Abstract
The epigenome changes that underlie cellular differentiation in developing organisms are poorly understood. To gain insights into how pancreatic beta-cells are programmed, we profiled key histone methylations and transcripts in embryonic stem cells, multipotent progenitors of the nascent embryonic pancreas, purified beta-cells, and 10 differentiated tissues. We report that despite their endodermal origin, beta-cells show a transcriptional and active chromatin signature that is most similar to ectoderm-derived neural tissues. In contrast, the beta-cell signature of trimethylated H3K27, a mark of Polycomb-mediated repression, clusters with pancreatic progenitors, acinar cells and liver, consistent with the epigenetic transmission of this mark from endoderm progenitors to their differentiated cellular progeny. We also identified two H3K27 methylation events that arise in the beta-cell lineage after the pancreatic progenitor stage. One is a wave of cell-selective de novo H3K27 trimethylation in non-CpG island genes. Another is the loss of bivalent and H3K27me3-repressed chromatin in a core program of neural developmental regulators that enables a convergence of the gene activity state of beta-cells with that of neural cells. These findings reveal a dynamic regulation of Polycomb repression programs that shape the identity of differentiated beta-cells.
Changes in chromatin structure, including the posttranslational modifications of histone tails, underlie the process of differentiation of pluripotent cells into all cell types of the body. The trimethylation of histone H3 lysine 4 and lysine 27 (H3K4me3 and H3K27me3, respectively) are among the best-characterized histone modifications. H3K4me3 is found at virtually all active transcriptional start sites of genes (Schneider et al. 2004; Barski et al. 2007). In contrast, H3K27me3 enrichment occurs in genes that are transcriptionally repressed by Polycomb group (PcG) proteins (Pietersen and van Lohuizen 2008; Schwartz and Pirrotta 2008). Large-scale studies revealed that PcG-dependent H3K27 methylation is not a universal mechanism for the repression of all inactive genes, but instead represses a restricted subset of genes, typically CpG island genes encoding for developmental regulators (Boyer et al. 2006; Lee et al. 2006). Further support that H3K27me3 plays a central role in developmental processes comes from knowledge that mutations in H3K27me3 methyltransferase and demethylase genes exhibit defects in stem-cell function, organogenesis, and differentiation (O'Carroll et al. 2001; Pasini et al. 2004; Lan et al. 2007; Ezhkova et al. 2009).
In vitro studies have provided insights into how cell-specific changes in H3K4 and H3K27 methylation shape cellular differentiation. In pluripotent embryonic stem (ES) cells, a major portion of genes that are methylated in H3K27 are also enriched in H3K4me3, a state termed bivalent (Bernstein et al. 2006; Mikkelsen et al. 2007). Lineage-specific differentiation has been associated to a resolution of this bivalent state, with a loss of either H3K27me3 or H3K4me3 leading to the concordant activation or repression of lineage-specific genes (Bernstein et al. 2006; Mikkelsen et al. 2007). A recent study showed that many genes also acquire H3K27me3 de novo after the ES cell stage during in vitro neuronal differentiation (Mohn et al. 2008). So far, however, H3K27me3 has been studied in a limited number of cell types, and has generally relied on cultured cellular models that may exhibit profound epigenetic differences with primary differentiated cells (Meissner et al. 2008). The H3 methylation changes that underlie lineage-specific differentiation in vivo thus remain elusive.
The insulin-producing beta-cell is a valuable model to study epigenetic regulation of development because of its implications for regenerative medicine. Beta-cells, and all other pancreatic epithelial cells including the exocrine acinar cells, originate from multipotent progenitors of the foregut endoderm that form the nascent pancreas (Oliver-Krasinski and Stoffers 2008; Zaret 2008). This developmental process can be potentially recapitulated from pluripotent ES cells to generate new beta-cells for replacement therapies in diabetes (Madsen and Serup 2006; Kroon et al. 2008; Zhou and Melton 2008) Alternately, beta-cells can be generated by transdifferentiation of pancreatic acinar cells or other tissues (Ferber et al. 2000; Baeyens et al. 2005; Zhou et al. 2008). However, the extent to which current protocols give rise to bona fide precursors and beta-cells is unknown, and their efficiency is limited (Madsen and Serup 2006; Zhou and Melton 2008). Knowledge of the epigenetic instructions underlying the in vivo generation of beta-cells and other cell types can uncover barriers that restrict lineage plasticity, and contribute to devise successful artificial beta-cell differentiation strategies.
We have now examined the H3K27me3 repression and gene activity programs of primary differentiated beta-cells. We studied pluripotent ES cells and multipotent pancreatic progenitors to understand how these programs are acquired during embryogenesis, and a panel of tissues to assess their unique properties. To our knowledge, this is the first large-scale analysis of Polycomb repression profiles in a cellular lineage of a developing organism or across multiple primary tissues.
Results
Beta-cells and neural tissues share a gene activity program
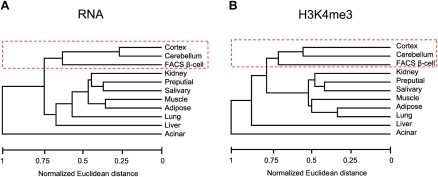
To study the transcriptional program of pancreatic islet cells, we first profiled mRNA from pancreatic FACS-purified beta-cells and 10 tissues from diverse developmental origins. While it is known that islet endocrine cells are electrically excitable and express many genes in common with neurons (Atouf et al. 1997; Rorsman 1997), we were surprised to find that the genome-wide mRNA profile of purified beta-cells or pancreatic islets is more similar to neural tissues than to tissues with a common endoderm origin, such as acinar or liver (Fig. 1A; for a principal component analysis including both purified beta-cells and islets, see Supplemental Fig. S1a). We profiled H3K4me3 as an alternate measure of gene activity and found an analogous resemblance with neural tissues (Fig. 1B; Supplemental Fig. S1b). Thus, despite their disparate developmental origins, beta-cells and neural tissues share a gene activity profile.
Figure 1.
Beta-cells and neural tissues share a gene activity program. Euclidean distances of genome-wide gene activity profiles, based on the presence or absence of mRNAs (A) or H3K4me3 (B) in each tissue. In both dendrograms, beta-cells are grouped with cerebral cortex and cerebellum. Similar conclusions were reached with principal component analysis using both purified beta-cells and islets (Supplemental Fig. S1).
Cell-specific H3K27me3 profiles are determined by gain and loss of histone methylation events
To assess how PcG-mediated mechanisms contribute to the unique gene activity program of beta-cells, we profiled H3K27me3 in ES cells, a panel of 10 tissues, and purified beta-cells. H3K27me3 enrichment is biased for the transcriptional start sites of genes (Barski et al. 2007; Mikkelsen et al. 2007). We thus classified genes according to the presence or absence of H3K27me3 and H3K4me3 enrichment at ±2 kb from the transcriptional start sites of 16,523 RefSeq genes.
