Abstract
The murine model for Turner Syndrome is the XO mouse. Unlike their human counterparts, XO mice are typically fertile, and their lack of a second sex chromosome can be transmitted from one generation to the next as an X-linked dominant trait with male lethality. The introduction of an X-linked coat-color marker (tabby) has greatly facilitated the maintenance of this useful mouse strain. XO mice can be produced in large numbers, generation after generation, and rapidly identified on the basis of their sex and coat color. Although this breeding scheme appears to be effective at the phenotype level, its utility has never been conclusively proved at the molecular or cytogenetic levels. Here, we clone and sequence the tabby deletion break point and present a multiplex polymerase chain reaction–based assay for the tabby mutation. By combining the results of this assay with whole-chromosome painting data, we demonstrate that genotype, phenotype, and karyotype all show perfect correlation in the publicly available XO breeding stock. This work lays the foundation for the use of this strain to study Turner Syndrome in particular and the X chromosome in general.
Women with Turner Syndrome typically have short stature, webbing of the posterior neck, an increased “carrying angle” at the elbows (cubitus valgus), and delayed or absent puberty (Turner 1938). Gonadal dysgenesis is seen in the vast majority of cases, and almost all affected individuals are infertile (Sybert and McCauley 2004). The classic cause of the disease is the complete absence of a second sex chromosome, leading to an abnormal 45,X karyotype, as opposed to the normal 46,XX or 46,XY karyotypes (Ford et al. 1959).
The murine model for Turner Syndrome is the XO mouse. Like their human counterparts, XO mice have a single X chromosome and no second sex chromosome. Note that the nomenclature differs between human and mouse. The International Standing Committee on Human Cytogenetic Nomenclature defines the human karyotype as “45,X,” whereas the International Committee on Standardized Genetic Nomenclature for Mice defines the mouse karyotype as “39,XO” or simply “XO,” where the letter “O” is a placeholder indicating the absence of a second sex chromosome; there is no “O” chromosome.
The existence of XO mice was first suspected after the discovery of mice that appeared to be homozygous for various X-linked traits but failed to transmit an allele of the trait to some of their female offspring. This finding was explained by the hypothesis that these unusual animals were not homozygous XX females but hemizygous XO females, and their female progenies who did not inherit an allele of the X-linked trait were also XO animals that failed to inherit an entire X chromosome from their mothers (Russell et al. 1959; Welshons and Russell 1959). Chromosome counts on a subset of these putative XO animals revealed 39 chromosomes in each case, as opposed to the normal 40, providing strong support for this hypothesis (Welshons and Russell 1959).
XO mouse stocks were subsequently developed using the tabby mutation as a coat-color marker. The tabby mutation (Ta) was the one of the first reported mouse traits to show a clear X-linked pattern of inheritance. On an agouti background, mice that are hemizygous or homozygous for the mutation have tan-colored fur and bald patches behind the ears, whereas heterozygous females have stripes of agouti fur and stripes of tan-colored fur, similar to that of a tabby cat (Falconer 1952). The mutation has been shown to be an ∼2-kb deletion at the 5′-end of the ectodysplasin-A gene (Eda) and has therefore been renamed EdaTa (Ferguson et al. 1997; Srivastava et al. 1997).
The first XO mouse stock was created by breeding female mice that were believed to be hemizygous for the wild-type ectodysplasin-A allele (Eda+/O) to male mice that were hemizygous for the EdaTa mutation (EdaTa/Y). Three kinds of offspring resulted: females with striped fur, who were presumably XX and heterozygous for the EdaTa mutation (Eda+/EdaTa); males with agouti fur, who were presumably XY and hemizygous for the wild-type Eda allele (Eda+/Y); and females with tan-colored fur and bald patches behind the ears, who were presumably XO and hemizygous for the EdaTa mutation (EdaTa/O). These presumed XO (EdaTa/O) females were then bred to their XY (Eda+/Y) brothers, and again 3 kinds of offspring resulted: females with striped fur (presumably XX, Eda+/EdaTa); males with tan-colored fur and bald spots behind the ears (presumably XY, EdaTa/Y); and females with agouti fur (presumably XO, Eda+/O). Brother–sister matings between XY and XO individuals could then be used to propagate the stock indefinitely (Cattanach 1962). The expected YO chromosome complement has been seen in 2-celled embryos but not thereafter, suggesting that it results in early embryonic lethality; YO mice are never seen in the live offspring (Morris 1968; Luthardt 1976).
