizing and nucleating the HRB three helix bundle (Russell et al., 2003; Yin et al., 2006).
Conclusions
The paramxyovirus F proteins fold to a metastable conformation, refolding at the right time and place to a lower energy post fusion form to drive membrane fusion and virus entry. Structural studies of a secreted fragment of the NDV Q F protein provided an initial model for F protein directed fusion based on the interpretation of that fragment structure representing a pre-fusion conformational state (Chen et al., 2001; Colman and Lawrence, 2003). However, the initial NDV Q F structure lacked electron density for key elements of the protein, including portions of the HRA helix, HRB and the fusion peptide. Subsequent studies of the secreted hPIV3 F protein revealed a similar conformation to the NDV Q F structure, but also demonstrated that the secreted protein forms the post-fusion 6HB with its HRA and HRB segments (Yin et al., 2005).
The pre-fusion form of the PIV5 F protein could be stabilized and crystallized by fusing a GCN-trimerization domain (Harbury et al., 1994) in frame with the HRB segment (Yin et al., 2006). EM studies of the PIV5 and hPIV3 F proteins reveal two distinct shapes, a lollipop pre-fusion and golf tee-like post-fusion form (Connolly et al., 2006). The pre-fusion PIV5 F can be converted to a post-fusion form by heat and biochemical studies support the conclusion that heat conversion leads to the exposure of the fusion peptide and membrane-insertion activity (Connolly et al., 2006). Similar distinct structures have been observed in the EM analysis of the RSV F glycoprotein (Calder et al., 2000), although a direct correspondence to the PIV5 pre-fusion and hPIV3 post-fusion structures has not been established.
The NDV Q F structure left open questions regarding the stability of its post-fusion stalk region and whether this differs from that observed for the corresponding hPIV3 protein. In addition, it has been important to demonstrate that other secreted F proteins can be stabilized in a pre-fusion form by the addition of a trimerization segment, in order to determine the generalizability of this approach to obtaining pre-fusion forms of other paramxyovirus F proteins.
Here we demonstrate by EM analysis that the NDV AV F protein can be expressed in two conformations consistent with the folding of the protein to distinct pre-and post-fusion states similar to those observed for the PIV5 F-GCNt and hPIV3 solF proteins. The pre-fusion NDV AV F-GCNt protein exists as a somewhat heterogeneous population consisting of both pre- and post-fusion forms and could not be crystallized. However, the post-fusion NDV solF could be crystallized and its structure determined. In contrast to the NDV Q F structure, the new NDV AV F structure reveals the presence of the anticipated 6HB, consistent with the structure of the hPIV3 solF protein and with the negative stain EM images. These results confirm that the secreted F ectodomains lacking a C-terminal trimerization domain adopts the post-fusion conformation.
Global comparisons of the NDV AV and hPIV3 F structures show best correspondence between distal regions of the proteins, which include the DIII core structural elements and HRB. Interestingly, the relationship between HRA/HRB helices in the 6HB appear to be shifted slightly between the two structures in this global alignment. Additional rearrangements in the DI/DII and DI/DIII interfaces between these structures are consistent with the flexible repositioning of these domains during refolding, and with previous differences observed between the NDV Q and hPIV3 F structures.
Electrostatic analysis of the two post-fusion and one pre-fusion structures demonstrate a reasonably well conserved charge profile. The stalk regions of all three proteins, which display HRB at the protein surface in both states, exhibits substantial negative charge. The head domain lobes, formed primarily by DI and DII domains, also reveal a consistent patch of positive charge, despite low sequence conservation between the proteins. These charge distributions in the stalk and head regions may serve to orient the F protein relative to the two membranes during fusion, and it is interesting to note that other class I fusion proteins including influenza virus HA, Ebola GP2 and HIV gp41, also exhibit negatively charged rod-like structures in their post-fusion states.
Finally, a normalized B-factor analysis of the F protein structures reveals features relevant to the conformational transition. One surprising observation is the distinct difference in B-factor profiles at the junction of the HRB-linker and HRB in the pre- and post-fusion states of the F protein structures. In the two post-fusion structures, this region is consistently more disordered, while in the pre-fusion form the disordered linker transitions to a region of better than average order at the junction of the F head and HRB stalk. This observation is consistent with the postulated role of the head in stabilizing the pre-fusion three-helix bundle stalk and with these junctional interactions playing an important role in the energy barrier for activation of the F protein conformational change. Mutations of residues at this stalk-head junction can lead to hyperfusogenic F proteins (Russell et al., 2003; Yin et al., 2006) and the introduction of a disulfide crosslink in this region prevents the F protein conformational change (Lee et al., 2007). However, the observation that the junction is better ordered than average is somewhat surprising, as it is this junction which is thought to dissociate in the early steps of F refolding (Russell et al., 2003; Yin et al., 2006). The disorder and accessibility of the HRB-Linker preceding this junction might play an important role in allowing receptor-dependent activation of F, which occurs in most of the paramyxoviruses through receptor engagement by a separate viral attachment glycoprotein (Lamb and Jardetzky, 2007; Lamb and Parks, 2007). Further studies of F protein stabilization and activation are required to fully understand the mechanism by which these membrane fusion machines fold and re-fold in response to cellular receptor interaction.
