Abstract
We have performed a whole-genome-sequence survey for the gregarine, Ascogregarina taiwanensis and herein describe both features unique to this early diverging apicomplexan and properties that unite it with Cryptosporidium, the Coccidia, and the Apicomplexa. Phylogenetic trees inferred from a concatenated protein sequence comprised of 10,750 amino acid positions, as well as the large subunit rRNA genes, robustly support phylogenetic affinity of Ascogregarina with Cryptosporidium at the base of the apicomplexan clade. Unlike Cryptosporidium, Ascogregarina possesses numerous mitochondrion-associated pathways and proteins, including enzymes within the Krebs cycle and a cytochrome-based respiratory chain. Ascogregarina further differs in the capacity for de novo synthesis of pyrimidines and amino acids. Ascogregarina shares with Cryptosporidium a Type I fatty acid synthase and likely a polyketide synthase. Cryptosporidium and Ascogregarina possess a large repertoire of multidomain surface proteins that align it with Toxoplasma and are proposed to be involved in coccidian-like functions. Four families of retrotransposable elements were identified, and thus, retroelements are present in Ascogregarina and Eimeria but not in other apicomplexans that have been analyzed. The sum observations suggest that Ascogregarina and Cryptosporidium share numerous molecular similarities, not only including coccidian-like features to the exclusion of Haemosporidia and Piroplasmida but also differ from each other significantly in their metabolic capacity.
Keywords: Apicomplexa, Ascogregarina taiwanensis, genome-sequence survey (GSS), Cryptosporidium
Introduction
The phylum Apicomplexa comprises a broad group of protists of which many members are obligate parasites and span diverse genera such as the causative agent of malaria, Plasmodium, and pathogens that cause coccidiosis (Levine 1988). Among apicomplexans, the haematozoa (e.g., Plasmodium and Theileria), coccidia (e.g., Toxoplasma and Eimeria), and Cryptosporidium are extensively studied owing to their importance as pathogens of humans or animals of veterinary interest. Recently, complete or near-complete genome sequences have been determined for apicomplexans that represent almost all known major groups within the phylum, including Cryptosporidium (Abrahamsen et al. 2004), Plasmodium (Gardner et al. 2002), Theileria (Gardner et al. 2005), Toxoplasma (http://www.toxodb.org), and Eimeria (8X coverage, http://www.sanger.ac.uk/Projects/E_tenella). Comparison of these genomes reveals a complex picture of the divergent metabolisms and evolutionary histories of apicomplexans. As members of the group Alveolata, apicomplexans are evolutionarily related to dinoflagellates and ciliates, which are comprised of both free-living and parasitic species. The complete genome sequence has recently been determined for the ciliate, Tetrahymena (Eisen et al. 2006), and thus, it is possible to begin comparison of apicomplexans with a distantly related member of the group Alveolata. Moreover, recent discovery of Chromera velia, the closest known free-living, photosynthetic relative of the Apicomplexa also provides additional opportunity to study the evolution of apicomplexan parasitism (Moore et al. 2008).
To understand the origin of the apicomplexans within the Alveolata, it is important to initiate genome-sequencing projects for dinoflagellates, such as the parasite of mollusks, Perkinsus marinus, as well as to study apicomplexans that are proposed to branch at the base of the apicomplexan clade. Cryptosporidium spp. has been traditionally considered to be an intestinal coccidium, but recent molecular phylogenies suggest that this genus is evolutionarily divergent from other coccidia and is an early branch at the base of the phylum (Zhu et al. 2000; Barta and Thompson 2006). Genomic and biochemical data indicate that Cryptosporidium differs from other apicomplexans in that it has lost the apicoplast organelle, as well as genomes for both the plastid and the mitochondrion (Zhu et al. 2000; Abrahamsen et al. 2004). In addition, Cryptosporidium appears to have lost many de novo biosynthetic pathways, such as the capacity to synthesize amino acids and nucleotides. Parasites of the class Gregarinia are also considered to branch at the base of the Apicomplexa, and like Cryptosporidium, they appear to lack a plastid genome and might additionally share a monophyletic relationship (Carreno et al. 1999; Leander et al. 2003). The gregarines are predominantly comprised of species that solely parasitize invertebrates (Levine 1988) and as a whole are a poorly studied group for which there are no in vitro culture methods and few insights into their biology and position within the evolutionary history of the apicomplexans.
The evaluation of a predicted evolutionary relationship between Cryptosporidium and gregarines is hampered because little is known about gregarine metabolism, and otherwise, there is no foundation of genome-sequence data. A small-scale project of expressed sequence tags (ESTs) has been reported on a gregarine, Gregarina niphandrodes, from the mealworm, but this survey yielded limited genomic information (Omoto et al. 2004). To understand in greater detail the metabolism and cellular biology of gregarines, we have initiated a genome-sequence survey (GSS) project for a gregarine, Ascogregarina taiwanensis, which infects mosquito larvae such as Aedes albopictus. Because Ascogregarina spp. is a globally distributed parasite of mosquitoes (Munstermann and Wesson 1990; Garcia et al. 1994; Chen 1999; Reyes-Villanueva et al. 2003; Morales et al. 2005), it may also serve as a candidate for a biological agent to control mosquitoes and thus target the vectors of many deadly pathogens in humans and animals. It is therefore hoped that genome-sequence information will provide valuable insights into manipulating this parasite as a biological control agent. The GSS project utilized a phi29 polymerase-based whole-genome amplification (WGA) methodology that provided virtually unlimited amount of material for constructing high-quality genomic DNA libraries. To date, over 20,000 sequence reads have been performed and assembled into contigs.
We describe here a preliminary annotation of the resulting Ascogregarina genome sequence, which provides a snapshot of the breadth of the metabolic capacity of an “early branching” apicomplexan and robust phylogenetic reconstructions to reconfirm its evolutionary affinity with Cryptosporidium species at the base of the Phylum.
Materials and Methods
Maintenance of Gregarines and Mosquitoes
The Asian tiger mosquito, A. albopictus, which originated from Linkou in northwest Taiwan, was cultured and infected by the gregarine, A. taiwanensis (Taoyoun-1 isolate), and maintained in the laboratory as described (Huang et al. 2006). To propagate the gregarine, about 100–200 larval mosquitoes from the infected colony were ground in a 1.5-ml microfuge tube containing 1 ml of distilled water (Chen and Yang 1996). The homogenate was passed through a 75 mesh nylon cloth to remove tissue debris and was then fed to uninfected newly hatched mosquito larvae.
Purification of Gregarine Oocysts and Isolation of Genomic DNA
The method for purification of gregarine oocysts has been described (Huang et al. 2006). Briefly, infected A. albopictus larvae were ground in distilled water and then filtered to obtain a crude suspension containing oocysts. The suspensions were centrifuged at 5,500 rpm (4 °C) for 90 min over a gradient composed of 1 ml each layers of 30%, 50%, 70%, and 90% Percoll (Amersham Biosciences). The interface between the 50% and 70% Percoll layers was collected and diluted with an equal volume of distilled water and then centrifuged at 3,000 rpm (4 °C) for 15 min. This procedure was repeated three times. The pellet was then layered over 500 μl of a 58% Percoll solution and centrifuged at 5,500 rpm (4 °C) for 30 min. The content taken from the interface of the 58% Percoll solution was mixed with an equal volume of distilled water and centrifuged at 3,000 rpm (4 °C) for 15 min in order to replace the Percoll solution with water. Purified oocysts were treated with 10% bleach solution in water for 10 min on ice, in order to remove bacterial contamination, followed by centrifugation for five times with water. Oocysts were then suspended in a DNA lysis buffer and subjected to five freeze and thaw cycles for isolating gDNA using a DNeasy isolation kit (Qiagen, Valencia, CA).
