Abstract
Comparative genomics provides a facile way to address issues of evolutionary constraint acting on different elements of the genome. However, several important DNA elements have not reaped the benefits of this new approach. Some have proved intractable to current day sequencing technology. These include centromeric and heterochromatic DNA, which are essential for chromosome segregation as well as gene regulation, but the highly repetitive nature of the DNA sequences in these regions make them difficult to assemble into longer contigs. Other sequences, like dosage compensation X chromosomal sites, origins of DNA replication, or heterochromatic sequences that encode piwi-associated RNAs, have proved difficult to study because they do not have recognizable DNA features that allow them to be described functionally or computationally. We have employed an alternate approach to the direct study of these DNA elements. By using proteins that specifically bind these noncoding DNAs as surrogates, we can indirectly assay the evolutionary constraints acting on these important DNA elements. We review the impact that such “surrogate strategies” have had on our understanding of the evolutionary constraints shaping centromeres, origins of DNA replication, and dosage compensation X chromosomal sites. These have begun to reveal that in contrast to the view that such structural DNA elements are either highly constrained (under purifying selection) or free to drift (under neutral evolution), some of them may instead be shaped by adaptive evolution and genetic conflicts (these are not mutually exclusive). These insights also help to explain why the same elements (e.g., centromeres and replication origins), which are so complex in some eukaryotic genomes, can be simple and well defined in other where similar conflicts do not exist.
Keywords: Drosophila, positive selection, protein–DNA interactions, rapid evolution
As sequencing becomes easier, faster, and cheaper, more and more genomes are routinely combed for functional elements like genes, control elements, and micro-RNAs (Stark et al. 2007). Even in cases where function was not previously known, the ability to compare multiple genomes for patterns of conservation can reveal elements that were previously unknown or were difficult to recognize. For instance, we can define exon/intron boundaries more accurately or use nucleotide conservation to identify transcription factor binding sites upstream of genes. This is primarily because such sequences fit an a priori expected pattern. When there is no such pattern, our predictions and insights begin to falter. A prime example of where the lack of an appropriate, universal model begins to affect genomic predictions is in the case of long, noncoding RNAs. Although newer and more sophisticated models have begun to tackle this problem systematically (Dowell and Eddy 2006), consensus suggests that the majority of important noncoding RNAs remain unidentified even in well-curated eukaryotic genomes.
An even more daunting challenge is posed by DNA elements that serve key organizing features in eukaryotic genomes. These elements range in function from replicating DNA to segregating chromosomes. Although these elements carry out essential roles in the organization of the eukaryotic genome, they can be composed of repetitive, redundant, or evolutionarily fluid DNA sequences. This observation poses a conundrum for evolutionary biologists interested in studying these elements: How can we use genetics or comparative evolutionary methods when the DNA elements themselves do not fit any expected pattern of conservation?
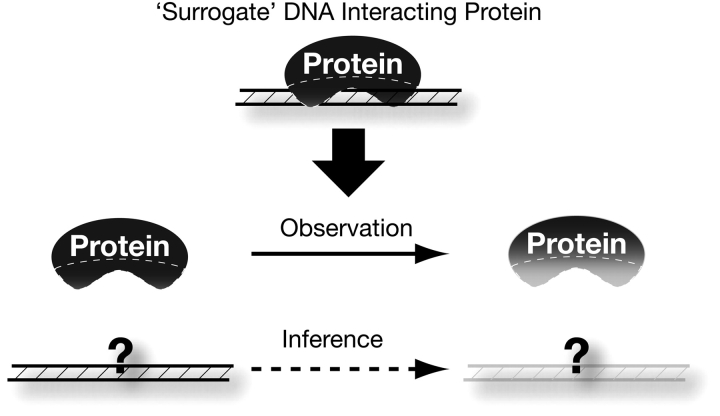
We have adopted an alternate “surrogate” approach to study the function and evolution of such DNA elements (Figure 1). Because we are unable to compare the DNA elements themselves, we instead study the evolution of the proteins that bind and, in many instances, epigenetically define the function of these elements. Selective pressure acting on these proteins, specifically in their putative DNA interfaces, therefore acts as a mirror image of the selective pressure on the DNA elements themselves. This approach provides us with unique insight as to what evolutionary pressures shape these DNA elements and the essential biological processes they carry out.
Figure 1.
A “surrogate” strategy to study noncoding DNA. In the schematic figure, the evolution of the underlying noncoding DNA is difficult to decipher because the sequence of the DNA is either unknown or difficult to describe. However, by studying the pattern of evolution acting specifically on the protein–DNA interfaces (hashed region) of proteins that specifically bind these DNAs, we can infer the types of changes that must have taken place on the underlying DNA, even in the absence of knowledge of what those DNA sequences are. Thus, the DNA-binding proteins serve as a surrogate allowing us to study the evolutionary constraints acting on important architectural noncoding DNAs that are hard to study otherwise.
In this review, we present 3 case studies of our surrogate approach, which includes the study of centromeres, origins of DNA replication, and dosage compensation X chromosomal binding sites. We will point out the difficulties associated with directly studying each of the DNA elements that serve these essential roles. Then, we will present our rationale for the selection of specific surrogate proteins. The study of evolutionary constraints acting on these surrogate proteins greatly increases our understanding of selective forces acting on the DNA elements that they bind. Although many of these surrogate studies are still in their infancy, they have already begun to reveal the remarkable biology that may shape some of the most important building blocks of eukaryotic genomes.
Centromeres: The Challenge of Unexplained Complexity
Centromeres are the sites on DNA that mediate proper chromosome segregation. Centromere evolution can have profound impacts on karyotype evolution within species (Pardo-Manuel de Villena and Sapienza 2001a), on the propensity of aneuploidy events in cancer (Lengauer et al. 1998), and even on the accuracy of male meiosis and therefore fertility in human males (Daniel 2002). Because of the importance of centromere function, proteins involved in chromosome segregation have been intensively studied by genetic and biochemical means in various organisms. On the other hand, the study of the underlying DNA sequence that scaffolds the assembly of these proteins and mediates chromosome segregation has lagged behind. The discrepancy between studies focused on centromeric proteins and DNA sequences exists chiefly because of the intractability of the highly repetitive sequences at the core of most plant and animal centromeric regions.
Centromeres range in size and complexity from the 125-bp point centromeres in Saccharomyces cerevisiae (Fitzgerald-Hayes et al. 1982) to the more complex centromeres in plants and animals that consist of hundreds of kilobases long arrays of satellite repeats (Copenhaver et al. 1999; Schueler et al. 2001). Centromeres are often flanked by heterochromatin; yet, the boundary between them is hard to define, making centromeric and heterochromatic sequences almost indistinguishable (Sun et al. 2003). Painstaking sequencing and assembly efforts have made only marginal progress on describing centromeric DNA complexity in diverse organisms (Schueler et al. 2001; Sun et al. 2003; Nagaki et al. 2004).
Studies on the human X chromosomal centromere support a simple mutation–recombination balance model where recombination (either unequal crossing over or gene conversion) is the underlying force that homogenizes centromeric repeats in the middle of an array, balanced by mutation and transposition in the flanks (McAllister and Werren 1999; Malik and Henikoff 2002). However, several theoretical studies have pointed out the inadequacy of mutation and recombination alone to explain increased array sizes, suggesting that selection must play a role in their evolution (Walsh 1987; Stephan 1989; Charlesworth et al. 1994; Stephan and Cho 1994). In keeping with this view, it has been demonstrated that pericentric satellites can contribute to a fitness difference between Drosophila melanogaster strains (Wu et al. 1989). In addition, a different pericentric satellite may contribute to hybrid inviability of Drosophila simulans/D. melanogaster interspecific hybrids (Sawamura and Yamamoto 1993; Sawamura et al. 1993).
Adding to the complexity of centromeric regions is the finding that satellite DNA sequences can change quite rapidly between closely related species. For instance, there has been a complete replacement of centromeric satellites between 2 closely related rice species, Oryza sativa and Oryza brachyantha (Lee et al. 2005). Similarly, the human X centromeric satellite appears to be only as old as the great apes (Schueler et al. 2001). These studies provide evidence that centromeric regions evolve rapidly between species. However, they do not provide evidence that directional selection drove this rapid evolution of large-scale accumulation of satellite repeats. Indeed, there is no a priori expectation that satellite repeats should evolve faster than nonrepetitive DNA in the absence of any mutational biases. Yet, comparison of centromeric to flanking heterochromatic repeats leads to the surprising finding that the centromeric arrays from different species are indeed more rapidly evolving than the pericentric units (Rudd et al. 2006). It is this paradoxical observation that leads to the idea that beneficial mutations repeatedly arise and fix, thereby increasing the substitution rates of the entire centromeric array. A process of biased gene conversion can, in theory, lead to dramatic turnover of satellite repeat arrays (Dover et al. 1982). Repeated innovation at the DNA level can recurrently fix “new satellite” repeats because of their advantage in recombinational processes.