Analysis of trypsin-Giemsa–banded chromosomes of these presumed XO mice has yielded 39 XO karyotypes at the gross cytogenetic level (Luthardt 1976), but more sophisticated chromosomal analysis, such as whole-chromosome painting, has never been reported. These studies would rule out other anomalies, such as cryptic translocations, which can be missed on routine karyotyping. Furthermore, although the genotype at the Eda locus is believed to correlate with the expected phenotype and karyotype of mice in an XO breeding stock, this has never been demonstrated. The genotype at the linked DXMit25 locus has previously been shown to correlate with the coat-color phenotype in one XO breeding stock, but this finding was not, in turn, shown to correlate with the animals’ karyotypes (Valdivia et al. 1996).
We describe here the cloning and sequencing of the EdaTa mutation and the development of a multiplex polymerase chain reaction (PCR)–based assay for genotyping at the Eda locus. We then use this assay to show that, for each kind of mouse seen in the publicly available XO mouse colony, an animal's genotype at the Eda locus correlates with its coat-color phenotype. Finally, we demonstrate that each mouse's genotype and phenotype also correlate with its karyotype, based on whole-chromosome painting of bone marrow cells. These results lay the groundwork for the use of this XO strain both as a murine model for Turner Syndrome and as a tool for the comprehensive functional analysis of the X chromosome.
Materials and Methods
Mice
The XO mouse breeding stock was obtained from The Jackson Laboratory (JAX, Bar Harbor, ME; JAX stock number 000314). The scheme for maintaining this stock has been described (Jackson Laboratory Staff 1990) and is explained in Figure 1. The animals were housed and cared for in the Transgenic Mouse Facility at Baylor College of Medicine (BCM). This is a specific pathogen-free mouse facility that is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). Animals were provided food and water ad libitum.
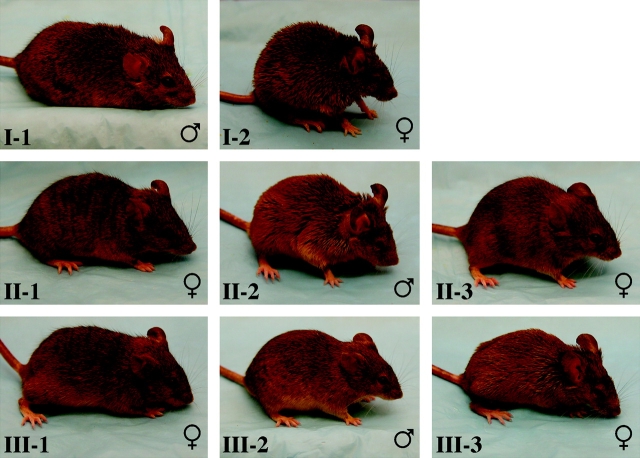
Figure 1.
Phenotypes of animals in the XO breeding stock. To maintain the XO breeding stock, an agouti male from an outside colony (I-1; Eda+/Y, XY) is bred to a presumed XO female (with tan-colored fur and bald spots behind the ears) from the XO breeding stock (I-2; presumed EdaTa/O, XO). Three types of offspring are seen: female animals with stripes of agouti fur and stripes of tan-colored fur (II-1; presumed Eda+/EdaTa, XX), male animals with tan-colored fur and bald spots behind the ears (II-2; presumed EdaTa/Y, XY), and female animals with agouti fur (II-3; presumed Eda+/O, XO). Animals II-2 and II-3 are interbred, and there are again 3 different kinds of progeny: female animals with stripes of agouti fur and stripes of tan-colored fur (III-1; presumed Eda+/EdaTa, XX), male animals with agouti fur (III-2; presumed Eda+/Y, XY), and female animals with tan-colored fur and bald spots behind the ears (III-3; presumed EdaTa/O, XO). Animal III-3 can then be bred to another agouti male from an outside colony to propagate the stock indefinitely. The sex of each animal is shown at the bottom right of each photograph (♂ = male, ♀ = female). All phenotypings (both sex and coat color) are done by simple visual inspection. The photographs also demonstrate that the hemizygous EdaTa mutants (animals I-2, II-2, and III-3) have a tendency to squint, which is likely secondary to well-documented deficiencies in the glands surrounding the eyes in mutant animals (Grüneberg 1971). Note that EdaTa/O, XO females (I-2 and III-3) are bred to Eda+/Y, XY males from an outside colony (I-1; a male from JAX stock number 001201) as opposed to Eda+/Y, XY males from within the XO breeding colony (III-2). This is done to promote fertility within the colony as the stock is difficult to maintain if brother–sister matings are used exclusively (Deckers and van der Kroon 1981).