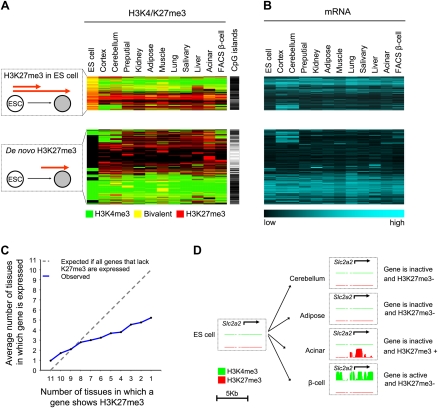
Because a global analysis of H3K27me3 across multiple cell types has not been reported, we first examined general tissue-specific patterns of trimethylated H3K27. We identified two major classes of H3K27me3 targets: (1) CpG-island genes that are already enriched in H3K27me3 in ES cells, and either maintain H3K27me3 or undergo tissue-selective loss of H3K27me3 during differentiation, and (2) genes in which H3K27me3 is not present in ES cells but instead arises de novo (Fig. 2A; Supplemental Fig. S2a). De novo H3K27me3 occurred in a remarkably large number of genes (69% of all genes showing H3K27me3 in at least one tissue) (Supplemental Fig. S2a) and was distinct in that it was highly tissue-selective (Supplemental Fig. S2b). Furthermore, unexpected from earlier analyses of H3K27me3 in fewer cell types (Mikkelsen et al. 2007; Mohn and Schubeler 2009), 48% of genes that showed de novo H3K27me3 in at least one tissue lacked CpG islands in their promoters (Supplemental Fig. S2a; for qPCR confirmations, see Supplemental Fig. S3a). These findings provided a framework to dissect the H3K27me3 patterns underlying beta-cell development.
Figure 2.
Tissue-specific loss and gain of H3K27 methylation. (A) Although many genes that show H3K27me3 enrichment in differentiated cells are also H3K27me3+ in ES cells, unexpectedly 69% of genes only show H3K27me3 enrichment in differentiated tissues (de novo H3K27me3). The cluster representation shows H3K27me3 (red), H3K4me3 (green), or both (yellow) in the 5248 genes that show H3K27me3 in at least one tissue. Genes with de novo H3K27me3 often lack CpG islands, which are indicated as a black line. (B) Most genes with H3K27me3 are inactive (see also Supplemental Fig. S2d). The mRNA heatmap maintains the same order as A. (C,D) H3K27me3 repression is both gene-specific and cell-specific. (C) Genes that are targeted by H3K27me3 in only few tissues are often also inactive in tissues where they do not show H3K27me3. This effect was more pronounced for non-CpG island genes (Supplemental Fig. S2c). (D) The beta-cell glucose transporter Slc2a2 shows H3K27me3 enrichment in acinar cells, yet is inactive in other tissues that lack H3K27me3. The graph shows posterior probability values ranging from 0–1 for the enrichment of H3K4me3 (green) and H3K27me3 (red).
PcG repression is employed in a cell context–dependent manner
To further assess the functional significance of tissue-specific H3K27 methylation, we examined the relationship between H3K27me3 and gene repression. As expected, most genes showed no mRNA expression in tissues where they were enriched in H3K27me3 (Fig. 2A; Supplemental Fig. S2d). Furthermore, as reported previously (Roh et al. 2006), bivalent H3K4me3 and H3K27me3 enrichment was found in differentiated tissues or purified beta-cells in numerous genes (Fig. 2A), which was confirmed in 12 genes by sequential ChIP (Supplemental Fig. S3b). Bivalent genes were generally also inactive (Supplemental Fig. S2d). Less expected, genes repressed by H3K27me3 in one tissue were often also inactive in other tissues where they lacked H3K27me3 (Fig. 2C,D; Supplemental Fig. S2c). For example, Slc2a2 was inactive in most tissues, yet underwent H3K27me3 repression only in acinar cells (Fig. 2D; for additional examples, see Supplemental Fig. S4). This was true regardless of whether we defined gene inactivity by the absence of detectable mRNA or H3K4me3 (Supplemental Fig. S4). This suggests that even among genes that are subject to PcG repression, this repressive mechanism is only employed in certain cellular contexts.
In beta-cells H3K27me3 represses genes important for their development and function
Next, we focused on the genes that are repressed by H3K27me3 in pancreatic beta-cells. Only 9% of all genes showed H3K27me3 in beta-cells, and these represented only about 19% of genes that are inactive in beta-cells.
As expected, H3K27me3 in beta-cells was primarily enriched in regulators of inappropriate developmental fates, such as skeletal, neural, and heart development (Fig. 3A; Supplemental Table S1, S2). However, among genes that showed a more selective H3K27me3 enrichment pattern in beta-cells, additional functional categories were enriched, including neurotransmitter receptors and cytokine activity (Supplemental Tables S3, S4), suggesting that H3K27me3 might repress genes that are not developmental regulators, but nevertheless must be repressed in differentiated beta-cells. To assess this possibility, we tested five genes that are known to be deleterious for mature beta-cells (Fig. 3B). These included (1) Fas, a mediator of cell death that is inactive in healthy beta-cells but is selectively induced during autoimmune destruction in type 1 diabetes (Hanafusa and Imagawa 2008); (2) the high-affinity hexokinases Hk1 and Hk2, whose expression would perturb the beta-cell-specific glycolytic glucose-sensing apparatus (Heimberg et al. 1993); (3) the monocarboxylic acid transporter Slc16a1, which leads to fatal insulin hypersecretion when it is inappropriately expressed in beta-cells (Ishihara et al. 1999; Otonkoski et al. 2007); and (4) Rest, a repressor of neuronal genes in non-neuronal tissues that also suppresses important beta-cell genes (Atouf et al. 1997; Johnson et al. 2007; Thiel and Schuit 2008). All these genes were selectively repressed by H3K27me3 in beta-cells (Fig. 3B; Supplemental Table S3). These results suggest that the H3K27me3 program of beta-cells is enriched in genes whose repression is specifically important for the development, function, or survival of beta-cells.
Figure 3.
H3K27me3 profiles suggest a central role in beta-cell identity. (A) Beta-cell H3K27me3 preferentially targets genes that promote alternate developmental fates. (B) H3K27me3 is also enriched in genes whose expression is known to be deleterious for mature beta-cells.
Beta-cell H3K27me3 is acquired through distinct developmental programs
To determine how PcG repression is established during the embryonic development of beta-cells, we examined H3K27me3 in pluripotent ES cells, Pdx1+ multipotent pancreatic progenitors of the E10.5 pancreas, and purified beta-cells. As anticipated, in pancreatic progenitors, H3K27me3 repressed alternate endoderm fates and other heterologous developmental programs, as well as genes that regulate pluripotency, while H3K27me3 was selectively absent in known regulators of the nascent embryonic pancreas (e.g., Foxa1, Foxa2, Gata4, and Pdx1) (for examples, see Supplemental Fig. S5a; for full lists of H3K27me3-repressed and derepressed genes in pancreatic progenitors, see Supplemental Tables S5, S6).