Construction of Genomic Libraries and GSS
Three plasmid-based genomic libraries were constructed from purified gDNA that was digested using the restriction enzymes EcoRI, BamHI, and XbaI and ligated into the corresponding restriction sites of pUC19. A fourth library was constructed from 100 ng of randomly sheared genomic DNA that was amplified via the WGA method using phi29 DNA polymerase (REPLI-g, Qiagen). The amplified DNA was nebulized, converted to blunt ends with T4 DNA polymerase, and size fractionated on a 1% agarose gel. The 1.5- to 2.5-kb-size fractions were isolated and cloned into pCR-Blunt II-TOPO or pCR4Blunt vector (Invitrogen). The resulting four plasmid libraries were transformed into XL10-gold Escherichia coli competent cells (Stratagene). Plasmids were isolated from individual bacterial colonies using an REAL Prep 96 Plasmid Kit (Qiagen). Nucleotide sequencing was performed at the Advanced Genetic Analysis Center at University of Minnesota. Approximately 20,000 sequence reads were obtained from the WGA library and an additional 3,000 from the restriction enzyme libraries. Sequence assembly was performed using Phrap following the masking of highly repeated sequences that were found to inhibit assembly. Two such examples of repeats were a 100-nt-long anonymous interspersed repeat and a contaminant bacterial tetracycline resistance gene that was enriched due to the tendency of the WGA method to preferentially amplify small circular molecules.
Sequence Analyses
For homologue searches, all contigs and singlets were used as queries to search the nonredundant protein and nucleotide databases at the National Center for Biotechnology Information using Blast algorithms. Ribosomal RNA (rRNA) genes were identified by BlastN searches using other apicomplexan rRNA genes as queries. The DNA region between the 28S- and 18S-coding region was subjected to Rfam (http://rfam.janelia.org) to define the boundaries to the 5S RNA–coding region. Repeats within individual sequences were identified with Tandem Repeats Finder (http://tandem.bu.edu/trf/trf.html). The transfer RNA (tRNA) genes were mapped using a local version of the tRNAscan-SE program (http://lowelab.ucsc.edu/tRNAscan-SE/) with various search modes and the final number of 1,235 was based on the eukaryotic mode. Introns within regions yielding Blast hits were predicted using GENEWISE (Birney et al. 2004).
Phylogenetic Reconstructions
To evaluate the phylogenetic position of Ascogregarina, all contigs were used as queries to identify overlapping gene orthologs in the partially sequenced P. marinus genome, as a first step in the creation of a concatenated data set. These orthologs, typically fragmented in either Ascogregarina or Perkinsus, were then used as queries to search for orthologs from other available alveolates. A total of 66 protein fragments shared among 14 alveolates were recovered and individually aligned using a ClustalW algorithm embedded in the MacVector v10.6 program (MacVector, Inc.), followed by visual inspection to correct apparent alignment errors and remove gapped and ambiguous positions. These alignments were then joined to form a concatenated data set containing 10,753 amino acid (aa) positions for subsequent phylogenetic reconstructions by Bayesian inference (BI) (Huelsenbeck and Ronquist 2001), maximum likelihood (ML), and a site-heterogeneous mixture model (CAT) (Lartillot and Philippe 2004).
BI analysis used a parallel version of MrBayes program for Apple PowerPC (version 3.1.2) (http://mrbayes.csit.fsu.edu/) under the Pooch environment for high performance parallel computing (http://www.daugerresearch.com/). The program was allowed to “jump” among 10 amino acid substitution models implanted within the program. Among-site rate heterogeneity considered four-rate Γ-distribution [Γ(4)] and the fraction of invariance (Finv). In the present study, all posterior probability (PP) values in the final trees were derived from the RTrev model (Dimmic et al. 2002). A typical analysis consisted of at least 1 × 106 generations of searches with two independent runs, each containing four chains running simultaneously. The current trees were saved every 1,000 generations. PP values were obtained by calculating consensus trees from the last 751 BI trees (of a total of 1,001 trees) that were obtained after the runs converged. ML bootstrapping analysis from 400 replicates was performed using the Treefinder program (version October 2008) (http://www.treefinder.de) with a RTrev + Γ(4) + Finv model of amino acid substitutions. Site-heterogeneous CAT model analysis used PhyloBayes program (version 3.1f) (http://www.phylobayes.org) (Lartillot et al. 2009). Using CAT + Γ, four independent Markov chains were run under default settings for at least 30,000 cycles with trees saved in each cycle. Posterior analysis discarded data generated in the first 8,000 cycles and sampled remaining trees at a frequency of 1 for every 10 cycles. The quality of BI and CAT analyses was evaluated by the trends of their ML values, uncorrected potential scale reduction factor (PDRF) (in MrBayes), and the largest (maxdiff) and mean (meandiff) discrepancy observed across all bipartitions (in PhyloBayes).
In addition, trees were also inferred from a large subunit (LSU) rRNA data set under CAT + Γ (>40,000 cycles under default settings with PhyloBayes) or under a general time reversal (GTR + Γ(4) + Finv) model with BI (2 runs, 4 chains, 5,000,000 generations of searches with MrBayes) and ML (bootstrap analysis from 1,000 replicates with Treefinder). Under BI and CAT, consensus trees were obtained after the first 25% of trees were discarded.
The α- and β-tubulin trees were inferred from corresponding protein sequences using BI (MrBayes, 2,000,000 generations of runs) and ML bootstrapping analysis with 100 replicates (ProML program in the PHYLIP package) using the same amino acid substitution described above and rate heterogeneity models. For plant and bacterial-type genes, only the best protein ML trees were computed using ProML program for eight representative genes to illustrate their unique phylogenetic affinities. These trees are provided in Supplementary Material online with additional details described in the legends (supplementary fig. S1, Supplementary Material online).
Results and Discussion
General Analysis of the GSS Database and Estimate of Genome Size
Genome studies of gregarines are hampered by the inability to culture the parasites and the paucity of material that is obtainable from infected hosts. For this reason, we utilized WGA followed by GSS in order to characterize the genome for A. taiwanensis. We obtained roughly 23,000 sequence reads, of which 20,165 reads were assembled into 3,434 contigs and 2,936 singlets (http://cryptogenome.umn.edu:3300), yielding 6.15-Mb nucleotide sequence information (GENBANK accession ABJQ00000000). The remaining reads were not included in the final Phrap assembly and were comprised largely of two repeats and a 12-kb bacterial plasmid contamination. The latter plasmid contaminant was likely due to amplification by the multiple displacement amplification methodology, which increases the abundance of small circular molecules at the expense of linear termini. In summary, approximately 6 Mb of A. taiwanensis genome-sequence information was obtained, with a median contig size of 1,131 nt and 49% G + C (table 1). The G + C content is considerably higher than that found for Cryptosporidium parvum (30%) and Plasmodium falciparum (19%).
Table 1.
Summary of the Ascogregarina taiwanensis Genome Survey Data.
| Sequence Information | |
| Contigs | 3,434 Contigs; 4,727,332 nt |
| Singlets | 1,945 Singlets; 1,422,079 nt |
| Median contig size | 1,131 nt |
| Largest contig | 11,727 nt |
| Total sequence | 6,149,411 nt |
| G/C content | 49% |
| Sequence quality | |
| Possible mosquito contaminant | 0 rRNA; 0 retrotransposons |
| Possible bacterial contaminant | 0.8% (contigs); 4% (singlets) |
| Information content | |
| rRNA | 21 Contigs |
| tRNA | 121 Contigs |
| Retrotransposons | 124 Contigs |
| XBlast hits | 1,340 Contigs; 1,250 singlets (Blast score ≥ 10) |
| Intron-containing sequences | 743 Contigs; 574 singlets |
| ORFs | 3,433 Contigs (stop to stop > 40 amino acids) |
| Median intron size | 62 nt (range: 13–1,010 nt) |
| Intergenic region | 69–300 nt between tRNA (e.g., 275 and 276 nt between α-tubulins) |
Note.—nt: nucleotides; XBlast: search against the NR database 12/07, E value cutoff = 5; introns: GENEWISE2.2 with GT/AG as the only requirement; possible bacterial contaminants: >75% nucleotide identity over >200-nt region.