The EdaTa mutation is known to cause tooth defects in hemizygous and homozygous animals, which appears to lead to impaired feeding and poor weight gain (Grüneberg 1965). In our hands, this effect is more pronounced in hemizygous female animals than in hemizygous males. Thus, cages containing EdaTa/O female mice were provided high-fat mouse chow (PicoLab Rodent Diet 20, #5058; LabDiet, Richmond, IN) as this chow is softer than regular mouse chow and has a higher caloric content. In addition, a quarter cup of powdered high-fat mouse chow (ground in a household kitchen blender) was added to these animals’ cages once a week. The rest of the animals were fed regular mouse chow (PicoLab Rodent Diet 20, #5053).
This work was approved by the Institutional Animal Care and Use Committee at BCM (Protocol #AN-1757).
Genomic DNA Preparation
Mouse genomic DNA was prepared from tail biopsies as described (Nagy et al. 2003), with minor modifications. The DNA was quantified on an ultraviolet (UV) spectrophotometer and diluted to a concentration of 25 ng/μl with Tris EDTA (TE) buffer (Sambrook and Russell 2001).
Amplification and Sequencing of the EdaTa Mutation
Southern blotting has previously shown that the EdaTa mutation is an ∼2-kb deletion at the 5′-end of the Eda gene (Ferguson et al. 1997; Srivastava et al. 1997). The deletion break point was amplified with the following primers: 5′-GAG GAC CTC CTC CCT CAT TC-3′ and 5′-CCA AAA ATC CTT GAC CAC CA-3′. This PCR product was then sequenced via standard protocols (Sambrook and Russell 2001) (GenBank accession number EU178100). Comparison of the break point sequence to the wild-type sequence of the mouse X chromosome (Kent et al. 2002) revealed the lesion to be a pure 1073-bp deletion that removes the entire first exon of the Eda gene (based on GenBank accession number AF016628).
Eda Multiplex PCR
Based on the sequence of the Eda+ and EdaTa alleles, we devised and optimized the following multiplex PCR reaction to amplify both the Eda+ and EdaTa alleles from genomic DNA. Primers were designed using the Primer3 algorithm (Rozen and Skaletsky 2000). Each 25-μl reaction consisted of 22-pmol primer A (5′-GTG AGG ACC TCC TCC CTC AT-3′), 10-pmol primer B (5′-CCT CTG CCT CAG TCC TTC AC-3′), 2.15-pmol primer C (5′-CAG GCT CCT CTT TGG AGT TG-3′), 13.4-pmol primer D (5′-CGC AGA AGA GAA TCC GTC TC-3′), 5-nm dNTPs, 1× standard Taq reaction buffer, 2.5 U Taq polymerase (New England Biolabs; Ipswich, MA), and 25 ng of genomic DNA. The thermocycler profile used was 94 °C for 3 min; 30 cycles of 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s; 72 °C for 10 min; and 4 °C until the PCR products were ready to be analyzed. The PCR products were then run on a 4% MetaPhor agarose (Cambrex Bio Science; Rockland, ME), 0.5× Tris-borate-EDTA (TBE) gel at 100 V and visualized via UV transillumination (Sambrook and Russell 2001).
Chromosome Preparation
Chromosome preparations were performed on bone marrow cells via a slightly modified version of the protocol found at The Jackson Laboratory's Web site (Available at: http://www.jax.org/cyto/marrow_preps_alt.html). The final cell pellet was resuspended in 0.5–1 ml of fixative. Approximately 20 μl of each cell suspension was placed on a standard microscope slide for whole-chromosome painting.
Whole-Chromosome Painting
The above slides were baked at 45 °C for 10 min, rinsed with acetic acid, and then washed for 2 min each in 75%, 85%, and 95% ethanol. A premade mixture of the following was added to each slide: 12-μl cyanine 3–labeled chromosome X probe, 12-μl fluorescein isothiocyanate (FITC)-labeled chromosome Y probe, and 36-μl hybridization solution (Open Biosystems, Huntsville, AL; Catalog numbers SFP2281 and SFP2241). The slides were heated to 73 °C for 3 min to denature the probes and then hybridized at 37 °C overnight. The following morning, the slides were rinsed twice in 0.4× standard saline citrate (SSC), 0.3% NP-40 at 73 °C for 2 min and then rinsed once in 2× standard saline citrate, 0.1% Igepal for 1 min. Finally, the slides were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and examined via standard fluorescence microscopy.