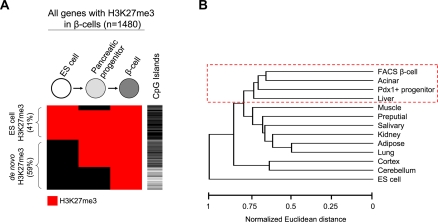
We next focused on all genes that are H3K27me3+ in beta-cells, and traced when this mark was established. We found that beta-cell H3K27me3 was acquired in a stepwise manner throughout development, consistent with stage-specific requirements to repress genomic targets (Fig. 4A; for illustrative genes, see Supplemental Fig. S6a). Overall, 59% of H3K27me3+ genes in beta-cells corresponded to de novo events (Fig. 4A). A more detailed analysis distinguished two major groups of genes that differed in when they acquired H3K27me3 enrichment relative to the pancreatic progenitor stage.
Figure 4.
The beta-cell H3K27me3 enrichment signature includes pancreatic endoderm and terminal differentiation methylation programs. (A) In 59% of the 1480 genes that are H3K27me3+ in beta-cells, the methylation is acquired de novo, either before the pancreatic progenitor stage or during subsequent differentiation. De novo events occurring during later stages of differentiation preferentially target genes that lack CpG islands. On the other hand, in 67% of genes with H3K27me3 enrichment in beta-cells, this mark is present in progenitors and is then maintained. The cluster diagram depicts H3K27me3 enrichment (in red) in the different stages, and the presence of CpG islands is indicated in black in the adjacent column. (B) The gene profile that is H3K27me3+ in beta-cells is most similar to that of acinar, pancreatic progenitors, and liver. This was determined by Euclidean distances of genome-wide H3K27me3 distributions, as described in Methods. Similar results were observed using principal component analysis (Supplemental Fig. S6b).
In nearly 400 genes, H3K27 methylation occurred de novo after the multipotent progenitor stage (Fig. 4A; Supplemental Fig. S6a). As anticipated from our multitissue analysis of de novo H3K27me3, these events were highly beta-cell-specific (data not shown). Furthermore, they occurred predominantly in genes that lacked CpG islands (71%, vs. 34% of all H3K27me3+ genes in beta-cells, P < 0.0001) (Fig. 4A). Thus, de novo H3K27 trimethylation in non-CpG island genes is common during terminal differentiation of beta-cells and is potentially linked to a stage-specific role of PcG repression in differentiated cells.
In 67% of all genes that are H3K27me3+ in beta-cells, however, this mark was already present in pancreatic progenitors (Fig. 4A). This suggested an epigenetic maintenance of repressive marks throughout pancreas organogenesis. In accordance, the beta-cell H3K27me3 repressive signature was most similar to acinar and pancreatic endoderm progenitors, and to a lesser extent to liver (Fig. 4B; for similar results using principal component analysis, see Supplemental Fig. S6b). Analogous conclusions were reached when we examined a set of 469 genes showing a beta-cell selective H3K27me3 pattern; of such genes 55% showed H3K27me3 in acinar cells and 40% in pancreatic endoderm progenitors, in contrast to an average of 13% in other tissues (P < 0.0001) (Supplemental Fig. S6c). Importantly, the shared H3K27me3 enrichment profile with acinar cells was not due to unrecognized cellular contamination, because known islet-specific genes that showed H3K27me3 enrichment in acinar cells did not exhibit H3K27me3 enrichment in beta-cells (e.g., Supplemental Fig. S7; Fig. 5B). Thus, the H3K27me3 repression signature in beta-cells is generated progressively throughout embryogenesis, and in sharp contrast to its gene activity signatures, it is preferentially shared with cells of common developmental origin.
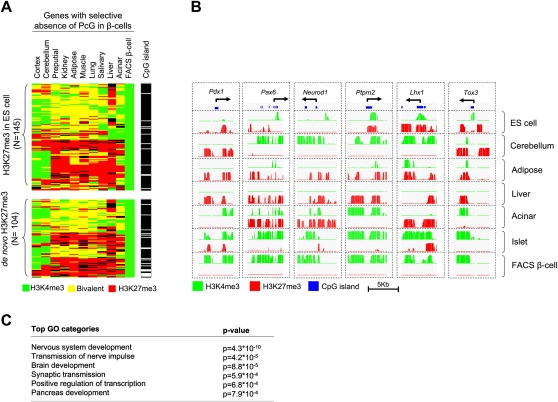
Figure 5.
A neural regulatory program shows selective absence of PcG repression in beta-cells. (A) A major fraction of genes with selective absence of PcG repression in beta-cells showed an identical pattern in cortex and cerebellum. The cluster diagram shows the 249 genes that are H3K27me3- H3K4me3+ in beta-cells but are H3K27me3+ in more than five other tissues. A quantitative analysis is shown in Supplemental Figure S6c. (B) Examples of genes with selective absence of PcG repression in beta-cells. (C) Most enriched nonredundant Gene Ontology biological process and molecular function terms (levels 4 and 5) among the 249 genes with beta-cell-selective absence of PcG repression. A more complete list is shown in Supplemental Table S8.
Beta-cells selectively lack PcG repression in a neural regulatory program
We next attempted to reconcile the paradox that in beta-cells a H3K27me3 repression signature shared with endoderm tissues is associated with a neural-like gene activity profile. We hypothesized that this could result from a selective loss of H3K27me3 repression in a small but functionally important set of regulators of neural gene activity.
We thus searched for genes that selectively lack H3K27me3 in beta-cells. We identified a core group of 249 genes that were H3K27me3+ in ≥50% of differentiated tissue samples, yet were H3K27me3− H3K4me3+ in beta-cells (Fig. 5A; for a full list, see Supplemental Table S7). We refer to these as genes with selective absence of PcG repression in beta-cells. As expected, mRNA expression in this gene set was highly enriched in beta-cells, islets, and tumor beta-cell lines (Supplemental Fig. S8a) and included many known beta-cell–enriched transcriptional regulators, including Pdx1, Neurod1, Pax6, Mnx1/Hlxb9, Nkx2-2, Glis3, and Isl1 (Fig. 5B; Supplemental Fig. S7; Supplemental Table S7). It also included many previously unrecognized putative regulators of beta-cells, such as Tox3, a calcium-dependent regulator of transcription in neurons (Yuan et al. 2009), and Lhx1, a LIM domain protein that is coexpressed with Pax6 and Isl1 in developing diencephalic neurons Fig. 5B; Supplemental Table S7; Mastick and Andrews 2001).
Remarkably, nearly 60% of genes with selective absence of PcG repression in beta-cells showed an identical histone methylation pattern in cortex and 46% in cerebellum, as opposed to 16% on average in other tissues (P < 0.001) (Fig. 5A; Supplemental Fig. S8b). In concordance, gene ontology analysis of this gene set showed a marked enrichment of functions related to neural development, in addition to regulators of pancreas development and transcription factors (Fig. 5C; Supplemental Table S8). In summary, although PcG repression in beta-cells is largely shared with other pancreatic cell types, beta-cells and neural tissues share a selective absence of PcG repression in a core set of developmental regulators. The transcriptional regulators that compose this comparatively small core program control target genes that are not necessarily subject to PcG regulation and therefore have the potential to activate broad transcriptional programs shared by beta-cells and neural tissues.