Because the A. taiwanensis material was purified from mosquito larvae, we were concerned about host tissue and midgut bacterial contamination. Mosquito contamination was estimated to be absent, because we did not find high-copy mosquito sequences such as rRNA or retrotransposons, the latter of which comprises as much as half of the mosquito genome. Bacterial contamination was also limited and included the 12-kb plasmid described above, rRNA, tRNA, and bacterial sequences. We estimate that bacterial contamination within the assembly is roughly 0.8% of contig DNA and 4% of the singlets and is readily identified and culled by evaluation of Blast hits.
An estimate of the size of the A. taiwanensis genome was determined by assessing the coverage of the ribosomal proteins and tRNA synthetase genes obtained by the survey (see below for a more detailed description of these genes) and extrapolation based upon the size of the GSS database. Thus, sequences for approximately 25% of the ribosomal proteins were identified, and roughly 5 Mb of assembled sequence was obtained, giving an estimate of at least 20 Mb for the A. taiwanensis genome. Molecular karyotype analysis was not performed due to the technical difficulties in obtaining sufficient purified Ascogregarina oocysts from infected mosquitoes. This minimal estimate of 20 Mb compares with ∼10 Mb in Theileria and Cryptosporidium, ∼22 Mb in Plasmodium, and ∼85 Mb in Toxoplasma.
Introns are found in apicomplexan genomes to varying degrees (supplementary table S1, Supplementary Materials online), although the average size and frequency in A. taiwanensis was not determined by the survey due to the lack of a sufficient number of matches between the GSS database and the available EST sequences in the public databases. To examine the genome of Ascogregarina, we relied upon Blast analysis of open reading frames (ORFs) and did not utilize gene-prediction programs. Blast hits were passed into GENEWISE to define introns yielding 1,804 introns in 55% of the Blast hit contigs, which suggested that roughly 55% of the genes have introns. The 55% estimate is likely low, as this search is dependent on good protein sequence matches. Consistent with an intron-rich genome, many genes had multiple introns and compilation of the splice junctions did not reveal preferences beyond the minimal GT/AG consensus sequence. The introns that were identified ranged up to 1,010 nt in length with a median size of 62 nt. Most of the contigs were short and encoded fragments of genes and were not useful for estimating gene spacing or gene density. Examples of intergenic distances include the tandem gene arrays of α-tubulin and tRNAs; specifically, 275 and 276 nt between the α-tubulins and 69–300 nt between the tRNA array.
The Ascogregarina sequences were compared with the 1,900 ESTs from Gregarina niphandrodes in the GenBank database. Only one transcript was identified with nucleotide similarity, C0636099, and had 74% nucleotide identities in the 28S rRNA sequence. Seven additional genes had amino acid sequence similarity, namely, ribonucleotide reductase, histone H2A, histone H2B, histone H4, ornithine-oxo-acid aminotransferase, tubulin β, and tubulin α. Additional searches against the NR database identified hits to DNA-dependent RNA polymerase subunit II from G. niphandrodes; and actin, myosin A, and myosin B from Gregarina plymorpha. All the protein-sequence similarities to gregarines were not best hits except for histone H3, RNA polymerase, and actin, suggesting that the Ascogregarina and Gregarina might exhibit great divergence. Following Blast analysis of GenBank, the majority of the best hits were against Cryptosporidium (310 best hits) versus Toxoplasma gondii (118 best hits) and P. falciparum (120 best hits). For comparison, there are only 13 top hits to human genes, 19 to Saccharomyces, 56 to Arabidopsis thaliana, and more significantly, only 12 top hits to proteins of the mosquito Aedes aegypti. These observations support an apicomplexan-type genome in A. taiwanensis with an apparent affinity to Cryptosporidium, as discussed in more detail in the following phylogenetic analysis.
Phylogenetic Position of Ascogregarina Inferred from Concatenated Protein Sequences and the LSU rRNA Genes
To confirm the relationship of Ascogregarina and Cryptosporidium with other apicomplexans, we established a large protein data set containing 10,753 concatenated aa positions that were recovered from 66 diverse gene fragments present in 14 partially or completely sequenced alveolate genomes. The gene fragments comprised, for example, ribosomal protein subunits, DNA and RNA polymerase subunits, DNA–RNA helicases, tRNA synthetases, various metabolic enzymes, protein kinases, and heat-shock proteins. This large data set permitted more robust tree reconstructions than a previously performed analysis based solely on SSU rRNA genes (Carreno et al. 1999).
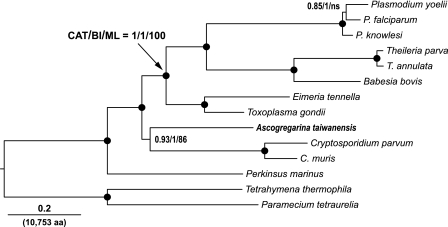
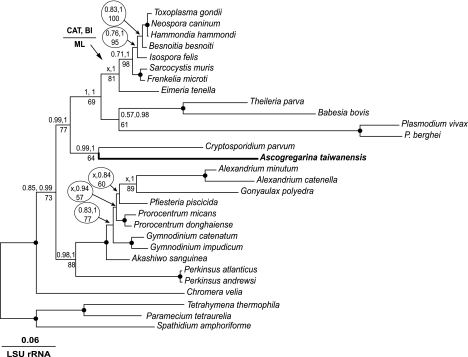
Trees inferred from this data set under three models CAT, BI, and ML displayed the same tree topology that united all apicomplexans as a monophyletic group, in which Ascogregarina and Cryptosporidium were grouped together at the base of the clade (fig. 1). As anticipated, Perkinsus was placed intermediate between the apicomplexans and ciliates. The tree was highly robust and, with the exception of two nodes, was supported by 100% bootstrap proportion (BP) and PP values. The monophyletic relationship between Ascogregarina and Cryptosporidium was uniformly supported by CAT (PP = 0.9), BI (PP = 1.0), and ML (BP = 86%). The high PP value by CAT is notable, as this site-heterogeneous model is well known to have a greater advantage than homogenous models in suppressing long-branch attraction artifacts (Lartillot et al. 2007). The phylogenetic affinity between Ascogregarina and Cryptosporidium was also firmly supported in LSU rRNA trees by CAT and BI (PP values = 0.99 and 1.0, respectively) and moderately by ML (BP = 64%) (fig. 2). An identical topology was also observed after the haematozoan sequences were removed in order to reduce possible long-branch effect (data not shown). The monophyly between the gregarine and Cryptosporidium observed in the concatenated protein tree and the LSU rRNA tree is in agreement with previous studies that used SSU rRNA and conserved proteins (Carreno et al. 1999; Zhu et al. 2000; Leander et al. 2003), but the present trees have much greater PP and bootstrap supports.
FIG. 1.
Phylogenetic relationships of the alveolates were inferred from concatenated protein sequences containing 10,753 amino acid positions by BI, ML and, exemplified here, a site-heterogeneous model (CAT). Under CAT, PP values were summarized from 2,400 trees after runs converged (sampled once per 100 cycles from a total number of >30,000 cycles for each of the four chains. Maxdiff = 0.0756 and meandiff = 0.0018). In BI analysis, PP values were derived from 751 trees after runs converged (sampled once per 1,000 generations from a total number of 1,000,000 generations of run for each of the four chains. Mean of the TL parameter = 3.259 with an uncorrected PDRF of 1.007). The RTrev amino acid substitution model contributed 100% to PP values. The BPs in the ML analysis were computed from 400 replicates. Solid circles indicate the nodes that are 100% supported by all three methods. ns indicates not supported under the majority ruling law.
FIG. 2.
Phylogenetic affinity between Ascogregarina and Cryptosporidium as inferred from LSU rRNA genes (29 taxa and 1,971-nt positions) using BI with MrBayes, ML bootstrapping analysis with Treefinder and site-heterogeneous CAT with PhyloBayes methods. Both BI and ML analyses used a GTR model with the consideration of four-rate gamma and the faction of invariance and yielded the same topology. The CAT method produced a similar topology except that the relationships of some species within the apicomplexan and dinoflagellate clusters were not fully resolved under the majority ruling law (as indicated by an “x”). Solid circles indicate nodes that are supported by all three methods at 95% level or higher.
