Abstract
The replication of a chromosomal region during S phase can be highly dynamic between cell types that differ in transcriptome and epigenome. Early replication timing has been positively correlated with several histone modifications that occur at active genes, while repressive histone modifications mark late replicating regions. This raises the question if chromatin modulates the initiating events of replication. To gain insights into this question, we have studied the function of heterochromatin protein 1 (HP1), which is a reader of repressive methylation at histone H3 lysine 9, in genome-wide organization of replication. Cells with reduced levels of HP1 show an advanced replication timing of centromeric repeats in agreement with the model that repressive chromatin mediates the very late replication of large clusters of constitutive heterochromatin. Surprisingly, however, regions with high levels of interspersed repeats on the chromosomal arms, in particular on chromosome 4 and in pericentromeric regions of chromosome 2, behave differently. Here, loss of HP1 results in delayed replication. The fact that these regions are bound by HP1 suggests a direct effect. Thus while HP1 mediates very late replication of centromeric DNA, it is also required for early replication of euchromatic regions with high levels of repeats. This observation of opposing functions of HP1 suggests a model where HP1-mediated repeat inactivation or replication complex loading on the chromosome arms is required for proper activation of origins of replication that fire early. At the same time, HP1-mediated repression at constitutive heterochromatin is required to ensure replication of centromeric repeats at the end of S phase.
Replication of eukaryotic genomes starts at origins of replication that are distributed throughout chromosomes and that can fire at different times during S phase (Gilbert 2002). Replication origins have been mapped at high resolution throughout the yeast genome, where they share a consensus motif (MacAlpine and Bell 2005; Nieduszynski et al. 2006). In metazoans, however, only a few origins of replication have been identified, and these lack a consensus sequence (MacAlpine et al. 2004; Lucas et al. 2007; Macalpine et al. 2010), suggesting that replication initiation in higher eukaryotes is determined not solely by DNA sequence but at least in part by epigenetic features such as chromatin structure.
Initiation of DNA replication can vary in timing and location, giving rise to highly cell type–specific replication timing programs. Early microscopic studies have shown that in most organisms, euchromatin replicates before heterochromatin (Gilbert 2002). Replication timing can be dynamic between different cell types, reflecting different epigenetic states (Hiratani et al. 2008, 2010; Pope et al. 2009; Schwaiger et al. 2009). Moreover, in higher eukaryotes, early replication correlates with a high probability of transcription and the presence of histone modifications characteristic of active chromatin such as histone acetylation (Schwaiger et al. 2009; Hansen et al. 2010). Histone hyperacetylation has been directly implicated to facilitate origin activation and thereby early replication timing (Aggarwal and Calvi 2004; Calvi et al. 2007; Goren et al. 2008) in agreement with a model of a function for open chromatin. If origin definition and activation would be purely defined by chromatin, one might expect that any relief of gene repression will lead to early replication at any chromosomal position. However, results in mammalian cells suggest that hyperactivation of a genomic region can also affect origin usage negatively (Mesner and Hamlin 2005; Gregoire et al. 2006). This suggests that transcription could also inhibit the firing of replication origins or fork progression, depending on the spatial relationship between transcription and DNA replication. To address the crosstalk between gene repression pathways and replication timing, we studied the consequences of reducing heterochromatin protein 1 [HP1; also referred to as HP1A, and as SU(VAR)205 in FlyBase] in the Drosophila genome.
The HP1 gene was discovered as a suppressor of position effect variegation (PEV) in Drosophila, and thus is referred to as Su(var)205 (James and Elgin 1986; Wustmann et al. 1989; Eissenberg et al. 1990). PEV describes the phenomenon that euchromatic genes acquire a variegated expression pattern when positioned close to or within heterochromatin (Muller and Altenburg 1930). This silencing requires heterochromatin functions and is suppressed by deletion of genes such as HP1 or Su(var)3-9 (Wustmann et al. 1989; Wallrath and Elgin 1995), the histone-methyltransferase that methylates lysine 9 on histone H3 (Schotta et al. 2002).
