Abstract
Alleles subject to strong, recent positive selection will be swept toward fixation together with contiguous sections of the genome. Whether the genomic signatures of such selection will be readily detectable in outbred wild populations is unclear. In this study, we employ haplotype diversity analysis to examine evidence for selective sweeps around knockdown resistance (kdr) mutations associated with resistance to dichlorodiphenyltrichloroethane and pyrethroid insecticides in the mosquito Anopheles gambiae. Both kdr mutations have significantly lower haplotype diversity than the wild-type (nonresistant) allele, with kdr L1014F showing the most pronounced footprint of selection. We complement these data with a time series of collections showing that the L1014F allele has increased in frequency from 0.05 to 0.54 in 5 years, consistent with a maximum likelihood–fitted selection coefficient of 0.16 and a dominance coefficient of 0.25. Our data show that strong, recent positive selective events, such as those caused by insecticide resistance, can be identified in wild insect populations.
Keywords: Anopheles gambiae, malaria, insecticide resistance, selection
Introduction
The sequencing of the Anopheles gambiae genome has opened up the possibility for genome-wide single nucleotide polymorphism (SNP)–based association mapping studies that have been successful in identifying positively selected loci in the human genome (Sabeti et al. 2002, 2007; Bersaglieri et al. 2004). The resolution of the association mapping approach is defined by the probability that recombination will have broken down the association between markers and a trait-associated functional polymorphism. Data from extensive resequencing of (primarily) detoxification genes in samples from wild populations of A. gambiae revealed a very high frequency of segregating sites (Wilding et al. 2009), consistent with high rates of recombination (Begun and Aquadro 1992; Begun et al. 2007) and/or a long history of outbreeding. In isofemale lab strains of Drosophila spp., it has been possible to observe selective sweeps around insecticide resistance–associated loci (Schlenke and Begun 2004; Aminetzach et al. 2005), but how long these signatures persist in wild populations is unknown. In this paper, we use linkage disequilibrium (LD)–based haplotype diversity analysis (Sabeti et al. 2006) to investigate the pattern of molecular genetic variation associated with insecticide resistance mutations at the pyrethroid and dichlorodiphenyltrichloroethane (DDT) knockdown resistance locus, kdr, in the African malaria mosquito A. gambiae s.s. Furthermore, as a corollary of this indirect genetic approach we demonstrate, using a series of temporal collections, a dramatic increase in kdr frequency in a population of A. gambiae s.s. over a period of approximately 72 generations. Data from these temporal collections are used to estimate the selection and dominance coefficients operating on kdr in the field to illustrate the potential levels of selection necessary to produce the patterns of LD we observe.
Insecticide-treated bed nets are the principal method for preventing malaria in sub-Saharan Africa. Currently, pyrethroids are the only class of insecticides licensed for use on nets, and there is concern that resistance will compromise control programs. To date the most commonly recorded resistance mechanism is termed “knockdown resistance” and results from single–base pair mutations in the voltage-gated sodium channel. The sodium channel gene, located within division 20C near the centromere of chromosome 2L, codes for a protein that is the target site of pyrethroid insecticides. Two alternative single–base pair mutations have been found in A. gambiae, and these kdr mutations can cause target-site insensitivity to pyrethroids as well as cross-resistance to DDT. The substitutions cause amino acid changes at codon 1014 within the transmembrane structure of segment 6 in domain II of the voltage-gated sodium channel (numbering according to the housefly para sequence, GenBank X96668). The L1014F mutation, a leucine to phenylalanine change, was first observed in West Africa (Martinez-Torres et al. 1998), and the same substitution has been observed in a diverse array of insects (Davies et al. 2007a). A second substitution, L1014S, was observed more recently in East African A. gambiae (Ranson et al. 2000) and involves the adjacent base of the same codon, resulting in a leucine to serine change.
There are two incipient species within the nominal taxon A. gambiae s.s. that are characterized by mutations on the X chromosome and are termed M and S form. The distribution of the kdr mutation is not uniform either within or between forms, although in general kdr alleles have been found at much higher frequencies in A. gambiae s.s. S-form samples compared with M-form samples (reviewed in Santolamazza et al. 2008). The reasons for the differences in distribution remain unclear because little is known about the origins of the kdr mutations and the selection pressures acting upon them in wild populations. In a sample from Benin, the L1014F was found in tight LD with two upstream intronic polymorphisms in both M- and S-form individuals. The two upstream polymorphisms associated with the L1014F variant were not found in wild-type M-form individuals but were common in wild-type S-form individuals, suggestive of an introgression event from S-form to M-form populations (Weill et al. 2000). This linkage between kdr and the intronic polymorphisms was not seen in M-form individuals from Bioko Island and was thought to indicate de novo mutation (Reimer et al. 2008). More recently, a study of S-form specimens from 15 countries suggested that the L1014F and L1014S mutations have both arisen independently on at least two separate occasions (Pinto et al. 2007).
Samples were obtained from three regions in sub-Saharan Africa; Kenya (East Africa) A. gambiae, S molecular form, kdr L1014S allele present; Ghana (West Africa) both M and S molecular form, kdr L1014F allele present; Gabon (Central Africa) S molecular form, both L1014S and L1014F kdr alleles present.
These population samples allow us to address a number of questions.
Available evidence suggests that the L1014S mutation has high penetrance for a DDT-resistant phenotype but lower penetrance for a pyrethroid-resistant phenotype than the L1014F mutation (Ranson et al. 2000). DDT was banned in Kenya in 1990, and we can investigate the signature of positive selection associated with weaker selection or recombination and relaxed selection.
The populations from central Africa are some of the few locations where both L1014F and L1014S alleles are observed sympatrically (Santolamazza et al. 2008). Indeed, in an earlier study, a significant, albeit marginal, L1014F/L1014S heterozygote excess was observed in samples from Libreville, Gabon (Pinto et al. 2006). By comparing patterns of LD around the three alleles, we investigate whether the unusually high frequency of the L1014S allele in these populations (63%; Pinto et al. 2006) is a result of a recent selective sweep.