Characterization of the Major Metabolic Pathways of Ascogregarina
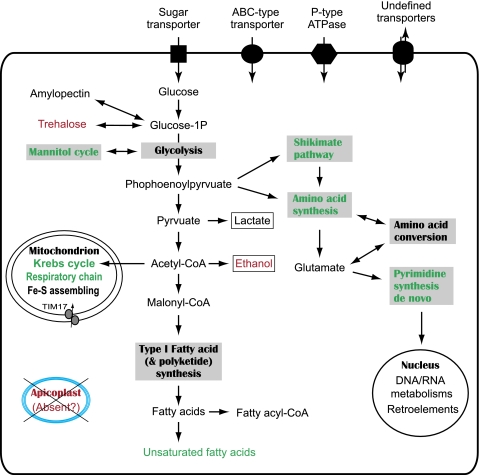
Numerous new genes encoding metabolic enzymes have been discovered by the GSS project (see supplementary table S1, Supplementary Material online, for a partial list), and this annotation provides a good snapshot with which to predict the major metabolic pathways in this gregarine (fig. 3). Among the Apicomplexa, Cryptosporidium is highly polar in the degree of its reduction in mitochondrion function and, indeed, it appears to altogether lack respiratory functions. In contrast, the Ascogregarina genome survey identified a number of mitochondrion-specific proteins and enzymes (table 2). These include genes encoding the Krebs cycle enzymes, components of the oxidative respiratory chain complexes, ferredoxin oxidoreductase, mitochondrial carrier proteins, chaperones, and a mitochondrial import apparatus; the sum of which is in agreement with the presence of typical mitochondrial organelles that have been observed in various gregarinal species (Chen et al. 1997; Toso and Omoto 2007). The presence of a Krebs cycle and respiratory chain suggests that gregarines are capable of generating energy via a complete oxidative pathway, similar to most other apicomplexans but differ from Cryptosporidium that solely relies on glycolysis (Thompson et al. 2005). Ascogregarina also possesses a cyanine-resistant mitochondrial alternative oxidase (AOX) that is present in Cryptosporidium (Abrahamsen et al. 2004) and ciliates but is absent from Toxoplasma and other apicomplexans for which whole-genome-sequence information is available. The survey did not detect the mitochondrial genome, but this is not unexpected because in other apicomplexans, it is typically present as small 5- to 6-kb linear elements, and thus, identification of a predicted genome awaits further studies.
FIG. 3.
The proposed major metabolic pathways in Ascogregarina taiwanensis as determined by annotations of the GSS. Pathways and features that are shared with Cryptosporidium to the exclusion of other apicomplexans are shown in red, whereas those shared with other apicomplexans to the exclusion of Cryptosporidium are shown in green. Two organic end products in the glycolytic pathway are boxed.
Table 2.
Putative Mitochondrial Proteins in Ascogregarina taiwanensis with Apparent Apicomplexan Affinity.
| Description | Contig or Singlet No. |
| Heat shock protein 60 (HSP60) | 3152 |
| Chaperon protein J (DnaJ homolog) | 1746; 2146 |
| Cytochrome C | 2828 |
| Cytochrome C oxidase subunit II | 1722 |
| Cytochrome C1, heme protein | CLRanP155-B03.b1.ab1; CLRanP155-B03.g1.ab1 |
| ATP synthase F1, β-subunit | 1526; CLRanP74-B10.g1.ab1 |
| ATPase subunit 9 | 2586 |
| Mitochondrial solute carrier | 2033 |
| Mitochondrial carrier protein | CLRanP94-G03.g1.ab1; CLEcoP1-A05.g1.033.ab1 |
| Mitochondrial import inner membrane translocase subunit | 747; 1433 |
| NAD(P) transhydrogenase | 3557 |
| Alternative NADH dehydrogenase 2 | CLRanP154-A08.b1.ab1 |
| Dihydrolipoamide dehydrogenase | CLRanP168-G08.g1.ab1 |
| Respiratory chain assembly protein (AFG3) | 3591 |
| GDP-forming succinate-CoA ligase α-subunit | 3091 |
| Cysteine desulfurase (nifS-like) | 474 |
| Mitochondrial Lon protease homolog 2 | 1730 |
| Mitochondrial ribosomal protein S15 | 1163 |
| Carbamoyl-phosphate synthetase II | 1867 |
| Branched-chain α-keto acid dehydrogenase E1 | CLRanP79-C01.g1.ab1 |
| 2-Oxoglutarate dehydrogenase e1 component | LRanP168-G08.g1.ab1 |
| Dihydrolipoamide succinyltransferase component | CLRanP117-G11.b1.ab1 |
| Mitochondrial succinate dehydrogenase flavoprotein subunit | CLRanP141-B09.b1.ab1 |
| Mitochondrial succinate dehydrogenase iron–sulfur subunit | CLRanP110-H04.g1.ab1 |
| Prohibitin | CLRanP154-G11.g1.ab1 |
| AOX | CLRANP132-G11.g1.ab1 |
Cryptosporidium possesses a highly streamlined repertoire of metabolic pathways and is largely devoid of metabolic functions beyond the core glycolytic enzymes, likely the result of its specialized parasitism of gut epithelial cells (Abrahamsen et al. 2004). Contrary to an expectation of similar streamlining in Ascogregarina, it instead appears to have a large metabolic repertoire. Ascogregarina is capable of synthesizing some amino acids de novo or from intermediates, including glutamate, alanine, aspartate, cysteine, phenylalanine, tyrosine, and tryptophan. Enzymes in the shikimate pathway are also present in the genome, which include 3-deoxy-7-phosphoheptulonate synthase, 3-dehydroquinate synthase, shikimate kinase, 5-enolpyruvylshikimate-3-phosphate synthase, and chorismate synthase. Ascogregarina possesses enzymes responsible for de novo synthesis of pyrimidines (e.g., orthologs for carbamoyl-phosphate synthase, dihydroorotate dehydrogenase, and orotate phosphoribosyl-transferase) and probably purines from glutamate (e.g., orthologs for phosphoribosyl-glycinamide formyltransferase, adenylosuccinate lyase, and bifunctional phosphoribosyl-aminoimidazole carboxamide formyltransferase/inosine monophosphate cyclohydrolase). The former pathway is present in all apicomplexans except for Cryptosporidium, whereas the latter is unique and has not yet been seen in other apicomplexans. We identified genes that are related to the de novo synthesis of purines, but they appear to be of bacterial origin, and further experiments are needed to determine if they represent contaminants or are examples of lateral gene transfer.
Like Cryptosporidium and Toxoplasma, A. taiwanensis possesses a Type I fatty acid synthase and/or a polyketide synthase (Zhu 2004). Other enzymes involved in fatty acid metabolism include a long chain fatty acid elongase, a fatty acid desaturase, and an array of long chain fatty acid–CoA ligases, which suggest that this gregarine might be unable to synthesize fatty acids de novo but is capable of elongating fatty acids and making unsaturated fatty acids. Another surprising feature is that A. taiwanensis possesses mannitol-1-phosphate dehydrogenase and trehalose-6-phosphate synthase (T6PS), suggesting that the mannitol cycle and trehalose syntheses coexist in this apicomplexan. Among other apicomplexans, the mannitol cycle is only seen in Eimeria (Schmatz 1997), whereas trehalose synthesis is found in Cryptosporidium, Toxoplasma, and piroplasmids (Theileria and Babesia) based on the presence of T6PS orthologs in their genomes. Ascogregarina is also likely capable of using trehalose by virtue of possessing a trehalose-6-phosphatase that is absent in other apicomplexans. Moreover, the presence of enzymes for producing lactate and ethanol as organic end products suggests that glycolysis and fermentation may play an important role in the energy metabolism in this gregarine. Among apicomplexans, the ability to produce ethanol is otherwise only present in Cryptosporidium (Abrahamsen et al. 2004; Thompson et al. 2005), and thus, this capacity is shared with Ascogregarina. In summary, although both Cryptosporidium and Ascogregarina are monoxenous and parasitize gut epithelial cells, Ascogregarina apparently has a much broader metabolic capacity. It is possible that the metabolic robustness of Ascogregarina reflects plesiomorphy within the Apicomplexa, and in its parasitic niche, it has not undergone specialization resulting in loss of, say, either the mannitol cycle or trehalose synthesis as have other apicomplexans.
