Abstract
Proteins of the green fluorescent protein family represent a convenient experimental model to study evolution of novelty at the molecular level. Here, we focus on the origin of Kaede-like red fluorescent proteins characteristic of the corals of the Faviina suborder. We demonstrate, using an original approach involving resurrection and analysis of the library of possible evolutionary intermediates, that it takes on the order of 12 mutations, some of which strongly interact epistatically, to fully recapitulate the evolution of a red fluorescent phenotype from the ancestral green. Five of the identified mutations would not have been found without the help of ancestral reconstruction, because the corresponding site states are shared between extant red and green proteins due to their recent descent from a dual-function common ancestor. Seven of the 12 mutations affect residues that are not in close contact with the chromophore and thus must exert their effect indirectly through adjustments of the overall protein fold; the relevance of these mutations could not have been anticipated from the purely theoretical analysis of the protein's structure. Our results introduce a powerful experimental approach for comparative analysis of functional specificity in protein families even in the cases of pronounced epistasis, provide foundation for the detailed studies of evolutionary trajectories leading to novelty and complexity, and will help rational modification of existing fluorescent labels.
Keywords: epistasis, complexity, ancestral reconstruction, comparative analysis, functional specificity, red fluorescent protein, kaede
Introduction
Reef Anthozoa, and reef-building corals (order Scleractinia) in particular, represent the largest natural repository of spectral features achievable within a green fluorescent protein (GFP)-like protein (Matz et al. 1999; Alieva et al. 2008). Four color classes of Anthozoan GFP-like proteins have been originally identified, according to spectral properties and chromophore structure: cyan–green fluorescent, yellow fluorescent, red fluorescent, and nonfluorescent chromoproteins (Matz et al. 1999; Labas et al. 2002). The recent additions to the original classification are three more types of red fluorescent chromophores: Kaede type with a very narrow orange–red fluorescence (Ando et al. 2002), kindling fluorescent protein type with broad deep red fluorescence (Quillin et al. 2005), and a novel chromophore observed in artificially generated mutant variant called mOrange (Shu et al. 2006). In corals, the green fluorescence is ancestral, whereas the present-day color diversity evolved from it on several independent occasions (Ugalde et al. 2004; Alieva et al. 2008), starting as early as the first appearance of modern coral reefs in late Triassic—early Jurassic (Kelmanson and Matz 2003). Autocatalytic synthesis of the green fluorescent chromophore involves two consecutive stages, whereas in red, purple, and some yellow GFP-like proteins, there is an additional processing step, which provides a more extensive p-orbital conjugation (Wachter 2006). Emergence of these nongreen colors from the ancestral green in evolution, therefore, represents a transition in functional complexity (Matz et al. 2002; Shagin et al. 2004; Ugalde et al. 2004), defined as the number of functions (in this case, stages of autocatalysis) performed by a biological entity (McShea 2000). Here, we aimed at identifying mutations that contributed to the color transition in one of the known independent cases of red fluorescence evolution: emergence of Kaede-type red proteins characteristic of corals of the Faviina suborder (Kelmanson and Matz 2003; Ugalde et al. 2004; Alieva et al. 2008).
A variety of computational methods have been developed to identify the specific amino acid sites that determine the functional differences within protein families by looking at the patterns of variation at individual sites in multiple-sequence alignments (Chakrabarti and Lanczycki 2007; Capra and Singh 2008). The basic idea of these methods is to compare the sequences of proteins endowed with the new function to other extant members of the protein family, exhibiting another function. However, comparison of extant proteins may overlook a number of important mutations because the common ancestor of the compared proteins may have possessed a dual function (fig. 1). This reflects a situation in which the new function originated within a single gene product and was subsequently followed by gene duplication that precipitated specialization. Such a mechanism of functional diversification, termed “escape from adaptive conflict” (Piatigorsky and Wistow 1991; Hughes 1994), appears to be more common than it was previously thought (Des Marais and Rausher 2008). In our particular case, ancestral reconstructions confirmed that the common ancestor of green and red fluorescent proteins (“r/g,” fig. 1D) indeed had an intermediate orange phenotype (Ugalde et al. 2004). Under such a scenario, the extant bearers of the ancestral function (green fluorescence) are expected to retain a number of latent sequence features that are relevant for the novel function (red fluorescence), due simply to a recent shared ancestry. These features will not be revealed through comparison of extant proteins. For a comprehensive analysis, we therefore had to compare extant red proteins with a deeper ancestral node (“a,” fig. 1D) for which the pure ancestral green phenotype was confirmed (Ugalde et al. 2004). Such a “vertical” comparative analysis (between the extant and resurrected ancestral proteins) is also more efficient than classical “horizontal” (between the extant proteins with different functions), because one has to deal with neutral mutations accumulated along only one of the two lines of descent connecting the extant representatives (fig. 1C).
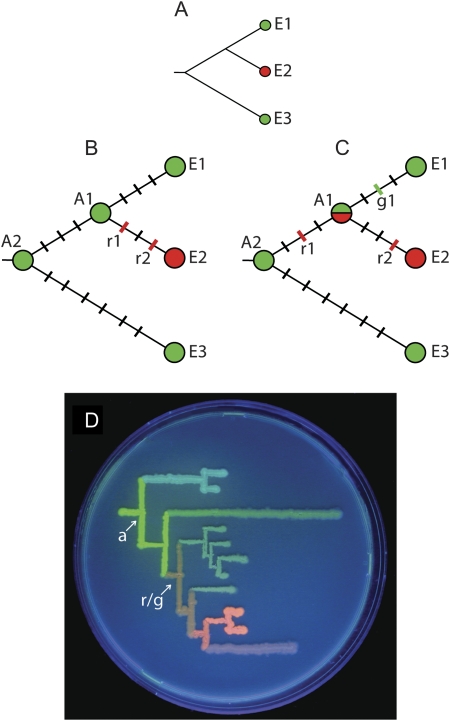
FIG. 1.
