Abstract
Identifying fertilization molecules is key to our understanding of reproductive biology, yet only a few examples of interacting sperm and egg proteins are known. One of the best characterized comes from the invertebrate archeogastropod abalone (Haliotis spp.), where sperm lysin mediates passage through the protective egg vitelline envelope (VE) by binding to the VE protein vitelline envelope receptor for lysin (VERL). Rapid adaptive divergence of abalone lysin and VERL are an example of positive selection on interacting fertilization proteins contributing to reproductive isolation. Previously, we characterized a subset of the abalone VE proteins that share a structural feature, the zona pellucida (ZP) domain, which is common to VERL and the egg envelopes of vertebrates. Here, we use additional expressed sequence tag sequencing and shotgun proteomics to characterize this family of proteins in the abalone egg VE. We expand 3-fold the number of known ZP domain proteins present within the VE (now 30 in total) and identify a paralog of VERL (vitelline envelope zona pellucida domain protein [VEZP] 14) that contains a putative lysin-binding motif. We find that, like VERL, the divergence of VEZP14 among abalone species is driven by positive selection on the lysin-binding motif alone and that these paralogous egg VE proteins bind a similar set of sperm proteins including a rapidly evolving 18-kDa paralog of lysin, which may mediate sperm–egg fusion. This work identifies an egg coat paralog of VERL under positive selection and the candidate sperm proteins with which it may interact during abalone fertilization.
Keywords: reproductive proteins, gamete recognition, reproductive isolation
Introduction
A major focus of reproductive biology is identifying how sperm and egg proteins interact during fertilization. Understanding these molecular interactions is of broad biological importance, but cognate sperm and egg molecules have been identified in just a few cases (Swanson and Vacquier 1997; Kamei and Glabe 2003; Harada et al. 2008). One of the best characterized involves fertilization proteins from abalone (Haliotis spp.). Abalone sperm contain two principle acrosomal proteins, lysin (Lewis et al. 1982) and an 18-kDa protein (Swanson and Vacquier 1995a), which are released upon binding of sperm to the elevated abalone egg envelope (the vitelline envelope [VE]). Lysin creates a hole in the VE, allowing sperm to cross this fertilization barrier by a nonenzymatic process involving binding to a large VE glycoprotein (the vitelline envelope receptor for lysin [VERL]) (Swanson and Vacquier 1997). VERL contains a tandemly repeated array of an ∼150 amino acid motif that is believed to be the lysin-binding sequence (Swanson and Vacquier 1997; Galindo et al. 2002). Though 18 kDa is ineffective at dissolving the VE, this fusagenic sperm protein coats the sperm acrosomal process and localizes to a second egg coat, structurally very similar to the VE, that overlies the plasma membrane at the egg surface (Mozingo et al. 1995). The 18-kDa sperm protein may bind a receptor at the egg plasma membrane, mediating sperm–egg fusion (Swanson and Vacquier 1995a). The similar structure and physical characteristics of lysin and 18 kDa support paralogy between these two sperm proteins, although they have evolved different functions in fertilization (Kresge et al. 2001a). Crystallography shows that these paralogs have the same alpha-helical bundle structure but strikingly different surface properties (Kresge et al. 2001b).
As is generally true for reproductive proteins (Swanson and Vacquier 2002; Clark et al. 2006), rapid adaptive divergence under positive selection is a common feature among abalone sperm and egg fertilization proteins. Lysin is among the most rapidly evolving proteins known (∼1 amino acid substitution among species per 105 years) (Metz et al. 1998) and 18 kDa evolves at least 2-fold faster (Metz et al. 1998), consistent with even stronger selection at the sperm–egg fusion stage of fertilization. Adaptive divergence is also prevalent among egg VE proteins (Aagaard et al. 2006), including lysin’s receptor VERL, for which positive selection is restricted to the two N-terminal repeats where lysin is believed to bind (Swanson et al. 2001; Galindo et al. 2003). Although the specific forces driving the evolution of abalone fertilization proteins are not yet clear (Swanson and Vacquier 2002; Clark et al. 2006, 2007), evidence from lysin and VERL demonstrates that the consequences of adaptive divergence can be significant. Positive selection targets lysin residues known to be important in species-specific dissolution of the VE (Lyon and Vacquier 1999; Yang, Swanson, and Vacquier 2000), and positive selection on the N-terminal VERL repeats may contribute to a rate-limiting step in VE dissolution by heterospecific lysins (Kresge et al. 2001a; Galindo et al. 2003). Because the species-specific interaction between lysin and the VE alone is known to constitute a significant barrier to hybridization (Leighton 2000), lysin and VERL have become prominent examples of sperm and egg recognition proteins that contribute to reproductive isolation and possibly speciation (Noor and Feder 2006).
Remarkably, despite rapid divergence of their constituent proteins (Swanson et al. 2003; Turner and Hoekstra 2006; Calkins et al. 2007), animal egg coats share a common molecular basis. In addition to its repeat array, VERL also contains a motif of approximately 260 amino acids known as the zona pellucida (ZP) domain. The ZP domain is found in diverse eukaryotic structures including the egg coats of vertebrates, where it is believed to play an important structural role in the formation of intermolecular fibers (Jovine et al. 2005). Mammalian egg coats contain a core set of 3–4 glycoproteins with a ZP domain (Lefievre et al. 2004), for which there is evidence of phylogenetic relatedness with a similar set of egg coat proteins of other vertebrates (Smith et al. 2005). ZP domain proteins are also prominent components of the egg envelopes of invertebrates, including abalone, for which at least nine additional ZP domain VE proteins were previously identified (Aagaard et al. 2006). Thus, although specific interactions between sperm and egg are probably species specific (Wassarman et al. 2004, 2005), the shared molecular features of egg coat structures suggest that invertebrate model systems such as abalone can provide insight into general features of animal reproduction (Evans 2000).
Previously, we identified ten of the ZP domain proteins that, along with VERL, are components of the abalone VE, more than half of which evolve under positive selection (Aagaard et al. 2006). Such pervasive adaptive divergence raises the intriguing possibility that, like VERL, these other vitelline envelope zona pellucida domain proteins (VEZPs) might also mediate interactions with rapidly evolving abalone sperm proteins during fertilization. The purpose of our current work is to characterize more fully this class of abalone egg coat proteins as well as test for binding between VEZPs and acrosomal sperm proteins. Toward these goals, we sequenced ∼5,000 randomly selected expressed sequence tags (ESTs) from a red abalone (Haliotis rufescens) ovary cDNA library and used tandem mass spectrometry (MS/MS) for more detailed characterization of red and green (Haliotis fulgens) abalone VEZPs. We then used lysin and 18-kDa affinity columns to identify several candidate VE receptors for these abalone sperm proteins, including a VERL paralog that contains a putative lysin-binding motif.