Unusual Tubulin Genes in Ascogregarina
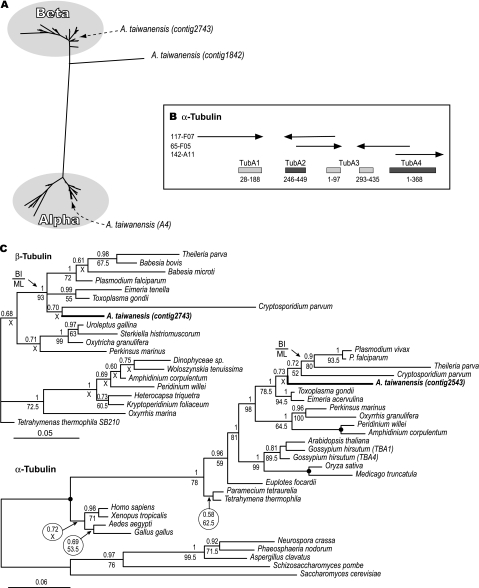
The genome survey identified two β-tubulin isoforms that shared 54% amino acid identity but were highly divergent (75% and 50% amino acid identity, respectively) from a G. niphandrodes β-tubulin (CO636144). One β-tubulin gene (contig 2743) encodes a typical apicomplexan protein that is phylogenetically clustered together with the Cryptosporidium ortholog (fig. 4). However, the phylogenetic position of the second one (contig 1842) could not be firmly determined. It was clearly placed at the base of the β-subunit clade in trees containing α- and β-subunits (fig. 4A and C) or as a long branch within the β-subunit clade when α-, β-, γ-, and ϵ-tubulins from animals, plants, and protists were used in the analysis (data not shown). These observations indicate that Ascogregarina possesses a novel type of β-tubulin that is under specific selection pressure. At least, five α-tubulin isoforms were identified that came from three plasmid clones, which allowed deduction of the arrangement of the α-tubulins in the genome as a tandem array (fig. 4B). The tandem arrangement is an anomaly among the Apicomplexa but is often the rule in other eukaryotes. Among other apicomplexans, P. falciparum and T. parva have two genes on two chromosomes; T. gondii has three genes on two chromosomes with the two closest genes separated by 100 kb, whereas Cryptosporidium has only one gene. The gregarinal genes encode α-tubulin proteins that share highest identities with apicomplexan orthologs. The phylogeny of α-tubulins is less resolved at major taxonomic levels, and the isoform A4 in Ascogregarina was placed within the apicomplexan clade (fig. 4C).
FIG. 4.
Phylogeny and gene arrangement of Ascogregarina tubulins. (A) Phylogenetic relationship of α- and β-tubulin protein sequences from 63 taxa and 335 amino acid positions as determined by BI method. The typical Ascogregarina α- and β-tubulins were placed within their corresponding groups, whereas the unusual β-tubulin was placed at the base of the beta cluster. (B) Diagram of α-tubulin gene cluster. The arrows indicate the sequence reads and the template name as indicated in the left. The arrows and boxes are not drawn to scale, and the sizes of the gaps in the sequence are not mapped. (C) Phylogenetic positions of the Ascogregarina taiwanensis typical β-tubulin (contig 2743) and α-tubulins (contig 2543) as determined by the BI and ML methods. A solid circle indicates the node that is 100% supported by both PP and BP, whereas an “x” indicates the node with <50% bootstrap support.
Ascogregarina rRNA, tRNA, and tRNA Synthetase Genes
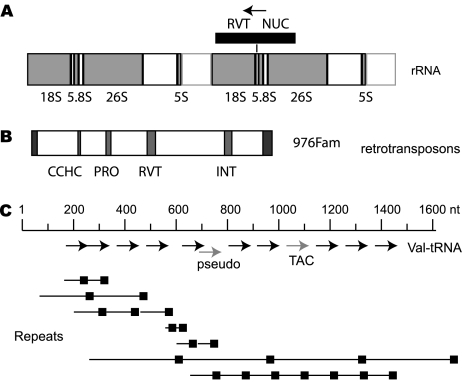
We examined the Ascogregarina rRNA genes in order to, firstly, ascertain the degree of contamination with mosquito and bacterial DNA and, secondly, to determine the arrangement of rRNA loci in the genome. As described above, only one bacterial rRNA sequence was identified, and no mosquito rRNA sequences, indicating that the GSS database harbored little contamination. The internal spacer region of rRNA corresponded to a known A. taiwanensis sequence, and no sequences in the GSS database were suggestive of coinfection by a closely related Ascogregarina species. Eukaryotes maintain large tandem repeats of rRNA genes and apicomplexans such as Plasmodium and Cryptosporidium have rRNA genes as dispersed telomeric repeats. The rRNA gene of A. taiwanensis was arranged as a 7-kb unit of 18S–5.8S–26S–5S (fig. 5A). The external spacer between 26S and 18S RNA–coding region contained 5S genes and two types of tandem repeats, nine copies of 40-nt repeats followed by seven copies of 35-nt repeats, presumed to function as the RNA polymerase I enhancer elements. Blast searches with the 5S sequence did not reveal any other 5S RNA sequences in the GSS database. Few exceptions to this organization were found, such as inverted repeats or many unique flanking sequences, suggesting that the rRNA genes are in a tandem array like a typical eukaryote. One exception was an insertion of a 3.3-kb sequence (Asta522Fam) inserted 18 nt from the 3′ end of the 18S-coding region (fig. 5A, right-hand panel). The Asta522Fam contained one ORF that encodes domains of reverse transcriptase and exonuclease and was similar to a LINE element. The same Asta522Fam sequence was also present in other contigs without the rRNA sequences indicating that this sequence is a dispersed retroelement. The presence of the Asta522Fam insertion in the rRNA sequence in the genome was confirmed by specific polymerase chain reaction amplification and sequencing.
FIG. 5.
Schematics of rRNA, tRNA, and retrotransposons. (A) Schematic of two tandem copies of the rRNA genes. The direction of the transcript is left to right. The insertion point of the retroelement 522fam is shown on the right-hand copy. The 522fam element has one ORF (as indicated by the arrow) including a nuclease domain, NUC, and a reverse transcriptase domain, RVT. (B) The gypsy/ty3 retroelement. The 976Fam element contains one contiguous ORF including a CCHC-type zinc finger (CCHC) domain; asparatic protease domain, PRO; reverse transcriptase domain, RVT; and an integrase/RNase H domain, INT. This element is bounded by 100-nt direct repeats. (C) Schematic of an array of tRNA genes. Each arrow represents a valine–tRNA gene. All tRNA found in this array had anticodon AAC except for two as indicated. Each line with a terminal box indicates tandem repeats found in this genomic segment.
The algorithm tRNAscan-SE identified 1,235 tRNA genes, in stark contrast to the 45 genes that were found in the completed nucleotide sequence of C. parvum. These tRNA genes were repeated 2–67 times (fig. 5C), a feature also found in Entamoeba histolytica. These tRNAs are unlikely to be SINE or LINE elements as the tRNAscan-SE is designed to discriminate against such pseudo-tRNAs. Moreover, the tRNA did not contain a 3′ CCA sequence, a signature of reverse transcriptase inception, and the tandem repeat analysis of the tRNA cluster failed to define a unit repeat consisting of tRNA plus flanking sequences common to the entire set. The tRNA synthetase genes for 7 of the 20 amino acids, phenylalanine (α- and β-subunits), glutamine, alanine, aspartate, lysine, valine, and tryptophan were found and all have introns.
