Abstract
Hermes are hAT transposons from Musca domestica that are very closely related to the hobo transposons from Drosophila melanogaster and are useful as gene vectors in a wide variety of organisms including insects, planaria, and yeast. hobo elements show distinct length variations in a rapidly evolving region of the transposase-coding region as a result of expansions and contractions of a simple repeat sequence encoding 3 amino acids threonine, proline, and glutamic acid (TPE). These variations in length may influence the function of the protein and the movement of hobo transposons in natural populations. Here, we determine the distribution of Hermes in populations of M. domestica as well as whether Hermes transposase has undergone similar sequence expansions and contractions during its evolution in this species. Hermes transposons were found in all M. domestica individuals sampled from 14 populations collected from 4 continents. All individuals with Hermes transposons had evidence for the presence of intact transposase open reading frames, and little sequence variation was observed among Hermes elements. A systematic analysis of the TPE-homologous region of the Hermes transposase-coding region revealed no evidence for length variation. The simple sequence repeat found in hobo elements is a feature of this transposon that evolved since the divergence of hobo and Hermes.
Keywords: Hermes, Herves, hobo, Musca domestica, P elements, transposable element
Hermes and hobo are closely related class II transposons of the hAT (hobo, Ac, Tam3) family of transposable elements, an ancient and diverse group of transposons found widely in plants and animals (Robertson 2002). The hobo transposon from Drosophila melanogaster is active and appears to have been introduced into this species on multiple occasions, whereas the Hermes transposon from the housefly, Musca domestica, is also active but with almost nothing known about its natural history (Simmons 1992; Warren et al. 1994; O'Brochta et al. 1996; Bonnivard et al. 2000; Boussy and Itoh 2004). The hobo transposon encodes a 661 amino acid transposase protein and has terminal inverted repeats of 12 bp (Streck et al. 1986; Calvi et al. 1991). The genome of M. domestica was analyzed for the presence of hAT elements using polymerase chain reaction (PCR) with degenerate primers to highly conserved coding regions of hAT transposase genes and found to contain only the Hermes element (Atkinson et al. 1993). Hermes contains a 612–amino acid open reading frame that encodes a 70-kD transposase protein 55% identical and 70% similar to hobo (Atkinson et al. 1993; O'Brochta et al. 1994; Warren et al. 1994). The 17-bp terminal inverted repeat sequences of Hermes are also very similar to those of hobo with 10 of the first 12 bp being identical (O'Brochta et al. 1994). In fact, hobo and Hermes are sufficiently similar to allow these elements to interact when present in the same cell (Atkinson et al. 1993; Sundararajan et al. 1999). Hermes elements are readily mobilized by hobo transposase, whereas hobo elements are inefficiently mobilized by Hermes transposase (Atkinson et al. 1993; Sundararajan et al. 1999).
hAT element transposases were initially thought to comprise a unique family of proteins, but recent biochemical and structural studies have demonstrated that these transposases are related to retroviral integrases (Zhou et al. 2004; Hickman et al. 2005). Retroviral integrases are characterized by the presence of an active site containing 3 highly conserved amino acids—2 aspartic acids (D) and a glutamic acid (E), forming the DDE motif (Mizuuchi and Baker 2002). Comparisons of hAT element transposases revealed at least 6 regions of sequence conservation referred to as domains A–F (Rubin et al. 2001) (Figure 1). More recently, the structure of a form of Hermes active in vitro and lacking the first 78 amino acids (Hermes79–612) was reported based on X-ray crystallography data and suggested the presence of 3 structural domains (Hickman et al. 2005). Residues 79–150 comprise what has been called the “N-terminal domain” and, according to Hickman et al. (2005), may be involved in site-specific DNA binding to the ends of the element. The “catalytic domain” consists of the characteristic integrase-like fold with the DDE motif and is formed from 2 stretches of amino acids interrupted by a long all-α helical domain referred to as the “insertion domain” (Hickman et al. 2005). The insertion domain has no known structural homologues and contains a highly conserved sequence domain (domain C) with an invariant tryptophan (Trp 319 in Hermes, Trp 370 in hobo) thought to play a crucial role in the formation and stabilization of DNA hairpin structures formed during element excision (Rubin et al. 2001; Hickman et al. 2005). Also, within the insertion domain and located between conserved sequence domains C and D is a rapidly evolving, poorly conserved region, referred to here as the 3′–nonconserved region (3′-NCR) (Rubin et al. 2001) (Figure 1).
Figure 1.