The binding pattern of HP1 throughout the sequenced part of the Drosophila genome has recently been studied in great detail using DamID and microarray technology in Drosophila cells (Greil et al. 2003). While HP1 binding to pericentric regions is stable throughout development, HP1 binds many, mostly long, genes in a developmental stage–specific way. HP1-bound regions often cover large, up to 100-kb-long regions in the genome, which do not overlap with polycomb target regions (de Wit et al. 2007). HP1 binds to H3K9 methylation independent of the enzyme that established the modification (Greil et al. 2003; de Wit et al. 2005, 2007; Seum et al. 2007; Tzeng et al. 2007). HP1 target genes that lie in pericentric heterochromatin (also termed heterochromatic genes) tend to be highly expressed and also have a different HP1 binding pattern than do euchromatic HP1 target genes, which show average expression levels (de Wit et al. 2007). Heterochromatic genes tend to be surrounded by repetitive sequences, which often reside within the introns of those genes (Devlin et al. 1990; Schulze et al. 2005). Upon loss of HP1 or translocation to euchromatin, the expression of heterochromatic genes is reduced in Drosophila (Wakimoto and Hearn 1990; Clegg et al. 1998; Lu et al. 2000; Schulze et al. 2005). It appears likely that this is due to the loss of heterochromatin-mediated silencing of associated repeats.
Interestingly, in Drosophila polytene chromosomes and Kc cells, HP1 target regions often overlap with regions bound by the Suppressor of Underreplication (SuUR) protein (Koryakov et al. 2006; Pindyurin et al. 2007). SuUR binds to underreplicated and late replicating regions on Drosophila polytene chromosomes (Makunin et al. 2002) and to late replicating regions in Kc cells (Pindyurin et al. 2007). SuUR might play a role in suppressing origin firing since it reduces the endoreplication of heterochromatic regions on polytene chromosomes (Belyaeva et al. 1998). HP1 has been shown to interact with SuUR and could be crucial for mediating chromatin binding of the SuUR protein (Pindyurin et al. 2008). Therefore it is conceivable that loss of HP1 from chromatin might induce an advance in replication timing of some heterochromatic regions. On the other hand, HP1 interacts with the origin recognition complex-2 (ORC2) protein (Pak et al. 1997), which is a member of the origin recognition complex (ORC) and important for origin activation (Bell and Dutta 2002). This might indicate a role for HP1 in recruiting the ORC to chromatin. In Schizosaccharomyces pombe, an organism with rather unusual early replication of heterochromatin (Kim et al. 2003), HP1 has recently been suggested to activate replication origins by recruiting the Dfp1 (Dbf4)-dependent kinase (DDK) to origins within pericentromeres and the silent mating-type locus (Hayashi et al. 2009).
To determine possible effects of HP1 on replication timing, we used RNA interference (RNAi) to reduce HP1 protein levels in Drosophila Kc cells. This resulted in advanced replication timing of centromeric repeats as measured by immunofluorescence and qPCR. In addition to changes at centromeric repeats, high-resolution tiling array analysis revealed that 5%–10% of the genome show altered replication timing upon knockdown of HP1. Unexpectedly, however, the majority of changes result in a replication timing delay. These delayed regions are direct targets of HP1, represent unique sequences embedded in repeats, and are particularly frequent on the fourth chromosome. Thus in Drosophila, HP1 functions in both delaying and advancing the replication timing of selected genomic regions.