Materials and Methods
Red Abalone Ovary ESTs
Our previous work (Aagaard et al. 2006) focused on ESTs from pink abalone (Haliotis corrugata). Because of conservation concerns, our current work employs only abalone taxa commonly found in aquaculture. We focus on red abalone (H. rufescens) as well as green abalone (H. fulgens) because of their predominance in previous studies of reproductive proteins and in order to span the phylogenetic range of the California abalone (Lee and Vacquier 1995). A new red abalone ovary library of ESTs was generated by randomly sequencing ovary cDNAs as previously done (Aagaard et al. 2006) with minor modifications. Briefly, this entailed isolation of messenger RNA from ovary, followed by cDNA synthesis and cloning using the CloneMiner kit (Invitrogen, Carlsbad, CA) employing the pDONR222 vector. Plasmid DNAs from ∼5,000 clones were directionally sequenced (M13 primer) using standard fluorescent sequencing methods and traces processed and assembled using default parameters in PHREDPHRAP (http://www.phrap.org). Assembled sequences were used to search the NCBI nonredundant protein database using the NCBI Blast client server BlastCl3 (http://www.ncbi.nih.gov) for similarity to ZP domain proteins based on BlastX E values less than 10−5. Full-length coding sequences of ESTs showing homology to ZP domain proteins were obtained using rapid amplification of cDNA ends (RACE) as in Aagaard et al. (2006), and the presence of the ZP as well as other domains was confirmed via simple modular architecture research tool (http://www.embl-heidelberg.de).
Phylogenetic Analysis of Abalone ZP Domain Genes
Phylogenetic relationships among newly and previously identified red abalone ZP domain genes, as well as egg coat proteins containing ZP domains from other marine gastropods (Tegula spp.), were determined from nucleotide alignments of the complete ZP domain. Analyses included all red abalone ovary ESTs newly identified in this study as having a ZP domain (22 in total), VERL (Galindo et al. 2002) and the ten ZP domain proteins we previously identified (Aagaard et al. 2006) from red abalone, and three vitelline coat proteins from Tegula pfeiferi (Haino-Fukushima et al. 2000), or the closely related Tegula funebralis (kindly provided by Dr M. Hellberg). Alignments were carried out initially based on the translated nucleotide (protein) sequence using the ClustalX algorithm implemented in BioEdit (T. Hall), followed by visual alignment. Areas with ambiguous alignments were excluded, and the nucleotide alignment (804 nt) used in phylogenetic analyses employing likelihood criteria is implemented in PAUP (Swofford 2002). Likelihood analyses used the general time reversible model with four rate categories, estimating the gamma shape parameter and the proportion of invariable sites (GTR + 1 + γ). Heuristic search criteria included tree-bisection-reconnection branch swapping with ten random addition replicates. Support for nodes was estimated by 100 bootstrap replicates using the maximum likelihood estimates of substitution parameters and heuristic search criteria.
Identification of ZP Domain Proteins in the Abalone VE
ZP domain proteins present within the VE of abalone egg were identified using a shotgun proteomics approach employing MS/MS. VE proteins from red as well as green abalone were fractionated from whole eggs (Lewis et al. 1982), solubilized (Swanson and Vacquier 1997), and prepared for MS/MS via trypsin digestion as previously done (Aagaard et al. 2006). Two replicates of both red and green VEs (50 μg each) were analyzed via multidimensional protein identification technology (MudPIT) as in Aagaard et al. (2006) but using a 13-step (0–5 M ammonium acetate) salt elution. In addition, red and green VEs (five technical replicates each) were analyzed by reversed-phase high-performance liquid chromatography. For reversed-phase analyses, digested proteins (∼5 μg) were injected into a 75-μm internal diameter capillary column packed with 30 cm of Jupiter C12 reversed-phase resin, peptides were eluted in a 4-h water:acetonitrile gradient, and mass spectra acquisition was handled exactly as for MudPIT analyses.
The acquired tandem mass spectra were searched against a database containing the red abalone EST 6 reading frame translations and full-length abalone ZP domain protein sequences, proteins of common contaminants (e.g., trypsin, keratin), and a shuffled decoy database using a parallelized implementation of Sequest (Eng et al. 1994). The program DTASelect (Tabb et al. 2002) was used to filter the peptide identifications and assemble peptides and proteins. DTASelect filters (≥2 tryptic peptides of ≥7 residues each per protein; XCorr > 1.8, 2.5, and 3.5 for +1, +2, and +3 peptides; deltaCN > 0.12) were selected to produce protein identifications with a false discovery rate of <5%. The relative abundance of each ZP domain protein present in the VE was then inferred using the spectral counting method of Florens et al. (2006) (normalized spectral abundance factor [NSAF]) and averaged across full (MudPIT) and technical (reversed-phase) replicates separately for red or green abalone VEs. The correlation in relative abundance of these ZP domain proteins between red and green abalone was assessed using a Spearman rank correlation test (Sokal and Rohlf 1981).
Tests of Positive Selection VEZP14
To test for evidence of positive selection on VEZP14 and make comparisons with homologous regions of VERL, VEZP14 orthologs were cloned from three additional abalone species. Using degenerate 3′-RACE primers designed from the red abalone VEZP14 sequence in combination with RACE cDNA libraries made previously (Aagaard et al. 2006) from ovary RNA of green, pink, and Japanese ezo (Haliotis discus hannai) abalone, portions of VEZP14 orthologs were polymerase chain reaction amplified, TOPO cloned (Invitrogen), and sequenced. Gene-specific primers were then designed from species-specific 3′-RACE sequences and used in 5′-RACE to obtain the complete sequence of VEZP14 orthologs.
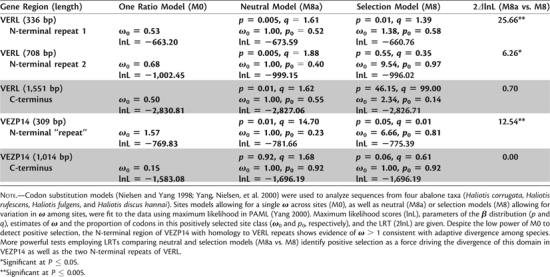
We tested for a signature of positive selection within two distinct regions of VEZP14 by comparing the proportion of amino acid changing nucleotide substitutions (dN) with the proportion of silent nucleotide substitutions (dS). The ratio (dN/dS or ω) can be used as an index of selection where ω < 1 is consistent with purifying selection, ω = 1 indicates neutral evolution, and ω > 1 is consistent with positive selection (i.e., adaptive diversification of orthologs). Codon substitution models employing maximum likelihood (Goldman and Yang 1994) including sites models (Nielsen and Yang 1998; Yang, Nielsen, et al. 2000) that allow for variable ω ratios among codons were used to individually analyze the N-terminal region of VEZP14 having homology to VERL repeats (309 nt; fig. 3) and the C-terminal region which includes the ZP domain (1,014 nt). For purposes of comparison, the homologous regions of VERL consisting of the nonhomogenized VERL repeats were analyzed in an identical fashion from the same taxa (N-terminal VERL repeats 1 and 2, accession numbers AF453553, AF490763, AF490764, AF490766; C-terminal region, accession numbers AF453553, DQ453750–DQ4537052). In addition to a model allowing for a single ω ratio among all codons (M0), several neutral (M1a, M7, M8a) and selection (M2a, M8) models were fit to the data using the computer program PAML (Yang 2000). All these codon substitution models assume a constant rate of silent substitution (dS). An unrooted phylogeny placing pink and Japanese ezo abalone as sisters (Coleman and Vacquier 2002) was used for all models, and those employing a β distribution (M7, M8a, and M8) included ten rate categories. Likelihood ratio tests (LRTs) of nested neutral and selection models were compared with χ2 distributions with one (M8a vs. M8) or two (M1a vs. M2a, M7 vs. M8) degrees of freedom to establish statistical support for the added parameters of the respective selection model.