The need for vertical comparative analysis. (A) A typical phylogenetic relationship suggesting that a new function (red) evolved from the ancestral (green). E1, E2, and E3 designate extant proteins. (B) A case when the new function evolved after gene duplication. The black hashes represent neutral mutations and the colored hashes mutations required for the new function. Comparison of E2 with the extant protein E1 captures both essential mutations, but comparison to their common ancestor (A1) is more efficient because it includes less neutral mutations. (C) A scenario when the new function evolved prior to gene duplication. In this case, comparing E1 and E2 does not capture all the essential mutations, unless g1 is a reversal of r1. Instead, E2 should be compared with the deeper ancestral node A2. (D) Phylogenetic tree of GFP-like proteins from Faviina corals drawn on a Petri dish using bacteria-expressing extant and ancestral proteins (Ugalde et al. 2004). The common ancestor of greens and reds (r/g) is a dual-function orange. The evolution of the red color, therefore, should be traced between the extant red and the common green ancestor of all coral colors (a).
The idea of looking at the ancestral protein as the reference point to infer potential function-transforming mutations is not new. This approach has been pioneered by Shi and Yokoyama (2003) and has been successfully applied to disentangle the structure–function relationships in such proteins as, for example, beta-lactamase (Weinreich et al. 2006), hormone receptors (Bridgham et al. 2006; Ortlund et al. 2007), and opsins (Yokoyama et al. 2008). In all these cases, site-directed mutagenesis of candidate sites in the ancestral and extant proteins provided the proof of the relevance of individual mutations for the phenotype. The application of the same approach to the evolution of red color in GFP-like proteins, however, presented an apparently insurmountable challenge: The least divergent pair of ancestral green and extant red proteins featured 37 amino acid substitutions, which meant that for an exhaustive search for the key combination of mutations, more than 100 billion combinatorial mutants needed characterization. Our earlier attempt to narrow down this vast search space by looking only at the mutations facilitated by positive natural selection specific to the red lineage was only partially successful: We identified three mutations that were necessary (i.e., severely impaired the red color when reverted to the ancestral state in an extant red protein) but clearly insufficient for red color evolution as their introduction into the green ancestral protein did not bring about even a trace of color change (Field et al. 2006). Here, we present a solution to our combinatorial problem, involving creation and analysis of the “transitional library” of possible evolutionary intermediates. This approach represents a general solution that can, in principle, be applied to any case of functional diversification in protein families, provided that it is feasible to test several hundred clones from the transitional library for the presence–absence of the novel function.
Materials and Methods
Preparation of Transitional Library
This work is based on results of the recreation of the common ancestor of all Faviina colors (Ugalde et al. 2004). This ancestral node was recreated as a combinatorial library of approximately equally probable sequence variants, according to the predictions of three different evolutionary models. All these variants demonstrated an identical green phenotype, and hence, any one of the clones from this combinatorial library could be used as a starting point for comparative analysis. When compared with the least divergent extant red protein, which was the clone designated R1–2 from the great star coral Montastrea cavernosa (Kelmanson and Matz 2003), these ancestral clones had at least 37 amino acid differences (35 substitutions, one insertion, and one deletion). All of these differences were incorporated into the degenerate gene synthesis, in all possible combinations, to create a bacterial expression library of the potential evolutionary intermediates connecting the ancestral green protein to the extant R1–2 protein. Additional variations were allowed at sites for which the ancestral state was reconstructed ambiguously. We followed the previously described procedures of oligonucleotide design and gene synthesis (Ugalde et al. 2004; Chang et al. 2005); see supplementary information, Supplementary Material online, for the gene-design overview and sequences of the ancestral gene, R1–2 gene, and oligonucleotides used in gene synthesis. The product of gene synthesis was ligated into pGEM-T vector (Promega) according to the manufacturer's protocol, transformed into Top10 Escherichia coli cells (Invitrogen), and plated onto Luria-Bertani agar supplemented with 50 μg/ml ampicillin and 1 mM Isopropyl β-D-1-thiogalactopyranoside.
Analysis of the Transitional Library
After incubating for 48 h at 37 °C, the plates were exposed to low-intensity UV-A light (“blacklight”) for 60 min to facilitate the beta-elimination reaction resulting in the emergence of the red chromophore (Mizuno et al. 2003; Nienhaus et al. 2005). The plates were then screened using a fluorescent stereomicroscope Leica MZ FL III equipped with a double-bandpass filter set (# 51004v2, Chroma Technology) allowing for simultaneous visualization of green and red fluorescence. After screening about 20,000 bacterial colonies, 28 clones exhibiting yellow or orange appearance were selected for further characterization, along with 67 purely green clones. The plasmids were isolated from the overnight cultures of these clones using QIAprep Spin Miniprep kit (Qiagen), and the inserts were sequenced using the Sanger method from the vector-specific primer to determine the state of each variable site in each clone. The counts in table 1 do not always sum up to 28 for yellow–orange and 67 for green clones, because of ambiguous sequence results and occasional artifacts stemming from the use of degenerate oligonucleotides to introduce variations, which led to the appearance of unplanned states at some sites. The representation of variable site states in the yellow–orange clones was compared with the purely green clones to detect associations of particular site states with the partial ability to synthesize the red chromophore using Fisher's exact test.
Table 1.