Results
Advanced replication of centromeric repeats after HP1 knockdown
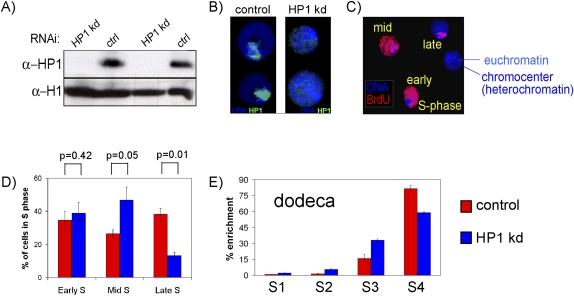
To reduce the levels of HP1 protein in Drosophila Kc cells by RNAi, we exposed cells for 8 d to double-stranded RNA (dsRNA) specific for the HP1 coding sequence. This treatment results in a strong depletion of HP1 protein, as detected by Western blot (Fig. 1A) and immunofluorescence staining (Fig. 1B), and does not lead to down-regulation of the expression of HP1B and HP1C (Supplemental Fig. 1D). To determine if HP1 reduction affects the temporal program of replication timing, we first measured the overall replication timing of constitutive heterochromatin, which in Drosophila clusters in a single chromocenter (Heitz 1934). We pulse-labeled cells with BrdU for 1 h, allowing us to detect replicating DNA by immunofluorescence with an antibody recognizing BrdU (Fig. 1C). Around 30% of all nuclei stained positive for BrdU, reflecting that they were in S phase at the time of the label. We observed, among labeled cells, three distinct patterns in the untreated control cells: labeling of only euchromatin, a characteristic of early S phase; labeling of both euchromatin and the chromocenter, a characteristic of mid S phase; and labeling of the chromocenter, a characteristic of late S phase (Fig. 1C). The same three patterns are also present in cells depleted for HP1, yet they occur at different frequencies. We observed a marked reduction of cells with chromocenter staining only, which, in untreated cells, is typical for late S phase. This decrease coincided with an increased number of cells showing the typical mid S-phase pattern (Fig. 1D). The cell cycle profiles measured by FACS were indistinguishable between HP1-depleted and untreated cells, strongly suggesting that this change does not reflect accumulation of cells in mid S phase (Supplemental Fig. 2); instead, it indicates advanced replication of the chromocenter DNA in cells with reduced HP1 levels, which leads to a pattern usually found more frequently in mid S phase.
Figure 1.
Depletion of HP1 by RNA interference. (A) Western blot detecting HP1 protein in untreated control cells (ctrl) and cells treated with dsRNA directed against HP1 (HP1 kd) for 7 d showing high efficiency of knockdown. H1 serves as a loading control for equal amounts of protein. (B) Cytological localization of HP1 by immunofluorescence with an antibody recognizing HP1 protein in Kc cells. HP1 localizes mainly to the chromocenter in control cells (left) but is detected only at very low levels in HP1 knockdown cells (right). (C) Cytological analysis of replication timing. Kc cell nuclei with three different patterns of BrdU incorporation after pulse-labeling are shown. (D) Quantification of the percentage of BrdU-positive nuclei with early, mid, and late S-phase pattern based on their BrdU staining in 486 control and 221 HP1-depleted BrdU-positive nuclei. Error bars, SEM between five biological repeats. P-values were calculated using the Wilcoxon rank sum test between five biological repeats. (E) Enrichments of BrdU containing DNA in four FACS sorted fractions (S1–S4) as quantified by real-time PCR. S1 represents the earliest and S4 the latest S-phase fraction as measured by DNA content. Here the dodeca pericentric repeat sequence is tested (see text). Error bars, SD between three biological repeats.
To study replication timing at higher resolution, we measured replication timing by labeling HP1 knockdown cells and control cells with BrdU for 1 h. Cells were then sorted into four different S-phase fractions based on DNA content using FACS (Supplemental Fig. 3A) as previously described (Schwaiger et al. 2009). Replicating DNA from each fraction was isolated by immunoprecipitation with an antibody detecting BrdU, and the abundance of replicating DNA in each S-phase fraction was compared using qPCR. Since the chromocenter represents mostly repetitive sequences within constitutive heterochromatin, we tested if the replication timing of centromeric repeats on chromosome 3 is advanced after HP1 RNAi. To this end, we used primers specific for the dodeca repeat sequence located in pericentric heterochromatin of chromosome 3 (Abad et al. 1992). In untreated cells, we detected the highest enrichment of replicating DNA in S4, representing the latest S-phase fraction (Fig. 1E). However, in HP1 RNAi cells, the enrichment in late S phase is reduced but increased in the two fractions representing mid S phase (S2 and S3) (Fig. 1E). This suggests that the dodeca repeat shows advanced replication timing after HP1 knockdown in agreement with the general behavior of heterochromatin. A small but reproducible advance in replication timing was also measured at the Bari1 transposon, which localizes preferentially into heterochromatin on chromosome 2 (Supplemental Fig. 3B; Caizzi et al. 1993). Based on cytological and repeat specific analysis, we conclude that depletion of HP1 results in a global advance of centromeric heterochromatin replication.