In many S-form populations in West Africa, including our collections from Ghana, the L1014F allele is close to fixation. In the absence of wild-type alleles, we are unable to control for local variation in recombination rates (Sabeti et al. 2007), and it is therefore impossible to ascribe patterns of LD to a positive selection event. Recently developed approaches such as cross-population extended haplotype homozygosity (EHH) have been developed to allow interpopulation comparisons in instances where alleles proceed to near fixation in some populations (Sabeti et al. 2007), but in our system resistance alleles may have multiple origins, presenting a confounding variable (Pinto et al. 2007). However, the presence of sympatric M-form populations in southern Ghana (Yawson et al. 2004, 2007) allows us to both document the increase in frequency of the same L1014F haplotype, following an introgression event, over a period of 5 years and estimate the selection and dominance coefficients associated with the signatures of positive selection.
Materials and Methods
Sample Sites, DNA Extraction, and Species Identification
Adult female A. gambiae s.s. mosquitoes used in this study were obtained from aspirator and pyrethroid knockdown collections from the field in various geographic locations (table 1). DNA was extracted from single female A. gambiae using either a modified Livak method or a phenol–chloroform method (Livak 1984; Ballinger-Crabtree et al. 1992). Species identification polymerase chain reaction (PCR) was carried out on A. gambiae s.l. according to the protocol (Scott et al. 1993). Reactions were then digested with CfoI restriction enzyme for 24 h at 37 °C in order to type A. gambiae s.s mosquitoes to M and S form (Fanello et al. 2002), and products visualized under UV light after electrophoresis on a 2% agarose Tris/borate/EDTA (TBE) gel with ethidium bromide. Kdr genotypes were determined by allele-specific PCR, heated oligonucleotide ligation assay (Lynd et al. 2005), or Taqman assay (Bass et al. 2007) depending upon year of collection.
Table 1.
Origin and kdr Genotype of Specimens Used in the Study.
| Population | Year Collected | Total N | Form | Number of Each kdr Genotype |
|||||
| L1014S/L1014S | L1014F/L1014F | L1014F/L1014S | L1014S/wt | L1014F/wt | Wt/wt | ||||
| Asembo Bay, Kenya, 00°10′S, 34°22′E | 20051 | 48 | S | 11 | — | — | 17 | — | 20 |
| Dienga, Gabon, 01°52′S, 12°40′E | 1999–20002 | 30 | S | — | — | — | 4 | 2 | 24 |
| Bakoumba, Gabon, 01°49′S, 13°01′E | 1999–20002 | 42 | S | — | 5 | 8 | 5 | 7 | 17 |
| Libreville, Gabon, 00°22′N, 09°26′E | 1999–20002 | 73 | S | 34 | 8 | 31 | — | — | — |
| Okyereko and Accra area, Ghana, 05°24.9′N, 00°36.6′W, 05°38′N, 00°15′E | 20023 | 35 | S | — | 33 | — | — | 2 | — |
| Okyereko, Ghana, 05°24.9′N, 00°36.6′W | 20023 | 30 | M | — | — | — | — | 2 | 28 |
NOTE.—The population name and total numbers of each DNA sample utilized. Molecular form is indicated, and the numbers of each kdr genotype are shown. Additional information on the collection sites may be obtained from the publications where the specimens are originally described: 1Müller et al. (2008), 2Pinto et al. (2006), and 3Yawson et al. (2004); wt, wild type.
Sodium Channel SNP Identification
The voltage-gated sodium channel gene is nearly 74 kbp in length and is composed of 35 exons including two duplicate exons (Davies et al. 2007a). Ten regions of the sodium channel were amplified by PCR for direct sequencing. Where possible, primers were designed to bind within exons to produce amplicons that spanned an intron with a maximum size of 1.5 kbp. Exons (numbering as Davies et al. 2007a) 1–2, 3, 4, 7–9, 13–14, 15–17, 20c, 23–24, 28–30, and 32–33 were selected as targets for sequencing. Primer and amplification details are provided (supplementary table 1, Supplementary Material online). Sequencing for SNP detection was carried out on up to 12 individuals of known kdr genotype from Ghana, São Tomé, Gabon, Angola, Mozambique, Malawi, and Kenya, from a susceptible laboratory strain (KISUMU), and from a permethrin tolerant resistant laboratory strain (reduced susceptibility to permethrin), both originating from Kenya. PCR products were cleaned using a Mini Elute PCR Purification kit (Qiagen) and then sequenced in both directions. Sequences were aligned using Bioedit software version 7.0.5.2 (Hall 1999) and then manually annotated for polymorphisms and ambiguities.
In addition, seven M-form individuals from Accra, Ghana, homozygous for the L1014F allele were bidirectionally sequenced across PCR amplicons 13–14, 15–17, and 21 to determine the associated haplotype of the kdr allele in this population.
SNP Screening
SNPs discovered through resequencing were screened in the large-scale SNP detection study using the SNPStart Primer Extension Kit on the Beckman CEQ 8000 Genetic Analysis System. Details of SNPs both included and excluded from the SNP screening are given in supplementary table 2, Supplementary Material online. Multiplex PCR was carried out to amplify the regions of DNA containing SNPs of interest, including a region of exon 20 and the preceding intron to allow high-throughput detection of the kdr mutation and three other well-characterized SNPs (Weill et al. 2000; Diabate et al. 2004; Pinto et al. 2006) (primers and reaction conditions detailed in supplementary table 3, Supplementary Material online). Products were visualized on a 2% TBE agarose gel. Successfully multiplexed samples were prepared for subsequent SNP extension by ExoI/shrimp alkaline phosphatase (SAP) enzymatic digestion. Interrogation primers were then designed for each individual SNP chosen for investigation according to the manufacturers’ recommendations (supplementary table 4, Supplementary Material online). Single-base extension to the 3′ end of the interrogation primer by a dye terminator molecule, corresponding to the nucleotide found at the SNP location, was carried out using a GenomeLab SNPStart Primer Extension Kit (Beckman Coulter, Amersham, UK). The SAP-digested product was then scored on the Beckman CEQ 8000 Genetic Analysis System.