Association of Site States with Color in the Transitional Library.
| State in |
Yellow–Orange Clones |
Green Clones |
|||||
| Site #a | Ancestral Green | Extant Red | Ancestral | Extant | Ancestral | Extant | P-Valueb |
| 11 | D | V | 17 | 8 | 20 | 47 | 0.001 |
| 21 | T | R | 13 | 10 | 42 | 25 | 0.627 |
| 26 | K | N | 14 | 12 | 32 | 35 | 0.649 |
| 28 | V | L | 11 | 7 | 30 | 37 | 0.290 |
| 30 | E | V | 14 | 12 | 45 | 22 | 0.242 |
| 43 | T | S | 15 | 13 | 34 | 33 | 0.826 |
| 45 | N | D | 17 | 9 | 37 | 29 | 0.485 |
| 47 | K | T | 11 | 16 | 28 | 37 | 0.501 |
| 60 | L | M | 16 | 12 | 37 | 30 | 1.000 |
| 63 | A | V | 23 | 5 | 29 | 38 | 0.001 |
| 65 | Q | H | 0 | 23 | 55 | 12 | 0.000 |
| 72 | T | A | 4 | 23 | 33 | 34 | 0.002 |
| 77 | D | H | 10 | 17 | 40 | 27 | 0.067 |
| 87 | S | M | 1 | 10 | 18 | 7 | 0.001 |
| 93 | S | F | 14 | 14 | 37 | 30 | 0.659 |
| 99 | T | N | 16 | 10 | 36 | 31 | 0.642 |
| 109 | T | R | 9 | 19 | 38 | 29 | 0.042 |
| 110 | S | N | 4 | 24 | 40 | 27 | 0.000 |
| 111 | D | E | 12 | 17 | 31 | 36 | 0.823 |
| 114 | L | M | 14 | 14 | 28 | 39 | 0.503 |
| 121 | Y | N | 1 | 28 | 46 | 21 | 0.000 |
| 122 | E | K | 16 | 13 | 30 | 37 | 0.381 |
| 123 | I | V | 16 | 13 | 39 | 28 | 0.825 |
| 162 | M | T | 5 | 24 | 34 | 33 | 0.003 |
| 165 | V | I | 9 | 22 | 34 | 33 | 0.051 |
| 182 | K | R | 12 | 18 | 35 | 32 | 0.282 |
| 186 | K | R | 18 | 12 | 35 | 32 | 0.515 |
| — | DEL | K | 10 | 20 | 23 | 44 | 1.000 |
| 194 | Q | E | 20 | 10 | 41 | 26 | 0.655 |
| 204 | R | C | 3 | 27 | 33 | 34 | 0.000 |
| 217 | N | K | 13 | 15 | 33 | 34 | 0.826 |
| 225 | V | E | 10 | 18 | 38 | 29 | 0.074 |
| 227 | R | H | 7 | 21 | 36 | 31 | 0.013 |
| — | Y | DEL | 5 | 23 | 38 | 29 | 0.001 |
| 228 | M | G | 3 | 14 | 18 | 11 | 0.005 |
| 231 | S | R | 13 | 15 | 36 | 31 | 1.000 |
| 232 | Q,L | V | 6,5 | 10 | 15,9 | 24 | 1.000 |
According to GFP from Aequorea victoria.
Two-tailed Fisher's exact test.
Mutagenesis of the Ancestral Protein
The transitional mutations were ranked according to the increasing P value of Fisher's exact test and introduced in that order into the ancestral gene. For this mutagenesis, we chose a clone from the ancestral combinatorial library that was most similar to the R1–2 protein (a total of 37 amino acid differences). Then, each of the mutations was individually reversed to the ancestral state, to double-check for its relevance for the red color. The site-directed mutagenesis experiments were performed using QuickChange II kit (Stratagene, La Jolla, CA). To evaluate the phenotypic effect of the mutations, the emission spectra of the mutants were recorded from bacterial colonies using a USB2000 spectrometer (Ocean Optics, Dunedin, FL) attached to an MZ FL III stereomicroscope (Leica, St Gallen, Switzerland), using the filter set BL/VIO (#11003, Chroma Technology, Rockingham, VT), and compared with the fluorescence of the extant R1-2 protein expressed in bacteria grown on the same agar plates, after exposure of the plates to UV-B light for 180 min.
Results and Discussion
Transitional Library Method
We present a novel method that is designed to find the combination of mutations minimally sufficient to transform the phenotype in evolution, despite a possibly astronomical number of combinations of potentially functionally relevant residues. We create a “transitional library” of possible evolutionary intermediates, in which the clones have every position differing between the ancestor and extant protein in either ancestral or derived state, with approximately equal probability (barring the biases of oligonucleotide synthesis and gene assembly). Each clone, therefore, contains about half of the sites in the derived state. To identify the phenotypically relevant sites, we test whether the clones exhibiting a trend toward novel phenotype preferentially contain certain sites in the derived state, using Fisher's exact test.
It is clear that the approach would work perfectly for the mutations the effect of which is predominantly additive (i.e., independent of the presence of other mutations). Suppose that the set of mutations includes one that visibly converts the phenotype irrespective of the others. In this case, every clone in the transitional library with the corresponding position in the derived state (i.e., about 50% of all clones) will exhibit the partially transformed phenotype. The unique strength of our method, however, lies in identifying combinations of mutations the effect of which strongly depends on each other's presence. This situation is termed positive epistasis and in extreme cases may result in no detectable phenotypinc effect until the whole combination of interacting mutations is assembled. To illustrate the power of our method for identifying such combinations, we simulated transitional library analysis with different numbers of phenotypically characterized and sequenced clones, under different epistasis scenarios (fig. 2). The simulations assumed that in each clone, each variable site is found in ancestral or derived state with equal probability. After simulating a random selection of such clones, we sorted them into ancestral and derived phenotypes based on the known mutations and epistasis scenario, counted the number of times a particular site is found in each of the states in phenotypically ancestral and derived clones, and analyzed the counts with Fisher's exact test to see if we can successfully recover the causal mutations. We investigated four epistasis scenarios (fig. 2).