Implementation of the beta-cell derepression program during differentiation
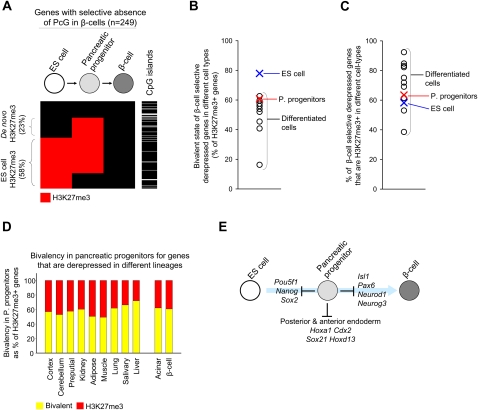
We next dissected the developmental steps that lead to the program of genes that exhibits selective absence of PcG repression in beta-cells. This gene set was frequently H3K27me3+ in pluripotent ES cells (58%, compared with 31% of all H3K27me3 targets in beta-cells, P < 0.0001), and often contained CpG islands (88% vs. 61% for all genes, P < 0.0001) (Fig. 6A). As expected from earlier studies in ES cells (Bernstein et al. 2006), H3K27me3 was bivalent in 80% of cases (Fig. 6B). Thus, beta-cell-specific gene activity is to a large extent determined by the selective loss of H3K27me3 in genes that show a bivalent chromatin state in ES cells.
Figure 6.
Pancreatic progenitors suppress regulators of beta-cell differentiation. (A) genes with selective absence of PcG repression in beta-cells often exhibit H3K27me3 enrichment in ES cells or pancreatic progenitors and have CpG islands. The cluster representation shows H3K27me3 (red) in the 249 genes with selective absence of PcG repression in beta-cells in different stages. In the adjacent column, the presence of CpG islands is indicated in black. (B) As expected, genes with selective absence of PcG repression in beta-cells are frequently bivalent in ES cells. In pancreatic progenitors, bivalency in such genes is similar to other differentiated cell types. (C) Genes with selective absence of PcG repression in beta-cells show H3K27me3 repression in both differentiated cell types of distant lineages as well as in pancreatic progenitors. (D) Pancreatic progenitors do not preferentially show a bivalent H3K27me3+ H3K4me3+ state (yellow) for genes with selective absence of PcG repression in their progeny (i.e., acinar and beta-cells), compared with genes with selective absence of PcG repression in distant lineages. (E) Pancreatic multipotent progenitors exhibit H3K27me3 repression of genes involved in pluripotency and alternate endoderm fates, but also in regulators of their progeny.
To assess when H3K27me3 repression is lost in genes with selective absence of PcG repression in beta-cells, we examined multipotent pancreatic progenitors. In numerous examples, progenitors exhibit anticipatory chromatin changes that signal transcriptional competence to activate genes during subsequent differentiation (Gualdi et al. 1996; Kontaraki et al. 2000; Palstra et al. 2003). However, 63% of genes with selective absence of PcG repression in beta-cells showed H3K27me3 enrichment at the pancreatic progenitor stage (Fig. 6C). which was not different than the frequency of H3K27me3 enrichment in these genes in nonpancreatic differentiated tissues (Fig. 6C). Analogous results were obtained for genes that showed a selective absence of PcG repression in acinar cells, an alternate fate of pancreatic progenitors (data not shown). Thus, the core beta-cell gene activity program remains largely suppressed by H3K27me3 in pancreatic progenitors and is selectively derepressed in the pancreatic endocrine lineage during later stages of differentiation.
Does bivalency in pancreatic progenitors signal transcriptional competence?
Because bivalent chromatin state has been linked to transcriptional potential in ES cells, we next asked if this is also true in pancreatic stem cells (Bernstein et al. 2006). Earlier studies (Roh et al. 2006) and our own analysis (Fig. 2A; Supplemental Figs. S2, S3b) show that bivalency is not uncommon in inactive genes of differentiated cells, although the frequency is lower than in pluripotent cells. We reasoned that if bivalency in pancreatic stem cells reflects a potential for gene activation, then it should be specifically present at genes that are activated in their cellular progeny.
We observed a bivalent state in many genes in pancreatic progenitors, yet not preferentially in genes that were activated in beta-cells and acinar cells compared with genes that are selectively active in distant lineages (Fig. 6D). Conversely, pancreatic progenitors and unrelated differentiated tissues had a comparable frequency of bivalency at genes that are selectively activated in beta-cells (Fig. 6B). In several genes that were bivalent in ES cells, progenitor cells retained H3K27me3 but not H3K4me3 enrichment and then became active in mature beta-cells (for examples, see Supplemental Fig. S5b). In summary, pancreatic progenitors use PcG-dependent mechanisms not only to suppress genes that promote pluripotency and alternate endoderm fates but also to control lineage-selective transcriptional programs of their cellular progeny (Fig. 6E). Furthermore, in pancreatic stem-cells, bivalent chromatin does not appear to be indicative of lineage-specific transcriptional competence.
Discussion
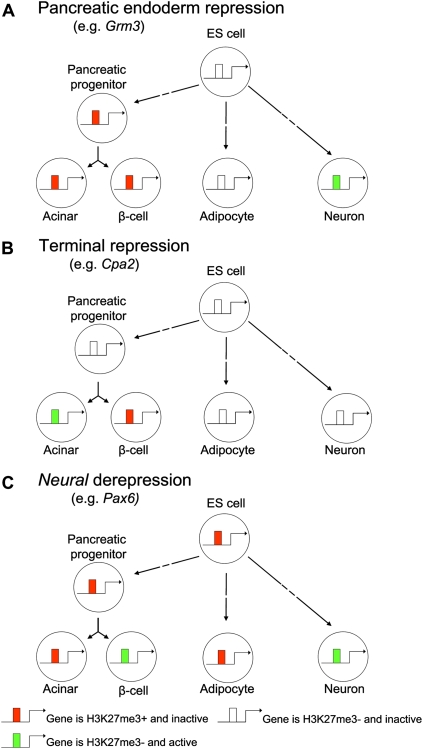
In the current study, we have examined the dynamics of PcG repression during the embryonic development of pancreatic beta-cells. Unlike earlier profiles of PcG-repressed chromatin, we examined primary progenitors and differentiated cells rather than cultured cell models, and simultaneously integrated gene expression and histone modifications profiles across diverse lineages. We have shown that throughout embryonic development PcG repression is placed or removed according to context-dependent requirements for this specific mode of repression. We traced the embryonic origin of PcG marks in beta-cells and decomposed three distinguishable PcG repression programs that underlie beta-cell identity (Fig. 7). First, we identified a repressive program that can be linked to the endodermal origin of beta-cells. Furthermore, we discovered two major epigenetic events that take place during the differentiation process that follows the pancreatic endoderm progenitor stage, namely, a wave of cell-specific de novo H3K27 methylation at non-CpG island genes and a selective removal of PcG repressive marks in a core developmental program that enables beta-cells to acquire a neural gene activity state.
Figure 7.