Data Analysis
As reviewed exhaustively by Sabeti et al. (2006), there are numerous statistical tests of positive selection which differ in their ability to detect selection events on different timescales. For the present SNP data set, it is not possible to use the suite of sequence-based tests that compare synonymous/nonsynonymous differences or detect an excess of rare alleles. We are therefore fortunate that on the timescales in which the emergence, and selection, of insecticide resistance is likely to occur, estimates of interpopulation divergence (e.g., based on F statistics) and screens of LD around selected versus wild-type alleles are likely to be the two most powerful analytical approaches. With the sample sizes available in our study, single-marker analyses based on F-statistic estimates would perform better as indicators of selection when markers can be typed at a more coarse scale, with consequently enhanced signal:noise ratio. However, with sample size constraints the signal would be difficult to localize. By contrast, long-range haplotype analyses, such as EHH (Sabeti et al. 2002) analysis, perform very well at a fine physical scale in identifying narrow candidate regions (Sabeti et al. 2006).
EHH analysis was carried out to assess the patterns of LD associated with wild type and the two kdr alleles. EHH can be defined as the probability that two random chosen chromosomes carrying the core (e.g., the wild-type or kdr allele) haplotype of interest are identical by descent. This approach first identifies core haplotypes surrounding the locus of interest and then examines the decay in LD from these core haplotypes to the surrounding loci. The resulting EHH can be used as evidence of recent positive selection at a locus in haplotypes that have high frequency and high EHH (Sabeti et al. 2002). EHH analysis requires haplotype information that cannot be empirically determined from the genotype data gathered by the methods used in this study. Therefore, haplotypes were inferred using PHASE software version 2.1.1 using default parameters (Stephens et al. 2001; Stephens and Scheet 2005). PHASE utilizes a Bayesian coalescent–based approach to determine phase and allows for varying rates of recombination at each SNP interval. The method is based on the idea that an unresolved haplotype is more likely to be the same or be similar to a previous haplotype. This approach was found to outperform other methods available for autosomal human data sets (Stephens et al. 2001; Stephens and Scheet 2005). Data were analyzed together rather than as separate subpopulations because 1) previous studies found this to be more accurate and 2) haplotype determination methods of this nature are relatively insensitive to departures from Hardy–Weinberg equilibrium so are fairly robust to population substructuring. This approach is also more conservative than determining haplotypes for individual populations because the latter is liable to lead to an underestimation in differences in haplotype frequencies (Stephens and Scheet 2005). Phase reconstruction was executed ten times upon the total data set, and differences in counts of best haplotypes were noted.
The estimated haplotypes obtained from PHASE were used as input for EHH analysis implemented by SWEEP version 2.1.1 (Sabeti et al. 2002). Core haplotypes were selected manually to include only the two adjacent kdr-causing loci. Significance of EHH values is usually assigned through comparison to an empirically generated null distribution from other regions of the genome. However, given that we had already identified the causal mutations of interest, we were able to make a comparison of patterns of LD around wild-type and resistant cores. The primary advantage of this approach is that it is not subject to the genome-wide variations in recombination rate which can affect the null distribution approach in species lacking detailed recombination maps. Significant differences in EHH values were determined in two ways: 1) Within country samples, at individual SNP positions with nonoverlapping 95% confidence intervals (CIs). These CIs were calculated at each SNP position using a bootstrapping procedure, carried out in SAS version 9 software. Resampling was carried out 1,000 times. 2) Across all SNPs within and among country samples, the diversity of the different kdr allele–bearing haplotypes was compared using sign tests, implemented by SPSS 14. Where exact sign test probabilities could not be calculated, a Monte Carlo procedure with 10,000 permutations was performed. The sequential Bonferroni procedure was applied to determine statistical significance following correction for multiple testing (Holm 1979). Although our data—EHH values at each SNP position—are not independent, it is this nonindependence caused by LD that will cause departure from the null hypothesis of equality of median EHH values. Therefore, the null hypothesis remains that there is no difference in median EHH between kdr and wild-type alleles. Bifurcation plots were also created using the SWEEP software. In a bifurcation plot, the core haplotype is represented as a black circle. Each SNP, moving out from the core both upstream and downstream of the kdr locus, is a potential site for a bifurcation that would result from the presence of two segregating alleles. Therefore, the diagram provides a means of displaying the breakdown in LD at increasing distance from the core haplotypes. The radius of the circle at each node is proportional to the number of individuals with that haplotype.
Calculation of Selection and Dominance Coefficients
The spread of the L1014F allele was modeled using the standard recursive population genetic formula:
| (1) |
where p is the frequency of the L1014F allele, p′ is the frequency in the next generation, s is the selective coefficient of the resistance mutation, h is the dominance coefficient (1 = complete dominance, 0 = complete recessivity), and W is the normalizing factor (Maynard-Smith 1998).
Tracking allele frequencies over time requires three input parameters: initial allele frequency at time zero, s, and h. Estimates of all three unknown parameters were obtained by maximum likelihood assuming a binomial distribution of observed allele frequencies around the predicted frequency. The analysis was performed in R (http://www.r-project.org) using maximum likelihood functions and optimizing routines. The generation time was set at the standard of one generation per calendar month (Lehmann et al. 1998).
Results
SNP Discovery and Screening
Ten genomic regions of a combined length of ≈6.5 kb of DNA, spanning a region of ≈73 kb of the voltage-gated sodium channel, were amplified and sequenced in A. gambiae s.s. individuals from seven countries across sub-Saharan Africa. A total of 62 potential SNPs were found, of which 14 were exonic (supplementary table 1, Supplementary Material online). Six intronic indels were observed, usually in poly-A or tandem AT repeats (supplementary table 1, Supplementary Material online). On average, there was one SNP every 106 bp, which represents a low SNP frequency for A. gambiae, but similar to other genes in the same genomic locality (chromosome 2L division 20; Wilding et al. 2009). Thirty-two SNPs, including the two kdr mutations, were selected for screening in 258 individuals. In S-form individuals, the SNP adjacent to the core in the upstream (centromeric) direction was excluded from further analysis as it was found to be monomorphic. Details of the populations and associated kdr genotypes are given in table 1. The genotypic data were resolved into haplotypes with ten runs of the analysis. In only one instance, did the replicate runs resolve a novel estimated haplotype, which in a subsequent comparative analysis was found to exert no qualitative effect on the results. Therefore, all analyses reported here are based upon the haplotypes resolved in the vast majority of the phasing runs.