We consider the possibility that the structure and tempo of centromeric satellite evolution may be a result of selective pressures. One form of selection could be simply purifying selection to maintain an uninterrupted, homogeneous array of a minimum size, so that it can form a functional centromere. However, this by itself is not an adequate explanation because it would lead to lower, not higher, rates of fixation of mutations in the centromere. An alternate selective force may be the transmission advantages of larger centromeres in female meiosis (Henikoff and Malik 2002; Malik and Henikoff 2002), which could result in both larger array sizes and rapid evolution of centromeric satellites as we detail below.
Centromeres: Centromeric Histones as Surrogates for Centromere Evolution
By looking at the selective constraints acting on proteins that bind centromeric DNA, we can infer information on centromere evolution. Centromeric H3s (CenH3s) are excellent surrogates for this purpose (Figure 2A). CenH3s are variant members of the histone H3 family of proteins, substituting for canonical H3 in centromere-specific nucleosomes (Sullivan et al. 1994; Yoda et al. 2000, 2004). Initially discovered in mammals (Palmer et al. 1987), CenH3s are now found to be encoded by every eukaryotic genome studied so far (Malik and Henikoff 2003) and are essential for accurate chromosome segregation (Stoler et al. 1995; Buchwitz et al. 1999; Blower and Karpen 2001). Localization of CenH3 can discriminate between the centromere and the surrounding heterochromatin (Takahashi et al. 2000), which provides a faithful marker of centromere identity throughout the entire range of centromere sizes.
Figure 2.
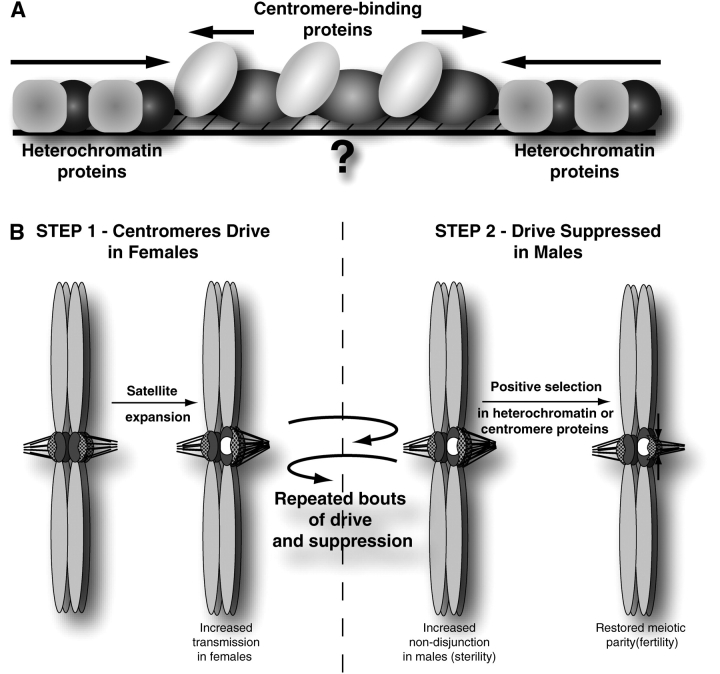
Surrogate strategy to study centromeric DNA. (A) “Surrogate” centromere proteins. Proteins like centromeric histones intimately associate with centromeric DNA sequence to directly identify the centromere. Heterochromatin proteins, on the other hand, indirectly define the centromere by establishing a boundary to the centromere. Evolutionary studies on either set of proteins provide novel hypotheses regarding the evolutionary dynamics and function of the underlying centromeric/pericentromeric DNA. (B) “Centromere-drive” model. Paired homologous chromosomes in metaphase I are illustrated. Centromeres (dark gray) are shown with an assembled kinetochore (stippled) bound to microtubules. In this example, the size of the kinetochore is dependent on the amount of centromeric sequence. In step 1, a novel centromeric satellite (white) expands and the new, larger centromere can drive in female meiosis by attracting more microtubules and gaining a favorable position in asymmetric female meiosis. This results in a selective advantage for this particular centromere, which will quickly fix in the population. In step 2, pairing of centromeres in male meiosis with unequal strengths results in unequal tension across the centromeres. Unequal tension can potentially increase the rate of nondisjunction in males, resulting in sterility. Selection acts on the DNA-binding affinity of proteins to restore parity across the centromeres and suppresses drive and sterility effects (shown here as restricting 1 centromere). Repeated bouts of drive and suppression will be observed as positive selection among centromere or heterochromatin proteins and fixation of new centromeric sequences (Henikoff and Malik 2002; Malik and Bayes 2006).
The centromere/heterochromatin boundary is not clearly defined but fluid. For instance, overexpression of heterochromatin proteins can encroach onto centromeric DNA and affect chromosome segregation (Halverson et al. 1997, 2000). Also, the packaging of centromeric sequences is heterogeneous such that smaller domains of contiguous CenH3-containing nucleosomes are interspersed with domains of canonical H3 nucleosomes (Ahmad and Henikoff 2002; Blower et al. 2002). This is significant because the proportion of centromeric DNA packaged by CenH3 nucleosomes appears to be determined by the dynamics and affinity of CenH3 versus canonical H3 nucleosomes for these sequences (Blower et al. 2002; Nagaki et al. 2004; Lam et al. 2006). Therefore, the centromere can be highly dynamic, and its identity is dependent on the relative DNA-binding affinities of CenH3s, canonical histones, as well as satellite-binding proteins (Figure 2A). Modulating the DNA-binding affinity of any one of these entities may affect centromere size and strength.
Centromeric histones differ from canonical H3 histones in 3 key sequence features (Sullivan et al. 1994; Shelby et al. 1997; Malik and Henikoff 2003). First, whereas canonical H3s in all eukaryotes have a well-conserved N terminal tail, the N terminal tails of CenH3s vary in both length and sequence and cannot be aligned across different lineages. Second, in a comparison of just the core histone fold domains (HFD), we found that CenH3s appear to have evolved more rapidly in contrast to canonical histone H3 (Henikoff et al. 2001; Malik and Henikoff 2003). Third, all CenH3s have a longer loop1 region than canonical H3s. Loop1 is one of the principal DNA interaction domains for H3 (Luger et al. 1997), and the longer loop1 of CenH3s has been inferred to allow them a greater DNA-binding specificity (Shelby et al. 1997).
In an attempt to infer selective pressure acting on centromeric DNA, we investigated the molecular evolution of Cid, the Drosophila CenH3. Histones are among the most conserved eukaryotic proteins; yet, we found that Cid, rather than evolving under purifying selection, evolves under positive selection (Malik and Henikoff 2001). Remarkably, both the N terminal tail and the HFD, which functions in wrapping the centromeric DNA, show a signature of positive selection. Comparing replacement (nonsynonymous) and synonymous polymorphisms in the Cid gene between D. melanogaster and D. simulans, we found that 18 replacement changes had been fixed between the 2 species, instead of the ∼3 expected in the absence of positive selection (Malik and Henikoff 2001), using the McDonald–Kreitman test (McDonald and Kreitman 1991). Positive selection is specifically found in the loop1 of the HFD, suggesting that changes in DNA-binding specificity are strongly selected for. Indeed, we showed that amino acid changes in loop1 are responsible for affecting centromeric targeting: the D. melanogaster loop1 region of Cid was necessary and sufficient to restore correct centromeric targeting to an otherwise mislocalized Cid protein from the distantly related Drosophila species, Drosophila bipectinata (Vermaak et al. 2002). These findings of positive selection in CenH3s have been extended to centromeric proteins from a variety of animal and plant taxa, using a variety of methods that compare the rates of nonsynonymous to synonymous changes. One notable exception is budding yeasts like S. cerevisiae, whose centromeres are simple 125-bp elements, which have centromeric proteins that are not found to be evolving under positive selection (Talbert et al. 2004).
Centromeres: Putting It Together in a New “Centromere-Drive Model”
Finding positive selection in our surrogate (Cid) allows us to formulate a model to explain centromere complexity and evolution (Henikoff et al. 2001; Henikoff and Malik 2002; Malik and Bayes 2006) (Figure 2B). The asymmetric nature of female meiosis in plants and animals can lead to genetic elements subverting this process for their own advantage. Under this model, centromeres compete via microtubule attachments for preferential transmission in female meiosis in animals and plants because only 1 of 4 meiotic products becomes the egg. This competition confers a selfish advantage to chromosomes that make attachments to the set of microtubules responsible for retention in the egg (Figure 2B). This selective advantage can quickly drive changes in satellite DNA sequence that (for instance) favor the recruitment of centromeric proteins, as well as expansions or contractions of preexisting satellite DNAs. It is worth noting that centromeres in budding yeasts, which lack an asymmetric meiosis, are devoid of this selfish opportunity, which likely explains their optimization to simple, point centromeres (Malik and Henikoff 2002).