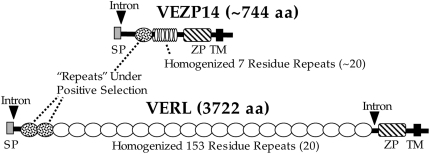
FIG. 3.
Gene structure supports paralogy between VERL and VEZP14. In addition to phylogenetic analysis based on the ZP domain (fig. 1), paralogy between VERL and VEZP14 is supported by several other shared features. Both genes have computationally predicted (SMART, http://www.embl-heidelberg.de) signal peptide (SP) and transmembrane (TM) domains followed by a furin cleavage site and both contain an intron at exactly the same location near the 5′-end of gene (following residue D26 or T26 for VERL or VEZP14, respectively). Significantly, VEZP14 also has a short (∼110 residue) region identified via Blast as having homology to the VERL repeats and which is thought to be important in binding of the lysin sperm protein (Swanson and Vacquier 1997). As with VERL, this N-terminal “repeat” of VEZP14 evolves under positive selection (table 1).
In order to test the codon model assumptions of a constant rate of silent substitution implemented in PAML, we constructed alternative likelihood models allowing for variable dS using the computer package HyPhy (Pond et al. 2005). Comparable likelihood models to those above (Muse and Gaut 1994) were constructed that either constrained dS among branches of the phylogeny to be constant (excluding the one internal branch without substitutions; a null model) or allowed for variation in dS among branches (the alternative model). LRTs of nested null and alternative models were compared with χ2 distributions with the appropriate degrees of freedom to establish statistical support for whether dS varied significantly among branches of the phylogeny.
Affinity Purification of Lysin and 18-kDa VE-Binding Partners
To identify additional VE proteins that might function in sperm–egg binding during abalone fertilization, we employed an affinity purification approach used previously to identify VERL as a receptor of lysin (Swanson and Vacquier 1997). We focused our biochemical studies on green abalone because of the ease of isolating large quantities of purified sperm proteins from this taxon. Briefly, this entailed purification of the green abalone sperm acrosomal protein lysin as well as 18 kDa using ion exchange column chromatography as in Swanson and Vacquier (1995b, 1997), construction of Affi-Gel 10 sperm protein (5 mg of lysin or 18 kDa) or negative control (5 mg chicken lysozyme or blank) affinity columns (1 ml Affi-Gel) followed by addition of 0.5 mg of solubilized green abalone VE protein and washing as in Swanson and Vacquier (1997), and finally three sequential elutions of bound VE proteins using 1 ml of 100 mM glycine, pH 2.8. The eluates were prepared and analyzed via reversed-phase MS/MS exactly as above for total solubilized VE material, and the relative abundance of each VE protein present in the eluate was then inferred using the spectral counting method of Florens et al. (2006) (NSAF).
Results
Diversity of Abalone Egg VEZPs
We identified 22 novel abalone ovary-expressed ZP domain genes by sequencing ∼5,000 clones from a red abalone ovary cDNA library and then used RACE to obtain full-length transcripts (accession numbers GQ851903–GQ851924). These include orthologs of the four pink abalone ovary ESTs containing a ZP domain identified previously from a pink abalone ovary EST library but for which we were unable to obtain full-length transcripts (Aagaard et al. 2006). These 22 genes represent all the ESTs in our current red abalone library identified by BlastX as having homology to ZP domain proteins. These sequences are all well diverged from VERL (Galindo et al. 2002) and the ten ZP domain genes that we identified previously (Aagaard et al. 2006) based on nucleotide alignment of the ZP domain (fig. 1). SMART computational prediction of functional domains for these 22 genes confirmed the presence of a canonical ZP domain with ten conserved cysteine residues. Each protein also had a predicted signal peptide. ZP proteins are often secreted (Jovine et al. 2005), and the presence of a signal peptide and a poly-A tail in each cDNA suggests that we recovered full-length coding sequences for all 22 genes. For one gene (ZPC, fig. 1), a premature stop codon suggests that it may have been pseudogenized, consistent with results from our MS/MS studies (see below). Six of the 22 genes have predicted transmembrane domains, but in most cases, there is little evidence of homology among these proteins outside of the ZP domains (though see below).
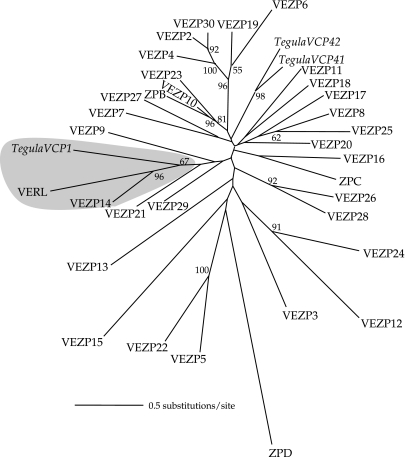
FIG. 1.
ZP domain genes identified from a red abalone (Haliotis rufescens) ovary EST library include a VERL paralog. RACE was used to extend ESTs predicted to have homology to ZP domain proteins, identifying 22 novel ovary-expressed genes in addition to VERL and the ten ZP domain genes identified previously (Aagaard et al. 2006). Phylogenetic analysis of the ZP domain (∼804 nt) from these abalone genes along with vitelline coat proteins (VCPs) from Tegula pfeifferi (Haino-Fukushima et al. 2000) and the closely related Tegula funebralis employing likelihood was performed under the general time reversible model with four rate categories using PAUP (Swofford 2002). Terminal branches in the tree are labeled with a proposed gene name specifying whether the protein was found within solubilized VE material via MS/MS (VERL, VEZP2–30; fig. 2) or not (ZPB–D), and bootstrap support based on 100 bootstrap replicates is shown for nodes with ≥50% support. VERL and VEZP14 cluster together with strong support and are co-orthologous to a ZP domain VCP from T. funebralis.
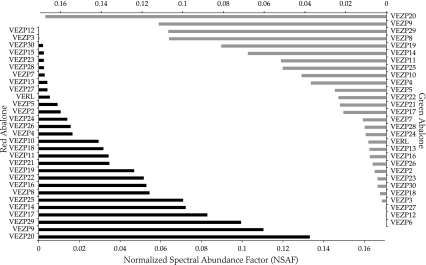
Shotgun proteomics identified peptides corresponding to a majority (19/22) of the newly identified ZP domain genes as constituents of the VE of abalone eggs. MS/MS spectra were matched to at least two peptides for each of these proteins in solubilized VEs from red or green abalone eggs, as well as for VERL and the nine VEZPs we identified previously (Aagaard et al. 2006). Peptides were also found for ZPA, the single ZP domain gene for which VE peptides were not found previously (Aagaard et al. 2006). In two cases (VEZP6 and VEZP15), peptides for low abundance VEZPs were identified in VEs of only one taxon (red or green, respectively; fig. 2). No peptides were identified for ZPC, which contains a premature stop codon within the ZP domain. In summary, our current study shows that the VE of abalone eggs contains at least 30 ZP domain proteins (fig. 2). Following the prior naming convention, all ZP domain proteins found within the VE are designated as VEZPs and are given a numeric designation based on the order in which each was found (e.g., ZPA is now VEZP11). ZP domain genes for which evidence of peptides within solubilized VEs is lacking are given an alphabetical designator (ZPB–ZPD; fig. 1). The relative abundance of VEZPs as inferred from the NSAF, a method that accounts for some aspects of detection bias among proteins and allows for comparison over separate experiments (Florens et al. 2006), differs markedly among VEZPs but is broadly consistent between red and green abalone (fig. 2; Spearman rank correlation rs = 0.80, P = 10−6) and between experimental procedures (replicates of MudPIT vs. reversed phase).