PcG repression programs that shape beta-cell identity. Schematic illustrating how PcG-dependent modifications are placed or removed in a highly context-dependent manner during beta-cell differentiation. (A) A subset of H3K27me3+ genes in beta-cells have acquired this mark by the pancreatic endoderm progenitor stage, which is then epigenetically inherited in their cellular progeny. (B) Another subset of genes undergoes de novo H3K27me3 repression during late differentiation. These often lack CpG islands and are highly cell-type-specific H3K27me3 events. In some of these genes, repression is essential for the differentiated function of beta-cells. (C) The selective loss of PcG repression after the pancreatic progenitor stage in a core set of developmental regulators enables a convergence of transcriptional profiles in beta-cells with neuroectoderm-derived tissues.
Distinct classes of lineage-specific H3K27 trimethylation
The analysis of a broad spectrum of differentiated tissues provided novel insights into how H3K27me3 generates cell type specificity. Consistent with earlier in vitro studies, we observed that a subset of CpG island genes that are highly enriched in transcription factors show H3K27me3 enrichment in ES cells and are then derepressed in a lineage-selective manner, thereby contributing to cell-specific gene activation (Bernstein et al. 2006; Boyer et al. 2006; Lee et al. 2006; Mikkelsen et al. 2007).
Also in keeping with a recent report (Mohn et al. 2008), we observed a high frequency of H3K27 methylation events that occurred de novo during differentiation. Interestingly, a significant subset of de novo events occurred in non-CpG island promoters, which was unanticipated from previous studies indicating a nearly exclusive occurrence of H3K27 trimethylation in CpG island genes (Mikkelsen et al. 2007; Mohn and Schubeler 2009). H3K27 trimethylation in non-CpG island genes was highly tissue-specific. It also had a marked preference to occur de novo during late stages of differentiation, and unlike H3K27 trimethylation in CpG island genes, it did not preferentially occur in genes encoding transcriptional regulators (Supplemental Tables S9, S10). These findings thus point to a previously unrecognized role of PcG repression in non-CpG island genes during cellular differentiation.
Tissue-specific H3K27me3 as a context-dependent repressive mechanism
Only a subset of genes is repressed by H3K27me3-dependent mechanisms. Interestingly, even among these genes there was no H3K27me3 enrichment in the majority of cell types or developmental stages in which they were transcriptionally inactive. This means that H3K27me3 repression profiles are not mirror images of gene activity but instead reflect which genes have a context-dependent requirement for inactivation through PcG-dependent mechanisms. It is plausible that this active repressive mechanism is unnecessary in cellular contexts in which the transcription of a gene is unlikely due to the lack of activators, or in which its transcription has no significant consequences. Additional epigenetic modifications that can support PcG-independent repression in different cell types include DNA methylation, dimethylated and trimethylated K9, or a void chromatin state (Mikkelsen et al. 2007; Meissner et al. 2008; Schwartz et al. 2010). For example, our analysis of a recently reported data set (Mohn et al. 2008) showed that of all genes that were PcG-repressed selectively in beta-cells, 23% were repressed by DNA methylation without H3K27me3 enrichment in differentiated neurons. The diversity of epigenetic states associated to gene activity and repression highlights fundamental differences in the information provided by epigenetic and transcriptional profiles.
Gene repression programs and beta-cell identity
In beta-cells, H3K27 trimethylation was primarily observed in genes that promote alternate developmental fates, as well as in several genes whose function is known to be deleterious for mature beta-cells. This suggests that PcG-dependent repression is central to the identity of the pancreatic beta-cell. Conditional ablation studies have shown that the Ezh2 H3K27 methyltransferase gene regulates the cell cycle suppressor Cdkn2a (also known as Ink4a or Arf) in beta-cells (Chen et al. 2009). However, beta-cells express Ezh1, another H3K27 methyltransferase (Margueron et al. 2008; Chen et al. 2009), and therefore, a complete understanding of the role of H3K27me3 in the development and mature phenotype of beta-cells will require a full abrogation of this repressive mechanism at different developmental stages.
PcG repressive marks reflect the developmental history of beta-cells
PcG repression has been proposed to mediate inheritance of epigenetic states (Orlando 2003; Ringrose and Paro 2007; Schwartz and Pirrotta 2008). Our findings point to a transmission of PcG repressive marks throughout embryonic development. They suggest that a subset of repressive events that are acquired during organogenesis to suppress alternate fates are epigenetically maintained and inherited by more differentiated cells. The shared H3K27me3 program of pancreatic beta-cells and acinar cells may thus reflect their common developmental history of PcG repressive events, irrespective of the similarity of their transcriptional activity states.
PcG derepression allows beta-cells to adopt a neural program
In sharp contrast to the H3K27me3 repression program of beta-cells, gene activity in beta-cells resembled that of ectoderm-derived neural tissues. The neuroendocrine nature of beta-cells has long been recognized (Atouf et al. 1997; Rorsman 1997), and although their endodermal origin is currently unchallenged, several early studies had proposed a neural crest origin (Pearse 1969; Le Douarin and Teillet 1973; Pictet et al. 1976). Our study now highlights the magnitude of the similarity of beta-cell and neural programs. We show that beta-cells and neural tissues share a selective lack of PcG repression in a core gene program that is small in comparison with the total number of beta-cell PcG-repressed genes, yet because it is highly enriched in transcriptional regulators, it can significantly impact beta-cell gene activity, including genes that are not directly regulated by PcG-dependent mechanisms. Our findings, therefore, suggest that beta-cells use PcG-dependent mechanisms to co-opt a neural program during late pancreatic differentiation, thus enabling a convergence of genetic programs of cells from disparate developmental origins.
What triggers the neural PcG derepression program in beta-cells? Previous studies showed that REST (also known as NRSF), a repressor of neuronal genes in non-neuronal cells, is inactive in beta-cells, and that misexpression of REST in beta-cells reduces the expression of selected REST targets (Atouf et al. 1997; Martin et al. 2008). The deactivation of Rest in beta-cells is thus a candidate mediator of the loss of PcG repression in neural genes in beta-cells. In strong support for this proposal, we find that (1) REST binds in ES cells to genes that form the beta-cell selective lack of PcG repression program (Supplemental Fig. S9a,b); (2) the expression of REST is extinguished in Neurog3+ pancreatic endocrine progenitors (Supplemental Fig. S9c); and (3) in keeping with this finding, the Rest gene acquires the H3K27me3 repressive mark after the pancreatic precursor stage during beta-cell differentiation, coinciding with the activation of the core beta-cell derepression program (Supplemental Fig. S9d). It is thus likely that induction of the neural beta-cell program is dependent on the repression of REST in concert with other changes in transcriptional regulators, notably the activation of Neurog3, an inducer of pancreatic endocrine differentiation (Gradwohl et al. 2000).