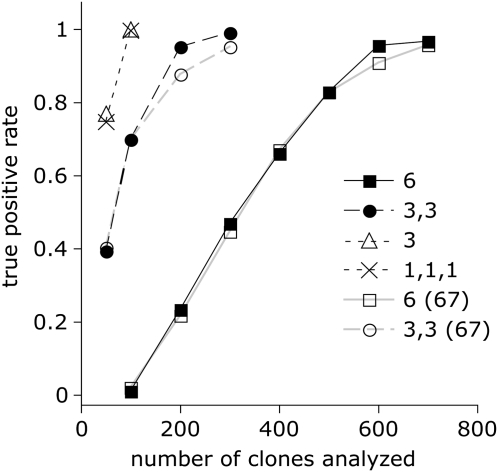
FIG. 2.
Simulated analysis of transitional libraries under different epistasis scenarios. Horizontal axis: number of clones characterized from the transitional library, vertical axis: causal mutation discovery rate, averaged over 100 replicates. The epistasis scenarios were as follows. “6”: a single group of six epistatically interacting mutations, that is, all six are required for the phenotype change. “3,3”: two groups of three epistatically interacting mutations with no epistasis between the groups, that is, the phenotype changes if all members of any one group are present. “3”: one group of three interacting mutations. “1,1,1”: three mutations with purely additive effects (no epistasis). We also performed a “sequencing budget” flavor of the analysis, where we used only 67 of all clones exhibiting ancestral phenotype in Fisher's test, as in our real study. These additional curves are designated “6 (67)” and “3,3 (67)”. The “sequencing budget” curves for other scenarios were identical to the full-analysis ones.
The simulation shows that even when the phenotype depends on six epistatically interacting mutations, there is a 95% or better chance of finding such a combination after phenotypic characterization of only 600–700 clones from the transitional library (fig. 2, “6” and “6 (67)”). For other cases of less pronounced epistasis, the analysis of 200–300 clones is sufficient. Importantly, limiting the sequencing effort of the ancestral-phenotype clones to 67, as in our real study, does not affect the success rate, except slightly for the challenging epistatic scenarios (“6” and “3,3”) with high numbers of clones analyzed. In all the simulation trials, the false positive rate (we assumed a total of 37 transitional mutations to mimic our experiment) remained at or below 0.05, indicating that Fisher's exact test is fully appropriate for this case, making our method both highly sensitive and specific.
To implement this methodology in the study of GFP-like proteins, we used degenerate gene synthesis to create a transitional bacterial expression library of possible evolutionary intermediates between ancestral green and extant red proteins (Ugalde et al. 2004; Field et al. 2006). To avoid errors due to the inherently ambiguous nature of ancestral reconstructions (Thornton 2004), we selected an ancestral sequence that deviates the least from the extant protein of interest among approximately equally probable reconstructions (Ugalde et al. 2004). Selecting the least divergent ancestral sequence makes the analysis conservative because it reduces the probability of “finding” a functionally important difference between ancestor and descendant that is in fact an error of reconstruction. In the transitional library, we looked for clones exhibiting the derived (red) rather than ancestral (green) fluorescence, even if it constituted a minor component of the total emission. After surveying about 20,000 clones under a fluorescent stereomicroscope, we were able to isolate only 28 such “partially evolved” clones that appeared yellow or orange due to a considerable proportion of red fluorescence along with the original green (supplementary fig. S2, Supplementary Material online). As suggested by our simulation results (fig. 2), such rarity of the derived phenotype in the transitional library most likely is due to extensive epistatic interactions. The sequences of yellow–orange clones were compared with sequences of 67 purely green clones to detect associations of individual site states with the yellow–orange phenotype via Fisher's exact test. The test was significant at the 0.05 level for 12 mutations (table 1). Curiously, two of these (D11V and A63V) were strongly favored in their ancestral state.
Forward and Reverse Mutagenesis
The next step after analysis of the transitional library is testing whether the discovered mutations are indeed sufficient to evolve the new phenotype in the ancestral protein. Obviously, to do this, it is necessary to introduce the mutations into the ancestral protein (“forward mutagenesis”), but in which order? We reasoned that it makes sense to start with the mutations showing the strongest association with the new phenotype, because their effect is likely to be the most dramatic, or, under epistasis scenario, they must be the most essential as a background for other mutations to work. We therefore chose to introduce the candidate mutations in the order of decreasing association, that is, in the order of increasing Fisher's test P values. It should be noted that this order is essentially arbitrary with respect to the true historical order of mutations.
First, we introduced 11 mutations with P values below 0.05, resulting in a protein that was nearly as red as the extant R1–2 (fig. 3A and B). Notably, the first three introduced mutations did not result in any change of phenotype (fig. 3A), despite their strong association with the red color in the transitional library (table 1), indicating extensive epistasis. This is in accord with our previous experiment (Field et al. 2006), which failed to generate any red fluorescence in the ancestral green protein even when all the three positively selected mutations (each of them necessary for the red color in the extant red protein) were introduced.
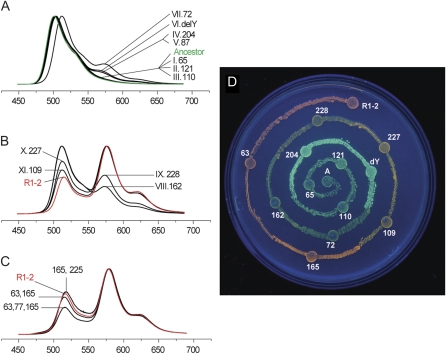
FIG. 3.