FIG. 2.
Relative abundance of ZP domain proteins in the VE of abalone eggs. Abalone eggs are surrounded by a raised VE separated from the egg (egg cytoplasm) by a perivitelline space. Isolated VEs from red (Haliotis rufescens) and green (Haliotis fulgens) abalone were solubilized and used for peptide identification in MS/MS experiments. Ovary-expressed ZP domain genes are designated as VEZPs if at least two peptides from red or green eggs were matched to a ZP domain gene. A spectral counting method (NSAF) (Florens et al. 2006) provides an estimate of the relative abundance of VEZPs in the VE of red (black bars) or green (gray bars) abalone eggs. Relative abundance of VEZPs is highly correlated between red and green abalone (Spearman rank correlation rs = 0.80, P = 10−6).
Gene Structure of VEZP14
Phylogenetic analyses of the ZP domain of abalone ovary-expressed genes identify one gene (VEZP14) that is sister to VERL with high confidence (fig. 1). Because this topology suggested that VERL and VEZP14 might be paralogs, we searched for evidence of sequence homology outside of the ZP domain or other shared features of gene structure. BlastP identified a region of ∼110 residues near the N-terminal end of VEZP14 (C187–P297) with sequence homology (E value = 5 × 10−11) to the N-terminal VERL repeat, as well as other repeats in the array. Sequencing of VEZP14 from genomic DNA of red abalone showed that although this gene lacks the 3′-intron found in VERL (Galindo et al. 2002), both genes do contain an intron in exactly the same location (following residue D26 or T26 for VERL or VEZP14, respectively) immediately following the signal peptide (fig. 3). Both genes also contain a furin cleavage site immediately C-terminal of the transmembrane domain. Finally, although the homogenized repeats of VERL (20 repeats of ∼153 residues) are distinct in sequence, VEZP14 shares this general feature of a homogenized repeat array (∼20 repeats of the simple canonical sequence TTTTTTP) N-terminal to the ZP domain.
Patterns of Positive Selection on VEZP14 Relative to VERL
Using RACE, we successfully identified orthologs of VEZP14 from green, pink, and Japanese ezo abalone (accession numbers GQ851925–GQ851927), allowing us to examine the evolutionary forces driving the divergence of this gene. To make comparisons with VERL, we focused our analyses of selection on regions with shared homology, including the N-terminal 110-residue VERL-like repeat and the 338 C-terminal residues that include the ZP domain (fig. 3). Nucleotide divergence among these taxa at silent sites in both regions of VEZP14 are similar (N-terminal dS = 0.19, C-terminal dS = 0.15) and do not differ significantly among branches of the abalone phylogeny (P = 0.43 and 0.40, respectively) as is seen for VERL among these taxa (N-terminal repeats 1 and 2 dS = 0.22 and 0.25, respectively, and C-terminal dS = 0.13) and are consistent with the recent divergence of the California abalone (<18 Ma) (Lee and Vacquier 1995; Metz et al. 1998). In contrast, nonsynonymous substitutions differ markedly between the VERL-like repeat of VEZP14 (dN = 0.31) and the C-terminus (dN = 0.02), a pattern similar to what is seen for VERL (N-terminal repeats 1 and 2 dN = 0.12 and 0.17, respectively, and C-terminal dN = 0.07).
A signature of positive selection on the N-terminal region of VEZP14 is suggested by the proportionally higher dN relative to dS, resulting in a dN/dS ratio (ω) > 1 (table 1). Powerful and robust likelihood-based selection models (M2a and M8) support this conclusion based on LRTs with corresponding nested neutral models (M1a, M7, or M8a, respectively) and demonstrate that positive selection is also a feature of the two N-terminal VERL repeats as has been shown previously among a broader group of taxa (Galindo et al. 2003). Table 1 presents results only for models M8a and M8, for which model comparisons using LRTs are the most conservative and robust (Wong et al. 2004), but results from other neutral and selection model comparisons are qualitatively similar. In contrast, there is no evidence of positive selection within the C-terminus of VEZP14 or VERL, consistent with strong purifying selection within these domains (table 1). Significantly, simulations show that the LRT applied to these model tests is robust and does not lead to an elevated type 1 error rate even for the small sample size used here (Anisimova et al. 2001).
Table 1.
Tests of Positive Selection among Homologous VERL and VEZP14 Domains.
 |
Identification of Egg VE Proteins That Bind to Lysin and 18 kDa
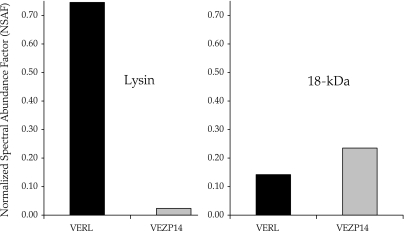
We used affinity purification to identify possible interactions among abalone sperm and egg coat proteins. Several abalone VE proteins (all VEZPs) in addition to VERL bound to the lysin and 18-kDa affinity columns as determined by MS/MS of the affinity column eluate. Whereas one protein (VEZP25) bound nonspecifically, repeatedly eluting under low-pH conditions from both negative control and sperm protein affinity columns, a total of five VEZPs bound specifically only to the lysin and/or 18-kDa columns. Three of these were found in the eluate of both lysin and 18-kDa columns, including VERL and its paralog VEZP14, as well as VEZP19. Peptide spectra corresponding to VEZP29 and VEZP9 were also found in the eluate from either the lysin or the 18-kDa column, respectively. No other ESTs were matched to spectra from any eluate. As expected from previous binding experiments (Swanson and Vacquier 1997), VERL was the predominant protein found in the eluate from the lysin column, but it also bound more strongly than other VEZPs to the18-kDa column based on spectral counts (NSAF). Figure 4 shows results for VERL relative to VEZP14 only from the third elution, representing the most tightly bound VEZPs, but this trend is equally apparent in elutions 1 and 2. Figure 4 also demonstrates an intriguing pattern of reduced binding between 18 kDa and VERL relative to lysin, whereas 18 kDa appears to bind more strongly to VERL’s paralog (VEZP14) relative to lysin.
FIG. 4.
VERL and VEZP14 paralogs both bind paralogous sperm proteins important in abalone fertilization. The paralogous acrosomal sperm proteins lysin and 18 kDa important in VE dissolution and sperm–egg fusion (Kresge et al. 2001b), respectively, were purified individually from acrosome-reacted green abalone (Haliotis fulgens) sperm and used for affinity purification of binding partners present within solubilized green abalone VEs. VEZPs binding with high affinity specifically to lysin or 18-kDa affinity columns were identified via MS/MS. A total of four VE proteins were found to elute from lysin and 18-kDA columns (pH 2.8 elution 3) but were absent from negative controls, including VERL and VEZP14. Shared binding interactions with lysin and 18 kDa are further support of paralogy between VERL and VEZP14, although the relative strength of binding may differ based on spectral abundance (NSAF) (Florens et al. 2006), consistent with functional divergence following duplication.