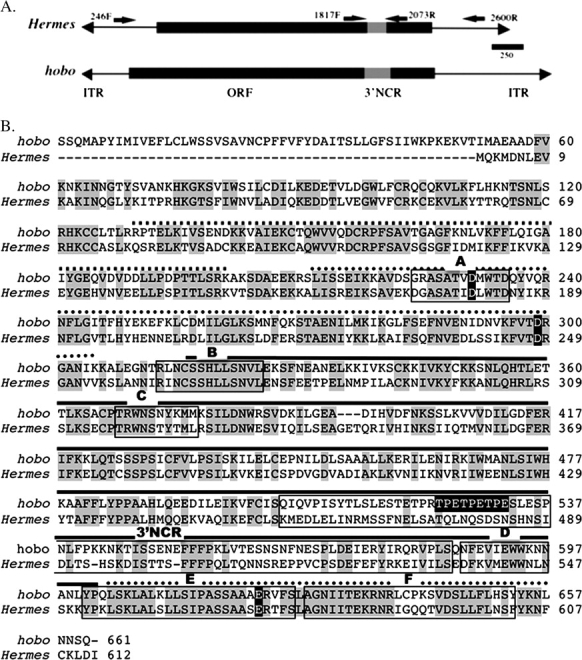
Structure and sequence of hobo and Hermes. (A) Diagrammatic representation of hobo and Hermes showing the location of the transposase-coding regions (open reading frame), the position of the 3′-NCR, the location of the priming sites, and the name of the primers. Primers 246F and 2600R were used to determine the structure of Hermes elements, and 1817F and 2073R were used to amplify the 3′-NCR in Hermes. The inverted terminal repeats (ITRs) are not drawn to scale. (B) An aligned amino acid sequence of hobo and Hermes transposases. Amino acids with a gray background indicate regions of amino acid identity. Sequences conserved within the hAT family (domains A–F) are outlined with a box and labeled A–F. The amino acids constituting the 3′-NCR between hobo and Hermes is outlined with a box and labeled 3′-NCR. The amino acids constituting the DDE motif are indicated with white letters on a black background. Residues (265–552) forming the α-helical domain in Hermes are indicated with a dashed line. The regions of the protein comprising the catalytic fold are indicated with a dotted line, whereas the insertion domain that splits the catalytic domain is indicated by a black line. The TPE repeat region within the 3′-NCR of hobo is shown in white letters on a black background.
In hobo, the 3′-NCR contains a simple sequence repeat of 9 nucleotides encoding the amino acids threonine, proline, and glutamic acid (TPE). This triamino acid repeat motif is polymorphic within and among populations of D. melanogaster with copy numbers varying from 1 to 10 (Bonnivard et al. 1997; Bonnivard et al. 2002). A 3-TPE repeat–containing hobo element is distributed worldwide and found in all natural populations of D. melanogaster. A 4-TPE–containing element is present in East African and South American populations, and a 5-TPE repeat is widely distributed in Europe with a frequency cline beginning in Western Europe (Bonnivard et al. 2000; Bonnivard et al. 2002). Furthermore, the mutation rate of these TPE repeats was observed to be higher than that of neutral microsatellites in experimental populations of D. melanogaster harboring 3- or 5-TPE repeats (Souames, Bazin, et al. 2003a). In experimental populations of D. melanogaster, the 5-TPE element was observed to decrease the activity of the 3-TPE element suggesting that these TPE repeats might be regulating the activity of the hobo transposase protein (Souames, Bazin, et al. 2003a).
Although much is known about the structure of Hermes transposase, the use of the element as a broad host range transgenic vector, and the mechanism of Hermes transposition, little is known about the natural history of the element and whether it shares some of the novel features of hobo elements such as a hypervariable transposase-coding region that might be involved in modulating element activity. In this study, we examine Hermes elements from populations of M. domestica from around the world to assess the frequency of this element in populations and whether this hobo-like element has a hypervariable region within the transposase insertion domain.
Materials and Methods
Insects
Fourteen populations of M. domestica were sampled from 4 continents and referred to as Korea; Lampang, Thailand; Nangpo, Thailand; Senegal; Uruguay; Panama; Lethbridge, Canada; Zimbabwe; Texas; Missouri; Pennsylvania; Maryland; and Cyromazine. The insects from Korea (n = 9), Thailand (n = 10), Senegal (n = 5), Uruguay (n = 5), Panama (n = 5), Zimbabwe (n = 5), and Canada (n = 5) were collected from the wild. Insects from Texas (n = 5), Missouri (n = 5), Pennsylvania (n = 6), Maryland (n = 6), Georgia (n = 6), and Cyromazine (n = 5) were collected from cage populations maintained in the laboratory for up to 20 years.