EHH analysis was carried out to assess the patterns of LD associated with the wild-type and the two kdr alleles. The intronic SNPs that have been used to identify the origin of the kdr mutations were the proximate SNPs in the centromeric direction (Weill et al. 2000; Pinto et al. 2007). LD decay was examined between these core haplotypes and the remaining 29 or 30 SNP loci (for S or M forms, respectively).
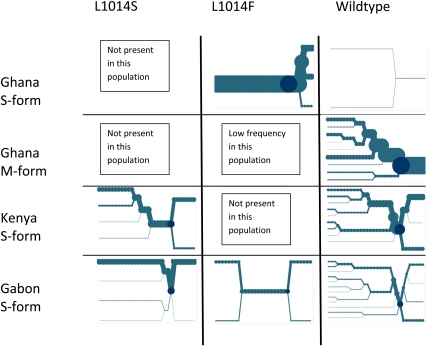
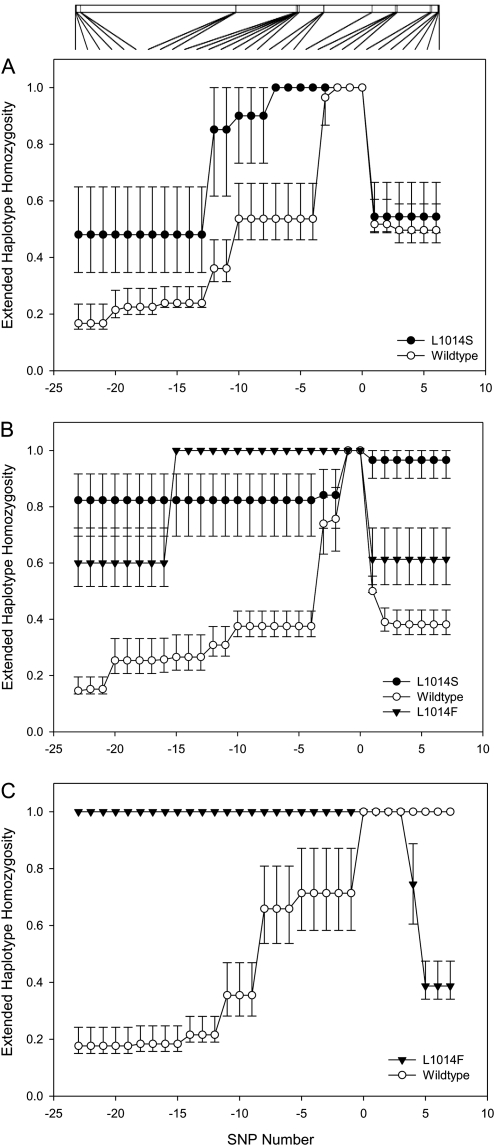
Only two core-alleles were present in the western Kenyan sample: wild type and L1014S. In the downstream telomeric direction, EHH decays at a similar rate for both wild type and L1014S, but there was a marked contrast between alleles in the centromeric direction, with entirely nonoverlapping confidence limits from just a few kilobases away from the core (figs. 1 and 2A). In the Gabonese collection, the difference between resistance-associated alleles and wild type was even more marked with significantly lower EHH in the wild type in both centromeric and telomeric directions less than 5 kb from the core (fig. 2B). Indeed, both the L1014F and the L1014S resistance mutations showed little haplotype bifurcation in the Gabon samples over the length of the sodium channel (fig. 1), suggesting a relatively recent origin for both these mutations accompanied by a strong selective sweep. The patterns of LD are most marked around the resistant L1014F haplotype in Ghanaian S-form samples in which the L1014F kdr allele was at very high frequency (figs. 1 and 2C), as would be expected given the near fixation of this allele in southern Ghana in the S molecular form (mean frequency = 0.96; 95% CI 0.95–0.97) (Yawson et al. 2004). The presence of only two wild-type haplotypes in the sample prevent any meaningful comparison of LD decay, but it should be noted that there was complete LD over the entire 64-kb length of the sodium channel in the centromeric direction. The wild-type allele, observed in the Ghanaian M-form populations (figs. 1 and 2C), showed marked LD, only in the telomeric direction, between exons 20 and 32, the opposite directional asymmetry to the L1014F mutation in Ghana S-form populations. Although simulation studies have shown that LD decay may be asymmetric even when rates of mutation and recombination are constant (Kim and Stephan 2002), it is possible that the LD observed in these samples may reflect the presence of one or more hitherto overlooked selectively advantageous mutants, although we cannot rule out recombination with unsampled haplotypes (supplementary table 5, Supplementary Material online). Davies et al. (2007b) have summarized that there are a number of additional nonsynonymous changes observed in a variety of taxa, and detailed association mapping studies are presently underway to investigate this phenomenon. Comparing overall levels of EHH for the whole 72.6-kb regions typed, it is interesting to note that median EHH values are statistically indistinguishable for the same allele typed in different populations (table 2) and that a clear hierarchy of evidence for selective sweeps emerged. Median EHH levels were highest for the L1014F resistance mutation, followed by those for the L1014S mutations, with the lowest for the wild-type allele (table 2). The only exception to this pattern was within the Gabonese sample, the only one in which both resistance alleles were present, where median EHH was equal for the two resistance alleles. Nevertheless, despite the possibilities of different origins of the same allele, and local variation in recombination rates, EHH levels across the genomic region investigated suggest some degree of commonality in selection across populations for each allele, although the actual rate of change in LD with distance can be quite complex and dependent on direction from the core (figs. 1 and 2).
FIG. 1.