Success in female meiosis may also negatively influence male meiosis. We present 2 examples of this duality of effects on female versus male meiosis. Robertsonian chromosomal fusions, which result from the fusion of 2 acrocentric chromosomes, provide the first example. Such fusion chromosomes have a transmission advantage through female but not male meiosis in humans (Pardo-Manuel de Villena and Sapienza 2001a, 2001b). Partly stemming from this transmission advantage, a significant proportion (0.12%) of the human population are carriers of a Robertsonian translocation (Nielsen and Wohlert 1991). There are no reports of any somatic (mitotic) effects but 3 quarters of male carriers of Robertsonian fusions appear to be partially to completely sterile (Daniel 2002). Thus, female meiotic success is balanced by the high cost to male fertility. This sterility likely results from a male meiotic checkpoint that monitors tension of microtubule attachment as described in mice (Eaker et al. 2001) and likely in Drosophila as well (McKee et al. 1998). Male meiosis is especially sensitive to such tension defects, and so there will be considerable selective pressure for mutations that can restore meiotic centromere parity and thus suppress the driving centromere.
A second example was recently elucidated by studies in Mimulus (monkeyflower) species. Dramatic (∼98%) segregation distortion was first observed in female interspecies hybrids of 2 closely related species, Mimulus guttatus and Mimulus nasutus (Fishman and Willis 2005). Such severe distortion could only result from either differential viability (which was ruled out) or due to distortion acting directly at the centromere in meiosis I (Zwick et al. 1999; Fishman and Willis 2005; Malik 2005). Subsequent studies revealed that even in intraspecies crosses of M. guttatus, 58:42 segregation distortion occurs in female meiosis due to divergence in centromere-associated repeat domains that can be cytologically visualized (Fishman and Saunders 2008). Intriguingly, like in the human Robertsonian cases, this female meiotic drive incurs a cost in male meiosis, as individuals homozygous for the driving allele suffer reduced pollen viability (Fishman and Saunders 2008).
Such meiotic drive systems can arise because of female meiotic asymmetries but are expected to be held in check (or eliminated) because of the detriment in male meiosis. Under such a situation, where meiotic drivers have thrived in a population but cannot drive to fixation, theory predicts that suppressor alleles may arise to alleviate the effects of the drive or to eliminate the drive itself (Sandler and Novitski 1957). These suppressor alleles would be unlinked from the drive locus so as to not reap the “benefits” of the drive (Hartl 1975). CenH3 is one such suppressor (Figure 2B). Its interaction surface with centromeric DNA is constantly under selection to change centromeric specificity and thus limit centromere size. Success of the suppressor alleles can lead to the degeneration of the drive system (in the absence of a transmission advantage), degeneration of the suppressor, and the retention of cryptic drive-suppressor systems (Tao et al. 2001). Typically, meiotic drivers and their suppressors are neomorphs (Merrill et al. 1999), and neither is essential for an organism. In the unusual scenario when essential elements act as drivers or suppressors, we could only uncover this cryptic genetic conflict by observing episodes of positive selection in them (Henikoff and Malik 2002). Thus, it was only possible from our study of surrogate proteins like CenH3s to uncover the interesting genetic conflicts that shape one of the most essential architectural DNA elements of eukaryotic genomes.
DNA Replication Origins: A Definition and Spacing Problem
There are intriguing analogies between DNA replication origins and centromeric DNA. Both can be simple and well-defined in some eukaryotes, like S. cerevisiae, but poorly defined in most others. Among eukaryotes studied so far, the sequence of genomic origins of replication is well-defined only in budding yeasts, where an 11 bp consensus-binding site embedded in 200 bp suffices as an autonomous origin of replication (Fangman et al. 1983; Brewer and Fangman 1987; Raghuraman et al. 2001). Three hundred such origins are spaced throughout the S. cerevisiae genome, although these do not all fire every cell cycle (Raghuraman et al. 2001). Origins of more complex eukaryotic genomes demonstrate no obvious sequence conservation and appear to be defined instead by epigenetic modification and/or transcriptional activity (MacAlpine and Bell 2005; Takeda and Dutta 2005). Studies of origin sequence specificity in some eukaryotes suggest that much or all euchromatin is competent to initiate replication if necessary (Mello et al. 1991; Kim et al. 1992; Coverley and Laskey 1994; Smith and Calos 1995).
Correctly defining origins of replication poses an evolutionary quandary. The transition to larger genome sizes and multiple chromosomes in eukaryotes necessitates the co-ordination of multiple origins of replication to ensure both faithful and efficient duplication of the genome. Pressure to replicate efficiently dictates that these origins cannot be spaced randomly because the largest continuous replicated regions of a chromosome (replicon) will be rate limiting for completion of replication. This origin spacing problem necessitates a more regular spacing of origins by some means (Blow et al. 2001). Even a regular spacing may not represent the optimal solution. In most complex eukaryotic genomes, efficient replication of eukaryotic genomes requires efficient replication of the (last-to-replicate) heterochromatic regions. Inability to do so efficiently can delay replication and normal S-phase progression (Quivy et al. 2008). It is unknown how complex eukaryotic genomes accomplish this task in the absence of cis-acting sequence information (Cvetic and Walter 2005), although it has been suggested that recruitment of replication proteins to heterochromatin via protein–protein interactions may be one solution (Quivy et al. 2008; Hayashi et al. 2009).
DNA Replication Origins: Cdc6 as a Surrogate
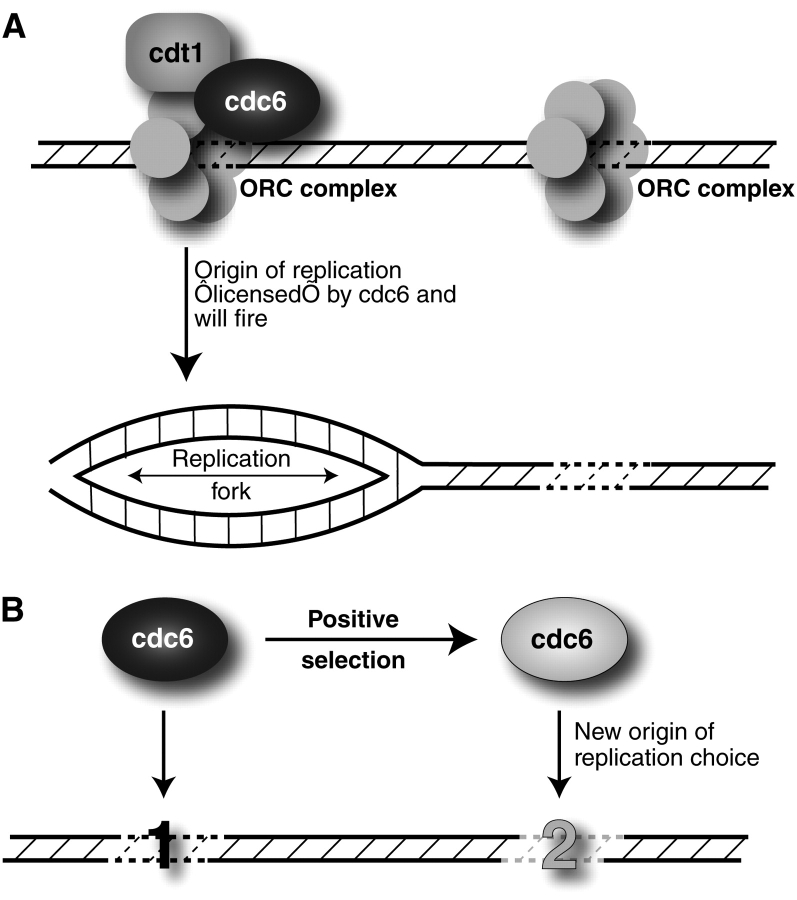
Due to the large amount of effort required to identify the compendium of replication origins in even a relatively simple eukaryotic genome, these questions about origin definition and spacing are unlikely to be suitably addressed by comparative genomics (Raghuraman et al. 2001). Therefore, we turned to the study of the Cdc6 protein, which serves a critical role in licensing DNA replication in eukaryotes. DNA licensing requires the ordered recruitment of a few highly conserved proteins at origins of replication to form the prereplication complex. This is followed by rapid removal of key components after initiation, thereby ensuring that each origin “fires” once and only once per cell cycle. Whereas many ORC complexes can be found on DNA and each ORC complex has the potential to initiate DNA replication, only a subset of them will be licensed through Cdc6 to allow replication to proceed (Figure 3A). Cdc6’s specificity of interaction with DNA sequences likely directly translates into specifying which origins will successfully fire based on 2 pieces of data. First, site-specific recruitment of Cdc6 to genomic DNA is sufficient to create an artificial origin of replication in mammalian cells (Takeda et al. 2005). Second, recent findings suggest that the Cdc6 ATPase activity may directly regulate the stability of the ORC–Cdc6 complex (Speck et al. 2005; Speck and Stillman 2007).
Figure 3.
Surrogate strategy to study origins of DNA replication. (A) “Surrogate” DNA replication proteins. Several proteins mediate the accurate, coordinated firing of multiple origins of DNA replication in eukaryotic genomes. For simplicity, we are only showing the ORC complex proteins that define origins, of which only a subset that recruits Cdc6 and Cdt1 will be “licensed” to result in productive replication origins. (B) Altering the genomic landscape of DNA replication. Cdc6 has undergone positive selection in 2 separate lineages of Drosophila. This leads to the model where adaptive evolution has changed the DNA-binding specificity of Cdc6 and thereby the pattern of replication origins. One mechanism by which this could occur is through changes in Cdc6-binding affinity altering the probability of replication licensing at different DNA sequences (Speck et al. 2005; Speck and Stillman 2007).