DNA Isolation
DNA from M. domestica was isolated using either the modified cetyl trimethyl ammonium bromide (CTAB) protocol of Shahjahan et al. (Shahjahan et al. 1995) or the phenol:chloroform extraction method described by Sambrook et al. (Sambrook et al. 1989). DNA from ethanol-preserved samples of M. domestica (Thailand only) was extracted using the DNA/RNA Isolation Kit from Amersham (Piscataway, NJ) according to manufacturer's specifications.
PCR, Cloning, and Sequencing
All reactions were done in a final volume of 40 μl containing 10 mM Tris–HCl, pH 8.3; 50 mM KCl; 2.5 mM MgCl2; 0.2 mM each of dATP, dTTP, dGTP, and dCTP; 2 units Taq DNA polymerase (Applied Biosystems, Foster City, CA); and 0.495 μM of each primer. A Perkin Elmer GeneAmp PCR System 9600 thermocycler was used for all the reactions. Primers for the analysis of Hermes in M. domestica were 246F (5′-TTT GGA GAG CGA AAA CAA CG-3′) and 2600R (5′-GTT TGA TGT TAA GAT CAC C-3′) (Figure 1). Reactions with these primers were performed at 95 °C (3 min); 30 cycles of 95 °C (15 s), 50 °C (15 s), and 72 °C (2 min); followed by 5 min at 72 °C. Reaction products were analyzed on a 1% agarose gel containing 5 μg/ml of ethidium bromide. The PCR was repeated for a few samples (Th16 from Thailand, Z93 from Zimbabwe, P52 from Panama, and Tx65 from Texas) in a separate 20-μl reactions containing 5× Phusion HF Buffer (NEB), 0.2 μM dNTPs (an equimolar mixture of dATP, dTTP, dCTP, dGTP), 0.4 units Finnzymes Phusion DNA Polymerase (New England Biolabs, Ipswich, MA; error rate = 4.4 × 10−7), and 24 pmoles of the above primers. Amplification reactions were performed under the following conditions: 98 °C (1min); 30 cycles of 98 °C (10 s), 57 °C (30 s), and 72 °C (1 min 30 s); followed by 10 min at 72 °C. The reaction products were visualized on a 1% agarose gel containing 5 μg/ml of ethidium bromide. The reaction products were cloned into pCR-Blunt II TOPO vector (Invitrogen, Carlsbad, CA). Clones were analyzed by colony PCR; the ones that showed the presence of the deleted forms of Hermes elements seen in the original PCR were sequenced. Six different deleted forms of the element from 4 individuals were sequenced.
The 3′-NCR of Hermes was amplified using the primers 1817F (5′-AAA TTA AAG AAT TTT GCT TAT CC-3′) and 2073R (5′-AAT TAA GAT TCC ACC ATT CC-3′) (Figure 1). Reaction conditions were the same as described above for the analysis of Hermes elements. Reaction products were analyzed on 2% MetaPhor agarose gel (FMC Bioproducts, Philadelphia, PA) containing 5 μg/ml of ethidium bromide. The PCR products were purified using the Wizard PCR DNA Purification System (Promega, Madison, WI) and were sequenced directly. DNA sequence analysis was done using the Wisconsin Package Version 7.1, Genetics Computer Group, Madison, WI.
DNA samples whose quality required verification (because of the failure to detect Hermes-specific PCR products, e.g.) were subjected to PCR using primers specific for the armadillo gene of M. domestica (Peifer and Wieschaus 1993). Primers arm1 (5′-GAC GAT ATG AAT CAA CAG C-3′) and arm2 (5′-AGG CTG TTG GGA GCC AGT AGT G-3′) were used in all reactions and resulted in a 1451-bp reaction product. Reaction conditions were as described above and were performed at 95 °C (3 min); 30 cycles of 95 °C (15 s), 56 °C (30 s), and 72 °C (1 min); followed by 5 min at 72 °C. Reaction products were analyzed on a 1% agarose gel containing 5 μg/ml of ethidium bromide.