Bifurcation plots showing patterns of recombination in the centromeric (5′ toward the left) and telomeric (3′ toward the right) directions. The core is marked by the dark circle, and each of the 29/30 SNPs is represented by a node and a recombination event is represented by a bifurcation. The diameter of the circle at each SNP node is proportional to the numbers of individuals with the same long-range haplotype at that position. No bifurcation plot is shown for the L1014F core in Ghanaian M-form populations as only a single haplotype was observed (see Results).
FIG. 2.
EHH analysis showing LD decay with increasing distance from the core (marked as the origin on the x axis). The 95% CIs were estimated by bootstrapping (see Materials and Methods). The x axis is ordinal, negative numbers are in the centromeric direction and positive numbers in the telomeric direction. The scale bar at the top of the figure is 72.6 kb in length and shows the physical distance between the SNPs. (A) Kenya data for L1014S and wild-type alleles; (B) Gabon data for L1014S, L1014F, and wild-type alleles, and (C) Ghana data for L1014F (S form) and wild type (M form).
Table 2.
Comparison of Median EHH Levels between Alleles at the kdr Loci.
| Kenya L1014S (S form) | Kenya Wild Type (S form) | Gabon L1014S (S form) | Gabon Wild Type (S form) | Gabon L1014F (S form) | Ghana Wild Type (M form) | |
| Kenya wild type (S form) | 0.0001 | |||||
| Gabon L1014S (S form) | 0.26 NS | 0.0001 | ||||
| Gabon wild type (S form) | 0.0001 | 0.026NS | 0.0001 | |||
| Gabon L1014F (S form) | 0.0001 | 0.0001 | 1.00NS | 0.0001 | ||
| Ghana wild type (M form) | 0.005 | 1.00NS | 0.005 | 0.86NS | 0.005 | |
| Ghana L1014F (S form) | 0.0005 | 0.0001 | 0.0001 | 0.0001 | 0.04NS | 0.0003 |
NOTE.—Probabilities from sign tests are shown. The values followed by NS were not significant after sequential Bonferroni corrections. Values that are underlined indicate that the EHH values were significantly higher for the sample given in the column heading; values that are in bold indicate that the EHH values were significantly higher for the sample given in the row heading.
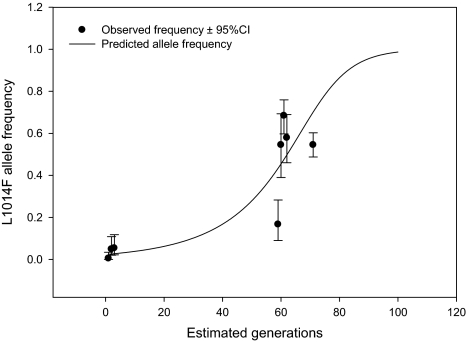
We investigated temporal change in the frequency of the L1014F allele and associated haplotype in sympatric populations of M-form individuals in a subset of the populations previously described by Yawson et al. (2004). Using the data reported in Yawson et al. (2004), we estimated the L1014F allele frequency in M-form populations from around Accra, southern Ghana (≈30 km diameter collection area), during 2002 (freqL1014F = 0.03; 95% CI 0.01–0.05). Additional screening in 2007 and 2008 from the same greater Accra regions revealed that within 5 years, this frequency had reached freqL1014F = 0.54 (95% CI 0.49–0.60; fig. 3). The data from years 2007 and 2008 are reported here for the first time. Phasing of the SNP genotypes of two M-form individuals with a wild-type/L1014F genotype showed that the L1014F-associated haplotype was identical to that found in the S form. This was confirmed by sequences obtained from seven M-form individuals collected from Accra, Ghana in 2008, which were homozygous for the L1014F allele (supplementary table 6, Supplementary Material online). Therefore, the L1014F allele, which has increased in frequency in M-form populations, is the same that has been putatively swept toward fixation in sympatric S-form populations. Introgression of kdr alleles between forms has been documented previously (Weill et al. 2000) and is unsurprising given that in southern Ghana there is a low but temporally stable level of interform matings (Yawson et al. 2004, 2007).
FIG. 3.
Observed and predicted changes in L1014F allele frequency in the Anopheles gambiae M-form populations from southern Ghana. Observed data obtained from surveys conducted in 2002, 2006, and 2007. First collection point (Generation 1) was June 2002. Data from 2002, first three data points, are taken from Yawson et al. (2004); all other data are novel. One generation per month is assumed following Lehmann et al. (1998). The 95% CIs for each observed data point were calculated according to Newcombe (1998). Expected data generated from simultaneous maximum likelihood estimates of initial frequency and selection and dominance coefficients (see Materials and Methods).
Using a maximum likelihood estimation procedure with random starting values for selection coefficient (s), dominance (h), and initial allele frequency (p0), the parameter estimates converged to s = 0.163 (standard deviation [SD] = 0.052), h = 0.249 (SD = 0.142), and initial frequency p0 = 0.025 (SD = 0.008) (fig. 3).
Discussion
These data show that there is marked LD around kdr mutations, loci exhibiting high penetrance, and, for L1014F at least, subject to strong recent positive selection. Despite similar median EHH levels, there were differences in the patterns of LD associated with the L1014S mutation in Kenya and Gabon. In Kenyan samples, the rate of dissipation of LD around the L1014S core was quite rapid suggesting that the mutation has not been subject to as recent or as strong a selective sweep as the same mutation in Gabon (or indeed as the L1014F mutation in Ghana). This is as predicted if the serine resistance allele was primarily selected by the use of DDT in the latter part of the 20th century rather than by the more recent use of pyrethroids in agriculture and insecticide control programs. In Culex mosquitoes, the equivalent L1014S mutation gives low levels of kdr to pyrethroids compared with the L1014F mutation but confers high levels of DDT resistance (Martinez-Torres et al. 1998; Ranson et al. 2000). Stump et al. (2004) investigated the change in allele frequency of the L1014S allele before and after the commencement of a large-scale ITN project in Asembo Bay, Western Kenya, the site of our collections (Stump et al. 2004). The frequency of the L1014S allele in the region approximately 10 years before bed net introduction was approximately 0.04 (95% CI 0.02–0.08). In 2002, 15 years after this initial survey and 5 years after the introduction of nets, the frequency of the L1014S allele had increased, nonsignificantly to only 0.075 (95% CI 0.05–0.12). This suggests that there is little selective advantage for this mutation in the present environment, although it should be noted that in a neighboring district in Uganda, a recent study reported that the L1014S mutation was at a frequency of 0.85 (95% CI 0.83–0.87) (Ramphul et al. 2009). An alternative explanation would be that in Uganda there is an epistatic interaction between L1014S and some, as yet unidentified locus, which may affect the selection, and indeed dominance coefficients, and thereby result in a higher L1014S frequency.