Replication timing changes around heterochromatic genes after HP1 knockdown
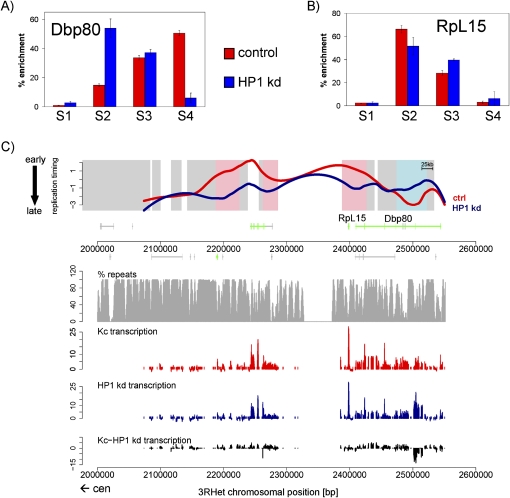
HP1 has been shown to bind to many unique genes that are embedded within dispersed repeats (Greil et al. 2003; de Wit et al. 2007). Proper transcription of these genes depends on their heterochromatic environment, since their translocation into euchromatin can eliminate their expression, as does the reduction of HP1 protein levels (Yasuhara and Wakimoto 2006). We tested if these genes change replication timing upon depletion of HP1. The light gene on chromosome 2L did not show a difference in enrichment for BrdU-IPd DNA in the four S-phase fractions between HP1 knockdown and control cells (Supplemental Fig. 5A,B). However, a tested region on chromosome 3, containing the Dbp80 and RpL15 genes, showed a strong effect on replication timing after HP1 knockdown. The large Dbp80 gene replicates late in Kc cells but showed advanced replication timing in HP1 knockdown cells (Fig. 2A), while the early replicating RpL15 gene replicated later after HP1 knockdown (Fig. 2B). This shows that modulating HP1 levels can have a strong but locus-specific effect on replication timing of heterochromatic genes.
Figure 2.
Differences in replication timing at heterochromatic genes. (A) Enrichments of BrdU containing DNA in four FACS sorted fractions (S1–S4) as quantified by real-time PCR with primers specific for the 3′ region of the Dbp80 gene. (B) Enrichments of BrdU containing DNA in four FACS sorted fractions (S1–S4) as quantified by real-time PCR with primers specific for the RpL15 gene. S1 represents the earliest and S4 the latest S-phase fraction as measured by DNA content. Error bars, SD between three biological repeats. (C) Replication timing profiles of control (red, ctrl) and HP1 knockdown (blue, HP1 kd) Kc cells for a representative region of pericentric heterochromatin on chromosome 3L (3LHet). X-axis represents 3LHet chromosomal position in base pairs; y-axis, log2 (early/late replication). Background coloring denotes regions that replicate earlier in HP1 kd cells (L:E, blue), regions that replicate earlier in control cells (E:L, pink), and regions replicating similarly in both cell types (white). Gene annotation is displayed below the profile (boxes, exons; lines, introns; small boxes, UTRs) and colored by expression status (for details, see Methods; green, expressed in control and HP1 kd cells; blue, expressed only in HP1 kd cells; red, expressed only in control cells; gray, not expressed in control and HP1 kd cells). The sequence analyzed in B overlaps with the Dbp80 exon and is located immediately to the right of the 2,500,000-bp marker. (Below) Transcription levels of control (red) and HP1 kd (blue) cells measured by tiling arrays are displayed, including transcription level differences (black). Repeat density is indicated as the percentage of repetitive element within 2-kb windows (gray; for details, see Methods). The direction toward the centromere (cen) is marked by an arrow.