Results
Distribution of Hermes in M. domestica
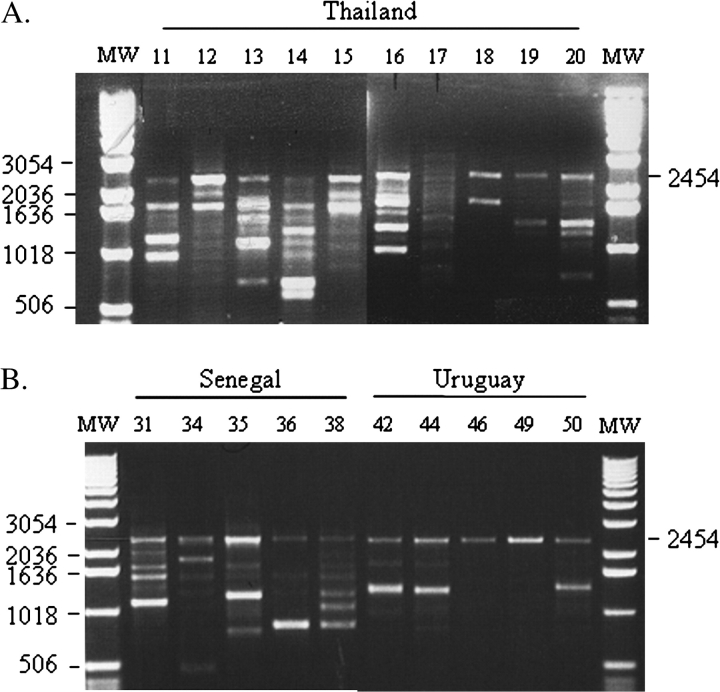
Seventy-seven individuals from 14 populations of M. domestica spanning 4 continents were sampled and analyzed for the presence of Hermes transposons. All populations sampled showed evidence of the presence of Hermes transposons (Figure 2). Seventy-seven individual houseflies were initially analyzed in this study, and all but one showed evidence for the presence of Hermes elements. A single individual collected in Lampang, Thailand (sample Th17), failed repeatedly (PCR conducted 3 times to confirm results) to show clear evidence of the element (Figure 3). The quality of this DNA sample was verified using primers specific for the M. domestica homologue of the D. melanogaster armadillo gene (Peifer and Wieschaus 1993). The expected armadillo PCR product was amplified, confirming that this DNA sample contained genomic DNA that could serve as PCR template. To test for further evidence of M. domestica lacking Hermes, 18 additional individual houseflies collected in Lampang, Thailand, and 15 collected in Nangpo, Thailand, were analyzed. All showed evidence for the presence of Hermes transposons (data not shown).
Figure 2.
Products of a PCR using Hermes primers 246F and 2600R using representatives of 11 of the 14 populations investigated in this study. MW refers to a molecular weight marker. Numbers along the left side of the figure refer to the size of the molecular weight markers measured in base pairs. The position of the 2454-bp PCR product resulting from the amplification of an intact element is indicated along the right side of the figure. K = Korea (sample K4); Th = Lampang,Thailand (sample Th16); S = Senegal (sample S38); U = Uruguay (sample U49); P = Panama (P52); Tx = Texas (sample Tx65); Mo = Missouri (sample M76); C = Lethbridge, Canada (sample L84); Z = Harare, Zimbabwe (sample Z93); G = Georgia (sample GA4); Md = Maryland (sample Md3).
Figure 3.
Products of a PCR using Hermes primers 246F and 2600R using samples from Thailand (A) and Senegal and Uruguay (B). MW refers to molecular weight markers. The size in base pairs of selected markers is indicated on the left side of each figure. The 2454-bp PCR product arising from intact elements is indicated. Numbers associated with each lane refer to sample numbers. Thailand sample 17 is referred to in the text.
Structure of Hermes Elements
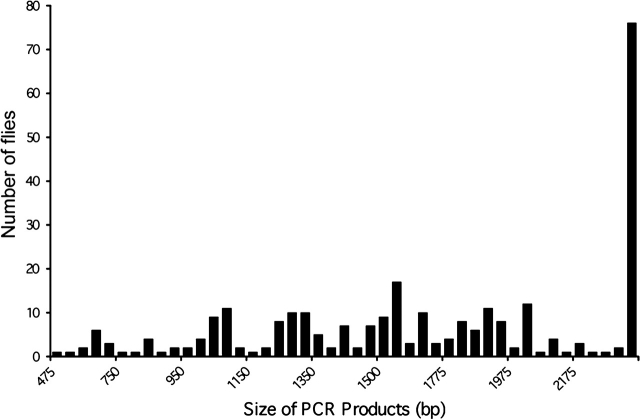
Of the individuals tested, approximately 90% had one or more internally deleted forms of Hermes. All insects with Hermes transposons had complete, nondeleted forms of the element as indicated by the presence of a 2454-bp PCR product (Figures 2 and 3). Few of the transposons detected had internal deletions that resulted in PCR products less than 1 kb in length, and none less than 475 bp in length were observed. One internally deleted form of Hermes 1550 bp in length was observed in 17 individuals originating from 8 populations, making it the most common deleted form of the element (Figure 4). Variations in the copy number and structure of Hermes transposons were also observed within populations (Figure 3). Individual houseflies collected in Thailand and Zimbabwe contained the highest number of internally deleted elements. Within the 5 individuals analyzed from insects collected in Zimbabwe, we observed 30 internally deleted elements of 18 different lengths of which 4 were unique to this location (Table 1). Within the houseflies collected in Thailand, we observed 31 internally deleted elements of 16 different lengths, 4 of which were present in only this location. Individuals collected in Missouri and Lethbridge, Canada, contained the fewest number of elements with both having only 4 different internally deleted forms of the element (Table 1). Some individuals within these populations had no evidence for internally deleted elements and appeared to contain only intact elements. Senegal and Korea had 3 forms unique to each location, Georgia had 2 forms, and Cyromazine, Texas, and Canada had 1 form unique to the location. Uruguay, Pennsylvania, Missouri, Panama, and Maryland shared the deleted forms with other locations.