When we examined Cdc6 evolution in 2 pairs of closely related species of Drosophila (D. melanogaster and D. simulans, Drosophila pseudoobscura and Drosophila miranda), we found that Cdc6 is subject to adaptive evolution (Wiggins and Malik 2007). Again, employing the McDonald–Kreitman test (McDonald and Kreitman 1991), we found an excess of fixed replacement changes over what is expected in the absence of positive selection (12 observed changes vs. 5 expected changes in mel/sim and 3 observed changes vs. 0 expected changes in pse/mir) (Wiggins and Malik 2007). In both independent species pairs, we found that adaptive evolution has specifically affected the C terminal domain, which contains the AAA-ATPase domain and is found in all eukaryotes. Additionally, the N terminal tail of Cdc6 is so variable among eukaryotes that this region was unalignable among species. It is worth pointing out the analogy of Cdc6 with CenH3s. N terminal tails in CenH3s also change rapidly and cannot be aligned, and it is the highly conserved HFD of CenH3 (like the AAA-ATPase domain of Cdc6) where we find the consistent action of positive selection.
DNA Replication Origins: Insights from Positive Selection of Cdc6
Our discovery of adaptive evolution in Cdc6 (Wiggins and Malik 2007) is one of the strongest pieces of evidence that origins of replication are acted on by natural selection, even in light of the fact that the selection coefficient associated with these adaptive changes may be relatively small. Under the model whereby DNA replication origins get defined epigenetically by the binding of replication proteins, this suggests that the adaptive evolution of replication proteins like Cdc6 might alter choice of DNA replication origins in order to optimize the placement and firing of multiple origins (Figure 3B). This may help explain the lack of any global sequence conservation of replication origins in higher eukaryotes.
What can this signature of positive selection on Cdc6 tell us about origin choice? First, changes in Cdc6 protein sequence might influence the pattern of replication initiation timing by affecting the subset of origins that successfully fire during replication (Speck et al. 2005; Takeda et al. 2005; Speck and Stillman 2007). Molecular details of how Cdc6 binding might alter the probability of origin firing have recently been elucidated. The ATPase activity of Cdc6 modulates the stability of the Cdc6–ORC complex specifically on certain DNAs and thereby determines which DNA sequences will successfully act as origins of replication. Amino acid replacements in Cdc6's ATPase domain may therefore alter the “preference” of Cdc6 for certain DNAs over others. The positive selection we have observed may be a result of selection for altering that pattern.
In most eukaryotic genomes, there is a compendium of competent origins bound by ORC proteins, but Cdc6 only licenses a smaller subset, enabling them to fire (Takeda and Dutta 2005). Thus, changes in Cdc6 might allow it to recognize a different subset of competent origins, thereby reshaping the replication landscape (Figure 3B), presumably to optimize the time required to finish DNA replication. Changes in the replication pattern would be especially necessary in the case of large-scale changes in the genome, like large expansions or deletions (Blumenthal et al. 1974). Heterochromatic regions of the genome are especially noteworthy because they are devoid of origins of replication, easily imposing a rate-limiting step in the replication of complex genomes. Large-scale changes in heterochromatin, by transposition, rearrangements, or recombination, thus provide the impetus to subsequently reorganize the landscape of replication origin firing. This might be achieved by a retargeting of Cdc6-binding preference especially at the “new” euchromatin–heterochromatin boundaries. Intriguingly, Cdc6 may not be unique in terms of its adaptive signature. Many of the ORC proteins also show signatures of adaptive evolution in genome-wide surveys of polymorphisms in McDonald–Kreitman analyses of D. melanogaster and D. simulans (Begun et al. 2007), suggesting that subtle pressure to reorganize and optimize replication landscapes subsequent to events like centromere drive may shape the evolution of essential DNA replication proteins. Thus far, these analyses have been primarily limited to Drosophila, but under our hypothesis, the expectation is that other complex eukaryotic genomes (animals and plants) will also be subject to such pressures, whereas “simple” heterochromatin-devoid genomes like budding yeast will not.
Dosage Compensation X Chromosomal Sites: Demarcating an Entire Chromosome
Centromeres and origins of DNA replication are examples of architectural DNA elements that are necessary for all eukaryotic chromosomes. Some other chromosome-organizing elements are only required in a subset of eukaryotic genomes. For instance, the evolutionary invention of sex chromosomes in animals presents a whole new problem; sex chromosomes require some mechanism of dosage compensation to provide parity between X: autosome gene expression in the 2 sexes (e.g., XX females vs. XY males). There are diverse means of accomplishing dosage compensation. In Drosophila, this is achieved by upregulating transcription from the single male X chromosome relative to the autosomes via members of a specialized protein–RNA complex (Hamada et al. 2005; Straub and Becker 2007). This complex is referred to as the male-specific lethal (MSL) complex because defects in any of the components result specifically in male inviability.
The MSL complex is present only in males and specifically binds to the X chromosome. Genetic translocation studies of any substantial part of X chromosome onto an autosome appear to be recognized and bound by the MSL complex, whereas autosomal translocations onto the X chromosome are generally not recognized or dosage compensated (Fagegaltier and Baker 2004; Oh et al. 2004). The hundreds of cis-acting X chromosomal DNA elements that recruit the MSL complex are referred to as the dosage compensation binding sites, and they are clearly important for the robust manifestation of dosage compensation and, therefore, viability in Drosophila males (Kelley et al. 1999; Meller et al. 2000). Targeting of the MSL complex to the X chromosome has been intensely studied; yet, the lack of distinguishing features of these X chromosomal DNA elements has stymied efforts to describe these sites. This is not due to lack of effort. Detailed chromatin immunoprecipitation efforts have led to an estimate of approximately 700 separable regions where the MSL complex is bound, covering roughly 25% of the X chromosome. These sites range in their capacity to recruit the MSL complex. Of these, a subset of 35–40 “high-affinity” sites are bound by the complex even in the absence of some of the MSL protein components (Kelley et al. 1999; Meller et al. 2000). Yet, despite a long list of DNA target sites, for a long period, no specific consensus DNA sequence had been defined (Alekseyenko et al. 2006; Dahlsveen et al. 2006; Gilfillan et al. 2006; Legube et al. 2006). Active transcription and histone modifications appeared to also play a role in attracting or maintaining the complex but could not explain the strong bias for binding to X chromosomal DNA (Schubeler 2006; Larschan et al. 2007; Bell et al. 2008). The combination of very detailed functional and computational analyses has identified features on the X chromosome that distinguishes it from the autosomes; however, extensive efforts at identifying common sequence predictors of MSL-binding sites have yielded limited prediction power at best (Stenberg et al. 2005; Gilfillan et al. 2006). These findings have led to the suggestion that degenerate and multiple weak signals may contribute to targeting (Alekseyenko et al. 2006; Dahlsveen et al. 2006; Gilfillan et al. 2006). Nonetheless, the dosage compensation machinery is able to function effectively for the sake of male survival.
Arguably, it is premature to suggest that comparative genomics methodology has had limited success to this problem because MSL-binding sites have not been mapped in divergent Drosophila species or even methodically in different D. melanogaster strains. Nevertheless, this degeneracy is observed even within D. melanogaster, in which multiple “entry” sites have been identified by chromatin immunoprecipitation studies (Alekseyenko et al. 2008; Straub et al. 2008). A consensus MSL recruitment site has been defined to encompass many, but not all, actual recruitment sites (Alekseyenko et al. 2008). One possibility emerges from these studies that a universal MSL-binding site consensus is hard to define because at least a subset of these motifs might be evolutionarily labile.