Ascogregarina Possesses a Number of Retrotransposons
Four distinct families of retrotransposable elements distributed in 11 contigs were identified in the Ascogregarina GSS database, and the sequence and structure of three of these elements were determined. The retroelements were confirmed to be of gregarinal origin and could be amplified from DNA that was isolated from purified oocysts or from mosquitoes infected with Ascogregarina but not from DNA that was isolated from uninfected mosquitoes (data not shown). Two elements are characteristic of the gypsy/ty3 family (Asta976Fam), and at least one appears to be active because it contains a 5.5-kb-long uninterrupted ORF for a gag-pol gene that encodes protease, reverse transcriptase, and integrase domains and is bounded by a 100-nt repeat (fig. 5B). One subset of the ribosomal genes has an 18S-coding region that is immediately followed by a retrotransposon. Among other apicomplexan genomes, only Eimeria has been reported to possess a reverse transcriptase (XP_001238615) associated with retrotransposons and retroviruses with long terminal repeats (Ling et al. 2007). It is likely that retroelements are absent in Haematozoa and Toxoplasma, and the few singlets containing partial retroelements that are present in the Plasmodium (e.g., PY07375) and Toxoplasma (e.g., 322.m00001 in contig TGG_994894) genome-sequence databases (ToxoDB.org and PlasmoDB.org, respectively) are contaminants, based on their sequence identities to gene fragments from mouse chromosomes 1 and 5 (i.e., GenBank # AC132854 and AC158548) or Brugia malayi (i.e, GenBank # DS237737), respectively. It is unclear if the sporadic presence of retroelements in the Apicomplexa, namely, within Eimeria and gregarines, is due to single or multiple examples of lineage-specific acquisition, or is due to widespread loss.
Plant- and Bacterial-Type Genes
Like Cryptosporidium and other apicomplexans (Striepen et al. 2004; Templeton et al. 2004), the A. taiwanensis genome possesses an expanded number of genes of predicted plant or bacterial origins. ML trees were derived from the sequences with reasonable lengths for effective phylogenetic analysis and are provided here as supplementary materials (supplementary fig. S1, Supplementary Material online). Examples of plant-type genes include α-glucan water kinase isoform 3, trehalose-6P synthase, NAD(P)H-dependent glutamate synthase (GS), aspartate aminotransferase, aspartate kinase (AspK) and a class III alcohol dehydrogenase. Among the bacterial-type genes, a few share extremely high identities to the bacterial orthologs at both protein and nucleotide levels, particularly to those from some α- and β-proteobacteria (e.g., contig 585 encoding a methylmalonyl-CoA mutase small subunit and two singlets encoding cytochrome C oxidase subunits I and III, respectively [vs. contig 1722 encoding cytochrome C oxidase subunit IIa that shares highest similarity to T. gondii ortholog]). Whether these genes are truly of Ascogregarina origin, or represent bacterial contaminants, requires experimental validation, but many of other genes share only moderate similarities to the bacterial homologues and/or significant identities to other apicomplexan orthologs, indicating that most of them are likely present within the Ascogregarina genome. Moreover, some bacterial-type genes possess apparent introns (e.g., contigs 2623, 3330, and 1226 that, respectively, encode homologues of phospho-2-dehydro-3-deoxyheptonate aldolase, propionyl-CoA carboxylase and a short chain dehydrogenase (see supplementary fig. S2, Supplementary Material online), indicative of a eukaryotic provenance rather than bacterial contamination. Additionally, we have observed that the base compositions and codon usages of the bacterial- and plant-type genes are not highly biased from the approximately 50% G + C content calculated from well-defined coding sequences within 101 gene fragments extracted from the GSS (see table 3 for a brief summary and supplementary table S2, Supplementary Material online, for details).
Table 3.
Base Composition and Codon Usage of Selected Genes with Bacterial or Plant Affinitya.
| Genes | No. Bases Analyzed | G/C Content (%) | G/C at Third Codon Positions (%) |
| Overall in Ascogregarina | 70,401 | 50.3 | 53.3 |
| Selected bacterial-type genesb | |||
| Alcohol dehydrogenase type E (adhE) | 573 | 49.5 | 51.2 |
| Phospho-2-dehydro-3-deoxyheptonate aldolase | 681 | 51.1 | 48.8 |
| Amidotransferase class-I anthranilate synthase (GAT1-AS) | 534 | 50.7 | 49.3 |
| TS | 1,059 | 51.9 | 51.9 |
| I3GPS | 501 | 46.4 | 48.6 |
| D-LDH | 279 | 49.5 | 53.3 |
| Overall in bacterial-type genes | 3,627 | 50.3 | 50.5 |
| Selected plant-type genesb | |||
| AspK | 783 | 51.4 | 57.8 |
| T6PS | 1,455 | 50.6 | 51.1 |
| GS | 876 | 50.7 | 50.1 |
| α-Glucan water kinase | 255 | 48.6 | 57.9 |
| Overall in plant-type genes | 3,369 | 50.7 | 52.9 |
Overall in Ascogregarina genome samples are calculated from the coding regions extracted from 101 gene fragments that encode highly conserved protein sequences to avoid possible ambiguity in defining codons. Please see supplementary table S2, Supplementary Material online, for details including compositions of individual bases, individual codon usages (frequencies).
The bacterial and plant-type genes are those with ML phylogenetic trees given in supplementary figure S1, Supplementary Material online.
Other bacterial-type genes include some of the enzymes involved in 1) glycolysis and fermentation, such as the type E alcohol dehydrogenase (adhE) and lactate dehydrogenase (LDH); 2) amino acid biosynthesis, including aspartate aminotransferase, glutamate synthetase, serine dehydratase, cysteine synthetase, and tryptophan synthase (TS); 3) purine biosynthesis; and 4) nutrient transporters, for example, several members of the ABC-type sugar transporters. One class of dehydrogenases shared a high number of bacterial homologues (supplementary table S1, Supplementary Material online), but it must be ruled out that these are not the results of bacterial contaminations. Plant and bacterial-type genes are commonly found in other apicomplexans including Cryptosporidium (Abrahamsen et al. 2004; Huang et al. 2004); the former likely originated from the ancient nuclear transfer of plastid genes (Huang et al. 2004), whereas the latter are suggestive of lateral gene transfer.
Ascogregarina possesses several bacterial-type enzymes indicating its capacity to synthesize tryptophan from chorismate, a pathway that among eukaryotes has only been described in plants and fungi. Specifically, gene fragments encoding glutamine amidotransferase class-I anthranilate synthase (GAT1-AS, contig 3220 and CLRanP166-H10), indole-3-glycerol-phosphate synthase (I3GP, contig 3208), and TS β-subunit (contig 652) were identified in the A. taiwanensis genome. These genes contained introns, indicating that they are unlikely to be bacterial contamination and do not resemble those found in fungi or plants. Among the alveolates, Tetrahymena and Paramecium lack the pathway, Plasmodium, Theileria, Babesia, and Toxoplasma have putative GAT1-AS (annotated as para-aminobenzoate [PABA] synthetase in some entries in the GenBank database) and I3GP-domain containing genes, whereas Cryptosporidium has only TS. GAT1-AS produces anthranilate and PABA from chorismate and glutamine, which differs from the “classic” AS (EC 4.1.3.27) that produces anthranilate and pyruvate from chorismate and ammonia. The fragmented distribution of the tryptophan synthesis pathway among the apicomplexans suggests that this pathway was either acquired early in the apicomplexan lineage and subsequently underwent lineage-specific gene loss, or that the individual enzymes were independently acquired multiple times. ML trees inferred from available amino acid positions support the bacterial affinity of these three enzymes from all apicomplexans but always separate the Ascogregarina orthologs from other apicomplexans as individual branches (supplementary fig. S1D–S1F, Supplementary Material online). Among other apicomplexans, Toxoplasma and Haematozoa were united together in the I3GPS tree (supplementary fig. S1E, Supplementary Material online) but separated from each other in the GAT1-AS tree (supplementary fig. S1F, Supplementary Material online). Thus, the apicomplexan tryptophan synthesis pathway appears complex, with possible vertical inheritance of some enzymes in some lineages, orthologous gene replacement of others, and perhaps examples of lineage-specific gene loss.