Figure 4.
Summary of PCR results. PCR products were placed in 1 of 45 size classes, and the frequency distribution of each class is shown. n = 77.
Table 1.
Hermes form polymorphisms
| Number of deleted forms |
||
| Location | Total | Unique |
| Thailand | 16 | 4 |
| Senegal | 13 | 3 |
| Uruguay | 4 | 0 |
| Cryomazine | 6 | 1 |
| Pennsylvania | 4 | 0 |
| Missouri | 6 | 0 |
| Georgia | 8 | 2 |
| Panama | 6 | 0 |
| Texas | 7 | 1 |
| Zimbabwe | 18 | 4 |
| Canada | 4 | 1 |
| Maryland | 4 | 0 |
| Korea | 9 | 3 |
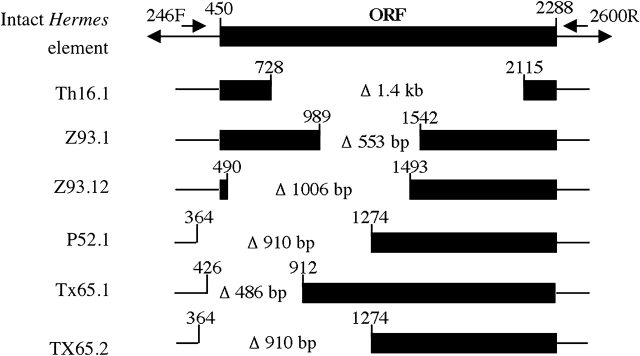
Six different forms of the element from 4 individuals were sequenced to confirm that they were Hermes elements and not artifacts of the PCR. Different regions of the element with parts of the open reading frame for Hermes transposase were found to be deleted (Figure 5). Two forms P52.1 and Tx65.2 from 2 different individuals, P52 from Panama, and Tx65 from Texas shared the same approximately 910-bp deletion (Figure 5).
Figure 5.
Structure of the Hermes elements from natural populations of Musca domestica. Diagrammatic representation of the Hermes elements isolated from the natural populations of M. domestica shown in comparison with the intact Hermes element. Size and the region of the deletion are shown. Th16.1 = isolated from Lampang, Thailand (sample Th16), Z93.1 and Z93.2 = isolated from Harare, Zimbabwe (sample Z93), P52.1 = isolated from Panama (sample P52), Tx65.1 and Tx65.2 = isolated from Texas (sample Tx65).
Structure of the 3′-NCR of Hermes Transposase
The 3′-NCRs from 65 individuals including representatives from 14 populations examined above were sequenced after amplification by PCR. The 3′-NCR contained few polymorphisms in its amino acid sequence. Nine individuals contained an FN (phenylalanine, asparagine) to VH (valine, histidine) diamino acid replacement starting at position 7 (Figure 6) and resulting in the conversion of a phenylalanine and asparagines to valine and histidine, respectively. This diamino acid replacement was due to 2 DNA sequence changes. Hermes transposons with both these sequences were always present in the same insect. A second polymorphism in the amino acid sequence was detected in only a single individual containing the nonconservative change D27H (aspartic acid 27 histidine) (Figure 6).
Figure 6.
Consensus nucleotide (A) and amino acid (B) sequence of the 3′-NCR of Hermes transposase. Boxed nucleotides and amino acids indicate sites of polymorphism. The observed polymorphisms and their frequency are shown above each sequence.