To quantify replication timing changes on a genome-wide scale, we obtained early and late replicating DNA by sorting BrdU-labeled cells into early and late S phase, followed by BrdU-IP, and hybridized it to a tiling array that covers the entire nonrepetitive genome at 35-bp resolution, as previously described (Schwaiger et al. 2009). The resulting replication timing profiles were highly reproducible (Supplemental Fig. 1A,B). At the level of the genome, replication timing was very similar between control and HP1 knockdown cells (Supplemental Fig. 1C), suggesting that only a subset of unique sequences change. Importantly, however, a significant fraction of probes on the array showed differences and, in most cases, replicate later in HP1 knockdown cells than in control cells (Supplemental Fig. 1C). Figure 2C shows a region within pericentric heterochromatin of chromosome 3L (chromosome 3LHet) containing the Dbp80 and RpL15 genes, which are differentially replicating according to qPCR analysis. In agreement with the qPCR data, we detect differences in replication timing using tiling arrays (Fig. 2C). The region of advanced replication timing in HP1 knockdown cells overlaps with the Dbp80 gene, while the RpL15 gene shows delayed replication timing after knockdown of HP1 (Fig. 2C). To determine if these changes in replication timing correlate with changes in gene expression, we hybridized RNA from control and HP1 knockdown cells to 3′ untranslated region (UTR) as well as chromosomal tiling arrays. Both the Dbp80 and RpL15 genes are active in Kc cells, even after depletion of HP1 (Fig. 2C). Tiling array analysis shows increased transcription within part of the Dbp80 gene in HP1 knockdown cells (Fig. 2C). It is possible that this detected RNA is derived from a small coding or noncoding transcript within Dbp80 or from the numerous repetitive elements in this region (Fig. 2C). Interestingly, the delay in replication timing around the RpL15 gene does not coincide with reduced transcript levels (Fig. 2C). This implies a direct role of HP1 in the early replication of this region. Alternatively, the transcription of repetitive elements not present on our tiling array could be up-regulated within this heterochromatic locus. This could interfere with origin selection, leading to the use of different origins of replication and thereby to different effects on replication timing.
Delayed replication timing after HP1 knockdown occurs at HP1 target sites
To determine genome-wide the regions of dynamic replication in an unbiased way, we employed a three-state hidden Markov model (HMM) to segment replication timing differences between control and HP1 knockdown cells (see Methods). In order to focus on robust changes, we excluded regional differences that are smaller than 20 kb or where the difference in timing extends over less than 1/12th of the total range of timing differences (delta log2 < 0.8). These stringent criteria reveal 52 regions, corresponding to 2.7% of the genome, that replicate earlier in control than in HP1 knockdown cells (E:L) and 38 regions, corresponding to 2.4% of the genome, that replicate earlier in HP1 RNAi than in control cells (L:E). These differentially replicating regions can be larger than 200 kb, have an average size of ∼70 kb (Supplemental Fig. 4; data not shown), and represent 5.1% of all sequences.
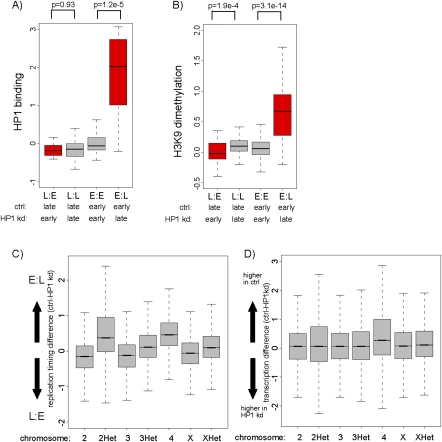
To determine if replication timing changes occur in regions that are actually bound by HP1, we compared differential replication timing to existing HP1 binding data for chromosomes 2 and 4 (de Wit et al. 2007). This analysis reveals that regions that replicate earlier upon HP1 reduction have low levels of HP1 binding in normal cells (Fig. 3A). In addition, chromosome 4, which is strongly bound by HP1 (de Wit et al. 2007), does not show earlier replicating regions in HP1 RNAi cells (Figs. 3C, 4). Together this suggests that advanced replication timing upon HP1 reduction on chromosomes 2 and 4 does not occur in regions bound by HP1. In contrast, however, regions that replicate later upon HP1 knockdown were frequently sites of actual HP1 binding (Fig. 3A). This suggests that their normal earlier replication timing depends directly on HP1 binding. Furthermore, the differences in replication timing were generally much higher in regions replicating later after HP1 knockdown than in those with advanced replication timing (Supplemental Fig. 4).
Figure 3.