Results and Discussion
Like humans with the classic 45,X Turner Syndrome karyotype, XO mice have only a single-sex chromosome in all their cells. Unlike their human counterparts, however, affected mice are usually fertile. The introduction of the tabby (EdaTa) mutation as an X-linked coat-color marker has allowed XO animals to be maintained for decades at The Jackson Laboratory by selecting for animals with the appropriate sex and coat color (Figure 1).
Genotyping by Eda multiplex PCR was performed on each kind of animal observed in this XO breeding stock and confirms that each animal has the expected genotype predicted by its observed coat-color phenotype (Figure 2). This assay conclusively demonstrates that XO females fail to transmit a copy of their Eda gene to their XO offspring, and it excludes the possibility that the predicted XO animals are Eda+/EdaTa, XX females with skewed X inactivation.
Figure 2.
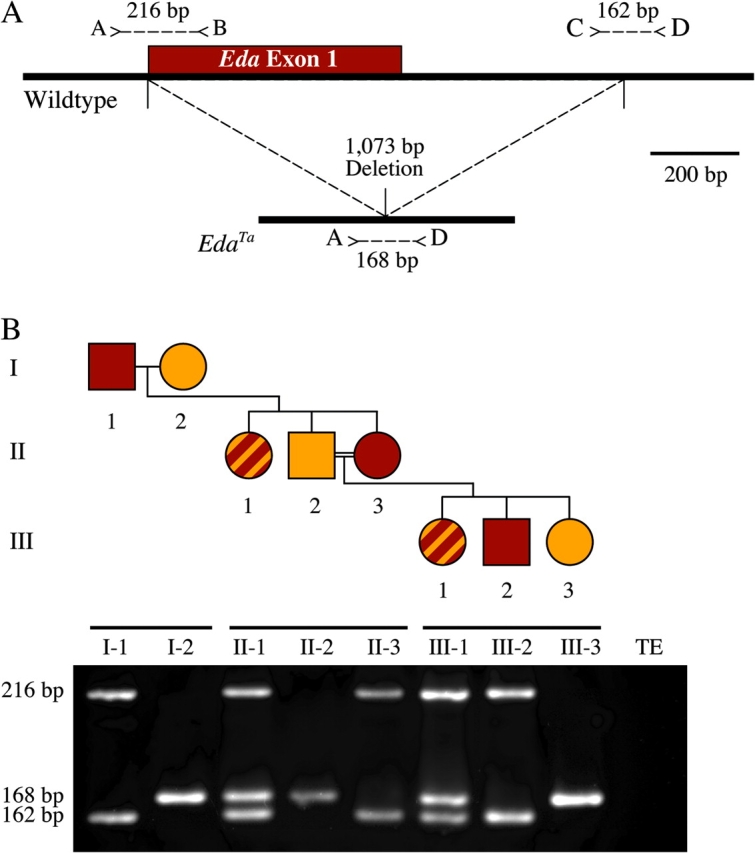
Eda genotyping. (a) The EdaTa mutation was cloned and sequenced and found to be a pure 1073-bp deletion that removes the entire first exon of the Eda gene (GenBank accession number EU178100). In a multiplex PCR reaction, primers A and B amplify a 216-bp product from wild-type DNA, whereas primers C and D amplify a 162-bp product. The EdaTa deletion removes the annealing sites for primers B and C and brings together the annealing sites for primers A and D, yielding a single 168-bp product. Thus, DNA from a homozygous or hemizygous wild-type animal yields 216-bp and 162-bp PCR products, DNA from a homozygous or hemizygous mutant animal yields a single band of 168 bp, and DNA from a heterozygote yields all 3 bands. The 3 different PCR products were all sequenced to confirm their identities (data not shown). (b) The pedigree shows a graphic representation of the breeding scheme shown in Figure 1. Males are shown as squares, and females are shown as circles. Dark brown shading indicates an agouti coat color, tan shading indicates a tan-colored coat color (and bald spots behind the ears), and stripes indicate a striped coat. The electrophoresis gel shows the results of the Eda multiplex PCR reaction for each of the respective animals in the pedigree from Figure 1. All animals have the expected genotypes, and there is perfect genotype–phenotype correlation. DNA from agouti animals yields 216-bp and 162-bp bands (Eda+), DNA from tan-colored animals (with bald spots behind the ears) yields a single band at 168 bp (EdaTa), and DNA from striped animals yields all 3 bands (Eda+/EdaTa). The assay is not quantitative and does not directly distinguish homozygotes (Eda+/Eda+ and EdaTa/EdaTa) from hemizygotes (Eda+/Y, Eda+/O, EdaTa/Y, and EdaTa/O). However, it demonstrates that animal I-2 fails to transmit her EdaTa allele to her female agouti offspring (II-3), and animal II-3 in turn fails to transmit her Eda+ allele to her offspring with tan-colored fur and bald spots behind the ears (III-3), which is consistent with all 3 animals having the XO karyotype.