Discussion
In this study, we examined the structure and natural history of Hermes transposons from 14 populations of M. domestica from 4 continents as part of an effort to characterize the distribution of Hermes in populations of M. domestica and whether it shares with hobo transposons a hypervariable transposase-coding region. Hermes was present in all individuals from all populations with the exception of a single individual collected in Lampang, Thailand. More extensive sampling failed to reveal any other individuals from that population that similarly lacked Hermes, making this individual quite unusual. Overall, individuals collected from all populations had evidence for the presence of Hermes transposons with intact or largely intact transposase open reading frames (±100 bp). The widespread occurrence of intact elements might be due to a number of reasons. First, Hermes might have recently been introduced into M. domestica and rapidly spread through this species. Such an event is expected to result in elements with low sequence diversity and a high frequency of autonomous (intact) elements. Second, the abundance of intact Hermes transposons may also indicate that Hermes may be less prone to accumulate internal deletions after element excision and transposition as has been seen with the well-studied P element of D. melanogaster (Engels et al. 1990). The widespread presence of intact elements within populations also seems to be a characteristic of hobo and other hAT elements such as Herves in Anopheles gambiae (Galindo 1995; Yamashita et al. 1999; Subramanian et al. 2007). In addition to the widespread presence of Hermes elements with transposase-coding regions largely intact, many individuals were also found to contain forms of Hermes with variable-size deletions within the coding region. The abundance of nondeleted forms of Hermes elements suggests that deleted forms may be produced more slowly during periods of active transposition, relative to P elements under the same conditions depending on the method of chromosome repair that is used after element excision. Note that hAT excision generally and Hermes excision specifically result in reaction products (excised elements and chromosomes with double-stranded gaps) that are chemically distinct from those produced during P-element excision, and this could influence the choice of post-excision gap repair pathways used subsequently (Zhou et al. 2004). Hermes excision results in double-stranded gaps in the donor chromosome with hairpin structures, initially sealing each end (Rio 2002; Zhou et al. 2004). This is not the case after P-element excision where the gapped chromosome ends in 5′-P and highly reactive 3′-OH (Rio 2002). That post-excision gap repair after hAT element excisions can follow repair pathways (e.g., nonhomologous end joining) less frequently used after P-element excision seems to also be reflected in excision “footprints” produced after P-element and hobo excision from plasmids introduced into developing D. melanogaster embryos (O'Brochta et al. 1991; Sundararajan et al. 1999; Gloor et al. 2000). We propose that nonhomologous end joining may be the preferred mode of gap repair after hAT element excision, whereas homology-dependent repair processes are used relatively more frequently after P-element excision (Coen et al. 1989; Rubin and Levy 1997; Yamashita et al. 1998, 1999).
This study also revealed variations in element form diversity as a function of geographic location. Populations of M. domestica sampled from Thailand, Zimbabwe, and Senegal had evidence of a greater number of forms of Hermes than other populations. Methods such as transposable element display could provide a more accurate estimate of Hermes copy number in these populations (Guimond et al. 2003).
The transposase of hobo is known to exist in nature in a number of forms as a consequence of a variable copy number of a simple sequence repeat consisting of 9 nucleotides encoding the amino acids T, P, and E (Bonnivard et al. 2000). This triamino acid repeat varies in copy number between 1 and 10 depending on the geographic origin of the D. melanogaster from which the elements were isolated (Bonnivard et al. 2002; Souames, Bonnivard, et al. 2003b). This correlation between transposase form and population location has been the basis for suggesting that these forms of transposase might have some functional significance such as different transposase or regulatory activities (Bonnivard et al. 2002; Souames, Bazin, et al. 2003a). That expansions and contractions of the TPE repeat in hobo transposase would lead to alterations in the activity or function of the protein would not be unusual as numerous similar examples have been described of simple sequence repeats modifying protein function (Kashi and King 2006). The region of hobo transposase in which the simple sequence repeat is located is the most rapidly evolving region of the protein. It is part of a much larger insertion domain that separates 2 conserved sequence domains that contributes to the formation of the integrase-like fold with the catalytic center of the transposase and the DDE motif (Warren et al. 1994; Rubin et al. 2001; Hickman et al. 2005; Kashi and King 2006). Despite the high degree of relatedness between Hermes and hobo, we found no evidence for the presence of a similar simple sequence repeat region or any length polymorphism in the insertion domain of Hermes transposase indicating that the simple sequence repeat present in hobo transposase has evolved relatively recently and after the divergence of the hobo and Hermes elements.
Funding
National Institutes of Health (GM48102).