Dosage Compensation X Chromosomal Sites: The MSL Complex as a Surrogate
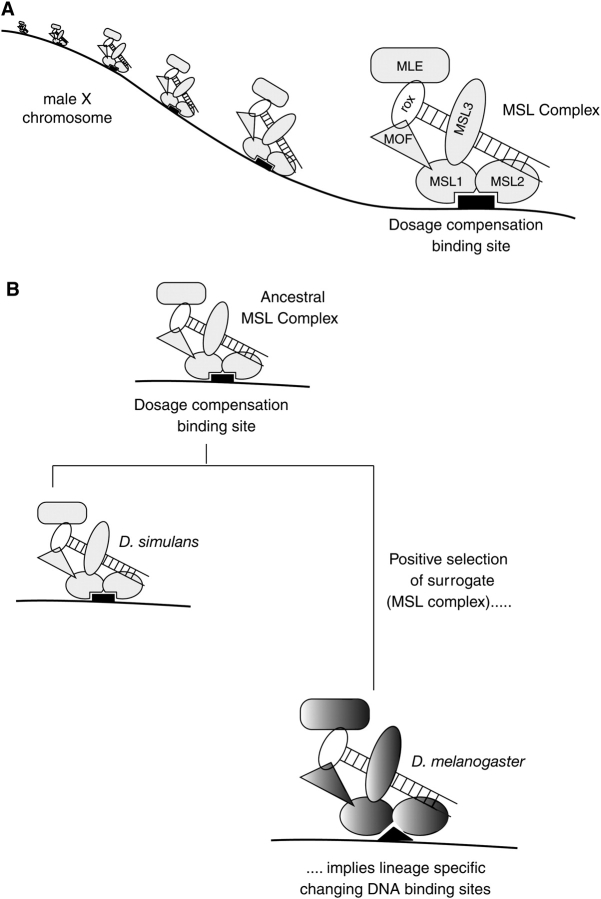
We explored the selective pressures shaping genes encoding MSL proteins as a surrogate to directly studying the MSL-binding sites. The MSL complex consists of 2 noncoding RNAs (rox1 and rox2) and 5 proteins, MLE (maleless), MOF (males absent on the fourth), and MSL1, MSL2, and MSL3 (Figure 4A). Targeting of the complex to the X chromosome is believed to enable MOF to specifically acetylate lysine 16 on histone H4 tails, a histone modification correlated with active transcription (Bone et al. 1994; Hilfiker et al. 1997; Akhtar and Becker 2000). However, this view is not universally held (Lavender et al. 1994; Hilfiker et al. 1997; Akhtar and Becker 2000; Bhadra et al. 2005). What is clear is that MSL1 and MSL2 play a central role in the assembly of the MSL complex and targeting to the X chromosome. MSL1 serves as a scaffold for the entire MSL complex. MSL1 binds to MSL2, and together with roX RNA, they bind to the X chromosome (Li et al. 2008). Mutational analyses of each MSL gene have shown that MSL1 and MSL2 complexed with roX RNA are capable of targeting high-affinity sites, independent of other known MSL components (Palmer et al. 1994; Lyman et al. 1997; Gu et al. 1998). Targeting requires an interaction between the N terminal domains of MSL1 and MSL2 and is abolished by deletion of the first 26 amino acids of MSL1 (Lyman et al. 1997; Copps et al. 1998; Scott et al. 2000; Li et al. 2005).
Figure 4.
Surrogate strategy to study dosage compensation X chromosomal sites. (A) “Surrogate” dosage compensation proteins. The MSL complex binds to many sites on the male X chromosome, and this binding is critical for dosage compensation and viability of Drosophila males. MSL1 and MSL2 play a key role in targeting of the complex to the X chromosome. (B) Alterations of dosage compensation X chromosomal sites. The MSL complex is evolving under positive selection in the Drosophila melanogaster lineage, but not in the Drosophila simulans lineage (Levine et al. 2007; Rodriguez et al. 2007). Positive selection appears to be concentrated in domains of MSL1 and MSL2 that are critical for targeting to the X chromosome (Rodriguez et al. 2007). This suggests that DNA-binding sites are rapidly changing in a lineage-specific manner, a prediction that has recently been confirmed (Bachtrog 2008).
In a comparison of D. melanogaster and D. simulans strains, all 5 protein coding genes of the MSL complex have evolved under positive selection (Levine et al. 2007; Rodriguez et al. 2007) using either McDonald–Kreitman (McDonald and Kreitman 1991) or Hudson–Kreitman–Aguade (Hudson et al. 1987) tests for adaptive evolution. Subsequent analyses found that this signature was largely confined to the D. melanogaster lineage (Levine et al. 2007; Rodriguez et al. 2007) (Figure 4B). This is a highly unexpected and remarkable finding given the essential function carried out by MSL proteins and suggests that strong, previously unappreciated selective forces are acting on the complex (see below). Interestingly, the positive selection in the D. melanogaster lineage maps to regions of MSL1 and MSL2 that are essential for targeting of the complex to the X chromosome (Rodriguez et al. 2007). This specific site of selection suggests that not only are the MSL protein components rapidly evolving but so are the DNA X chromosomal sites, at least on the D. melanogaster X chromosome (Figure 4B). This observation begins to support the possibility that although DNA sequence may be important for binding specificity, the elusive, evolutionarily fixed, “consensus”-binding site on the D. melanogaster X chromosome may not even exist.
Dosage Compensation X Chromosomal Sites: Implication of Rapid Evolution
By using a surrogate approach, we have found that instead of contributing little to recognition and evolving under relaxed constraints, DNA target sequences may be under strong selection to change. What might this selection be? One possibility is genetic conflict of the MSL complex with male-killing bacteria. For instance, Spiroplasma poulsonii specifically kill male D. melanogaster flies. Recent studies have directly implicated the presence of a functional MSL complex as a requirement for male-specific killing (Veneti et al. 2005). Under such a “genetic conflict” scenario, one could imagine bacterial proteins evolving to “detect” MSL components via direct binding, whereas MSL components could be under strong selective pressure to evolve away from this recognition. This “arms race” would result in changes in one or all the MSL components because fixation of slightly deleterious mutations in the MSL complex would be preferred over bacteria-induced male lethality.
Genetic conflict with male-killing bacteria could result in positive selection of any protein surface of the MSL complex but would not necessarily be predicted to impact its DNA-binding interface or drive the evolution of dosage compensation sites. Rather, an alternate (and not mutually exclusive) genetic conflict could be with MSL components and transposable elements (Lyon 2000; McDonald et al. 2005). One might imagine that transposable elements may benefit from increased expression through the recruitment of the MSL complex (Matyunina et al. 2008). This scenario would place the MSL complex under pressure to avoid these selfish DNA sequences on the X chromosome by avoiding certain MSL recruitment sites. This places an impetus for novel sites to become competent for MSL recruitment to restore compensation of X chromosomal genes. A scenario where sites are lost and born anew would result in positive selection of the MSL complex–DNA interface reflected in changes in binding affinity over time (Rodriguez et al. 2007). Intriguingly, a follow-up study found that, in a limited analysis of entry sites, these entry sites had evolved much faster in D. melanogaster as compared with D. simulans (Bachtrog 2008), consistent with our suggestion that cognate DNA entry sites would evolve rapidly in a species-specific manner.
Regardless of the selective force, the asymmetry in positive selection acting on the complex suggests that the X chromosomal binding sites are labile in at least D. melanogaster (Bachtrog 2008). This is of importance because dosage compensation has been intensely studied only in D. melanogaster; yet, the D. melanogaster genome may turn out to be an unsuitable genome to search for a consensus. Rapid evolution of MSL complex binding sites in D. melanogaster may be obscuring the identification of a consensus sequence in this species. On the other hand, consensus-binding sites may exist in related genomes like D. simulans and Drosophila yakuba where no evidence of positive selection acting on MSL1 and MSL2 has been found (Rodriguez et al. 2007).
Other Organizing DNAs Amenable to Surrogate Analysis
We have presented 3 case studies where the insights provided by surrogate proteins greatly clarify our knowledge of how these DNA elements function and evolve. Many other noncoding DNAs are likely to benefit from such insights. A specific example is that all the “completed” eukaryotic genomes represent only the euchromatic regions, and we are still largely missing assembled sequence data from heterochromatic regions. A central remaining question is whether all this heterochromatic DNA sequence is simply an inert aspect of a genome, subject to neutral evolutionary pressures, or does it have a function, subject to purifying or adaptive selective rationale?
Certainly, some functions of heterochromatin are appreciated. It is needed to support centromeric function, especially during meiosis (Bernard et al. 2001; Nonaka et al. 2002; Yamagishi et al. 2008). Some genes embedded in heterochromatin actually depend on the heterochromatic environment for proper expression (Weiler and Wakimoto 1995; Yasuhara and Wakimoto 2006). However, heterochromatin might act to silence otherwise destructive mobile elements via piwi-associated RNAs (piRNAs) that silence mobile elements in both male and female germ lines. These “piRNA clusters” embedded in heterochromatin have recently been shown to evolve under positive selection, presumably to expand the repertoire of silencing to include newly encountered mobile elements (Assis and Kondrashov 2009). Our surrogate approach turned out to be very useful in this case again. In flies, we found that an ovary-specific heterochromatin protein (rhino/HP1D) evolves under positive selection, hinting at a possible genetic conflict within heterochromatin (Vermaak et al. 2005). At this time, we did not know what the molecular basis of this genetic conflict was. However, recent work has shown that rhino is a piRNA transcription factor (Klattenhoff et al. 2009). Absence of rhino leads to impaired piRNA production and unleashing of mobile elements in the female germ line, resulting in female sterility (Volpe et al. 2001; Klattenhoff et al. 2009). The positive selection of rhino is predicted to expand the repertoire of piRNA clusters to ensure the silencing of newly encountered mobile elements (Vermaak et al. 2005; Klattenhoff et al. 2009). Thus, our study of a surrogate heterochromatin protein under positive selection revealed an unexpected but exciting genetic conflict. We suggest that other heterochromatic proteins may be under positive selection to act as suppressors of centromere drive (like Cid as discussed earlier), in silencing mobile elements (like rhino), or both.
Similarly, other examples of DNA elements that enable dosage compensation, such as in Caenorhabditis elegans (by transcriptional silencing) and in mammals (by X inactivation), may be especially suitable for analysis. In the latter case, it is known what events are required for choice and initiation of X inactivation, but the mechanism of how this initial inactivation is “spread” to the rest of the X chromosome is controversial (Lyon 1998, 2000). Protein components involved in these processes are beginning to be identified. Their evolutionary patterns may reveal yet more surprises in this arena of biology that is essential for sex-specific viability and may be inherently subject to genetic conflict (Haig 2006; Engelstadter and Haig 2008).