Discussion
We present three complimentary findings. First, the abalone egg VE contains a remarkable diversity of proteins with ZP domains (Aagaard et al. 2006). Second, one of these (VEZP14) is a paralog of lysin’s receptor, VERL, and the putative sperm-binding motif of VEZP14 has diverged among abalone species by positive selection. Third, paralogous acrosomal proteins from abalone sperm (lysin and 18 kDa) bind a similar set of VEZPs including VERL and VEZP14. Together, these results provide compelling insight into the molecular mechanisms of fertilization and the forces driving gamete recognition protein evolution in abalone.
Abalone VEZPs Are Extraordinarily Diverse
The current study expands our earlier results showing that, like the egg coat structures of vertebrates, ZP domain proteins are also a prominent component of the VE of abalone. Previously, we identified nine proteins from the abalone VE that, along with VERL, contain a ZP domain (VEZP2–10) (Aagaard et al. 2006). Here, we identify 22 additional ovary-expressed ZP domain genes (fig. 1), the majority of which encode proteins that our MS/MS studies confirm are present in the abalone VE. In total, we have now identified 30 abalone VEZPs (fig. 2). This is 3-fold greater than previous estimates and is dramatically greater than the diversity of vertebrate egg coat ZP domain proteins. Phylogenetic analyses of vertebrate ZP domain proteins suggest that five to six ancestral genes were present prior to diversification of the vertebrates (Smith et al. 2005). Subsequent lineage-specific loss or gain resulted in a core set of just a few orthologous proteins found within the egg coats of vertebrates (three to four in higher mammals). Thus, this family of egg coat proteins is severalfold more diverse in abalone.
The number of abalone VEZPs is remarkable when compared with the mammalian egg ZP, though neither the source nor the significance of this diversity is clear. Abalone VEZPs are highly diverged (fig. 1), similar to what is seen for vertebrate ZP genes (Clark et al. 2006), and there is evidence of homology with the egg coat proteins of another marine gastropod (Tegula spp., fig. 1) which diverged from the lineage leading to abalone >250 Ma (Tracey et al. 1993). Thus, ZP domain proteins are clearly ancient features of the egg coats of marine invertebrates, with recent gene duplication contributing to certain lineage-specific expansions (e.g., in the lineage leading to VERL; fig. 1 and see below). One possible explanation for these ancient and diverse abalone VEZPs relates to their mode of broadcast spawning external fertilization, a common and presumed ancestral state among marine invertebrates (Parker 1984; Wray 1995). Because their egg coat structures mediate a host of biotic and abiotic factors in addition to interactions with sperm proteins (Podolsky 2004), perhaps marine invertebrates maintain a greater diversity of structural proteins. Tests of selection among all 30 abalone VEZPs are beyond the scope of our current work, but adaptive divergence is known to be a prominent force driving the evolution of many of these molecules (Aagaard et al. 2006) consistent with their potentially dynamic functions. Addressing the question of egg coat protein diversity relative to mode of fertilization awaits further comparative studies, particularly more thorough characterization of proteins from nonvertebrate taxa. However, the extraordinary diversity of VEZPs we have identified clearly hints at the complexity of roles played by the egg coats of marine broadcast spawners such as abalone.
VERL and VEZP14 Are Paralogs
Phylogenetic analyses of abalone ZP domain proteins indicate that VEZP14, an abundant VE protein, is a paralog of abalone VERL. In our ZP domain phylogeny (fig. 1), an egg coat VERL-like protein identified from the marine gastropod Tegula is sister to a well-supported clade that includes both abalone VERL and VEZP14 (fig. 2). Based on this topology inferred from alignment of the ZP domain alone, VERL and VEZP14 arose by gene duplication sometime after the lineages leading to abalone and Tegula split more than 250 Ma (Tracey et al. 1993). The ZP domain of VERL and VEZP14 is well diverged (∼44% identity), and there is no evidence of recent polyploidy in abalone (Thiriot-Quievreux 2003; Gallardo-Escarate et al. 2004) suggesting that duplication was ancient. Comparison of the gene structure of VERL and VEZP14 further supports this relationship (fig. 3). In addition to a ZP domain and several short conserved motifs, both genes contain a 5′-intron in exactly the same location. Perhaps most significantly, VEZP14 also contains a region of ∼110 amino acids with homology to the VERL repeats. Taken together, this similarity of gene structure is an independent evidence of paralogy between VERL and VEZP14.
Sequence-based evidence supports the idea that VEZP14 may function as a sperm receptor similar to its paralog VERL. VEZP14 contains the ∼110 amino acid motif with homology to VERL repeats (fig. 3). Stoichiometry of lysin–VERL binding identifies individual VERL repeats as the lysin-binding motif (Kresge et al. 2001a). Although the motif in VEZP14 is highly diverged from VERL repeats at the amino acid level (∼11% mean identity), it too might facilitate binding with lysin or perhaps another related but similarly diverged sperm protein (see below). In addition, the pattern of positive selection discretely focused on N-terminal VERL repeats is mirrored in VEZP14. Although there is no evidence of positive selection on the C-terminus region of either gene, our statistical analyses provide strong evidence of adaptive divergence within the two N-terminal repeats of VERL as seen previously for a larger data set (Galindo et al. 2003) as well as the homologous motif in VEZP14 (table 1 and fig. 3). Positive selection on the putative lysin-binding motif in VEZP14 hints at the possibility of coevolution between sperm proteins and VERL’s paralog. Intriguingly, amino acid substitutions in this motif are at least 2-fold greater for VEZP14 (ω = 1.57) than for VERL (ω = 0.53 and 0.68 for repeats 1 and 2, respectively), consistent with stronger positive selection on VEZP14. If VEZP14 does indeed function as a sperm protein receptor, this suggests either tighter coevolution between ligand and receptor as compared with lysin–VERL or that another more rapidly evolving sperm protein serves as the primary ligand of VEZP14.
Sperm and Egg Protein Paralogs Bind Each Other
Abalone lysin and 18-kDa sperm proteins are ancient paralogs (Kresge et al. 2000) that are highly diverged in primary amino acid sequence (∼14% identity) (Kresge et al. 2001b) and have evolved specialized functions in fertilization. Lysin mediates sperm passage across the elevated abalone VE (Lewis et al. 1982), but 18 kDa is ineffective at VE dissolution and is thought to function in sperm–egg fusion at the plasma membrane surface (Swanson and Vacquier 1995a). Despite divergence, our affinity purification experiments demonstrate that these two sperm paralogs bind a similar repertoire of VEZPs. Although two VEZPs were identified from the eluate of only lysin or 18-kDa affinity columns (VEZP29 and VEZP9, respectively), the majority of proteins (three of five) were found in the eluate of both columns, including VEZP19 along with VERL and VEZP14. With the notable exception of VERL’s paralog VEZP14, there is no evidence of a lysin-binding motif (a region of homology to the VERL repeats) among these other three genes. Because the ZP domain can form intermolecular disulfide bonds with other egg coat proteins (Darie et al. 2004; Jovine et al. 2005) which should remain intact during our affinity purification experiments, this suggests that VEZP9, VEZP19, and VEZP29 might elute from columns as a result of interactions with either VERL or VEZP14, rather than by binding directly to lysin or 18 kDa. This is an intriguing result because of evidence from mouse that multiprotein complexes govern sperm binding to the egg coat based on gross three-dimensional structure (Hoodbhoy and Dean 2004). Strong positive selection is known to act on at least one of these additional abalone proteins (VEZP9) (Aagaard et al. 2006) which suggests the possibility of correlated evolution with rapidly evolving sperm proteins even in the absence of direct interactions. Taken together with results from our affinity purification studies, this suggests that future studies in abalone consider the possibility of coevolution among ZP domain egg coat proteins as another component of recognition between sperm and egg.