Changes in fluorescence of the ancestral protein as candidate color-changing mutations are introduced into it in the order of decreasing association with red color in the transitional library analysis. The graphs are normalized fluorescence spectra of expressing bacterial colonies after 120-min UV-B exposure; the horizontal axis is wavelength in nanometers. Green fluorescence corresponds to the peak at 500–520 nm and red to the peak at 575–580 nm. The numbering of mutated sites is according to the Aequorea victoria GFP sequence (Prasher et al. 1992) within a familywide alignment (Alieva et al. 2008); DelY stands for the deletion of a tyrosine between positions 227 and 228. (A,B) Introduction of mutations for which the association was significant at 0.05 level results in an 11-mutation clone that is almost as red as the extant red protein R1-2. (C) Adding different combinations of mutations that are significant at the 0.1 level meets and surpasses the red maturation efficiency of R1–2. (D) Visual appearance of bacteria-expressing mutant proteins that retrace the green-to-red evolutionary transition, under UV-A light.
Because in Kaede-type proteins, the maturation of the red chromophore is promoted by 400-nm light (Mizuno et al. 2003; Nienhaus et al. 2005), the fluorescence spectra of the bacterial colonies expressing mutant proteins were measured over the course of exposure to low-intensity UV-B (“blacklight”) for up to 180 min. All the mutants in which red fluorescence was detectable tended to accumulate it more or less linearly for at least 120 min of UV-B exposure, and then level off (supplementary fig. S3A, Supplementary Material online). There was a strong correlation (R2 = 0.93) between the initial maturation rate and the maximal achieved redness (supplementary fig. S3B, Supplementary Material online). This suggests either that the proportion of convertible protein and the rate of conversion are tightly interdependent, or, more likely, that all the mutants differ predominantly in the rate of maturation and for less efficient mutants the plateau is simply never reached (the level-off may be apparent or be the result of protein damage due to, e.g., photobleaching).
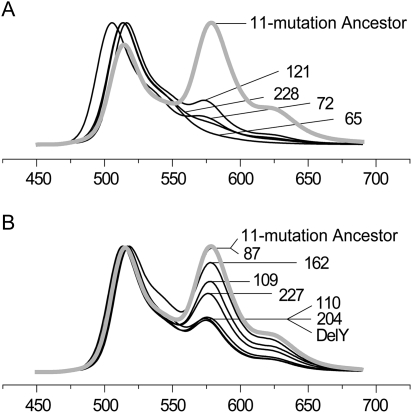
We then tested for effects of each of the mutations by reverting them individually in the newly “evolved” red protein (“reverse mutagenesis,” fig. 4). This experiment indicated that one of the mutations, S87M, was irrelevant for the red color (fig. 4B). Indeed, this mutation looks like a pure false positive in our data: Not only does it not affect the red color upon reversal, but it also does not lead to any phenotype change in the original forward mutagenesis (fig. 3A). It may be surprising that in the transitional library, S87M is apparently strongly associated with red color (table 1); however, the most likely reason for this is that the number of transitional library clones in which site 87 was successfully sequenced was only 36, potentially leading to spurious test results.
FIG. 4.
Effect of individual mutation reversals in the ancestral clone bearing 11 candidate mutations (“11-mutation ancestor”). The graphs are normalized fluorescence spectra; the horizontal axis is wavelength in nanometers. (A) Large-effect mutations. (B) Mutations with lesser effect (“fine tuning”). Note that reversal of mutation at site 87 does not affect fluorescence, and hence, this mutation is not required for the red color.
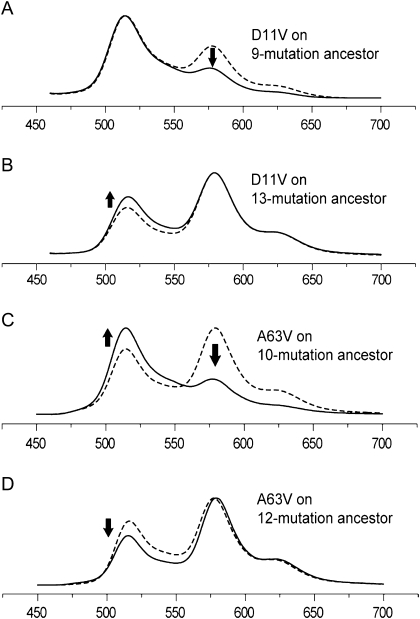
We then proceeded to investigate the phenotypic effect of additional mutations that were highlighted by Fisher's test including the two that were strongly preferred in their ancestral state (D11V and A63V). Mutation D11V was detrimental for the red color, as suggested by the association test (fig. 5A and B). Such deleterious mutations may be either introduced via neutral mechanisms or related to some other aspect of adaptation rather than color. Curiously, the A63V mutation was beneficial (fig. 3C), despite strong association with ancestral green color in the transitional library (table 1). Further analysis provided an explanation for this, by revealing that this mutation exhibits sign epistasis (Weinreich et al. 2005; Poelwijk et al. 2007): Its effect can be detrimental or beneficial depending on the sequence background, in such a way that it is likely to be detrimental at the earlier evolutionary stages but beneficial later (fig. 5C and B). Adding more mutations with P values between 0.05 and 0.1 (D77H, V165I, and V225E) enhanced the red fluorescence beyond the level of the extant red protein (fig. 3C), suggesting that in the extant red protein, they are compensating for the effect of deleterious mutations such as D11V.
FIG. 5.
Effect of mutations for which the association study strongly suggested preference toward the ancestral state in the redder clones. The graphs are normalized fluorescence spectra; the horizontal axis is wavelength in nanometers; dashed curves correspond to proteins with the mutation in question in the ancestral state. (A,B) mutation D11V is always deleterious for the red color, suppressing red fluorescence (575–580 nm) and promoting green (515–520 nm). (C,D) Mutation A63V exhibits sign epistasis (Weinreich et al. 2005): It is deleterious at less advanced evolutionary stage (C) but is beneficial later (D).