Concluding Comments about Adaptive Scenarios of Organizing DNA Elements
It is important to point out explicitly that there is not an expectation that all noncoding DNA elements, such as the ones we have described, will be shaped by adaptive evolution. Indeed, although we have focused our attention on a few cases where surrogates have been found to evolve under positive selection, most noncoding DNA–binding proteins might not evolve under positive selection. One must reemphasize that effectively nonadaptive, neutral evolution must remain a robust null hypothesis for the gain of complexity even in these essential DNA elements. Indeed, very compelling arguments have been presented that the reduced efficacy of selection in some eukaryotic lineages due to a lower effective population size could have easily resulted in this apparently unresolved and inexplicable complexity (Lynch 2007). However, we have presented 3 instances (4 including rhino) where closer examination has revealed not just adaptation in organizing DNA elements but an amazing level of biological organization and evolutionary lability stemming from recurrent genetic conflict. This biological viewpoint would have remained obscured were it not for the insights revealed from a surrogate approach, which provides a very useful tool to study the otherwise intractable components of complex genomes.
Funding
National Institute of Health (PHS NRSA T32 GM07270 to J.J.B.); National Institute of Health (R01-GM074108 to H.S.M.).
Acknowledgments
We thank Michael Lynch for his suggestion about writing this review, his encouragement, and patient suggestions to improve the clarity of our presentation. We also thank Mia Levine and members of the Malik laboratory for comments on the manuscript. This review is solely the responsibility of the authors and does not necessarily represent the official views of NIGMS or National Institutes of Health.
References
- Ahmad K, Henikoff S. Histone H3 variants specify modes of chromatin assembly. Proc Natl Acad Sci USA. 2002;99(Suppl 4):16477–16484. doi: 10.1073/pnas.172403699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar A, Becker PB. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol Cell. 2000;5:367–375. doi: 10.1016/s1097-2765(00)80431-1. [DOI] [PubMed] [Google Scholar]
- Alekseyenko AA, Larschan E, Lai WR, Park PJ, Kuroda MI. High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev. 2006;20:848–857. doi: 10.1101/gad.1400206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseyenko AA, Peng S, Larschan E, Gorchakov AA, Lee OK, Kharchenko P, McGrath SD, Wang CI, Mardis ER, Park PJ, et al. A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell. 2008;134:599–609. doi: 10.1016/j.cell.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assis R, Kondrashov AS. Rapid repetitive element-mediated expansion of piRNA clusters in mammalian evolution. Proc Natl Acad Sci USA. 2009; 106:7079–7082. doi: 10.1073/pnas.0900523106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D. Positive selection at the binding sites of the male-specific lethal complex involved in dosage compensation in Drosophila. Genetics. 2008;180:1123–1129. doi: 10.1534/genetics.107.084244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, Holloway AK, Stevens K, Hillier LW, Poh YP, Hahn MW, Nista PM, Jones CD, Kern AD, Dewey CN, et al. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 2007;5:e310. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell O, Conrad T, Kind J, Wirbelauer C, Akhtar A, Schubeler D. Transcription-coupled methylation of histone H3 at lysine 36 regulates dosage compensation by enhancing recruitment of the MSL complex in Drosophila melanogaster. Mol Cell Biol. 2008;28:3401–3409. doi: 10.1128/MCB.00006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- Bhadra MP, Bhadra U, Kundu J, Birchler JA. Gene expression analysis of the function of the male-specific lethal complex in Drosophila. Genetics. 2005;169:2061–2074. doi: 10.1534/genetics.104.036020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Gillespie PJ, Francis D, Jackson DA. Replication origins in Xenopus egg extract Are 5-15 kilobases apart and are activated in clusters that fire at different times. J Cell Biol. 2001;152:15–25. doi: 10.1083/jcb.152.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower MD, Karpen GH. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat Cell Biol. 2001;3:730–739. doi: 10.1038/35087045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal AB, Kriegstein HJ, Hogness DS. The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harb Symp Quant Biol. 1974;38:205–223. doi: 10.1101/sqb.1974.038.01.024. [DOI] [PubMed] [Google Scholar]
- Bone JR, Lavender J, Richman R, Palmer MJ, Turner BM, Kuroda MI. Acetylated histone H4 on the male X chromosome is associated with dosage compensation in Drosophila. Genes Dev. 1994;8:96–104. doi: 10.1101/gad.8.1.96. [DOI] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- Buchwitz BJ, Ahmad K, Moore LL, Roth MB, Henikoff S. A histone-H3-like protein in C. elegans. Nature. 1999;401:547–548. doi: 10.1038/44062. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Sniegowski P, Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994;371:215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- Copenhaver GP, Nickel K, Kuromori T, Benito MI, Kaul S, Lin X, Bevan M, Murphy G, Harris B, Parnell LD, et al. Genetic definition and sequence analysis of Arabidopsis centromeres. Science. 1999;286:2468–2474. doi: 10.1126/science.286.5449.2468. [DOI] [PubMed] [Google Scholar]
- Copps K, Richman R, Lyman LM, Chang KA, Rampersad-Ammons J, Kuroda MI. Complex formation by the Drosophila MSL proteins: role of the MSL2 RING finger in protein complex assembly. Embo J. 1998;17:5409–5417. doi: 10.1093/emboj/17.18.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coverley D, Laskey RA. Regulation of eukaryotic DNA replication. Annu Rev Biochem. 1994;63:745–776. doi: 10.1146/annurev.bi.63.070194.003525. [DOI] [PubMed] [Google Scholar]
- Cvetic C, Walter JC. Eukaryotic origins of DNA replication: could you please be more specific? Semin Cell Dev Biol. 2005;16:343–353. doi: 10.1016/j.semcdb.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Dahlsveen IK, Gilfillan GD, Shelest VI, Lamm R, Becker PB. Targeting determinants of dosage compensation in Drosophila. PLoS Genet. 2006;2:e5. doi: 10.1371/journal.pgen.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel A. Distortion of female meiotic segregation and reduced male fertility in human Robertsonian translocations: consistent with the centromere model of co-evolving centromere DNA/centromeric histone (CENP-A) Am J Med Genet. 2002;111:450–452. doi: 10.1002/ajmg.10618. [DOI] [PubMed] [Google Scholar]
- Dover GA, Strachan T, Coen ES, Brown SD. Molecular drive. Science. 1982;218:1069. doi: 10.1126/science.7146895. [DOI] [PubMed] [Google Scholar]
- Dowell RD, Eddy SR. Efficient pairwise RNA structure prediction and alignment using sequence alignment constraints. BMC Bioinformatics. 2006;7:400. doi: 10.1186/1471-2105-7-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaker S, Pyle A, Cobb J, Handel MA. Evidence for meiotic spindle checkpoint from analysis of spermatocytes from Robertsonian-chromosome heterozygous mice. J Cell Sci. 2001;114:2953–2965. doi: 10.1242/jcs.114.16.2953. [DOI] [PubMed] [Google Scholar]
- Engelstadter J, Haig D. Sexual antagonism and the evolution of X chromosome inactivation. Evolution Int J Org Evolution. Forthcoming 2008 doi: 10.1111/j.1558-5646.2008.00431.x. [DOI] [PubMed] [Google Scholar]
- Fagegaltier D, Baker BS. X chromosome sites autonomously recruit the dosage compensation complex in Drosophila males. PLoS Biol. 2004;2:e341. doi: 10.1371/journal.pbio.0020341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fangman WL, Hice RH, Chlebowicz-Sledziewska E. ARS replication during the yeast S phase. Cell. 1983;32:831–838. doi: 10.1016/0092-8674(83)90069-7. [DOI] [PubMed] [Google Scholar]
- Fishman L, Saunders A. Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science. 2008;322:1559–1562. doi: 10.1126/science.1161406. [DOI] [PubMed] [Google Scholar]
- Fishman L, Willis JH. A novel meiotic drive locus almost completely distorts segregation in mimulus (monkeyflower) hybrids. Genetics. 2005;169:347–353. doi: 10.1534/genetics.104.032789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald-Hayes M, Clarke L, Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982;29:235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- Gilfillan GD, Straub T, de Wit E, Greil F, Lamm R, van Steensel B, Becker PB. Chromosome-wide gene-specific targeting of the Drosophila dosage compensation complex. Genes Dev. 2006;20:858–870. doi: 10.1101/gad.1399406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Szauter P, Lucchesi JC. Targeting of MOF, a putative histone acetyl transferase, to the X chromosome of Drosophila melanogaster. Dev Genet. 1998;22:56–64. doi: 10.1002/(SICI)1520-6408(1998)22:1<56::AID-DVG6>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Haig D. Self-imposed silence: parental antagonism and the evolution of X-chromosome inactivation. Evolution Int J Org Evolution. 2006;60:440–447. [PubMed] [Google Scholar]