Binding of VEZPs by lysin and 18 kDa fits expectations from past experimental results as well as our data on sequence divergence among paralogs. In our affinity purification experiments, VERL is relatively more abundant in the eluate from lysin as compared with 18-kDa affinity columns (fig. 4). Binding between 18 kDa and VERL (net positive and negative charge, respectively) is unlikely to be an artifact of charge alone as one of our negative control columns contains a protein (lysozyme) of similar charge and size as both lysin and 18 kDa. Rather, the approximately 4-fold decrease in 18 kDa’s ability to bind VERL is consistent with divergence in amino acid sequence between 18 kDa and VERL’s primary ligand lysin and may help to explain the inability of 18 kDa to dissolve the abalone VE (Lewis et al. 1982). Despite overall conservation of structure between these paralogous acrosomal sperm proteins that allows for some residual binding to VERL, the surface properties of 18 kDa are distinct from lysin (Kresge et al. 2001b) apparently reducing its affinity for VERL.
Although the relative abundance of VEZP14 in the eluate from lysin and 18-kDA affinity columns is lower overall, possibly due to fewer copies of the putative sperm-binding motif in the molecule, this situation is mirrored for VERL’s paralog (fig. 4). VEZP14 is relatively more abundant in the eluate from the 18-kDa affinity column as compared with the lysin column (a 12-fold increase). Significantly, the preferential binding between 18 kDa and VEZP14 is consistent with the proportional increase in the intensity of positive selection on both 18 kDa and VEZP14’s sperm protein–binding motif relative to lysin and VERL (>2-fold in both cases). Divergence in primary amino acid sequence is the most likely cause of the differences we observed in specificity and strength of binding between these sperm and egg proteins, but patterns of glycosylation also could potentially play a role. Trypsin generated glycopeptides of VERL that have the carbohydrate moieties intact, but the peptide backbone destroyed, have no effect on lysin binding of VERL (Swanson WJ, unpublished data), whereas intact VERL is strongly inhibitory (Swanson and Vacquier 1997). Although the three-dimensional presentation of carbohydrates on trypsin-digested VERL is likely to be modified, this suggests that lysin–VERL interactions are protein–protein mediated. However, oligosaccharides are known to affect binding of mammalian sperm to the egg coat (Florman and Wassarman 1985; Bleil et al. 1988), and amino acid substitutions at and surrounding glycosylation sites of the likely binding domain of a candidate egg receptor of mouse sperm appear to mediate specificity (Bleil et al. 1988) and occur as a result of positive selection (Swanson et al. 2003). Among the abalone taxa we studied, 8 of 22 (VERL repeat 1), 38 of 58 (VERL repeat 2), and 10 of 21 (VEZP14) possible sites of O-linked (serine/threonine) oligosaccharides harbor amino acid substitutions. Thus, although the potential role of oligosaccharides in mediating binding among abalone sperm and egg proteins is beyond the scope of our present work, this is a topic deserving consideration in future work.
Is VEZP14 the 18-kDa Sperm Protein Receptor?
Two lines of evidence presented above are consistent with the hypothesis that VEZP14 may function as a receptor of 18 kDa during abalone fertilization. Briefly, these include the following: 1) the sperm protein–binding motif of VEZP14 exhibits the predicted intensity of positive selection given the very rapid divergence of 18 kDa and 2) 18 kDa binding of VEZP14 appears to be much greater relative to lysin. This is an appealing hypothesis given that VEZP14 is a paralog of VERL, the receptor of 18-kDa’s paralog lysin. More conclusive evidence will likely involve several distinct lines of investigation. Future studies focusing on pairwise interactions among sperm and egg proteins would allow direct tests of the affinities among sperm-binding motifs and 18 kDa, which our results suggest should be highest for VEZP14. Testing for correlated sequence divergence between VEZP14 and 18 kDa would similarly provide an indirect means of testing this hypothesis based on patterns of molecular evolution. Of particular importance are investigations of proteins at the egg plasma membrane as the18-kDa sperm protein’s role in sperm–egg fusion likely involves a plasma membrane–associated receptor (Swanson and Vacquier 1995a). Thus, a key prediction of our hypothesis is that in addition to the elevated abalone VE, VEZP14 should be present at the egg plasma membrane. Because ultrastructure studies show that the plasma membrane of abalone eggs is overlain by a second fibrous layer that shares physical characteristics with the VE, including fibers of similar size and dimensions (Lewis et al. 1982), it seems plausible that the VE and egg surface coat might be composed of similar structural proteins. Furthermore, almost all the characteristic egg coat molecules in our ovary EST library (ZP domain proteins) were matched to peptides from the VE, thus there are few other candidates for such a structure.
Why Do 18 kDa and VEZP14 Evolve So Rapidly?
Both 18 kDa and VEZP14 evolve at least twice as fast as lysin and VERL, their respective paralogs. If VEZP14 functions as a receptor of 18 kDa, this rapid adaptive divergence suggests that selection on both sperm and egg components of fusion is extremely strong, raising the question of what might drive their divergence. Gamete pathogens (Vacquier 1998) are one possibility, though this seems unlikely as pathogen attack should be focused principally on the external VE. Furthermore, although we cannot directly discount the possibility of reinforcement (Ortiz-Barrientos et al. 2004) on 18 kDa or VEZP14, it seems an unlikely explanation as lysin and VERL are known to undergo selective sweeps even in the absence of selection to avoid hybrid formation (Clark et al. 2007). The opportunity for intersexual conflict (Rice and Holland 1997) remains strong, however, and could also drive the divergence of fertilization proteins that function following passage of sperm through the VE. Multiple sperm penetrate the VE during abalone fertilization, providing for sperm competition (Clark et al. 1999) that can drive selection on sperm proteins (e.g., lysin and 18 kDa) because sperm compete individually for first access to the egg. This race can cause intersexual conflict (Rice and Holland 1997) because development will arrest if multiple sperm fuse with an egg (polyspermy), resulting in strong selection on eggs to slow sperm access or blocks to polyspermy (Gould-Somero and Jaffe 1984). Female receptors of sperm proteins (VERL and potentially VEZP14) could act in this capacity, resulting in a continual chase between sperm and egg that our results suggest might intensify immediately prior to fusion, at which point depolarization of the egg membrane signals the end of the race (Gould and Stephano 2003).