Materials and Methods
Expression and purification of NDV F constructs
The complete NDV F ectodomain construct, lacking the transmembrane domain and cytoplasmic tail region and containing mutation of the F protein furin cleavage site to a single R residue and a C-terminal (His)6 tag for purification purposes were cloned into a pFastBac baculovirus expression vector (Invitrogen). The pre-fusion NDV F construct contained an engineered GCN4 trimerization domain in frame with the heptad repeat of the HRB and a C-terminal (His)6 tag for purification purposes. High-titer baculovirus stocks were grown in Sf9 cells. Proteins were expressed by infecting High Five cells with virus and harvesting the media supernatant two days post infection.
Proteins were purified by two successive chelation purification steps. First, to separate the F protein from the abundant insect ferritin in the media, CuCl2 was added to the media supernatant to a final concentration of 500 micromolar. To each 800 mls of media, four milliliters of EDTA-stripped chelation resin (TALON Resin, BD Biosciences) was added and the slurry was rocked for 30 min at 4°C and separated by a gravity column. The resin was washed with ten-times column volume of equilibration buffer (25 mM Tris pH 7.5, 300 mM NaCl) and the F protein was eluted with ten-times column volume of elution buffer (equilibration buffer with 250 mM imidazole). The elution was dialyzed against 25 mM Tris buffer pH 7.5, and the resulting solution was loaded onto a 5 ml Hitrap chelation column charged with CoCl2 and eluted in 25 mM Tris pH 7.5, 300 mM NaCl and a gradient of imidazole. Fractions containing pure, uncleaved Fo constructs were collected, dialyzed against 25 mM Tris pH 7.5, 50 mM NaCl and were concentrated using Vivaspin columns to a concentration of approximately 10 mgs/ml.
EM of NDV constructs
Protein solutions of 50 micromolar NDV F pre-fusion and post-fusion constructs were absorbed onto glow-discharged carbon coated grids and were negatively stained with 2% sodium phosphotungstate (pH 6.6). The grids were observed on a JOEL 1230 transmission electron microscope (JEOL, Peabody, MA) operating at 100 kV.
Crystallization of the post-fusion NDV F
Crystals were grown at 4 degrees centigrade in 96 well sitting-drop plates (Corning), mixing equal volumes of protein solution (10 mg/ml) and precipitant [10% (wt/vol) polyethylene glycol 1000, 1.26 M ammonia sulfate and 0.1 M sodium acetate (pH 4.5)] as delivered by the Hydra II crystallization robot (Thermo Scientific). The crystals grew as hexagonal disks with the largest being approximately 100 micrometers wide and 20 micrometers deep and were harvested after reaching their observed largest dimensions between 3 and 5 days after tray setup. Crystals were harvested and subsequently transferred into 15% propylene glycol, 12% polyethylene glycol 1000, 1.26 M ammonia sulfate and 0.1 M sodium acetate (pH 4.5) for flash freezing in liquid nitrogen. The crystals belong to the space group H3 with two NDV F monomers (from two different trimers) in the asymmetric unit.
Structure determination and refinement
An NDV F data set was collected to 3.5 Å. Diffraction data were processed with HKL2000 (Otwinowski and Minor, 1997) and scaled and reduced to structure factor amplitudes by using the CCP4 suite of programs (CCP4, 1994). A molecular replacement phase solution was determined using the program PHASER (CCP4, 1994; Storoni et al., 2004) using the partial NDV F trimer (PDB ID code 1G5G). The resultant phases were refined using two-fold noncrystallographic symmetry averaging using the program REFMAC (Murshudov et al., 1997). The resulting electron-density maps revealed features not present in the starting partial NDV F trimer model, similar to what was observed for the hPIV3 post-fusion F model, and the 6-helix bundle stalk region was built into this density using the program COOT (Emsley and Cowtan, 2004). The final NDV AV F model was built and refined using several iterative runs of the REFMAC, Phenix Refine (Adams et al., 2002) and COOT. The final refinement and model statistics are collected in Table I. The NDV F coordinates and reflections have been deposited in the RCSB Protein Data Bank under PDB ID code 3MAW.