Delayed replication timing of HP1- and H3K9me2-positive chromatin. (A) Distribution of HP1 binding levels (de Wit et al. 2007) for regions with differential replication timing. The boxplots illustrate that regions with delayed replication after HP1 knockdown (E:L) show high levels of HP1 binding. L:E, regions replicating earlier in HP1 kd cells; L:L, regions replicating late in both; E:E, regions replicating early in both; E:L, regions replicating earlier in control cells. P-values were calculated using the Wilcoxon rank sum test. (B) Distribution of H3K9me2 for regions with differential replication timing. The boxplots illustrate that regions replicating later after HP1 knockdown (E:L) show high levels of H3K9me2. L:E, regions replicating earlier in HP1 kd cells; L:L, regions replicating late in both; E:E, regions replicating early in both; E:L, regions replicating earlier in control cells. P-values were calculated using the Wilcoxon rank sum test. (C) Distribution of replication timing differences (control-HP1 kd RNAi replication timing, log2 scale, y-axis) on different chromosomes. The individual boxplots represent the distribution of control minus HP1 kd replication timing of all array probes on the indicated chromosome. The boxplots illustrate that chromosome 4 (4, P-value < 2.2 × 10−16) and pericentric regions on chromosome 2 (2Het, P-value < 2.2 × 10−16), and to a lesser extent on chromosome 3 and X (3Het, XHet, P-value < 2.2 × 10−16), show delayed replication timing (E:L) more often than euchromatin on chromosome 2,3 and X (2,3, X). (D) Distribution of transcription differences (control-HP1 RNAi transcription, log2 scale, y-axis) within different chromosomes. The boxplots illustrate that chromosome 4 (4, P-value < 2.2 × 10−16) shows reduced transcription levels in HP1 knockdown cells (higher in control) more frequently than other chromosomes.
Figure 4.
Delayed replication timing of chromosome 4 after knockdown of HP1. Replication timing profiles of control (red, ctrl) and HP1 knockdown (blue, HP1 kd) Kc cells for a representative region on chromosome 4. X-axis, chromosomal position in base pairs; y-axis, log2 (early/late replication). Background coloring denotes regions that replicate earlier in HP1 kd cells (L:E, pink), regions that replicate earlier in control cells (E:L, blue), and regions replicating similarly in both cell types (white). Annotated genes are displayed below the profile (boxes, exons; lines, introns; small boxes, UTRs) and colored by their expression status (for details, see Methods; green, expressed in control and HP1 kd cells; blue, expressed only in HP1 kd cells; red, expressed only in control cells; gray, not expressed in control and HP1 kd cells). Transcription levels of control (red) and HP1 kd (blue) cells measured by tiling arrays are displayed on the same scale below, including transcription level differences (black). The direction toward the centromere (cen) is marked by an arrow.
To characterize the chromatin status of regions that change replication timing after HP1 knockdown, we performed chromatin immunoprecipitation with microarray hybridization (ChIP-chip) for dimethylation of lysine 9 and trimethylation of lysine 27 on histone H3. We find H3K27me3 associated with nontranscribed, late replicating regions (Supplemental Fig. 6E–G), while H3K9me2 generally marks regions containing dispersed repeats (Supplemental Fig. 6A). Similar to previous reports, we find additional H3K9me2 at coding regions of active genes (Supplemental Fig. 6H,I; Yasuhara and Wakimoto 2008). Importantly, regions of high H3K9me2 do not replicate as late in S phase as H3K27me3 domains (Supplemental Fig. 6G,J) and colocalize with HP1-bound regions (Supplemental Fig. 6B). HP1/H3K9me2 do not colocalize with H3K27me3 (Supplemental Fig. 6C,D), in agreement with previous reports for HP1 and PRC1 (de Wit et al. 2007). Importantly, like HP1, H3K9me2 is highly enriched at regions of differential replication timing (Fig. 3B), while H3K27me3 mostly occurs in regions that replicate late in control and knockdown cells (Supplemental Fig. 7A). Taken together, reduction of HP1 in Kc cells leads to a delay in replication timing of unique sequences, which are characterized by HP1 binding and H3K9 dimethylation.
Delayed replication timing of chromosome 4 and pericentric regions after HP1 knockdown
To ask how HP1-dependent changes in replication timing relate to repeat density, we measured the abundance of repeats in differentially replicating regions that are present on our tiling array. This reveals that regions on the chromosomal arms with high repeat content show frequently delayed replication timing after HP1 knockdown (Supplemental Fig. 7B). Chromosome 4 is particularly high in its repeat content and shows high abundance of HP1 binding and H3K9 dimethylation (Riddle and Elgin 2006; Riddle et al. 2009). When chromosomes were compared, we observed that delayed replication timing occurs mostly on repeat rich regions such as those on chromosome 2 (Fig. 3B; Supplemental Fig. 8A).