Conclusions
The VE of abalone eggs contains a remarkable diversity of ZP domain proteins. This study provides evidence of at least 30 of these VEZPs including VEZP14, a paralog of the VE receptor for abalone sperm lysin (VERL). VEZP14 contains a putative sperm-binding motif like VERL, and the ∼2-fold increased rate of divergence of this domain under positive selection among abalone taxa parallels our affinity purification studies demonstrating increased binding between VEZP14 and lysin’s paralog 18 kDa. In short, both sequence divergence and binding assays implicate VEZP14 as a receptor of the18-kDa abalone sperm fusion protein. If confirmed in future studies, our work suggests that abalone may prove a useful model system for deciphering the molecular basis of sperm–egg fusion as well as the evolutionary forces acting during this final step in fertilization.
Acknowledgments
We thank M. E. Hellberg for sharing the Tegula funebralis vitelline coat protein 1 ZP domain gene sequence prior to publication. We also thank the Swanson Laboratory and two anonymous reviewers for comments on the manuscript. This work was supported by the National Institutes of Health (NIH) grants HD042563, HD054631, and HD057974 and National Science Foundation grants DEB-0743539 and DEB-0918106 (to W.J.S.), NIH grants DK069386 and HG004263 (to M.J.M.), NIH grant HD12986 (to V.D.V.), and NIH grant FHD053185A (to J.E.A.).
References
- Aagaard JE, Yi X, MacCoss MJ, Swanson WJ. Rapidly evolving zona pellucida domain proteins are a major component of the vitelline envelope of abalone eggs. Proc Natl Acad Sci USA. 2006;103:17302–17307. doi: 10.1073/pnas.0603125103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M, Bielawski JP, Yang Z. Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol Biol Evol. 2001;18:1585–1592. doi: 10.1093/oxfordjournals.molbev.a003945. [DOI] [PubMed] [Google Scholar]
- Bleil JD, Greve JM, Wassarman PM. Identification of a secondary sperm receptor in the mouse egg zona pellucida: role in maintenance of binding of acrosome-reacted sperm to eggs. Dev Biol. 1988;128:376–385. doi: 10.1016/0012-1606(88)90299-0. [DOI] [PubMed] [Google Scholar]
- Calkins JD, El-Hinn D, Swanson WJ. Adaptive evolution in an avian reproductive protein: ZP3. J Mol Evol. 2007;65:555–563. doi: 10.1007/s00239-007-9034-8. [DOI] [PubMed] [Google Scholar]
- Clark AG, Begun DJ, Prout T. Female × male interactions in Drosophila sperm competition. Science. 1999;283:217–220. doi: 10.1126/science.283.5399.217. [DOI] [PubMed] [Google Scholar]
- Clark NL, Aagaard JE, Swanson WJ. Evolution of reproductive proteins from animals and plants. Reproduction. 2006;131:11–22. doi: 10.1530/rep.1.00357. [DOI] [PubMed] [Google Scholar]
- Clark NL, Findlay GD, Yi X, MacCoss MJ, Swanson WJ. Duplication and selection on abalone sperm lysin in an allopatric population. Mol Biol Evol. 2007;24:2081–2090. doi: 10.1093/molbev/msm137. [DOI] [PubMed] [Google Scholar]
- Coleman AW, Vacquier VD. Exploring the phylogenetic utility of its sequences for animals: a test case for abalone (haliotis) J Mol Evol. 2002;54:246–257. doi: 10.1007/s00239-001-0006-0. [DOI] [PubMed] [Google Scholar]
- Darie CC, Biniossek ML, Jovine L, Litscher ES, Wassarman PM. Structural characterization of fish egg vitelline envelope proteins by mass spectrometry. Biochemistry. 2004;43:7459–7478. doi: 10.1021/bi0495937. [DOI] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR., 3rd An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–986. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Evans JP. Getting sperm and egg together: things conserved and things diverged. Biol Reprod. 2000;63:355–360. doi: 10.1095/biolreprod63.2.355b. [DOI] [PubMed] [Google Scholar]
- Florens L, Carozza MJ, Swanson SK, Fournier M, Coleman MK, Workman JL, Washburn MP. Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods. 2006;40:303–311. doi: 10.1016/j.ymeth.2006.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florman HM, Wassarman PM. O-linked oligosaccharides of mouse egg ZP3 account for its sperm receptor activity. Cell. 1985;41:313–324. doi: 10.1016/0092-8674(85)90084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo BE, Moy GW, Swanson WJ, Vacquier VD. Full length sequence of VERL, the egg vitelline envelope receptor for abalone sperm lysin. Gene. 2002;288:111–117. doi: 10.1016/s0378-1119(02)00459-6. [DOI] [PubMed] [Google Scholar]
- Galindo BE, Vacquier VD, Swanson WJ. Positive selection in the egg receptor for abalone sperm lysin. Proc Natl Acad Sci USA. 2003;100:4639–4643. doi: 10.1073/pnas.0830022100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo-Escarate C, Alvarez-Borrego J, Del Rio Portilla M, Kober V. Karyotype of Pacific red abalone Haliotis rufescens using image analysis. J Shellfish Res. 2004;23:205–211. [Google Scholar]
- Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- Gould MC, Stephano JL. Polyspermy prevention in marine invertebrates. Microsc Res Tech. 2003;61:379–388. doi: 10.1002/jemt.10351. [DOI] [PubMed] [Google Scholar]
- Gould-Somero M, Jaffe LA. Control of cell fusion at fertilization by membrane potential. In: Beers RF, Bassett EG, editors. Cell fusion and transformation. New York: Raven Press; 1984. pp. 27–38. [Google Scholar]
- Haino-Fukushima K, Fan X, Nakamura S. Three new components contained in the vitelline coat of Tegula pfeifferi. Zygote. 2000;8:S61. [PubMed] [Google Scholar]
- Harada Y, Takagaki Y, Sunagawa M, Saito T, Yamada L, Taniguchi H, Shoguchi E, Sawada H. Mechanism of self-sterility in a hermaphroditic chordate. Science. 2008;320:548–550. doi: 10.1126/science.1152488. [DOI] [PubMed] [Google Scholar]
- Hoodbhoy T, Dean J. Insights into the molecular basis of sperm-egg recognition in mammals. Reproduction. 2004;127:417–422. doi: 10.1530/rep.1.00181. [DOI] [PubMed] [Google Scholar]
- Jovine L, Darie CC, Litscher ES, Wassarman PM. Zona pellucida domain proteins. Annu Rev Biochem. 2005;74:83–114. doi: 10.1146/annurev.biochem.74.082803.133039. [DOI] [PubMed] [Google Scholar]
- Kamei N, Glabe CG. The species-specific egg receptor for sea urchin sperm adhesion is EBR1, a novel ADAMTS protein. Genes Dev. 2003;17:2502–2507. doi: 10.1101/gad.1133003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kresge N, Vacquier VD, Stout CD. The high resolution crystal structure of green abalone sperm lysin: implications for species-specific binding of the egg receptor. J Mol Biol. 2000;296:1225–1234. doi: 10.1006/jmbi.2000.3533. [DOI] [PubMed] [Google Scholar]
- Kresge N, Vacquier VD, Stout CD. Abalone lysin: the dissolving and evolving sperm protein. Bioessays. 2001a;23:95–103. doi: 10.1002/1521-1878(200101)23:1<95::AID-BIES1012>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Kresge N, Vacquier VD, Stout CD. The crystal structure of a fusagenic sperm protein reveals extreme surface properties. Biochemistry. 2001b;40:5407–5413. doi: 10.1021/bi002779v. [DOI] [PubMed] [Google Scholar]