Calculation of normalized temperature factors
Individual normalized atomic temperature factors were calculated by subtracting the mean temperature facture calculated over all atoms (Bmean) from each refined value (Bobs), and dividing by the maximum observed value (Bmax) minus the mean (Bnorm = (Bobs-Bmean)/(Bmax-Bmean)). The values were averaged over independently observed chains in the crystallographic asymmetric unit, as initial calculations showed significant chain differences due to crystal packing interactions for the PIV5 F structure. Finally, the average temperature factor per residue was calculated by taking the average over all atoms in each residue.
ACKNOWLEDGEMENTS
R.A.L is an Investigator of the Howard Hughes Medical Institute. This research was supported in part by National Institutes of Health Research Grants R01-GM61050 to T.S.J. and R01-AI23173 to R.A.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Baker K, et al. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell. 1999;3:309–319. doi: 10.1016/s1097-2765(00)80458-x. [DOI] [PubMed] [Google Scholar]
- Baker NA, et al. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullough PA, et al. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- Calder LJ, et al. Electron microscopy of the human respiratory syncytial virus fusion protein and complexes that it forms with monoclonal antibodies. Virology. 2000;271:122–131. doi: 10.1006/viro.2000.0279. [DOI] [PubMed] [Google Scholar]
- CCP4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Chen J, et al. N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA(2) subunit to form an N cap that terminates the triple-stranded coiled coil. Proc. Natl. Acad. Sci. 1999;96:8967–8972. doi: 10.1073/pnas.96.16.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, et al. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure. 2001;9:255–266. doi: 10.1016/s0969-2126(01)00581-0. [DOI] [PubMed] [Google Scholar]
- Colman PM, Lawrence MC. The structural biology of type I viral membrane fusion. Nat. Rev. Mol. Cell Biol. 2003;4:309–319. doi: 10.1038/nrm1076. [DOI] [PubMed] [Google Scholar]
- Connolly SA, et al. Refolding of a paramyxovirus F protein from prefusion to postfusion conformations observed by liposome binding and electron microscopy. Proc. Natl. Acad. Sci. 2006;103:17903–17908. doi: 10.1073/pnas.0608678103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutch RE, et al. Paramyxovirus fusion protein: characterization of the core trimer, a rod-shaped complex with helices in anti-parallel orientation. Virology. 1999;254:147–159. doi: 10.1006/viro.1998.9532. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Harbury PB, et al. Crystal structure of an isoleucine-zipper trimer. Nature. 1994;371:80–83. doi: 10.1038/371080a0. [DOI] [PubMed] [Google Scholar]
- Joshi SB, et al. A core trimer of the paramyxovirus fusion protein: parallels to influenza virus hemagglutinin and HIV-1 gp41. Virology. 1998;248:20–34. doi: 10.1006/viro.1998.9242. [DOI] [PubMed] [Google Scholar]
- Lamb RA, Jardetzky TS. Structural basis of viral invasion: lessons from paramyxovirus F. Curr. Opin. Struct. Biol. 2007;17:427–436. doi: 10.1016/j.sbi.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb RA, Parks GD. Paramyxoviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. Fifth ed. Vol. 1. Lippincott WIlliams & Wilkins; Wolters Kluwer: 2007. pp. 1449–1496. 2 vols. [Google Scholar]
- Lee JK, et al. Reversible inhibition of the fusion activity of measles virus F protein by an engineered intersubunit disulfide bridge. J. Virol. 2007;81:8821–8826. doi: 10.1128/JVI.00754-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malashkevich VN, et al. Crystal structure of the simian immunodeficiency virus (SIV) gp41 core: conserved helical interactions underlie the broad inhibitory activity of gp41 peptides. Proc. Natl. Acad. Sci. 1998;95:9134–9139. doi: 10.1073/pnas.95.16.9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malashkevich VN, et al. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9 Å resolution. Proc. Natl. Acad. Sci. 1999;96:2662–2667. doi: 10.1073/pnas.96.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, et al. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Methods in Enzymology: Macromolecular Crystallography, part A. Vol. 276. Academic Press; 1997. Processing of X-ray Diffraction Data Collected in Oscillation Mode. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Russell CJ, et al. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 2001;20:4024–4034. doi: 10.1093/emboj/20.15.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell CJ, et al. A dual-functional paramyxovirus F protein regulatory switch segment: activation and membrane fusion. J. Cell Biol. 2003;163:363–74. doi: 10.1083/jcb.200305130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storoni LC, et al. Likelihood-enhanced fast rotation functions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- Weissenhorn W, et al. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol. Cell. 1998;2:605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- Weissenhorn W, et al. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- White JM, et al. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin HS, et al. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc. Natl. Acad. Sci. 2005;102:9288–93. doi: 10.1073/pnas.0503989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin HS, et al. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439:38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]