The high frequency and marked LD associated with L1014S in Gabon may be a result of the co-occurrence in genotypes, though not haplotypes (supplementary table 5, Supplementary Material online), with L1014F. A recent study from Cameroon showed that although L1014F/L1014S heterozygotes were significantly less resistant to permethrin than L1014F homozygotes, L1014F/L1014S heterozygotes were significantly more resistant to all insecticides tested than L1014F/L1014-wild type heterozygotes (Reimer et al. 2008). Repetitive mutation at the 1014 locus could, at least in part, be responsible for the patterns of LD around the kdr locus in the Gabonese data. Indeed, there is evidence for repeated mutations of kdr alleles across the species range of A. gambiae (Pinto et al. 2007). However, we argue that on the recent timescales on which kdr has arisen and spread it is more parsimonious to assume that recombination is the dominant influence on patterns of LD rather than high rates of repetitive mutations.
Although kdr is the best-documented resistance mechanism in A. gambiae, there are many other resistance-associated loci. Microarray and recombinant protein expression work has shown that resistant mosquitoes over express a small number of enzymes that catalyze insecticide degradation (Ortelli et al. 2003; Müller et al. 2007; Chiu et al. 2008; Müller et al. 2008). LD-based screens could be a powerful way of identifying regions of the genome carrying the scars of recent selection that regulate such overexpression. However, whether association mapping approaches will effectively identify genes subject to much older and comparatively weaker selection is currently unclear. The bounded estimate of the selection coefficient reported here is at the upper limit of estimates generated to date and of a similar magnitude to estimates generated for resistance alleles in the mosquito Culex pipiens (Labbe et al. 2009). In human populations, mutations associated with resistance to malaria infection such as G6PD and sickle cell trait have coefficients of selection of 0.02–0.05 (Tishkoff and Williams 2002) and 0.05–0.18 (Li 1975), respectively. In the third actor in the malaria transmission cycle of Plasmodium falciparum, a selection coefficient of 0.1 has been obtained for the locus dhfr that confers resistance to the chemotherapeutic agent, pyrimethamine (Nair et al. 2003).
Together with strong and recent positive selection, the major determinant of LD around selected loci will be the rate of recombination. Indications of dramatic variation in the recombination rate across the A. gambiae genome have already been reported (Pombi et al. 2006; Black et al. 2008), and it is possible that, being close to the centromere of chromosome 2L, the sodium channel locus is in an area of reduced recombination. However, our Kenyan data are consistent with rates of recombination sufficient to reduce the region hitchhiked with a selectively advantageous locus in a relatively short period of time. Indeed, detection of the signatures of selection for loci with low selection coefficients will be more logistically challenging in A. gambiae than humans because of much lower background levels of LD (Weetman D, Wilding CS, Steen K, Donnelly MJ, unpublished data). We attempted to amplify microsatellites from around the sodium channel to fully define the extent of the swept region as has been done for drug resistance loci in P. falciparum (Wootton et al. 2002; Nair et al. 2003). However, the sodium channel is situated in a region with an abundance of repetitive sequences and it was not possible to identify unique locus-specific microsatellite primer pairs.
Given the apparently high selection pressure on the L1014F mutation, it is curious that there are no studies, with adequate sample size, that have observed either of the kdr alleles at fixation (Santolamazza et al. 2008). One explanation would be that of overdominance; however, insecticide bioassays studies suggest that this is unlikely to be the case (Chandre et al. 2000; Reimer et al. 2008), and our estimate of the dominance coefficient shows the kdr L1014F allele to be partially recessive. Therefore, it is likely that there is some fitness cost to the L1014F allele and that this could be attributable to heterogeneity in exposure to pyrethroids in the environment or a consequence of an Hill–Robertson effect where selection at a kdr locus can interfere with the selection at nearby beneficial mutations (Hill and Robertson 1966).
The data presented herein show that it is possible to detect genomic signatures of strong positive selection in pest species with large effective population size and generally low levels of LD. We suggest that such approaches are likely to extremely powerful in many nonmodel taxa subject to similar selective events.
Supplementary Material
Supplementary tables 1–6 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
We thank Nelson Cuamba, Eveline Klinkenberg, and Pie Muller for providing wild caught specimens and the associate editor for constructive comments. This work was supported by the National Institutes for Health (U01 AI58542 to Dr Edward Walker, Michigan State University), the Innovative Vector Control Consortium, and the Royal Society.