Effects of Individual Mutations
As a result of our analysis, we have determined that a group of 12 historical substitutions are sufficient to recapitulate the evolution of red fluorescence (fig. 3D and table 1): A63V, Q65H, T72A, T109R, S110N, Y121N, M162T, V165I, R204C, R227H, delY[227–228], and M228G. There are a small number of mutations with a large effect that are most critical for the red color (fig. 4A), whereas the rest of the mutations are more of a “fine-tuning” type (fig. 4B), each bringing about a relatively small improvement once the large-effect mutations are in place. Interestingly, it is possible to name slightly different sets of fine-tuning mutations that would result in red fluorescence equivalent to the extant red protein, because their full complement in the absence of deleterious mutations such as D11V (fig. 5A and B) results in a more efficient protein than the extant one. For example, V225E and A63V are interchangeable within the list of 12 sufficient mutations (fig. 3C).
Mutation Q65H seems to be the only one that is absolutely essential for red fluorescence, as reflected both in the association analysis (table 1) and results of reverse mutagenesis (fig. 4A). The crucial role of Q65H is not surprising, because it provides the imidazole group that becomes incorporated into the red chromophore (Mizuno et al. 2003). Notably, because Q65H alone does not lead to any red fluorescence (Field et al. 2006) (fig. 3A), whereas all the other mutations must have Q65H as a background for protein chemistry reasons, it follows that the evolution of red color required more than one mutation before any color change was achieved. Therefore, it could not have occurred purely by natural selection for the new fluorescence color along a “selection-accessible mutation path” sensu Weinreich et al. (2006). Other evolutionary mechanisms, such as genetic drift, gene conversion, or selection for some other aspect of the protein function, must have also played a role. Another indication of a potential involvement of factors other than natural selection is the fact that only three of the color-converting mutations (Q65H, Y121N, and M228G) were previously identified as driven by positive selection in the red lineage (Field et al. 2006). Still, this may simply reflect insufficient power of the test for positive selection in our particular case.
Our results clearly illustrate the need for vertical comparative analysis: If we were to compare extant red proteins not with the ancestral green protein (vertical analysis), but with the extant green proteins (horizontal analysis), we would overlook five of the color-converting mutations (A63V, T72A, S110N, delY[227–228], and M228G). These mutations happened before the separation of green and red gene lineages, leading to the evolution of the dual-function common ancestor of greens and reds (fig. 1D).
Structural Basis of Red Fluorescence Evolution
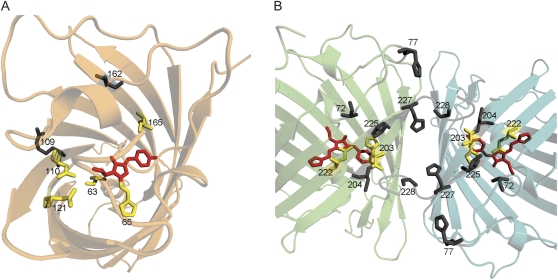
Only five of the identified 12 mutations affect side chains that are predictably located in the immediate vicinity of the chromophore (fig. 6A). The most obvious of these is Q65H, which contributes the histidine side chain that becomes incorporated into the red chromophore structure (Mizuno et al. 2003; Nienhaus et al. 2005). The other four mutations with a direct effect are S110N, Y121N, V165I, and possibly A63V. All these mutations are likely to affect fluorescence by modifying polar interactions of the chromophore and/or the size of the internal cavity in which it is located (fig. 6A). The remaining seven mutations must exert their effect indirectly, by adjusting the relative positions of functional groups within the protein through modifications of the overall fold. It is important to note that such influences are virtually impossible to predict solely on the basis of theoretical analysis of the protein structure, which highlights the value of an unbiased empirical analysis such as we undertook here. Sites 109 and 162, with side chains facing outside the globule but directed toward another monomer in the tetrameric structure, may aid precise positioning of sites 110 and 165, mentioned above, being their neighbors in their respective beta strands. There is a remarkable cluster of interacting residues close to the C-terminus of the protein, which affects the interface of adjacent subunits within the tetramer (fig. 6B). We hypothesize that these mutations act in a similar fashion to the ones at sites 109 and 162, that is, by tugging on the wall of the protein to readjust the functional groups inside, only in this case the groups being readjusted stayed invariant in evolution. These groups most likely are Glu-222 and His-203 (fig. 6B), which are known to be important for autocatalytic formation or modification of spectral features of the chromophore in GFPs (Ormo et al. 1996; Barondeau et al. 2003; Sniegowski et al. 2005). The mutation T72A, located within the chromophore-bearing helix, has a strong impact on the color, shifts the whole emission spectrum toward red by 15 nm, and appreciably dims the overall fluorescent output. It can be speculated that T72A mutation adjusts the conformation of the internal helix, resulting in alignment of the chromophore's phenolate ring with the His-203 side chain, which enables stacking of their conjugated pi-systems. This particular interaction may be responsible for the whole-spectrum wavelength shift and reduced brightness and may be one of the key facilitators of the extended autocatalysis resulting in synthesis of the red chromophore. In the future, we would like to study the recreated intermediates by X-ray crystallography to verify these hypotheses, because this knowledge is directly relevant for the ability to rationally design novel genetically encoded fluorescent labels based on GFP-like proteins.
FIG. 6.
Distribution of the sites responsible for the red fluorescence, as suggested by this study, in a Kaede-type red fluorescent protein EosFP (Nienhaus et al. 2005). (A) Side chains that directly contribute to the chromophore or its environment are shown in yellow. Two residues in gray, with outward-directed side chains, most likely exert their effects through adjusting the positions of the yellow ones. The invariant chromophore portion (not including the part contributed by His65) is shown in red. (B) Side chains of the residues with indirect effect (gray). The chromophore is shown in red. Yellow side chains belong to the two evolutionarily conserved residues (Glu-222 and His-203); it is likely that mutations at the gray sites result in adjustment of positions of these side chains relative to the chromophore.