Extracellular Proteins That Are Shared with Other Apicomplexans
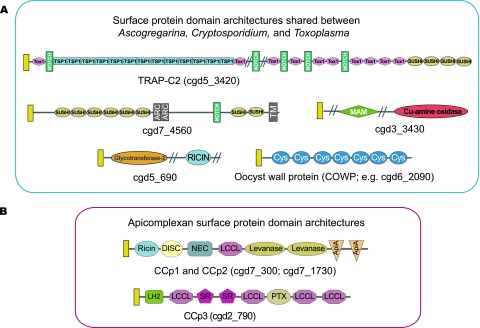
The Apicomplexa are unusual among the protozoans in that they posses repertoires of extracellular proteins with complex multidomain architectures that are reminiscent of the complexity that is found in metazoan extracellular proteins (Templeton et al. 2004; Templeton 2006, 2007; Anantharaman et al. 2007). To characterize the extracellular repertoire of Ascogregarina, we screened the GSS database for known apicomplexan extracellular proteins and domains and in particular comparison with the extracellular protein repertoire of Cryptosporidium. No Cryptosporidium extracellular proteins were found that were conserved in Ascogregarina to the exclusion of Toxoplasma, whereas numerous proteins that are conserved in Cryptosporidium and the Coccidia were also found in this gregarine (fig. 6A). For example, the MAM and Notch domains are found only in coccidians, Cryptosporidium and Ascogregarina, but are absent in Plasmodium and Theileria, both genera for which complete genome-sequence information is available. This supports the hypothesis that Cryptosporidium and Ascogregarina share extracellular proteins that group them within a “coccidian” natural history, and accordingly, perhaps these proteins function during the excreted oocyst stages. Survey of Ascogregarina also revealed two multidomain extracellular proteins that are likely conserved throughout the apicomplexan clade, namely, members of the LCCL domain–containing family which include in Cryptosporidium cgd7_300, cgd7_1730, and cgd2_790 (fig. 6B). Thus, the LCCL-domain proteins are perhaps the most conserved extracellular proteins in the apicomplexans in that their complex multidomain architectures arose prior to the split of all extant apicomplexans and their roles are likely similarly conserved in a fundamental aspect of apicomplexan biology. Ascogregarina possesses several predicted secreted proteins that contain repeats of a cysteine-rich domain, termed CPW–WPC, which is found only in the apicomplexan clade. The CPW–WPC domain has not been formally described other than during the course of genome annotations and was first noted as a repeated domain within an amplified protein family in P. falciparum (e.g., PFL1675c) and is conserved in Theileria (e.g., TP04_0183) and Toxoplasma (e.g., 50.m03161). The domain is notably absent in Cryptosporidium, and thus, the presence of CPW–WPC domain proteins in Ascogregarina demonstrates that this parasite harbors widely conserved apicomplexan-specific extracellular proteins to the exclusion of Cryptosporidium.
FIG. 6.
Domain organizations of extracellular proteins with architectures that are shared between Ascogregarina, Cryptosporidium, and Toxoplasma to the exclusion of Plasmodium and Theileria (A) or conserved among all apicomplexans (B). All proteins have a signal peptide, indicated by a yellow box at the amino terminus. Domains present within the protein architectures include: Tox1 (ShKT); Notch (NL); TSP1; Sushi (CCP); ARC; MAM; Cu-amine oxidase (Cu_amine_oxidase); Glycotransferase-1 (glycos_transf_1); Ricin; Cys; Discoidin (F5_F8_type_C); NEC; LCCL; Levanase; ApiECA; LH2 (PLAT_SR); SR; Pentraxin (PTX); and transmembrane domains (TM). Many of these domains are described further at one of the following browsers of domains: http://smart.embl-heidelberg.de/browse.shtml and http://pfam.janelia.org/browse.shtml. Not described in these browsers is the Cys domain found in the highly cysteine-rich coccidian oocyst–wall proteins and the ARC domain that is a small domain with characteristically spaced cysteine residues that is fused to a papain-like protease domain in the secreted protein AF1946 from the archaean, Archaeoglobus fulgidus.
Conclusions
We have performed a GSS for the gregarine, A. taiwanensis, using oocyst stage genomic DNA that was purified from infected mosquito larvae of A. albopictus. The GSS produced approximately 5 Mb of assembled sequences which we predict represents about 25% of an approximately 20-Mb-size genome sequence. Phylogenetic affinities between gregarines and Cryptosporidium at the base of the Apicomplexa were observed with strong BP and PP supports based on a large data set comprised of concatenated protein sequences and LSU rRNA data sets. Annotation of the GSS revealed many “apicomplexan” biological features, as well as indications that this monoxonous parasite has a greater metabolic capacity of synthetic pathways compared with other members of the clade.
Reconstructions of core metabolic pathways included the energy metabolism and biosyntheses of fatty acids, nucleosides, amino acids, and polysaccharides. A. taiwanensis possesses a number of features that are absent in Cryptosporidium but shared with some or many other apicomplexans. For example, the mannitol cycle is only present in the coccidium Eimeria, whereas the shikimate pathway, synthesis of select amino acids, and the de novo pyrimidine synthetic capacity are widespread in apicomplexans except for Cryptosporidium. Thus, the extremely streamlined nature of the Cryptosporidium metabolism, combined with the phylogenetic relationship with the gregarines, indicates that during adaptation Cryptosporidium underwent widespread gene loss. Nonetheless, Ascogregarina and Cryptosporidium possess features that unite them with the Coccidia, including an environmental oocyst stage; a typical gut location in the respective hosts, particularly for sexual development; metabolic pathways such as the Type I fatty acid and polyketide synthetic enzymes; and a number of conserved extracellular-protein-domain architectures. The genome of Ascogregarina notably contains an array of retroelements that are not seen in other apicomplexans other than Eimeria. The acquisition of complete-genome-sequence database for Ascogregarina will undoubtedly provide a rich trove of information with which to compare the metabolic capacities, lineage-specific parasitic adaptations, and gene loss in the apicomplexans.
The lack of an apicoplast has been confirmed in Cryptosporidium (Zhu et al. 2000; Abrahamsen et al. 2004), and organelle loss was recently proposed for another gregarinal species, G. niphandrodes (Toso and Omoto 2007). Our genome survey also failed to detect plastid genome sequences and plastid-associated metabolic pathways, such as those involved in the Type II fatty acid synthesis and isoprenoid metabolism. However, due to the limited breadth of this survey, these negative data are more of not rejecting, rather than confirming, the hypothesis of plastid loss in gregarines. The apicoplast is a significant organelle because it is not only an attractive drug target but is also critical to understanding apicomplexan evolutionary history. It is anticipated that a future completion of the genome sequence for Ascogregarina will provide new insight into the evolutionary history of this unique organelle as well as other important metabolic pathways among apicomplexans.
Supplementary Material
Supplementary tables S1 and S2 and supplementary figures S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
We are very gratefully to Dr Hsuan-Wien Chen for his technical assistance at the early stage of this project. This project is in part supported by grants from National Institute of Allergy and Infectious Diseases at the National Institutes of Health, USA (R01 AI44594 to G.Z.) and National Science Council of Taiwan (NSC96-2320-B-182-016-MY3 to W-J.C.). This Whole Genome Shotgun project has been deposited at DNA Data bank of Japan (DDBJ)/European Molecular Biology Laboratory (EMBL)/Genbank databases under the project accession ABJQ00000000. The version described in this paper is the first version, ABJQ01000000.