References
- Atkinson PW, Warren WD, O'Brochta DA. The hobo transposable element of Drosophila can be cross-mobilized in houseflies and excises like the Ac element of maize. Proc Natl Acad Sci U S A. 1993;90:9693–9697. doi: 10.1073/pnas.90.20.9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnivard E, Bazin C, Denis B, Higuet D. A scenario for the hobo transposable element invasion, deduced from the structure of natural populations of Drosophila melanogaster using tandem TPE repeats. Genet Res. 2000;75:13–23. doi: 10.1017/s001667239900395x. [DOI] [PubMed] [Google Scholar]
- Bonnivard E, Bazin C, Higuet D. High polymorphism of TPE repeats within natural populations of Drosophila melanogaster: a gradient of the 5TPE hobo element in Western Europe. Mol Biol Evol. 2002;19:2277–2284. doi: 10.1093/oxfordjournals.molbev.a004051. [DOI] [PubMed] [Google Scholar]
- Bonnivard E, Higuet D, Bazin C. Characterization of natural populations of Drosophila melanogaster with regard to the hobo system: a new hypothesis on the invasion. Genet Res. 1997;69:197–208. doi: 10.1017/s0016672397002826. [DOI] [PubMed] [Google Scholar]
- Boussy IA, Itoh M. Wanderings of hobo: a transposon in Drosophila melanogaster and its close relatives. Genetica. 2004;120:125–136. doi: 10.1023/b:gene.0000017636.08925.55. [DOI] [PubMed] [Google Scholar]
- Calvi BR, Hong TJ, Findley SD, Gelbart WM. Evidence for a common evolutionary origin of inverted repeat transposons in Drosophila and plants: hobo, activator, and Tam3. Cell. 1991;66:465–471. doi: 10.1016/0092-8674(81)90010-6. [DOI] [PubMed] [Google Scholar]
- Coen ES, Robbins TP, Almeida J, Hudson A, Carpenter R. Consequences and mechanisms of transposition in Antirrhinum majus. In: Berg DE, Howe MM, editors. Mobile DNA. American Society for Microbiology Biology, Washington; 1989. pp. 413–436. [Google Scholar]
- Engels WR, Johnson-Schlitz DM, Eggleston WB, Sved J. High-frequency P element loss in Drosophila is homologue dependent. Cell. 1990;62:515–525. doi: 10.1016/0092-8674(90)90016-8. [DOI] [PubMed] [Google Scholar]
- Galindo MI, Ladeveze V, Lemeunier F, Kalmes R, Periquet G, Pascual L. Spread of the autonomous transposable element hobo in the genome of Drosophila melanogaster. Mol. Biol. Evol. 1995;12:723–734. doi: 10.1093/oxfordjournals.molbev.a040251. [DOI] [PubMed] [Google Scholar]
- Gloor GB, Moretti J, Mouyal J, Keeler KJ. Distinct P element excision products in somatic and germline cells of Drosophila. Genetics. 2000;155:1821–1830. doi: 10.1093/genetics/155.4.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimond N, Bideshi DK, Pinkerton AC, Atkinson PW, O'Brochta DA. Patterns of Hermes transposition in Drosophila melanogaster. Mol Gen Genet. 2003;268:779–790. doi: 10.1007/s00438-002-0800-4. [DOI] [PubMed] [Google Scholar]
- Hickman AB, Perez ZN, Zhou LQ, Musingarimi P, Ghirlando R, Hinshaw JE, Craig NL, Dyda F. Molecular architecture of a eukaryotic DNA transposase. Nat Struct Mol Biol. 2005;12:715–721. doi: 10.1038/nsmb970. [DOI] [PubMed] [Google Scholar]
- Kashi Y, King DG. Simple sequence repeats as advantageous mutators in evolution. Trends Genet. 2006;22:253–259. doi: 10.1016/j.tig.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K, Baker TA. Chemical mechanisms for mobilizing DNA. In: Craig NL, Craige R, Gellert M, editors. Mobile DNA II. ASM Press, Washington; 2002. pp. 12–23. [Google Scholar]
- O'Brochta DA, Gomez SP, Handler AM. P-element excision in Drosophila melanogaster and related drosophilids. Mol Gen Genet. 1991;225:387–394. doi: 10.1007/BF00261678. [DOI] [PubMed] [Google Scholar]
- O'Brochta DA, Warren WD, Saville KJ, Atkinson PW. Interplasmid transpostion of Drosophila hobo elements in non-drosophilid insects. Mol Gen Genet. 1994;244:9–14. doi: 10.1007/BF00280181. [DOI] [PubMed] [Google Scholar]