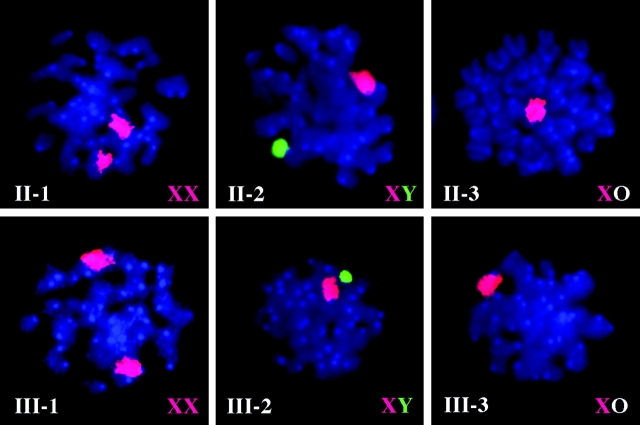
Figure 3.
Whole-chromosome painting. Whole-chromosome painting of the X and Y chromosomes was performed on chromosome spreads from bone marrow cells of the animals from generations II and III in Figures 1 and 2. The DAPI counterstain gives all chromosomes a blue fluorescent background color. The cyanine 3–labeled X chromosome probe produces a red fluorescent signal, and the fluorescein isothiocyanate-labeled Y chromosome probe produces a green fluorescent signal. Cells from animals II-1 and III-1 have 2 chromosomes with red signals (XX), and cells from animals II-2 and III-2 have 1 chromosome with a red signal and 1 with a green signal (XY), whereas cells from animals II-3 and III-3 have 1 chromosome with a red signal and no second sex chromosome signal (XO).
In order to confirm that the predicted XO animals have a pure XO karyotype and not a cryptic translocation or other subtle chromosomal anomaly, whole-chromosome painting was performed on the animals from generations II and III of the pedigree shown in Figures 1 and 2. These results show that animals II-3 and III-3 inherited a single X chromosome and no second sex chromosome. Animals II-1 and III-1 have the expected XX karyotype, and animals II-2 and III-2 have the expected XY karyotype. No evidence of a complex rearrangement involving the X or Y chromosomes was seen in any of the cells examined (Figure 3). This confirms that this stock is truly segregating the absence of a second sex chromosome as an X-linked dominant trait with male lethality. Taken together, these results show that this breeding stock permits the maintenance of XO mice with perfect genotype, phenotype, and karyotype correlation.
Progeny counts on animals in the stock from both our laboratory at Baylor College of Medicine (BCM) and from The Jackson Laboratory (JAX) show a clear deviation from the expected 1:1:1 ratio of XX, XY, and XO animals (Tables 1 and 2). This likely occurs for several reasons. First, it has been previously demonstrated that there is nonrandom segregation of the single X chromosome during oogenesis in XO mice, with the X chromosome being transmitted to the oocyte about two-thirds of the time (as opposed to the predicted one-half) (Kaufman 1972), so XO embryos are expected to occur only half as frequently as XX and XY embryos (i.e., the expected XX:XY:XO ratio immediately after fertilization is 2:2:1). Second, some XO embryos that implant near the cervix develop extremely poorly. It is not clear why this phenomenon occurs, but it could presumably reduce the number of XO progenies that are born (Burgoyne et al. 1983). Third, the EdaTa mutation is not simply a coat-color mutation. It is a pleiotropic mutation that affects a number of organ systems and has a significant deleterious effect on homozygous and hemizygous mutant animals (Grüneberg 1965, 1971).