References
- Abrahamsen MS, Templeton TJ, Enomoto S, et al. (20 co-authors) Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science. 2004;304:441–445. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- Anantharaman V, Iyer LM, Balaji S, Aravind L. Adhesion molecules and other secreted host-interaction determinants in Apicomplexa: insights from comparative genomics. Int Rev Cytol. 2007;262:1–74. doi: 10.1016/S0074-7696(07)62001-4. [DOI] [PubMed] [Google Scholar]
- Barta JR, Thompson RC. What is Cryptosporidium? Reappraising its biology and phylogenetic affinities. Trends Parasitol. 2006;22:463–468. doi: 10.1016/j.pt.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Birney E, Clamp M, Durbin R. GeneWise and genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreno RA, Martin DS, Barta JR. Cryptosporidium is more closely related to the gregarines than to coccidia as shown by phylogenetic analysis of apicomplexan parasites inferred using small-subunit ribosomal RNA gene sequences. Parasitol Res. 1999;85:899–904. doi: 10.1007/s004360050655. [DOI] [PubMed] [Google Scholar]
- Chen WJ. The life cycle of Ascogregarina taiwanensis (Apicomplexa:Lecudinidae) Parasitol Today. 1999;15:153–156. doi: 10.1016/s0169-4758(99)01418-0. [DOI] [PubMed] [Google Scholar]
- Chen WJ, Chow CY, Wu ST. Ultrastructure of infection, development and gametocyst formation of Ascogregarina taiwanensis (Apicomplexa: Lecudinidae) in its mosquito host, Aedes albopictus (Diptera: Culicidae) J Eukaryot Microbiol. 1997;44:101–108. doi: 10.1111/j.1550-7408.1997.tb05945.x. [DOI] [PubMed] [Google Scholar]
- Chen WJ, Yang CH. Developmental synchrony of Ascogregarina taiwanensis (Apicomplexa: Lecudinidae) within Aedes albopictus (Diptera: Cuclicidae) J Med Entomol. 1996;33:212–215. doi: 10.1093/jmedent/33.2.212. [DOI] [PubMed] [Google Scholar]
- Dimmic MW, Rest JS, Mindell DP, Goldstein D. RArtREV: an amino acid substitution matrix for inference of retrovirus and reverse transcriptase phylogeny. J Mol Evol. 2002;55:65–73. doi: 10.1007/s00239-001-2304-y. [DOI] [PubMed] [Google Scholar]
- Eisen JA, Coyne RS, Wu M, et al. (53 co-authors) Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006;4:e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia JJ, Fukuda T, Becnel JJ. Seasonality, prevalence and pathogenicity of the gregarine Ascogregarina taiwanensis (Apicomplexa: Lecudinidae) in mosquitoes from Florida. J Am Mosq Control Assoc. 1994;10:413–418. [PubMed] [Google Scholar]
- Gardner MJ, Bishop R, Shah T, et al. (44 co-authors) Genome sequence of Theileria parva, a bovine pathogen that transforms lymphocytes. Science. 2005;309:134–137. doi: 10.1126/science.1110439. [DOI] [PubMed] [Google Scholar]
- Gardner MJ, Hall N, Fung E, et al. (45 co-authors) Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CG, Tsai KH, Wu WJ, Chen WJ. Intestinal expression of H+ V-ATPase in the mosquito Aedes albopictus is tightly associated with gregarine infection. J Eukaryot Microbiol. 2006;53:127–135. doi: 10.1111/j.1550-7408.2005.00086.x. [DOI] [PubMed] [Google Scholar]
- Huang J, Mullapudi N, Lancto CA, Scott M, Abrahamsen MS, Kissinger JC. Phylogenomic evidence supports past endosymbiosis, intracellular and horizontal gene transfer in Cryptosporidium parvum. Genome Biol. 2004;5:R88. doi: 10.1186/gb-2004-5-11-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Leander BS, Clopton RE, Keeling PJ. Phylogeny of gregarines (Apicomplexa) as inferred from small-subunit rDNA and beta-tubulin. Int J Syst Evol Microbiol. 2003;53:345–354. doi: 10.1099/ijs.0.02284-0. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Brinkmann H, Philippe H. Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. MBC Evol Biol. 2007;7(suppl 1):S4. doi: 10.1186/1471-2148-7-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S. PhyloBayes 3. A Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25:2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- Levine ND. Progress in taxonomy of the Apicomplexan protozoa. J Protozool. 1988;35:518–520. doi: 10.1111/j.1550-7408.1988.tb04141.x. [DOI] [PubMed] [Google Scholar]
- Ling KH, Rajandream MA, Rivailler P, et al. (33 co-authors) Sequencing and analysis of chromosome 1 of Eimeria tenella reveals a unique segmental organization. Genome Res. 2007;17:311–319. doi: 10.1101/gr.5823007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore RB, Obornik M, Janouskovec, et al. (14 co-authors) A photosynthetic alveolate closely related to apicomplexan parasites. Nature. 2008;451:959–963. doi: 10.1038/nature06635. [DOI] [PubMed] [Google Scholar]
- Morales ME, Ocampo CB, Cadena H, Copeland CS, Termini M, Wesson DM. Differential identification of Ascogregarina species (Apicomplexa: Lecudinidae) in Aedes aegypti and Aedes albopictus (Diptera: Culicidae) by polymerase chain reaction. J Parasitol. 2005;91:1352–1357. doi: 10.1645/GE-442R.1. [DOI] [PubMed] [Google Scholar]
- Munstermann LE, Wesson DM. First record of Ascogregarina taiwanensis (Apicomplexa: Lecudinidae) in North American Aedes albopictus. J Am Mosq Control Assoc. 1990;6:235–243. [PubMed] [Google Scholar]
- Omoto CK, Toso M, Tang K, Sibley LD. Expressed sequence tag (EST) analysis of Gregarine gametocyst development. Int J Parasitol. 2004;34:1265–1271. doi: 10.1016/j.ijpara.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Reyes-Villanueva F, Becnel JJ, Butler JF. Susceptibility of Aedes aegypti and Aedes albopictus larvae to Ascogregarina culicis and Ascogregarina taiwanensis (Apicomplexa: Lecudinidae) from Florida. J Invertebr Pathol. 2003;84:47–53. doi: 10.1016/s0022-2011(03)00119-8. [DOI] [PubMed] [Google Scholar]
- Schmatz DM. The mannitol cycle in Eimeria. Parasitology. 1997;114:S81–S89. [PubMed] [Google Scholar]
- Striepen B, Pruijssers AJ, Huang J, Li C, Gubbels MJ, Umejiego NN, Hedstrom L, Kissinger JC. Gene transfer in the evolution of parasite nucleotide biosynthesis. Proc Natl Acad Sci USA. 2004;101:3154–3159. doi: 10.1073/pnas.0304686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton TJ. Toxoplasma gondii: the model apicomplexan: perspectives and methods. Netherlands (Amsterdam): Elsevier; 2006. The surface protein repertoire of Toxoplasma gondii and other Apicomplexa. [Google Scholar]
- Templeton TJ. Whole-genome natural histories of apicomplexan surface proteins. Trends Parasitol. 2007;23:205–212. doi: 10.1016/j.pt.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Templeton TJ, Iyer LM, Anantharaman V, Enomoto S, Abrahante JE, Subramanian GM, Hoffman SL, Abrahamsen MS, Aravind L. Comparative analysis of apicomplexa and genomic diversity in eukaryotes. Genome Res. 2004;14:1686–1695. doi: 10.1101/gr.2615304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RC, Olson ME, Zhu G, Enomoto S, Abrahamsen MS, Hijjawi NS. Cryptosporidium and cryptosporidiosis. Adv Parasitol. 2005;59:77–158. doi: 10.1016/S0065-308X(05)59002-X. [DOI] [PubMed] [Google Scholar]
- Toso MA, Omoto CK. Gregarina niphandrodes may lack both a plastid genome and organelle. J Eukaryot Microbiol. 2007;54:66–72. doi: 10.1111/j.1550-7408.2006.00229.x. [DOI] [PubMed] [Google Scholar]
- Zhu G. Current progress in the fatty acid metabolism in Cryptosporidium parvum. J Eukaryot Microbiol. 2004;51:381–388. doi: 10.1111/j.1550-7408.2004.tb00384.x. [DOI] [PubMed] [Google Scholar]
- Zhu G, Keithly JS, Philippe H. What is the phylogenetic position of Cryptosporidium? Int J Syst Evol Microbiol. 2000;50:1673–1681. doi: 10.1099/00207713-50-4-1673. [DOI] [PubMed] [Google Scholar]
- Zhu G, Marchewka MJ, Keithly JS. Cryptosporidium parvum appears to lack a plastid genome. Microbiology. 2000;146:315–321. doi: 10.1099/00221287-146-2-315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.