Our Results and Real Evolution
There are four aspects in which our experiment is an approximation, rather than literal recreation, of the real evolutionary process. First, because the sequence of a reconstructed ancestral protein is unavoidably ambiguous (Thornton 2004), the mutations identified here comprise a likely, but not necessarily the true list. We expect, however, that the true list would include more rather than fewer mutations, because we deliberately selected the pair of ancestral and extant sequences showing the least possible divergence. Second, our method, just as any other technique substituting heuristics for exhaustive search, does not guarantee that the obtained solution is unique. Theoretically, there may be other combinations of mutations leading to the same phenotype. We expect that in our particular case, the alternative solutions may only be possible with respect to the “fine-tuning” mutations (fig. 3C); a separate study would be required to prove it rigorously. Third, the rate and extent of red fluorescence development in the recreated intermediates and mutants in E. coli may be affected, for example, by the lack of binding partners that may be present in the coral (Field et al. 2006); hence, our estimates of how mutations change the color may not be an adequate modeling of their effect in the coral. It may be argued, however, that such a bias due to heterologous expression is likely to be the same for all our mutant proteins, as well as for the extant protein R1–2, because they are all still very similar in sequence. Therefore, it is reasonable to assume that the relative (but maybe not the absolute) contributions of individual mutations to the red fluorescence measured in bacteria reflect the situation in the coral correctly. Encouragingly, there is also an indication that there may not be too much heterologous expression bias in the first place: The photoconversion of M. cavernosa Kaede-like protein from green to red in vivo under blacklight may take on the order of 3 h (Leutenegger et al. 2007), which is quite similar to what we observed in E. coli (supplementary fig. S3, Supplementary Material online). Fourth, as we mentioned above, the order in which we introduced the mutations into the ancestral protein is essentially arbitrary. Evaluating which mutation paths are more likely to have been realized through evolution by natural selection (Weinreich et al. 2006; Ortlund et al. 2007) would be a logical follow-up project; however, to properly operate in terms of natural selection, such a study would require a much better understanding of the biological function of coral fluorescence than we possess at the moment (Matz et al. 2006). It is quite possible that the red color evolution might have been largely facilitated by selection for other aspects of the protein's function, such as the tentative abilities to detoxify reactive oxygen species (Bou-Abdallah et al. 2006) or donate electrons upon excitation (Bogdanov et al. 2009).
Supplementary Material
Supplementary figures S1-S3 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
This work was supported by the NIH grants R01-GM066243 and R01-GM078247 to M. V. M.
References
- Alieva NO, Konzen KA, Field SF, Meleshkevitch EA, Beltran-Ramirez V, Miller DJ, Salih A, Wiedenmann J, Matz MV. Diversity and evolution of coral fluorescent proteins. PLoS ONE. 2008;3:e2680. doi: 10.1371/journal.pone.0002680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc Natl Acad Sci USA. 2002;99:12651–12656. doi: 10.1073/pnas.202320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barondeau DP, Putnam CD, Kassmann CJ, Tainer JA, Getzoff ED. Mechanism and energetics of green fluorescent protein chromophore synthesis revealed by trapped intermediate structures. Proc Natl Acad Sci USA. 2003;100:12111–12116. doi: 10.1073/pnas.2133463100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov AM, Mishin AS, Yampolsky IV, Belousov VV, Chudakov DM, Subach FV, Verkhusha VV, Lukyanov S, Lukyanov KA. Green fluorescent proteins are light-induced electron donors. Nat Chem Biol. 2009;5:459–461. doi: 10.1038/nchembio.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bou-Abdallah F, Chasteen ND, Lesser MP. Quenching of superoxide radicals by green fluorescent protein. Biochim Biophys Acta. 2006;1760:1690–1695. doi: 10.1016/j.bbagen.2006.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgham JT, Carroll SM, Thornton JW. Evolution of hormone–receptor complexity by molecular exploitation. Science. 2006;312:97–101. doi: 10.1126/science.1123348. [DOI] [PubMed] [Google Scholar]
- Capra JA, Singh M. Characterization and prediction of residues determining protein functional specificity. Bioinformatics. 2008;24:1473–1480. doi: 10.1093/bioinformatics/btn214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S, Lanczycki CJ. Analysis and prediction of functionally important sites in proteins. Prot Sci. 2007;16:4–13. doi: 10.1110/ps.062506407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang BSW, Ugalde JA, Matz MV. Applications of ancestral protein reconstruction in understanding protein function: GFP-like proteins. Meth Enzymol. 2005;395:652–670. doi: 10.1016/S0076-6879(05)95034-9. [DOI] [PubMed] [Google Scholar]
- Des Marais DL, Rausher MD. Escape from adaptive conflict after duplication in an anthocyanin pathway gene. Nature. 2008;454:U762–U785. doi: 10.1038/nature07092. [DOI] [PubMed] [Google Scholar]