Interestingly, we detected a small, but global, delay of replicating timing of the repeat-rich, early replicating chromosome 4 in HP1-depleted cells (Figs. 3C, 4). This suggests that high levels of HP1 present on chromosome 4 might be important for maintaining its early replication. We also observed a weak reduction in transcription levels throughout this chromosome in HP1-depleted cells (Fig. 3D). Interestingly, however, while many genes showed a small decrease in transcript levels, only a few genes showed a complete loss of gene expression. Furthermore, not all differentially replicating regions also showed a reduction in transcription (Fig. 4). These data indicate that the effect of HP1 on the timing of chromosome 4 replication is independent from its effect on transcription at some loci, while at others, replication and transcription are both affected.
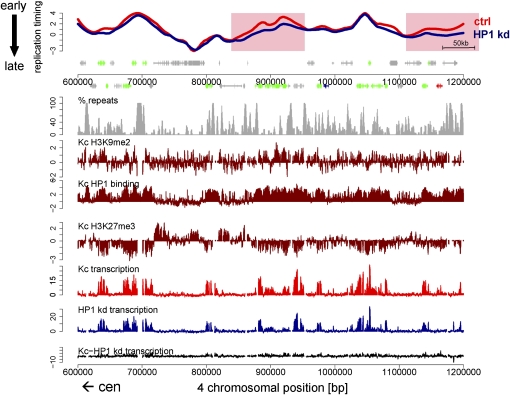
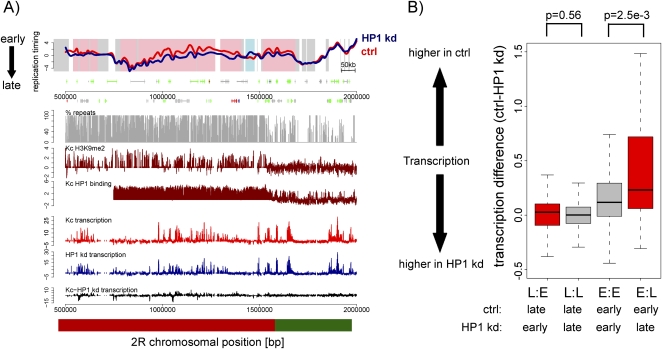
Several regions on chromosome 2 and 3 are differentially replicating after depletion of HP1. Most of these regions lie within centromere proximal regions rich in dispersed repeats (Fig. 3C), which also show high levels of HP1 binding (de Wit et al. 2007). Figure 5A displays a region that spans part of the pericentric heterochromatin and part of euchromatin on chromosome 2R. The region closer to the centromere (Fig. 5A, left) shows high levels of HP1 binding and H3K9 dimethylation and is repeat-dense throughout (Fig. 5A). Interestingly, it contains many regions with subtle replication timing differences, mostly replicating later in HP1 RNAi cells than in control cells (Fig. 5A, left). The overall transcription differences within chromosome 2 heterochromatin tend toward reduced expression after HP1 knockdown (Fig. 5A). However, some of those differentially replicating regions do not contain any differences in transcriptional levels (Fig. 5A). Pericentric regions of other chromosomes are similarly affected in replication timing and transcription, yet to a lesser extent (Fig. 3B,D; Supplemental Fig. 8A).
Figure 5.
Differences in replication timing do frequently but not always coincide with transcription differences. (A) Replication timing profiles of control (red, ctrl) and HP1 knockdown (blue, HP1 kd) Kc cells for a representative region on chromosome 2R. X-axis, chromosomal position in base pairs; y-axis, log2 (early/late replication). Background coloring denotes regions that replicate earlier in HP1 kd cells (L:E, pink), regions that replicate earlier in control cells (E:L, blue), and regions replicating similarly in both cell types (white). Annotated genes are displayed below the profile (boxes, exons; lines, introns; small boxes, UTRs) and colored by their expression status (for details, see Methods; green, expressed in control and HP1 kd cells; blue, expressed only in HP1 kd cells; red, expressed only in control cells; gray, not expressed in control and HP1 kd cells). Transcription levels of control (red) and HP1 kd (blue) cells measured by tiling arrays are displayed on the same scale below, including transcription level differences (black). The direction toward the centromere (cen) is marked by an arrow, and the parts of the figure that we refer to in the main text as pericentric heterochromatin (red bar) and euchromatin (green bar) are indicated. (B) Distribution of transcription differences (control-HP1 kd transcription levels) for regions with differential replication timing. The boxplots illustrate that on average differences in replication timing coincide with changes in transcription. L:E, regions replicating earlier in HP1 kd cells; L:L, regions replicating late in both; E:E, regions replicating early in both; E:L, regions replicating earlier in control cells. P-values were calculated using the Wilcoxon rank sum test.