- Lee Y-H, Vacquier VD. Evolution and systematics in Haliotidae (Mollusca:Gastropoda): inferences from DNA sequences of sperm lysin. Mar Biol. 1995;124:267–268. [Google Scholar]
- Lefievre L, Conner SJ, Salpekar A, et al. (12 co-authors) Four zona pellucida glycoproteins are expressed in the human. Hum Reprod. 2004;19:1580–1586. doi: 10.1093/humrep/deh301. [DOI] [PubMed] [Google Scholar]
- Leighton DL. The biology and culture of the California abalones. Pittsburgh (PA): Dorrance Publishing Co; 2000. [Google Scholar]
- Lewis CA, Talbot CF, Vacquier VD. A protein from abalone sperm dissolves the egg vitelline layer by a nonenzymatic mechanism. Dev Biol. 1982;92:227–239. doi: 10.1016/0012-1606(82)90167-1. [DOI] [PubMed] [Google Scholar]
- Lyon JD, Vacquier VD. Interspecies chimeric sperm lysins identify regions mediating species-specific recognition of the abalone egg vitelline envelope. Dev Biol. 1999;214:151–159. doi: 10.1006/dbio.1999.9411. [DOI] [PubMed] [Google Scholar]
- Metz EC, Robles-Sikisaka R, Vacquier VD. Nonsynonymous substitution in abalone sperm fertilization genes exceeds substitution in introns and mitochondrial DNA. Proc Natl Acad Sci USA. 1998;95:10676–10681. doi: 10.1073/pnas.95.18.10676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozingo NM, Vacquier VD, Chandler DE. Structural features of the abalone egg extracellular matrix and its role in gamete interaction during fertilization. Mol Reprod Dev. 1995;41:493–502. doi: 10.1002/mrd.1080410412. [DOI] [PubMed] [Google Scholar]
- Muse SV, Gaut BS. A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome. Mol Biol Evol. 1994;11:715–724. doi: 10.1093/oxfordjournals.molbev.a040152. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Yang Z. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics. 1998;148:929–936. doi: 10.1093/genetics/148.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor MA, Feder JL. Speciation genetics: evolving approaches. Nat Rev Genet. 2006;7:851–861. doi: 10.1038/nrg1968. [DOI] [PubMed] [Google Scholar]
- Ortiz-Barrientos D, Counterman BA, Noor MA. The genetics of speciation by reinforcement. PLoS Biol. 2004;2:e416. doi: 10.1371/journal.pbio.0020416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker GA. Sperm competition and the evolution of animal mating strategies. In: Smith RL, editor. Sperm competition and the evolution of animal mating systems. Orlando (FL): Academic Press; 1984. pp. 1–60. [Google Scholar]
- Podolsky RD. Life-history consequences of investment in free-spawned eggs and their accessory coats. Am Nat. 2004;163:735–753. doi: 10.1086/382791. [DOI] [PubMed] [Google Scholar]
- Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Rice WR, Holland B. The enemies within: intergenomic conflict, interlocus contest evolution (ICE), and the intraspecific Red Queen. Behav Ecol Sociobiol. 1997;41:1–10. [Google Scholar]
- Smith J, Paton IR, Hughes DC, Burt DW. Isolation and mapping the chicken zona pellucida genes: an insight into the evolution of orthologous genes in different species. Mol Reprod Dev. 2005;70:133–145. doi: 10.1002/mrd.20197. [DOI] [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ. Biometry. 2nd ed. New York: W H Freeman and Company; 1981. [Google Scholar]
- Swanson WJ, Aquadro CF, Vacquier VD. Polymorphism in abalone fertilization proteins is consistent with the neutral evolution of the egg's receptor for lysin (VERL) and positive Darwinian selection of sperm lysin. Mol Biol Evol. 2001;18:376–383. doi: 10.1093/oxfordjournals.molbev.a003813. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Nielsen R, Yang Q. Pervasive adaptive evolution in Mammalian fertilization proteins. Mol Biol Evol. 2003;20:18–20. doi: 10.1093/oxfordjournals.molbev.a004233. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD. Liposome fusion induced by a M(r) 18,000 protein localized to the acrosomal region of acrosome-reacted abalone spermatozoa. Biochemistry. 1995a;34:14202–14208. doi: 10.1021/bi00043a026. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD. Extraordinary divergence and positive Darwinian selection in a fusagenic protein coating the acrosomal process of abalone spermatozoa. Proc Natl Acad Sci USA. 1995b;92:4957–4961. doi: 10.1073/pnas.92.11.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD. The abalone egg vitelline envelope receptor for sperm lysin is a giant multivalent molecule. Proc Natl Acad Sci USA. 1997;94:6724–6729. doi: 10.1073/pnas.94.13.6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD. Rapid evolution of reproductive proteins. Nat Rev Genet. 2002;3:137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- Swofford D. PAUP*: phylogenetic analysis using parsimony (and other methods) Sunderland (MA): Sinauer Associates; 2002. [Google Scholar]
- Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiriot-Quievreux C. Advances in chromosomal studies of gastropod molluscs. J Moll Stud. 2003;69:187–201. [Google Scholar]
- Tracey S, Dodd JA, Erwin H. Mollusca:Gastropoda. In: Benton MJ, editor. The fossil record 2. London: Chapman and Hall; 1993. pp. 131–167. [Google Scholar]
- Turner LM, Hoekstra HE. Adaptive evolution of fertilization proteins within a genus: variation in ZP2 and ZP3 in deer mice (Peromyscus) Mol Biol Evol. 2006;23:1656–1669. doi: 10.1093/molbev/msl035. [DOI] [PubMed] [Google Scholar]
- Vacquier VD. Evolution of gamete recognition proteins. Science. 1998;281:1995–1998. doi: 10.1126/science.281.5385.1995. [DOI] [PubMed] [Google Scholar]
- Wassarman PM, Jovine L, Litscher ES, Qi H, Williams Z. Egg-sperm interactions at fertilization in mammals. Eur J Obstet Gynecol Reprod Biol. 2004;115(Suppl 1):S57–S60. doi: 10.1016/j.ejogrb.2004.01.025. [DOI] [PubMed] [Google Scholar]
- Wassarman PM, Jovine L, Qi HY, Williams Z, Darie C, Litscher ES. Recent aspects of mammalian fertilization research. Mol Cell Endocrinol. 2005;234:95–103. doi: 10.1016/j.mce.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Wong WS, Yang Z, Goldman N, Nielsen R. Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics. 2004;168:1041–1051. doi: 10.1534/genetics.104.031153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA. Evolution of larvae and developmental modes. In: McEdward L, editor. Ecology of marine invertebrate larvae. Boca Raton (FL): CRC Press; 1995. pp. 412–448. [Google Scholar]
- Yang Z. Phylogenetic analysis by maximum likelihood (PAML) London: University College London; 2000. [Google Scholar]
- Yang Z, Nielsen R, Goldman N, Pedersen AM. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Swanson WJ, Vacquier VD. Maximum-likelihood analysis of molecular adaptation in abalone sperm lysin reveals variable selective pressures among lineages and sites. Mol Biol Evol. 2000;17:1446–1455. doi: 10.1093/oxfordjournals.molbev.a026245. [DOI] [PubMed] [Google Scholar]