Progenitors use H3K27me3 to suppress beta-cell selective genes
Our studies allowed us to examine the chromatin state of beta-cell–specific genes in pancreatic stem cells. In contrast to our expectations, the data suggested that transcriptional competence in pancreatic progenitors is not dependent on a bivalent state or on a premature loss of H3K27me3 in genes that will become activated in acinar or beta-cells. Instead, pancreatic progenitors employ H3K27me3 to suppress differentiation programs of their progeny. These results are consistent with a recent report where the Ezh2 H3K27me3 methyltransferase was shown to specifically repress terminal differentiation of skin progenitors to keratinocytes (Ezhkova et al. 2009). They are also conceptually concordant with data showing that genes that are activated during in vitro differentiation of neurons undergo de novo H3K27 trimethylation at the neuronal progenitor stage (Mohn et al. 2008). However, it should be noted that our data indicate that de novo K27 trimethylation in pancreatic progenitors does not occur in a preferential manner in beta-cell–specific genes.
The model implied by these findings is that pancreatic differentiation entails a controlled lineage-specific release of PcG repressive mechanisms in selected gene targets. These changes occur concomitantly with the progressive de novo acquisition of H3K27me3 in genes to repress alternate cell fates, and at terminal stages in genes that are deleterious for the differentiated phenotype of beta-cells (Fig. 7). More generally, our results underscore the importance of examining endogenous progenitors and a broad representation of cell types to understand how PcG-dependent mechanisms shape a lineage-specific differentiation program.
Implications for pancreatic beta-cell disease
Our findings are relevant for current efforts to generate insulin-producing beta-cells to treat diabetes. Concerning transdifferentiation strategies, our data raise the hypothesis that cell types with similar epigenomes might share similar cell fate restrictions and may thus have an increased ability to undergo interconversions compared with epigenetically distant cell types. It also suggests that manipulating PcG repression (rather than solely gene activity) could be used to promote cell fate changes. Knowledge of the PcG-repression and derepression programs of pancreatic progenitors and beta-cells can furthermore provide benchmarks to direct and evaluate ES cell–based beta-cell differentiation protocols. Finally, these and related genome-scale chromatin studies in islet cells (Gaulton et al. 2010) provide a baseline to investigate how perturbations of the beta-cell epigenome can contribute to the development of diabetes.
Methods
Cell preparation
All experiments were approved by the Institutional animal care committees of the University of Barcelona and Vrije Universiteit Brussel. Tissues were isolated from 10- to 16-wk-old C57Bl/6J male mice. Mouse pancreatic islets and hepatocytes were isolated according to the method described by Luco et al. (2008); other tissues, according to the method described by Antal et al. (2007). Murine ES cells (CGR8) were maintained as described previously (Skoudy et al. 2004).
FACS purification
Pdx1-expressing progenitors were obtained by dissecting embryonic (E10.5) pancreatic buds of Pdx1-eGFP mice. Buds were dissociated to single cells using collagenase (0.5 mg/mL) and trypsin (10 μg/mL, Sigma), filtered (30 μm), incubated with propidium iodide (2 μg/mL, Sigma) and TSQ (2 μg/mL, Molecular Probes) for 15 min, and sorted on a FACSAria (Becton Dickinson) as described (Xu et al. 2008). FACS purification of beta-cells was according to the method described by Stange et al. (2003). Purity of cell preparations was > 90% by immunofluorescence stainings for insulin and DAPI.
RNA analysis
Total RNA was extracted using TRIzol (Invitrogen). RNA integrity was verified with a 2100 Bioanalyzer (Agilent), amplified through cDNA synthesis, and labeled cRNA from biological duplicates was hybridized to Affymetrix Mouse Genome 430 2.0 arrays. RNA expression data were RMA normalized using the Affymetrix Expression Console software. Presence/absence P-values were obtained using the MAS5 algorithm. Transcripts were considered present if both replicate probes showed a presence call with P-value < 0.065 and an average RMA expression of at least 100. For genes with multiple probes, we selected the single probe with the highest expression value.
Chromatin immunoprecipitation
Chromatin immunoprecipitations (ChIPs) were performed essentially as described (Luco et al. 2008), with some modifications. For tissues, we used ∼4 × 106 cells. For pancreatic islets, we used 0.5 × 106 cells for H3K4me3 and 1 × 106 cells for H3K27me3. For FACS-purified beta-cells, we employed 1.5 × 105 cells for H3K4me3 and 3 × 105 cells for H3K27me3. For pancreatic progenitors, we employed 0.25 × 105 cells for H3K4me3 and 0.75 × 105 cells for H3K27me3. Tissues were first finely chopped with razor blades in ice-cold PBS. All cells were fixed in 1% formaldehyde and sonicated using Bioruptor (Diagenode) to a length of 200–1000 bp. Samples were precleared with protein A+G-Sepharose (1:1) and immunoprecipitated with anti-H3K4me3 (Upstate, 05-745), anti-H3K27me3 (Upstate, 07-449), mouse monoclonal anti-REST (12C11 (Dr. David J. Anderson, California Institute of Technology, Pasadena) (Chen et al. 1998), or anti-mouse IgG (Abcam, ab6709) overnight at 4°C. Precipitations on FACS purified beta-cells and progenitors were performed in the presence of 2.5 mg/mL BSA and 25 μg/mL tRNA. Immune complexes were collected by adsorption to protein A+G-Sepharose for 2 h at 4°C. Beads were washed and immunocomplexes eluted prior to DNA purification with Qiaquick columns (Qiagen).
ChIP and input DNA were amplified using either the Sigma GenomePlex WGA2 kit or the WGA4 single cell kit for FACS-purified cells. Amplification was performed as described (O'Geen et al. 2006), except that dUTPs were added to a final concentration of 0.4 mM during the amplification reaction to enable subsequent fragmentation. We fragmented 6–7.5 μg DNA, labeled it using the Affymetrix GeneChip WT Double-Stranded DNA terminal Labeling Kit, and hybridized to GeneChip Mouse Promoter 1.0R Arrays covering about −6.5 kb to +2.5 kb of transcription start sites of RefSeq mRNAs (NCBI GenBank April 7, 2005).
Significant enrichment in ChIP experiments relative to input DNA was determined using Cisgenome (Ji et al. 2008). We applied a hidden Markov model as described (Ji and Wong 2005) and used a posterior probability cut-off of 0.5 in at least five continuous probes. Using identical criteria, <0.05% of sites detected with histone antibodies were observed in IgG control ChIP experiments. Enriched intervals were mapped to Refseq genes when located within 2000 bp of transcriptional start sites in the case of histone methylations and 7000 bp for REST.
Sequential ChIP was performed as described above except that during the elution of the first immunoprecipitation (for H3K4me3) DDT was added to a final concentration of 30 mM. Five percent of this eluted chromatin was set aside to assess enrichment of targets in the first and second (H3K27me3) immunoprecipitations. The remaining chromatin was concentrated to a volume of 50 μL using Micocon centrifugal columns (Ultracel YM-3; Millipore), diluted 30 times in sonication buffer, and used for the second immunoprecipitation with anti-H3K27me3 in the presence of 2.5 mg/mL BSA and 25 μg/mL tRNA. DNAs from input and both immunoprecipitations were amplified with the Sigma GenomePlex WGA2 kit and quantified with a ND-1000 spectrophotometer, and equal amounts of total DNA were then used for quantitative PCR analysis.