- Halverson D, Baum M, Stryker J, Carbon J, Clarke L. A centromere DNA-binding protein from fission yeast affects chromosome segregation and has homology to human CENP-B. J Cell Biol. 1997;136:487–500. doi: 10.1083/jcb.136.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halverson D, Gutkin G, Clarke L. A novel member of the Swi6p family of fission yeast chromo domain-containing proteins associates with the centromere in vivo and affects chromosome segregation. Mol Gen Genet. 2000;264:492–505. doi: 10.1007/s004380000338. [DOI] [PubMed] [Google Scholar]
- Hamada FN, Park PJ, Gordadze PR, Kuroda MI. Global regulation of X chromosomal genes by the MSL complex in Drosophila melanogaster. Genes Dev. 2005;19:2289–2294. doi: 10.1101/gad.1343705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL. Modifier theory and meiotic drive. Theor Popul Biol. 1975;7:168–174. doi: 10.1016/0040-5809(75)90012-x. [DOI] [PubMed] [Google Scholar]
- Hayashi MT, Takahashi TS, Nakagawa T, Nakayama J, Masukata H. The heterochromatin protein Swi6/HP1 activates replication origins at the pericentromeric region and silent mating-type locus. Nat Cell Biol. 2009;11:357–362. doi: 10.1038/ncb1845. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Malik HS. Centromeres: selfish drivers. Nature. 2002;417:227. doi: 10.1038/417227a. [DOI] [PubMed] [Google Scholar]
- Hilfiker A, Hilfiker-Kleiner D, Pannuti A, Lucchesi JC. mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. Embo J. 1997;16:2054–2060. doi: 10.1093/emboj/16.8.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguade M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley RL, Meller VH, Gordadze PR, Roman G, Davis RL, Kuroda MI. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell. 1999;98:513–522. doi: 10.1016/s0092-8674(00)81979-0. [DOI] [PubMed] [Google Scholar]
- Kim CS, Preer JR, Jr., Polisky B. Bacteriophage lambda DNA fragments replicate in the Paramecium macronucleus: absence of active copy number control. Dev Genet. 1992;13:97–102. doi: 10.1002/dvg.1020130202. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana J, Schultz N, Koppetsch BS, Nowosielska A, Seitz H, et al. The Drosophila HP1 homologue Rhino is required for transposon silencing and piRNA production by dual strand clusters. Cell. Forthcoming 2009 doi: 10.1016/j.cell.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam AL, Boivin CD, Bonney CF, Rudd MK, Sullivan BA. Human centromeric chromatin is a dynamic chromosomal domain that can spread over noncentromeric DNA. Proc Natl Acad Sci USA. 2006;103:4186–4191. doi: 10.1073/pnas.0507947103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan E, Alekseyenko AA, Gortchakov AA, Peng S, Li B, Yang P, Workman JL, Park PJ, Kuroda MI. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell. 2007;28:121–133. doi: 10.1016/j.molcel.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Lavender JS, Birley AJ, Palmer MJ, Kuroda MI, Turner BM. Histone H4 acetylated at lysine 16 and proteins of the Drosophila dosage compensation pathway co-localize on the male X chromosome through mitosis. Chromosome Res. 1994;2:398–404. doi: 10.1007/BF01552799. [DOI] [PubMed] [Google Scholar]
- Lee HR, Zhang W, Langdon T, Jin W, Yan H, Cheng Z, Jiang J. Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc Natl Acad Sci USA. 2005;102:11793–11798. doi: 10.1073/pnas.0503863102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legube G, McWeeney SK, Lercher MJ, Akhtar A. X-chromosome-wide profiling of MSL-1 distribution and dosage compensation in Drosophila. Genes Dev. 2006;20:871–883. doi: 10.1101/gad.377506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- Levine MT, Holloway AK, Arshad U, Begun DJ. Pervasive and largely lineage-specific adaptive protein evolution in the dosage compensation complex of Drosophila melanogaster. Genetics. 2007;177:1959–1962. doi: 10.1534/genetics.107.079459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Parry DA, Scott MJ. The amino-terminal region of Drosophila MSL1 contains basic, glycine-rich, and leucine zipper-like motifs that promote X chromosome binding, self-association, and MSL2 binding, respectively. Mol Cell Biol. 2005;25:8913–8924. doi: 10.1128/MCB.25.20.8913-8924.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Schiemann AH, Scott MJ. Incorporation of the noncoding roX RNAs alters the chromatin-binding specificity of the Drosophila MSL1/MSL2 complex. Mol Cell Biol. 2008;28:1252–1264. doi: 10.1128/MCB.00910-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Lyman LM, Copps K, Rastelli L, Kelley RL, Kuroda MI. Drosophila male-specific lethal-2 protein: structure/function analysis and dependence on MSL-1 for chromosome association. Genetics. 1997;147:1743–1753. doi: 10.1093/genetics/147.4.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. The origins of genome architecture. Sinauer Associates; 2007. [Google Scholar]
- Lyon MF. X-chromosome inactivation: a repeat hypothesis. Cytogenet Cell Genet. 1998;80:133–137. doi: 10.1159/000014969. [DOI] [PubMed] [Google Scholar]
- Lyon MF. LINE-1 elements and X chromosome inactivation: a function for “junk” DNA? Proc Natl Acad Sci USA. 2000;97:6248–6249. doi: 10.1073/pnas.97.12.6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine DM, Bell SP. A genomic view of eukaryotic DNA replication. Chromosome Res. 2005;13:309–326. doi: 10.1007/s10577-005-1508-1. [DOI] [PubMed] [Google Scholar]
- Malik HS. Mimulus finds centromeres in the driver's seat. Trends Ecol Evol. 2005;20:151–154. doi: 10.1016/j.tree.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Malik HS, Bayes JJ. Genetic conflicts during meiosis and the evolutionary origins of centromere complexity. Biochem Soc Trans. 2006;34:569–573. doi: 10.1042/BST0340569. [DOI] [PubMed] [Google Scholar]
- Malik HS, Henikoff S. Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics. 2001;157:1293–1298. doi: 10.1093/genetics/157.3.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik HS, Henikoff S. Conflict begets complexity: the evolution of centromeres. Curr Opin Genet Dev. 2002;12:711–718. doi: 10.1016/s0959-437x(02)00351-9. [DOI] [PubMed] [Google Scholar]
- Malik HS, Henikoff S. Phylogenomics of the nucleosome. Nat Struct Biol. 2003;10:882–891. doi: 10.1038/nsb996. [DOI] [PubMed] [Google Scholar]
- Matyunina LV, Bowen NJ, McDonald JF. LTR retrotransposons and the evolution of dosage compensation in Drosophila. BMC Mol Biol. 2008;9:55. doi: 10.1186/1471-2199-9-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister BF, Werren JH. Evolution of tandemly repeated sequences: what happens at the end of an array? J Mol Evol. 1999;48:469–481. doi: 10.1007/pl00006491. [DOI] [PubMed] [Google Scholar]
- McDonald JF, Matzke MA, Matzke AJ. Host defenses to transposable elements and the evolution of genomic imprinting. Cytogenet Genome Res. 2005;110:242–249. doi: 10.1159/000084958. [DOI] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- McKee BD, Wilhelm K, Merrill C, Ren X. Male sterility and meiotic drive associated with sex chromosome rearrangements in Drosophila. Role of X-Y pairing. Genetics. 1998;149:143–155. doi: 10.1093/genetics/149.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller VH, Gordadze PR, Park Y, Chu X, Stuckenholz C, Kelley RL, Kuroda MI. Ordered assembly of roX RNAs into MSL complexes on the dosage-compensated X chromosome in Drosophila. Curr Biol. 2000;10:136–143. doi: 10.1016/s0960-9822(00)00311-0. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. Embo J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill C, Bayraktaroglu L, Kusano A, Ganetzky B. Truncated RanGAP encoded by the segregation distorter locus of Drosophila. Science. 1999;283:1742–1745. doi: 10.1126/science.283.5408.1742. [DOI] [PubMed] [Google Scholar]
- Nagaki K, Cheng Z, Ouyang S, Talbert PB, Kim M, Jones KM, Henikoff S, Buell CR, Jiang J. Sequencing of a rice centromere uncovers active genes. Nat Genet. 2004;36:138–145. doi: 10.1038/ng1289. [DOI] [PubMed] [Google Scholar]
- Nielsen J, Wohlert M. Chromosome abnormalities found among 34,910 newborn children: results from a 13-year incidence study in Arhus, Denmark. Hum Genet. 1991;87:81–83. doi: 10.1007/BF01213097. [DOI] [PubMed] [Google Scholar]
- Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SI, Watanabe Y. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- Oh H, Bone JR, Kuroda MI. Multiple classes of MSL binding sites target dosage compensation to the X chromosome of Drosophila. Curr Biol. 2004;14:481–487. doi: 10.1016/j.cub.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Palmer DK, O'Day K, Wener MH, Andrews BS, Margolis RL. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J Cell Biol. 1987;104:805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer MJ, Richman R, Richter L, Kuroda MI. Sex-specific regulation of the male-specific lethal-1 dosage compensation gene in Drosophila. Genes Dev. 1994;8:698–706. doi: 10.1101/gad.8.6.698. [DOI] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena F, Sapienza C. Female meiosis drives karyotypic evolution in mammals. Genetics. 2001a;159:1179–1189. doi: 10.1093/genetics/159.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena F, Sapienza C. Nonrandom segregation during meiosis: the unfairness of females. Mamm Genome. 2001b;12:331–339. doi: 10.1007/s003350040003. [DOI] [PubMed] [Google Scholar]