Table 1.
Progeny counts: Eda+/Y × EdaTa/O
| Laboratory | Number of litters | Average litter size at weaning | Striped females (Eda+/EdaTa, XX) | Tan males (EdaTa/Y, XY) | Agouti females (Eda+/O, XO) |
| BCM | 36 | 4.5 | 68 (42%) | 65 (40%) | 30 (18%) |
| JAX | — | — | 272 (46%) | 249 (42%) | 76 (13%) |
Table 2.
Progeny counts: EdaTa/Y × Eda+/O
| Laboratory | Number of litters | Average litter size at weaning | Striped females (Eda+/EdaTa, XX) | Agouti males (Eda+/Y, XY) | Tan females (EdaTa/O, XO) |
| BCM | 133 | 4.3 | 267 (47%) | 236 (42%) | 65 (11%) |
| JAX | — | — | 257 (41.5%) | 239 (39%) | 121 (19.5%) |
Two of the most clinically important features of Turner Syndrome are cardiac disease and renal malformations (Sybert and McCauley 2004). There are no published reports of studies on XO mice for either of these conditions. Our multiplex PCR assay allows the rapid and easy identification of XO mice at any stage of development, so studies of the hearts and kidneys of these animals can now be performed to determine if cardiac or renal disease is a common finding in XO mice.
Finally, the XO mouse can be a powerful tool for the functional analysis of the mouse X chromosome. Previously, phenotype-driven mutagenesis screens have focused on genome-wide autosomal dominant mutations (Hrabe de Angelis et al. 2000; Nolan et al. 2000) or region-specific autosomal recessive mutations (Kile et al. 2003). A phenotype-driven mutagenesis screen for X-linked recessive mutations has never been reported. The XO mouse provides an obvious avenue to accomplish this. ENU-mutagenized males (Eda+/Y, XY) can be bred to EdaTa/O, XO females, and the Eda+/O animals that result from such a cross can be identified via our multiplex PCR assay. These mice will be hemizygous for an N-ethyl-N-nitrosourea (ENU)-mutagenized X chromosome and will therefore express any X-linked recessive traits that were induced by the mutagen. Thus, the use of the XO mouse allows for a one-generation screen for X-linked recessive mouse mutants. Furthermore, the Eda+/EdaTa, XX females that result from such a cross can also be examined for any deviation from the expected striped-fur phenotype as these animals may represent mutations that cause skewing of X inactivation. Clearly, the XO mouse is valuable not only just as a murine model for Turner Syndrome but also as a tool for the study of all the genes on the X chromosome.
Funding
Burroughs Wellcome Fund (Career Award for Medical Scientists #1006860 to F.J.P.); National Institutes of Health (F32HG003942 to F.J.P. and R01CA115503 to M.J.J.).
Acknowledgments
We thank Curtis Clark of California State Polytechnic University for designing the male and female symbols used in the text and figures, Julie Soukup and Muriel Davisson of The Jackson Laboratory for providing progeny counts from their XO colony, and Muriel Davisson of The Jackson Laboratory for her suggestions for improving the manuscript.
References
- Burgoyne PS, Tam PPL, Evans EP. Retarded development of XO conceptuses during early pregnancy in the mouse. J Reprod Fertil. 1983;68:387–393. doi: 10.1530/jrf.0.0680387. [DOI] [PubMed] [Google Scholar]
- Cattanach BM. XO mice. Genet Res. 1962;3:487–490. [Google Scholar]
- Deckers JFM, van der Kroon PHW. Some characteristics of the XO mouse (Mus musculus L.) I. Vitality: growth and metabolism. Genetica. 1981;55:179–185. [Google Scholar]
- Falconer DS. A totally sex-linked gene in the house mouse. Nature. 1952;169:664–665. doi: 10.1038/169664b0. [DOI] [PubMed] [Google Scholar]
- Ferguson BM, Brockdorff N, Formstone E, Ngyuen T, Kronmiller JE, Zonana J. Cloning of tabby, the murine homolog of the human EDA gene: evidence for a membrane-associated protein with a short collagenous domain. Hum Mol Genet. 1997;6:1589–1594. doi: 10.1093/hmg/6.9.1589. [DOI] [PubMed] [Google Scholar]