- O'Brochta DA, Warren WD, Saville KJ, Atkinson PW. Hermes, a functional non-Drosophilid insect gene vector from Musca domestica. Genetics. 1996;142:907–914. doi: 10.1093/genetics/142.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M, Wieschaus E. The product of the Drosophila melanogaster segment polarity gene armadillo is highly conserved in sequence and expression in the housefly Musca domestica. J Mol Evol. 1993;36:224–233. doi: 10.1007/BF00160477. [DOI] [PubMed] [Google Scholar]
- Rio DC. P transposable elements in Drosophila melanogaster. In: Craig NL, Craige R, Gellert M, Lambowitz AM, editors. Mobile DNA II. ASM Press, Washington; 2002. p. 1204. [Google Scholar]
- Robertson HM. Evolution of DNA transposons in eukaryotes. In: Craig NL, Craige R, Gellert M, Lambowitz AM, editors. Mobile DNA II. ASM Press, Washington; 2002. pp. 1093–1110. [Google Scholar]
- Rubin E, Levy A. Abortive gap repair: underlying mechanism for Ds element formation. Mol Cell Biol. 1997;17:6294–6302. doi: 10.1128/mcb.17.11.6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin E, Lithwick G, Levy AA. Structure and evolution of the hAT transposon superfamily. Genetics. 2001;158:949–957. doi: 10.1093/genetics/158.3.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 1989. Molecular cloning: a laboratory manual. [Google Scholar]
- Shahjahan RM, Hughes KJ, Leopold RA, DeVault JD. Lower incubation temperature increases yield of insect genomic DNA isolated by the CTAB method. Biotechniques. 1995;19:333–334. [PubMed] [Google Scholar]
- Simmons GM. Horizontal transfer of hobo transposable elements within the Drosophila melanogaster species complex: evidence from DNA sequencing. Mol Biol Evol. 1992;9:1050–1060. doi: 10.1093/oxfordjournals.molbev.a040774. [DOI] [PubMed] [Google Scholar]
- Souames S, Bazin C, Bonnivard E, Higuet D. Behavior of the hobo transposable element with regard to TPE repeats in transgenic lines of Drosophila melanogaster. Mol Biol Evol. 2003a;20:2055–2066. doi: 10.1093/molbev/msg221. [DOI] [PubMed] [Google Scholar]
- Souames SM, Bonnivard E, Bazin C, Higuet D. High mutation rate of TPE repeats: a microsatellite in the putative transposase of the hobo element in Drosophila melanogaster. Mol Biol Evol. 2003b;20:1826–1832. doi: 10.1093/molbev/msg194. [DOI] [PubMed] [Google Scholar]
- Streck RD, MacGaffey JE, Beckendorf SK. The structure of hobo transposable elements and their insertion sites. EMBO J. 1986;5:3615–3623. doi: 10.1002/j.1460-2075.1986.tb04690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian RA, Arensburger P, Atkinson P, O'Brochta DA. Transposable element dynamics of the hAT element Herves in the human malaria vector, Anopheles gambiae, s.s. Genetics. 2007;176:2477–2487. doi: 10.1534/genetics.107.071811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundararajan P, Atkinson PW, O'Brochta DA. Transposable element interactions in insects: crossmobilization of hobo and Hermes. Insect Mol Biol. 1999;8:359–368. doi: 10.1046/j.1365-2583.1999.83128.x. [DOI] [PubMed] [Google Scholar]
- Warren WD, Atkinson PW, O'Brochta DA. The Hermes transposable element from the housefly, Musca domestica, is a short inverted repeat-type element of the hobo, Ac, and Tam3 (hAT) element family. Genet Res. 1994;64:87–97. doi: 10.1017/s0016672300032699. [DOI] [PubMed] [Google Scholar]
- Yamashita S, Mikami T, Kishima Y. Tam3 in Antirrhinum majus is an exceptional transposon in resistance to alteration by abortive gap repair: identification of nested transposons. Mol Gen Genet. 1998;259:468–474. doi: 10.1007/s004380050837. [DOI] [PubMed] [Google Scholar]
- Yamashita S, TakanoShimizu T, Kitamura K, Mikami T, Kishima Y. Resistance to gap repair of the transposon Tam3 in Antirrhinum majus: a role of the end regions. Genetics. 1999;153:1899–1908. doi: 10.1093/genetics/153.4.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Mitra R, Atkinson PW, Hickman AB, Dyda F, Craig NL. Transposition of hAT elements links transposable elements and V(D)J recombination. Nature. 2004;432:995–1001. doi: 10.1038/nature03157. [DOI] [PubMed] [Google Scholar]