- Quivy JP, Gerard A, Cook AJ, Roche D, Almouzni G. The HP1-p150/CAF-1 interaction is required for pericentric heterochromatin replication and S-phase progression in mouse cells. Nat Struct Mol Biol. 2008;15:972–979. doi: 10.1038/nsmb.1470. [DOI] [PubMed] [Google Scholar]
- Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL. Replication dynamics of the yeast genome. Science. 2001;294:115–121. doi: 10.1126/science.294.5540.115. [DOI] [PubMed] [Google Scholar]
- Rodriguez MA, Vermaak D, Bayes JJ, Malik HS. Species-specific positive selection of the male-specific lethal complex that participates in dosage compensation in Drosophila. Proc Natl Acad Sci USA. 2007;104:15412–15417. doi: 10.1073/pnas.0707445104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd MK, Wray GA, Willard HF. The evolutionary dynamics of alpha-satellite. Genome Res. 2006;16:88–96. doi: 10.1101/gr.3810906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler L, Novitski E. Meiotic drive as an evolutionary force. Am Nat. 1957;41:105–110. [Google Scholar]
- Sawamura K, Yamamoto MT. Cytogenetical localization of Zygotic hybrid rescue (Zhr), a Drosophila melanogaster gene that rescues interspecific hybrids from embryonic lethality. Mol Gen Genet. 1993;239:441–449. doi: 10.1007/BF00276943. [DOI] [PubMed] [Google Scholar]
- Sawamura K, Yamamoto MT, Watanabe TK. Hybrid lethal systems in the Drosophila melanogaster species complex. II. The Zygotic hybrid rescue (Zhr) gene of D. melanogaster. Genetics. 1993;133:307–313. doi: 10.1093/genetics/133.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler D. Dosage compensation in high resolution: global up-regulation through local recruitment. Genes Dev. 2006;20:749–753. doi: 10.1101/gad.1423006. [DOI] [PubMed] [Google Scholar]
- Schueler MG, Higgins AW, Rudd MK, Gustashaw K, Willard HF. Genomic and genetic definition of a functional human centromere. Science. 2001;294:109–115. doi: 10.1126/science.1065042. [DOI] [PubMed] [Google Scholar]
- Scott MJ, Pan LL, Cleland SB, Knox AL, Heinrich J. MSL1 plays a central role in assembly of the MSL complex, essential for dosage compensation in Drosophila. Embo J. 2000;19:144–155. doi: 10.1093/emboj/19.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby RD, Vafa O, Sullivan KF. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JG, Calos MP. Autonomous replication in Drosophila melanogaster tissue culture cells. Chromosoma. 1995;103:597–605. doi: 10.1007/BF00357686. [DOI] [PubMed] [Google Scholar]
- Speck C, Chen Z, Li H, Stillman B. ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA. Nat Struct Mol Biol. 2005;12:965–971. doi: 10.1038/nsmb1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck C, Stillman B. Cdc6 ATPase activity regulates ORC x Cdc6 stability and the selection of specific DNA sequences as origins of DNA replication. J Biol Chem. 2007;282:11705–11714. doi: 10.1074/jbc.M700399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Lin MF, Kheradpour P, Pedersen JS, Parts L, Carlson JW, Crosby MA, Rasmussen MD, Roy S, Deoras AN, et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007;450:219–232. doi: 10.1038/nature06340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenberg P, Pettersson F, Saura AO, Berglund A, Larsson J. Sequence signature analysis of chromosome identity in three Drosophila species. BMC Bioinformatics. 2005;6:158. doi: 10.1186/1471-2105-6-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan W. Tandem-repetitive noncoding DNA: forms and forces. Mol Biol Evol. 1989;6:198–212. doi: 10.1093/oxfordjournals.molbev.a040542. [DOI] [PubMed] [Google Scholar]
- Stephan W, Cho S. Possible role of natural selection in the formation of tandem-repetitive noncoding DNA. Genetics. 1994;136:333–341. doi: 10.1093/genetics/136.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler S, Keith KC, Curnick KE, Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- Straub T, Becker PB. Dosage compensation: the beginning and end of generalization. Nat Rev Genet. 2007;8:47–57. doi: 10.1038/nrg2013. [DOI] [PubMed] [Google Scholar]
- Straub T, Grimaud C, Gilfillan GD, Mitterweger A, Becker PB. The chromosomal high-affinity binding sites for the Drosophila dosage compensation complex. PLoS Genet. 2008;4:e1000302. doi: 10.1371/journal.pgen.1000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan KF, Hechenberger M, Masri K. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J Cell Biol. 1994;127:581–592. doi: 10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Le HD, Wahlstrom JM, Karpen GH. Sequence analysis of a functional Drosophila centromere. Genome Res. 2003;13:182–194. doi: 10.1101/gr.681703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Chen ES, Yanagida M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- Takeda DY, Dutta A. DNA replication and progression through S phase. Oncogene. 2005;24:2827–2843. doi: 10.1038/sj.onc.1208616. [DOI] [PubMed] [Google Scholar]
- Takeda DY, Shibata Y, Parvin JD, Dutta A. Recruitment of ORC or CDC6 to DNA is sufficient to create an artificial origin of replication in mammalian cells. Genes Dev. 2005;19:2827–2836. doi: 10.1101/gad.1369805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Bryson TD, Henikoff S. Adaptive evolution of centromere proteins in plants and animals. J Biol. 2004;3:18. doi: 10.1186/jbiol11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Hartl DL, Laurie CC. Sex-ratio segregation distortion associated with reproductive isolation in Drosophila. Proc Natl Acad Sci USA. 2001;98:13183–13188. doi: 10.1073/pnas.231478798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veneti Z, Bentley JK, Koana T, Braig HR, Hurst GD. A functional dosage compensation complex required for male killing in Drosophila. Science. 2005;307:1461–1463. doi: 10.1126/science.1107182. [DOI] [PubMed] [Google Scholar]
- Vermaak D, Hayden HS, Henikoff S. Centromere targeting element within the histone fold domain of Cid. Mol Cell Biol. 2002;22:7553–7561. doi: 10.1128/MCB.22.21.7553-7561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaak D, Henikoff S, Malik HS. Positive selection drives the evolution of rhino, a member of the heterochromatin protein 1 family in Drosophila. PLoS Genet. 2005;1:96–108. doi: 10.1371/journal.pgen.0010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe AM, Horowitz H, Grafer CM, Jackson SM, Berg CA. Drosophila rhino encodes a female-specific chromo-domain protein that affects chromosome structure and egg polarity. Genetics. 2001;159:1117–1134. doi: 10.1093/genetics/159.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh JB. Persistence of tandem arrays: implications for satellite and simple-sequence DNAs. Genetics. 1987;115:553–567. doi: 10.1093/genetics/115.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler KS, Wakimoto BT. Heterochromatin and gene expression in Drosophila. Annu Rev Genet. 1995;29:577–605. doi: 10.1146/annurev.ge.29.120195.003045. [DOI] [PubMed] [Google Scholar]
- Wiggins BL, Malik HS. Molecular evolution of Drosophila Cdc6, an essential DNA replication-licensing gene, suggests an adaptive choice of DNA replication origins. Fly. 2007;1:155–163. doi: 10.4161/fly.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CI, True JR, Johnson N. Fitness reduction associated with the deletion of a satellite DNA array. Nature. 1989;341:248–251. doi: 10.1038/341248a0. [DOI] [PubMed] [Google Scholar]
- Yamagishi Y, Sakuno T, Shimura M, Watanabe Y. Heterochromatin links to centromeric protection by recruiting shugoshin. Nature. 2008;455:251–255. doi: 10.1038/nature07217. [DOI] [PubMed] [Google Scholar]
- Yasuhara JC, Wakimoto BT. Oxymoron no more: the expanding world of heterochromatic genes. Trends Genet. 2006;22:330–338. doi: 10.1016/j.tig.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Yoda K, Ando S, Morishita S, Houmura K, Hashimoto K, Takeyasu K, Okazaki T. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc Natl Acad Sci USA. 2000;97:7266–7271. doi: 10.1073/pnas.130189697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda K, Morishita S, Hashimoto K. Histone variant CENP-A purification, nucleosome reconstitution. Methods Enzymol. 2004;375:253–269. doi: 10.1016/s0076-6879(03)75017-4. [DOI] [PubMed] [Google Scholar]
- Zwick ME, Salstrom JL, Langley CH. Genetic variation in rates of nondisjunction: association of two naturally occurring polymorphisms in the chromokinesin nod with increased rates of nondisjunction in Drosophila melanogaster. Genetics. 1999;152:1605–1614. doi: 10.1093/genetics/152.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]