- Ford CE, Jones KW, Polani PE, De Almeida JC, Briggs JH. A sex-chromosome anomaly in a case of gonadal dysgenesis (Turner's syndrome) Lancet. 1959;273:711–713. doi: 10.1016/s0140-6736(59)91893-8. [DOI] [PubMed] [Google Scholar]
- Grüneberg H. Genes and genotypes affecting the teeth of the mouse. J Embryol Exp Morphol. 1965;14:137–159. [PubMed] [Google Scholar]
- Grüneberg H. The glandular aspects of the tabby syndrome in the mouse. J Embryol Exp Morphol. 1971;25:1–19. [PubMed] [Google Scholar]
- Hrabe de Angelis MH, Flaswinkel H, Fuchs H, Rathkolb B, Soewarto D, Marschall S, Heffner S, Pargent W, Wuensch K, Jung M, et al. Genome-wide, large-scale production of mutant mice by ENU mutagenesis. Nat Genet. 2000;25:444–447. doi: 10.1038/78146. [DOI] [PubMed] [Google Scholar]
- Jackson Laboratory Staff. Tabby strains available from The Jackson Laboratory. 1990. JAX Notes. 440a. Available at http://jaxmice.jax.org/jaxnotes/archive/440a.html. [Google Scholar]
- Kaufman MH. Non-random segregation during mammalian oogenesis. Nature. 1972;238:465–466. doi: 10.1038/238465a0. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kile BT, Hentges KE, Clark AT, Nakamura H, Salinger AP, Liu B, Box N, Stockton DW, Johnson RL, Behringer RR, et al. Functional genetic analysis of mouse chromosome 11. Nature. 2003;425:81–86. doi: 10.1038/nature01865. [DOI] [PubMed] [Google Scholar]
- Luthardt FW. Cytogenetic analysis of oocytes and early preimplantation embryos from XO mice. Dev Biol. 1976;54:73–81. doi: 10.1016/0012-1606(76)90287-6. [DOI] [PubMed] [Google Scholar]
- Morris T. The XO and OY chromosome constitutions in the mouse. Genet Res. 1968;12:125–137. doi: 10.1017/s0016672300011745. [DOI] [PubMed] [Google Scholar]
- Nagy A, Gertsenstein M, Vintersten K, Behringer R. Manipulating the mouse embryo: a laboratory manual. 3rd ed. Cold Spring Harbor (NY): Cold Spring Harbor Press; 2003. [Google Scholar]
- Nolan PM, Peters J, Strivens M, Rogers D, Hagan J, Spurr N, Gray IC, Vizor L, Brooker D, Whitehill E, et al. A systematic, genome-wide, phenotype-driven mutagenesis programme for gene function studies in the mouse. Nat Genet. 2000;25:440–443. doi: 10.1038/78140. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics methods and protocols: methods in molecular biology. Totowa (NJ): Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Russell WL, Russell LB, Gower JS. Exceptional inheritance of a sex-linked gene in the mouse explained on the basis that the X/O sex-chromosome constitution is female. Proc Natl Acad Sci USA. 1959;45:554–560. doi: 10.1073/pnas.45.4.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3rd ed. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Srivastava AK, Pispa J, Hartung AJ, Du Y, Ezer S, Jenks T, Shimada T, Pekkanen M, Mikkola ML, Ko MS, et al. The tabby phenotype is caused by mutation in a mouse homologue of the EDA gene that reveals novel mouse and human exons and encodes a protein (ectodysplasin-A) with collagenous domains. Proc Natl Acad Sci USA. 1997;94:13069–13074. doi: 10.1073/pnas.94.24.13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sybert VP, McCauley E. Turner's syndrome. N Engl J Med. 2004;351:1227–1238. doi: 10.1056/NEJMra030360. [DOI] [PubMed] [Google Scholar]
- Turner HH. A syndrome of infantilism, congenital webbed neck, and cubitus valgus. Endocrinology. 1938;23:566–574. [PubMed] [Google Scholar]
- Valdivia RP, Kunieda T, Hattori M, Toyoda Y. Typing of X chromosomes bearing tabby allele in mouse preimplantation embryos by detection of a microsatellite marker. Exp Anim. 1996;45:395–398. doi: 10.1538/expanim.45.395. [DOI] [PubMed] [Google Scholar]
- Welshons WJ, Russell LB. The Y-chromosome as the bearer of male determining factors in the mouse. Proc Natl Acad Sci USA. 1959;45:560–566. doi: 10.1073/pnas.45.4.560. [DOI] [PMC free article] [PubMed] [Google Scholar]