Quantitative PCR analysis
Quantitative PCR was performed on a 7300 Realtime PCR System (Applied Biosystems) using the Power SYBR Green reagent (Applied Biosystems). A dilution of genomic input DNA served to establish a standard curve. Threshold cycles of ChIP samples were projected on this curve to generate quantitative values in arbitrary units. Enrichments were then visualized relative to positive or negative controls. A full list of primers used is provided as Supplemental Table S11.
Immunofluorescence
Immunofluorescence in E15.5 embryos was performed as described previously (Maestro et al. 2003) Guinea-pig anti-NEUROG3 (Michael S. German, University of California San Francisco) was used at 1:1000, and mouse anti-REST (12C11) (Chen et al. 1998) was used at a 1:20 with TSA amplification (TSA Fluorescence Systems, PerkinElmer).
Integrated data analysis
CpG islands were downloaded from UCSC (table name “cpgIslandExt”). The criteria to define CpG islands was thus as follows: GC content of ≥50%, length >200 bp, ratio >0.6 of observed number of CG dinucleotides to the expected number on the basis of the number of Gs and Cs in the segment. All RefSeq genes with a CpG island within 2000 bp of transcriptional start sites were classified as CpG island genes, corresponding to 61% of genes in this study.
To generate dendrograms and gene cluster representations, tissues were normalized for the total number of present/absent genes for that histone methylation and then clustered based on their similarity according to the Euclidian distance using Cluster3.0. Clusters were visualized in Java Treeview1.1.1.
The DAVID functional annotation tool (http://david.abcc.ncifcrf.gov/) was used to study the overrepresentation of annotation terms, using all RefSeq genes as the background. Nonredundant Gene Ontology categories from the top “Molecular Function” and “Biological Process” term nodes 4 and 5 were selected.
Principal component analysis (Pearson 1901) was performed using a binary classification of all genes based on the presence/absence of mRNA, H3K27me3, or H3K4me3. After performing PCA for each data set, the two components with maximum Eigen values were plotted in pairs to show the maximum data variance.
Overrepresentation of transcription factor binding sites was determined using oPOSSUM (Ho Sui et al. 2005). The top 30% conserved regions located within 10 kb of transcription start sites were analyzed for overrepresentation of known transcription factor motifs with a matrix match threshold of 80%.
Statistical analysis
Comparisons of expression levels between sets of genes were analyzed with SPSS 16 by two-sided unpaired Student's t-test or one-way ANOVA and post-hoc Fisher's least significant difference test, as appropriate. Fractions of genes with methylation patterns in different tissues were compared with a two-sided χ2 test.
Acknowledgments
We thank Adrian Bracken, Duncan Odom, Miguel Vidal, and Ken Zaret for insightful comments; Finn Cilius Nielsen, Rehannah Borup, and Susanne Smed (RH Microarray Center, Rigshospitalet, Copenhagen) for array hybridizations; Mohammad Massumi for maintenance of ES cells; Carlos Castaño and Vanessa Grau for mouse colony maintenance; Carme Sanahuja for administrative assistance; Michael S. German and David J. Anderson for antibodies; Tatiana Konovolova and Alexander Kel (BIOBASE) for help with initial data analysis; and funding from Ministerio de Ciencia e Innovación (SAF2008-03116) and the European Union VI Framework Programme (LSHB-CT-2005-512145).
Footnotes
[Supplemental material is available online at http://www.genome.org. The microarray data for RNA expression and ChIP experiments from this study have been submitted to ArrayExpress (http://www.ebi.ac.uk/microarray-as/ae/) under accession nos. E-TABM-877 and E-TABM-906, respectively.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.101709.109.
References
- Antal C, Teletin M, Wendling O, Dgheem M, Auwerx J, Mark M 2007. Tissue collection for systematic phenotyping in the mouse. Curr Protoc Mol Biol Chapter 29: Unit A.4 doi: 10.1002/0471142727.mb29a04s80 [DOI] [PubMed] [Google Scholar]
- Atouf F, Czernichow P, Scharfmann R 1997. Expression of neuronal traits in pancreatic beta cells. Implication of neuron-restrictive silencing factor/repressor element silencing transcription factor, a neuron-restrictive silencer. J Biol Chem 272: 1929–1934 [DOI] [PubMed] [Google Scholar]
- Baeyens L, De BS, Lardon J, Mfopou JK, Rooman I, Bouwens L 2005. In vitro generation of insulin-producing beta cells from adult exocrine pancreatic cells. Diabetologia 48: 49–57 [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Chen ZF, Paquette AJ, Anderson DJ 1998. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat Genet 20: 136–142 [DOI] [PubMed] [Google Scholar]
- Chen H, Gu X, Su IH, Bottino R, Contreras JL, Tarakhovsky A, Kim SK 2009. Polycomb protein Ezh2 regulates pancreatic beta-cell Ink4a/Arf expression and regeneration in diabetes mellitus. Genes Dev 23: 975–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, Tarakhovsky A, Fuchs E 2009. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell 136: 1122–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferber S, Halkin A, Cohen H, Ber I, Einav Y, Goldberg I, Barshack I, Seijffers R, Kopolovic J, Kaiser N, et al. 2000. Pancreatic and duodenal homeobox gene 1 induces expression of insulin genes in liver and ameliorates streptozotocin-induced hyperglycemia. Nat Med 6: 568–572 [DOI] [PubMed] [Google Scholar]
- Gaulton KJ, Nammo T, Pasquali L, Simon JM, Giresi PG, Fogarty MP, Panhuis TM, Mieczkowski P, Secchi A, Bosco D, et al. 2010. A map of open chromatin in human pancreatic islets. Nat Genet 42: 255–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradwohl G, Dierich A, LeMeur M, Guillemot F 2000. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci 97: 1607–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualdi R, Bossard P, Zheng M, Hamada Y, Coleman JR, Zaret KS 1996. Hepatic specification of the gut endoderm in vitro: Cell signaling and transcriptional control. Genes & Dev 10: 1670–1682 [DOI] [PubMed] [Google Scholar]
- Hanafusa T, Imagawa A 2008. Insulitis in human type 1 diabetes. Ann N Y Acad Sci 1150: 297–299 [DOI] [PubMed] [Google Scholar]
- Heimberg H, De VA, Vandercammen A, Van SE, Pipeleers D, Schuit F 1993. Heterogeneity in glucose sensitivity among pancreatic beta-cells is correlated to differences in glucose phosphorylation rather than glucose transport. EMBO J 12: 2873–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Sui SJ, Mortimer JR, Arenillas DJ, Brumm J, Walsh CJ, Kennedy BP, Wasserman WW 2005. oPOSSUM: Identification of over-represented transcription factor binding sites in co-expressed genes. Nucleic Acids Res 33: 3154–3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara H, Wang H, Drewes LR, Wollheim CB 1999. Overexpression of monocarboxylate transporter and lactate dehydrogenase alters insulin secretory responses to pyruvate and lactate in beta cells. J Clin Invest 104: 1621–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Wong WH 2005. TileMap: Create chromosomal map of tiling array hybridizations. Bioinformatics 21: 3629–3636 [DOI] [PubMed] [Google Scholar]
- Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH 2008. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol 26: 1293–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B 2007. Genome-wide mapping of in vivo protein–DNA interactions. Science 316: 1497–1502 [DOI] [PubMed] [Google Scholar]
- Kontaraki J, Chen HH, Riggs A, Bonifer C 2000. Chromatin fine structure profiles for a developmentally regulated gene: Reorganization of the lysozyme locus before trans-activator binding and gene expression. Genes Dev 14: 2106–2122 [PMC free article] [PubMed] [Google Scholar]
- Kroon E, Martinson LA, Kadoya K, Bang AG, Kelly OG, Eliazer S, Young H, Richardson M, Smart NG, Cunningham J, et al. 2008. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat Biotechnol 26: 443–452 [DOI] [PubMed] [Google Scholar]
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, et al. 2007. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449: 689–694 [DOI] [PubMed] [Google Scholar]
- Le Douarin NM, Teillet MA 1973. The migration of neural crest cells to the wall of the digestive tract in avian embryo. J Embryol Exp Morphol 30: 31–48 [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. 2006. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125: 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Maestro MA, Sadoni N, Zink D, Ferrer J 2008. Targeted deficiency of the transcriptional activator Hnf1α alters subnuclear positioning of its genomic targets. PLoS Genet 4: e1000079 doi: 10.1371/journal.pgen.1000079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen OD, Serup P 2006. Towards cell therapy for diabetes. Nat Biotechnol 24: 1481–1483 [DOI] [PubMed] [Google Scholar]
- Maestro MA, Boj SF, Luco RF, Pierreux CE, Cabedo J, Servitja JM, German MS, Rousseau GG, Lemaigre FP, Ferrer J 2003. Hnf6 and Tcf2 (MODY5) are linked in a gene network operating in a precursor cell domain of the embryonic pancreas. Hum Mol Genet 12: 3307–3314 [DOI] [PubMed] [Google Scholar]
- Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D 2008. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell 32: 503–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D, Allagnat F, Chaffard G, Caille D, Fukuda M, Regazzi R, Abderrahmani A, Waeber G, Meda P, Maechler P, et al. 2008. Functional significance of repressor element 1 silencing transcription factor (REST) target genes in pancreatic beta cells. Diabetologia 51: 1429–1439 [DOI] [PubMed] [Google Scholar]
- Mastick GS, Andrews GL 2001. Pax6 regulates the identity of embryonic diencephalic neurons. Mol Cell Neurosci 17: 190–207 [DOI] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454: 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Schubeler D 2009. Genetics and epigenetics: Stability and plasticity during cellular differentiation. Trends Genet 25: 129–136 [DOI] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D 2008. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell 30: 755–766 [DOI] [PubMed] [Google Scholar]
- O'Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T 2001. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol 21: 4330–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Geen H, Nicolet CM, Blahnik K, Green R, Farnham PJ 2006. Comparison of sample preparation methods for ChIP-chip assays. Biotechniques 41: 577–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver-Krasinski JM, Stoffers DA 2008. On the origin of the beta cell. Genes Dev 22: 1998–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando V 2003. Polycomb, epigenomes, and control of cell identity. Cell 112: 599–606 [DOI] [PubMed] [Google Scholar]
- Otonkoski T, Jiao H, Kaminen-Ahola N, Tapia-Paez I, Ullah MS, Parton LE, Schuit F, Quintens R, Sipila I, Mayatepek E, et al. 2007. Physical exercise-induced hypoglycemia caused by failed silencing of monocarboxylate transporter 1 in pancreatic beta cells. Am J Hum Genet 81: 467–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, W.de L 2003. The beta-globin nuclear compartment in development and erythroid differentiation. Nat Genet 35: 190–194 [DOI] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, Lazzerini DE, Helin K 2004. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J 23: 4061–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearse AG 1969. The cytochemistry and ultrastructure of polypeptide hormone-producing cells of the APUD series and the embryologic, physiologic and pathologic implications of the concept. J Histochem Cytochem 17: 303–313 [DOI] [PubMed] [Google Scholar]
- Pearson K 1901. On lines and planes of closest fit to systems of points in space. Philos Mag 2: 559–572 [Google Scholar]
- Pictet RL, Rall LB, Phelps P, Rutter WJ 1976. The neural crest and the origin of the insulin-producing and other gastrointestinal hormone-producing cells. Science 191: 191–192 [DOI] [PubMed] [Google Scholar]
- Pietersen AM, van Lohuizen M 2008. Stem cell regulation by polycomb repressors: postponing commitment. Curr Opin Cell Biol 20: 201–207 [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R 2007. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development 134: 223–232 [DOI] [PubMed] [Google Scholar]
- Roh TY, Cuddapah S, Cui K, Zhao K 2006. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci 103: 15782–15787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorsman P 1997. The pancreatic beta-cell as a fuel sensor: An electrophysiologist's viewpoint. Diabetologia 40: 487–495 [DOI] [PubMed] [Google Scholar]
- Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T 2004. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol 6: 73–77 [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V 2008. Polycomb complexes and epigenetic states. Curr Opin Cell Biol 20: 266–273 [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Stenberg P, Ohno K, Bourgon R, Pirrotta V 2010. Alternative epigenetic chromatin states of polycomb target genes. PLoS Genet 6: e1000805 doi: 10.1371/journal.pgen.1000805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoudy A, Rovira M, Savatier P, Martin F, Leon-Quinto T, Soria B, Real FX 2004. Transforming growth factor (TGF)β, fibroblast growth factor (FGF) and retinoid signalling pathways promote pancreatic exocrine gene expression in mouse embryonic stem cells. Biochem J 379: 749–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stange G, Van De Casteele M, Heimberg H 2003. Purification of rat pancreatic B-cells by fluorescence-activated cell sorting. Methods Mol Med 83: 15–22 [DOI] [PubMed] [Google Scholar]
- Thiel G, Schuit F 2008. No REST for healthy beta cells. Diabetologia 51: 1343–1346 [DOI] [PubMed] [Google Scholar]
- Xu X, D'Hoker J, Stange G, Bonne S, De LN, Xiao X, Van De Casteele M, Mellitzer G, Ling Z, Pipeleers D, et al. 2008. Beta cells can be generated from endogenous progenitors in injured adult mouse pancreas. Cell 132: 197–207 [DOI] [PubMed] [Google Scholar]
- Yuan SH, Qiu Z, Ghosh A 2009. TOX3 regulates calcium-dependent transcription in neurons. Proc Natl Acad Sci 106: 2909–2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS 2008. Genetic programming of liver and pancreas progenitors: Lessons for stem-cell differentiation. Nat Rev Genet 9: 329–340 [DOI] [PubMed] [Google Scholar]
- Zhou Q, Melton DA 2008. Pathways to new beta cells. Cold Spring Harb Symp Quant Biol 73: 175–181 [DOI] [PubMed] [Google Scholar]
- Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA 2008. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature 455: 627–632 [DOI] [PMC free article] [PubMed] [Google Scholar]