References
- Aminetzach YT, Macpherson JM, Petrov DA. Pesticide resistance via transposition-mediated adaptive gene truncation in Drosophila. Science. 2005;309:764–767. doi: 10.1126/science.1112699. [DOI] [PubMed] [Google Scholar]
- Ballinger-Crabtree ME, Black WC, IV, Miller BR. Use of genetic polymorphisms detected by the random-amplified polymorphic DNA polymerase chain reaction (RAPD-PCR) for differentiation and identification of Aedes aegypti subspecies and populations. Am J Trop Med Hyg. 1992;47:893–901. doi: 10.4269/ajtmh.1992.47.893. [DOI] [PubMed] [Google Scholar]
- Bass C, Nikou D, Donnelly MJ, Williamson MS, Ranson H, Ball A, Vontas J, Field LM. Detection of knockdown resistance (kdr) mutations in Anopheles gambiae: a comparison of two new high-throughput assays with existing methods. Malar J. 2007;6e:111. doi: 10.1186/1475-2875-6-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, Aquadro C. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature. 1992;356:519–520. doi: 10.1038/356519a0. [DOI] [PubMed] [Google Scholar]
- Begun DJ, Holloway AK, Stevens K, et al. (13 co-authors) Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 2007;5:e310. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersaglieri T, Sabeti PC, Patterson N, Vanderploeg T, Schaffner SF, Drake JA, Rhodes M, Reich DE, Hirschhorn JN. Genetic signatures of strong recent positive selection at the lactase gene. Am J Hum Genet. 2004;74:1111–1120. doi: 10.1086/421051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black WC, IV, Gorrochetegui-Escalante N, Randle NP, Donnelly MJ. The yin and yang of linkage disequilibrium: mapping of genes and nucleotides conferring insecticide resistance in insect disease vectors. Transgenesis and the Management of Vector-Borne Disease. 2008;627:71–83. doi: 10.1007/978-0-387-78225-6_6. [DOI] [PubMed] [Google Scholar]
- Chandre F, Darriet F, Duchon S, Finot L, Manguin S, Carnevale P, Guillet P. Modifications of pyrethroid effects associated with kdr mutation in Anopheles gambiae. Med Vet Entomol. 2000;14:81–88. doi: 10.1046/j.1365-2915.2000.00212.x. [DOI] [PubMed] [Google Scholar]
- Chiu T, Wen Z, Rupasinghe S, Schuler M. Comparative molecular modeling of Anopheles gambiae CYP6Z1, a mosquito P450 capable of metabolizing DDT. Proc Natl Acad Sci U S A. 2008;105:8855–8860. doi: 10.1073/pnas.0709249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies TGE, Field LM, Usherwood PNR, Williamson MS. A comparative study of voltage-gated sodium channels in the Insecta: implications for pyrethroid resistance in Anopheline and other Neopteran species. Insect Mol Biol. 2007a;16:361–375. doi: 10.1111/j.1365-2583.2007.00733.x. [DOI] [PubMed] [Google Scholar]
- Davies TGE, Field LM, Usherwood PNR, Williamson MS. DDT, pyrethrins, pyrethroids and insect sodium channels. IUBMB Life. 2007b;59:151–162. doi: 10.1080/15216540701352042. [DOI] [PubMed] [Google Scholar]
- Diabate A, Brengues C, Baldet T, et al. (11 co-authors) The spread of the Leu-Phe kdr mutation through Anopheles gambiae complex in Burkina Faso: genetic introgression and de novo phenomena. Trop Med Int Health. 2004;9:1267–1273. doi: 10.1111/j.1365-3156.2004.01336.x. [DOI] [PubMed] [Google Scholar]
- Fanello C, Santolamazza F, della Torre A. Simultaneous identification of species and molecular forms of the Anopheles gambiae complex by PCR-RFLP. Med Vet Entomol. 2002;16:461–464. doi: 10.1046/j.1365-2915.2002.00393.x. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hill WG, Robertson A. The effect of linkage on limits to artificial selection. Genet Res. 1966;8:269–294. [PubMed] [Google Scholar]
- Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat. 1979;6:65–70. [Google Scholar]
- Kim Y, Stephan W. Detecting a local signature of genetic hitchhiking along a recombining chromosome. Genetics. 2002;160:765–777. doi: 10.1093/genetics/160.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbe P, Sidos N, Raymond M, Lenormand T. Resistance gene replacement in the mosquito Culex pipiens: fitness estimation from long-term cline series. Genetics. 2009;182:303–312. doi: 10.1534/genetics.109.101444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann T, Hawley WA, Grebert H, Collins FH. The effective population size of Anopheles gambiae in Kenya: implications for population structure. Mol Biol Evol. 1998;15:264–276. doi: 10.1093/oxfordjournals.molbev.a025923. [DOI] [PubMed] [Google Scholar]
- Li W. The first arrival time and mean age of a deleterious mutant gene in a finite population. Am J Hum Genet. 1975;27:274–286. [PMC free article] [PubMed] [Google Scholar]
- Livak KJ. Organization and mapping of a sequence on the Drosophila melanogaster X and Y chromosomes that is transcribed during spermatogenesis. Genetics. 1984;107:611–634. doi: 10.1093/genetics/107.4.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynd A, Ranson H, McCall PJ, Randle NP, Black WC, Walker ED, Donnelly MJ. A simplified high-throughput method for pyrethroid knock-down resistance (kdr) detection in Anopheles gambiae. Malar J. 2005;4:16. doi: 10.1186/1475-2875-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Torres D, Chandre F, Williamson MS, Darriet F, Berge JB, Devonshire AL, Guillet P, Pasteur N, Pauron D. Molecular characterization of pyrethroid knockdown resistance (kdr) in the major malaria vector Anopheles gambiae s.s. Insect Mol Biol. 1998;7:179–184. doi: 10.1046/j.1365-2583.1998.72062.x. [DOI] [PubMed] [Google Scholar]
- Maynard-Smith J. Evolutionary genetics. Oxford: Oxford University Press; 1998. [Google Scholar]
- Müller P, Chouaibou M, Pignatelli P, Etang J, Walker ED, Donnelly MJ, Simard F, Ranson HW. Pyrethroid tolerance is associated with elevated expression of antioxidants and agricultural practice in Anopheles arabiensis sampled from an area of cotton fields in Northern Cameroon. Mol Ecol. 2008;17:1145–1155. doi: 10.1111/j.1365-294X.2007.03617.x. [DOI] [PubMed] [Google Scholar]