- Field SF, Bulina MY, Kelmanson IV, Bielawski JP, Matz MV. Adaptive evolution of multicolored fluorescent proteins in reef-building corals. J Mol Evol. 2006;62:332–339. doi: 10.1007/s00239-005-0129-9. [DOI] [PubMed] [Google Scholar]
- Hughes AL. The evolution of functionally novel proteins after gene duplication. Proc R Soc Lond Ser B-Biol Sci. 1994;256:119–124. doi: 10.1098/rspb.1994.0058. [DOI] [PubMed] [Google Scholar]
- Kelmanson I, Matz M. Molecular basis and evolutionary origins of color diversity in great star coral Montastraea cavernosa (Scleractinia: Faviida) Mol Biol Evol. 2003;20:1125–1133. doi: 10.1093/molbev/msg130. [DOI] [PubMed] [Google Scholar]
- Labas YA, Gurskaya NG, Yanushevich YG, Fradkov AF, Lukyanov KA, Lukyanov SA, Matz MV. Diversity and evolution of the green fluorescent protein family. Proc Natl Acad Sci USA. 2002;99:4256–4261. doi: 10.1073/pnas.062552299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leutenegger A, D'Angelo C, Matz MV, Denzel A, Oswald F, Salih A, Nienhaus GU, Wiedenmann J. It's cheap to be colorful—Anthozoans show a slow turnover of GFP-like proteins. FEBS J. 2007;274:2496–2505. doi: 10.1111/j.1742-4658.2007.05785.x. [DOI] [PubMed] [Google Scholar]
- Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- Matz MV, Labas YA, Ugalde J. Evolution of functions and color in GFP-like proteins. In: Chalfie M, Kain SR, editors. Green fluorescent protein: properties, applications and protocols. 2nd ed. 2006. New York: Wiley-Interscience. [Google Scholar]
- Matz MV, Lukyanov KA, Lukyanov SA. Family of the green fluorescent protein: journey to the end of the rainbow. Bioessays. 2002;24:953–959. doi: 10.1002/bies.10154. [DOI] [PubMed] [Google Scholar]
- McShea DW. Functional complexity in organisms: parts as proxies. Biol Phil. 2000;15:641–668. [Google Scholar]
- Mizuno H, Mal TK, Tong KI, Ando R, Furuta T, Ikura M, Miyawaki A. Photo-induced peptide cleavage in the green-to-red conversion of a fluorescent protein. Mol Cell. 2003;12:1051–1058. doi: 10.1016/s1097-2765(03)00393-9. [DOI] [PubMed] [Google Scholar]
- Nienhaus K, Nienhaus GU, Wiedenmann J, Nar H. Structural basis for photo-induced protein cleavage and green-to-red conversion of fluorescent protein EosFP. Proc Natl Acad Sci USA. 2005;102:9156–9159. doi: 10.1073/pnas.0501874102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- Ortlund EA, Bridgham JT, Redinbo MR, Thornton JW. Crystal structure of an ancient protein: evolution by conformational epistasis. Science. 2007;317:1544–1548. doi: 10.1126/science.1142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatigorsky J, Wistow G. The recruitment of crystallins—new functions precede gene duplication. Science. 1991;252:1078–1079. doi: 10.1126/science.252.5009.1078. [DOI] [PubMed] [Google Scholar]
- Poelwijk FJ, Kiviet DJ, Weinreich DM, Tans SJ. Empirical fitness landscapes reveal accessible evolutionary paths. Nature. 2007;445:383–386. doi: 10.1038/nature05451. [DOI] [PubMed] [Google Scholar]
- Prasher DC, Eckenrode VK, Ward WW, Prendergast FG, Cormier MJ. Primary structure of the Aequorea victoria green-fluorescent protein. Gene. 1992;111:229–233. doi: 10.1016/0378-1119(92)90691-h. [DOI] [PubMed] [Google Scholar]
- Quillin ML, Anstrom DM, Shu X, O'Leary S, Kallio K, Chudakov DM, Remington SJ. Kindling fluorescent protein from Anemonia sulcata: dark-state structure at 1.38 Å resolution. Biochemistry. 2005;44:5774–5787. doi: 10.1021/bi047644u. [DOI] [PubMed] [Google Scholar]
- Shagin DA, Barsova EV, Yanushevich YG, et al. (13 co-authors) GFP-like proteins as ubiquitous Metazoan superfamily: evolution of functional features and structural complexity. Mol Biol Evol. 2004;21:841–850. doi: 10.1093/molbev/msh079. [DOI] [PubMed] [Google Scholar]
- Shi Y, Yokoyama S. Molecular analysis of the evolutionary significance of ultraviolet vision in vertebrates. Proc Natl Acad Sci USA. 2003;100:8308–8313. doi: 10.1073/pnas.1532535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu X, Shaner NC, Yarbrough CA, Tsien RY, Remington SJ. Novel chromophores and buried charges control color in mFruits. Biochemistry. 2006;45:9639–9647. doi: 10.1021/bi060773l. [DOI] [PubMed] [Google Scholar]
- Sniegowski JA, Lappe JW, Patel HN, Huffman HA, Wachter RM. Base catalysis of chromophore formation in Arg(96) and Glu(222) variants of green fluorescent protein. J Biol Chem. 2005;280:26248–26255. doi: 10.1074/jbc.M412327200. [DOI] [PubMed] [Google Scholar]
- Thornton JW. Resurrecting ancient genes: experimental analysis of extinct molecules. Nat Rev Genet. 2004;5:366–375. doi: 10.1038/nrg1324. [DOI] [PubMed] [Google Scholar]
- Ugalde JA, Chang BSW, Matz MV. Evolution of coral pigments recreated. Science. 2004;305:1433. doi: 10.1126/science.1099597. [DOI] [PubMed] [Google Scholar]
- Wachter RM. The family of GFP-like proteins: structure, function, photophysics and biosensor applications. Photochem Photobiol. 2006;82:339–344. doi: 10.1562/2005-10-02-IR-708. [DOI] [PubMed] [Google Scholar]
- Weinreich DM, Delaney NF, DePristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- Weinreich DM, Watson RA, Chao L. Perspective: sign epistasis and genetic constraint on evolutionary trajectories. Evolution. 2005;59:1165–1174. [PubMed] [Google Scholar]
- Yokoyama S, Tada T, Zhang H, Britt L. Elucidation of phenotypic adaptations: molecular analyses of dim-light vision proteins in vertebrates. Proc Natl Acad Sci USA. 2008;105:13480–13485. doi: 10.1073/pnas.0802426105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.