- Müller P, Donnelly MJ, Ranson H. Transcription profiling of a recently colonised pyrethroid resistant Anopheles gambiaestrain from Ghana. BMC Genomics. 2007;8:e36. doi: 10.1186/1471-2164-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller P, Warr E, Stevenson BJ, et al. (12 co-authors) Field-caught permethrin-resistant Anopheles gambiae overexpress CYP6P3, a P450 that metabolises pyrethroids. PLoS Genet. 2008;4:e1000286. doi: 10.1371/journal.pgen.1000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S, Williams JT, Brockman A, et al. (12 co-authors) A selective sweep driven by pyrimethamine treatment in southeast asian malaria parasites. Mol Biol Evol. 2003;20:1526–1536. doi: 10.1093/molbev/msg162. [DOI] [PubMed] [Google Scholar]
- Newcombe RG. Two-sided confidence intervals for the single proportion: comparison of seven methods. Stat Med. 1998;17:857–872. doi: 10.1002/(sici)1097-0258(19980430)17:8<857::aid-sim777>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Ortelli F, Rossiter LC, Vontas J, Ranson H, Hemingway J. Heterologous expression of four glutathione transferase genes genetically linked to a major insecticide-resistance locus from the malaria vector Anopheles gambiae. Biochem J. 2003;373:957–963. doi: 10.1042/BJ20030169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto J, Lynd A, Elissa N, Donnelly MJ, Costa C, Gentile G, Caccone A, Do Rosario VEI. Co-occurrence of East and West African kdr mutations suggests high levels of resistance to pyrethroid insecticides in Anopheles gambiae from Libreville, Gabon. Med Vet Entomol. 2006;20:27–32. doi: 10.1111/j.1365-2915.2006.00611.x. [DOI] [PubMed] [Google Scholar]
- Pinto J, Lynd A, Vicente JL, et al. (13 co-authors) Multiple origins of knockdown resistance mutations in the Afrotropical mosquito vector Anopheles gambiae. PLoS ONE. 2007;2:e1243. doi: 10.1371/journal.pone.0001243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombi M, Stump AD, Della Torre A, Besansky NJ. Variation in recombination rate across the X chromosome of Anopheles gambiae. Am J Trop Med Hyg. 2006;75:901–903. [PubMed] [Google Scholar]
- Ramphul U, Boase T, Bass C, Okedi LM, Donnelly MJ, Müller P Forthcoming. Insecticide resistance and its association with target-site mutations in natural populations of Anopheles gambiaefrom eastern Uganda. Trans R Soc Trop Med Hyg. 2009;103:1121–1126. doi: 10.1016/j.trstmh.2009.02.014. [DOI] [PubMed] [Google Scholar]
- Ranson H, Jensen B, Vulule JM, Wang X, Hemingway J, Collins FHI. Identification of a point mutation in the voltage-gated sodium channel gene of Kenyan Anopheles gambiae associated with resistance to DDT and pyrethroids. Insect Mol Biol. 2000;9:491–497. doi: 10.1046/j.1365-2583.2000.00209.x. [DOI] [PubMed] [Google Scholar]
- Reimer L, Fondjo E, Patchoke S, et al. (11 co-authors) Relationship between kdr mutation and resistance to pyrethroid and DDT insecticides in natural populations of Anopheles gambiae. J Med Entomol. 2008;45:260–266. doi: 10.1603/0022-2585(2008)45[260:rbkmar]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Reich DE, Higgins JM, et al. (17 co-authors) Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Schaffner SF, Fry B, Lohmueller J, Varilly P, Shamovsky O, Palma A, Mikkelsen TS, Altshuler D, Lander ESW. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Varilly P, Fry B, et al. (12 co-authors) Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santolamazza F, Calzetta M, Etang J, et al. (11 co-authors) Distribution of knock-down resistance mutations in Anopheles gambiaemolecular forms in west and west-central Africa. Malar J. 2008;7:e74. doi: 10.1186/1475-2875-7-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenke TA, Begun DJ. Strong selective sweep associated with a transposon insertion in Drosophila simulans. Proc Natl Acad Sci U S A. 2004;101:1626–1631. doi: 10.1073/pnas.0303793101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JA, Brogdon WG, Collins FH. Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am J Trop Med Hyg. 1993;49:520–529. doi: 10.4269/ajtmh.1993.49.520. [DOI] [PubMed] [Google Scholar]
- Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet. 2005;76:449–462. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stump AD, Atieli FK, Vulule JM, Besansky NJ. Dynamics of the pyrethroid knockdown resistance allele in western Kenyan populations of Anopheles gambiae in response to insecticide-treated bed net trials. Am J Trop Med Hyg. 2004;70:591–596. [PubMed] [Google Scholar]
- Tishkoff SA, Williams SM. Genetic analysis of African populations: human evolution and complex disease. Nat Rev Genet. 2002;3:611–621. doi: 10.1038/nrg865. [DOI] [PubMed] [Google Scholar]
- Weill M, Chandre F, Brengues C, Manguin S, Akogbeto M, Pasteur N, Guillet P, Raymond M. The kdr mutation occurs in the Mopti form of Anopheles gambiae s.s. through introgression. Insect Mol Biol. 2000;9:451–455. doi: 10.1046/j.1365-2583.2000.00206.x. [DOI] [PubMed] [Google Scholar]
- Wilding CS, Weetman D, Steen K, Donnelly MJ. High, clustered, nucleotide diversity in the genome of Anopheles gambiaerevealed by SNP discovery through pooled-template sequencing: implications for high-throughput genotyping protocols. BMC Genomics. 2009;10:e320. doi: 10.1186/1471-2164-10-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton JC, Feng XR, Ferdig MT, Cooper RA, Mu JB, Baruch DI, Magill AJ, Su XZI. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature. 2002;418:320–323. doi: 10.1038/nature00813. [DOI] [PubMed] [Google Scholar]
- Yawson AE, McCall PJ, Wilson MD, Donnelly MJ. Species abundance and insecticide resistance of Anopheles gambiae in selected areas of Ghana and Burkina Faso. Med Vet Entomol. 2004;18:372–377. doi: 10.1111/j.0269-283X.2004.00519.x. [DOI] [PubMed] [Google Scholar]
- Yawson AE, Weetman D, Wilson MD, Donnelly MJ. Ecological zones rather than molecular forms predict genetic differentiation in the malaria vector Anopheles gambiae s.s. in Ghana. Genetics. 2007;175:751–761. doi: 10.1534/genetics.106.065888. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.