A genome-wide comparison of average transcription differences within differentially replicating regions to replication timing (Fig. 5B) reveals that regions, which replicate earlier in control cells (E:L) show higher transcription in control cells (Fig. 5B). Notably, in many cases, these transcriptional changes only occur at a low percentage of genes in each differentially replicating region (data not shown). At the same time, only ∼25% of differentially replicating regions show transcription differences, which are significantly higher than in most regions replicating at the same time (Fig. 5B). This suggests that HP1 can affect replication timing and transcription independently at several regions within euchromatin and heterochromatin. Of note, we cannot formally exclude the possibility of differential transcription of repetitive elements, which due to their nonuniqueness are not represented on the tiling array.
In summary, we show that depletion of HP1 from Drosophila tissue culture cells results in distinct changes in the temporal program of replication, most of which lie within heterochromatin and on the fourth chromosome. While many replication timing changes correlate with transcriptional differences, many do not, suggesting that HP1 modulates replication timing via its role in chromatin organization.
Discussion
We demonstrate that HP1 modulates the temporal organization of DNA replication in Drosophila cells in a locus-specific fashion. Reduction of HP1 levels causes an advance of replication timing of late replicating heterochromatic centromeric repeats. Surprisingly, repeat-rich regions that are embedded within or close to euchromatin such as pericentromeric heterochromatin and the fourth chromosome show the opposite behavior. Their replication timing is often delayed after knockdown of HP1. This suggests that repeat silencing is required for the organization of both very early as well as very late replication. Since ORC and HP1 have been shown to interact, the differential timing effect could also be the result of a potential loss of ORC recruitment to origins due to absence of HP1 in pericentric regions, which in turn could result in a delay of replication timing. In the case of advanced replication timing, an increased accessibility of chromatin for the replication machinery in centromeric heterochromatin due to absence of HP1 might be the predominant mechanism.
Replication timing of centromeric repeats is advanced after HP1 knockdown
Several lines of evidence support that HP1 is important for heterochromatic silencing. It binds the repressive H3K9me2 histone modification (Bannister et al. 2001; Lachner et al. 2001), and loss of HP1 results in suppression of PEV, while an increase in HP1 protein can enhance PEV in Drosophila (Locke et al. 1988; Wustmann et al. 1989; Eissenberg et al. 1990). HP1 is furthermore required for proper expression of some heterochromatic genes (Wakimoto and Hearn 1990; Clegg et al. 1998; Lu et al. 2000; Schulze et al. 2005). Interaction of HP1 with CAF-1 is required for replication of heterochromatin in mouse cells, suggesting that compact chromatin mediated by HP1 needs to be alleviated to allow replication fork progression through S phase (Quivy et al. 2008). Depletion of HP1 could result in the loss of the requirement for this mechanism and therefore allow slightly earlier replication of centromeric repetitive heterochromatin. This advanced replication timing of repeats is consistent with observations in mouse fibroblasts, where replication timing of centromeric repeats is advanced in cells depleted of the mouse homologs of the Drosophila histone methyltransferase SU(VAR)3-9 that methylate lysine 9 on histone H3 (Wu et al. 2006). Interestingly, however, embryonic stem cells lacking the SUV39H1 and SUV39H2 H3K9 methyltransferases showed delayed replication timing of centromeric repeats (Jorgensen et al. 2007), underscoring the context dependency of the interaction between chromatin and the replication timing program. Studies of endoreplication of Drosophila polytene chromosomes have suggested a role for HP1 in the maintenance of underreplicated regions in connection with the SuUR protein (Makunin et al. 2002; Koryakov et al. 2006; Pindyurin et al. 2007, 2008). It has been suggested that open chromatin plays a role in replication initiation (Aggarwal and Calvi 2004; Goren et al. 2008; Schwaiger et al. 2009; Hansen et al. 2010; Macalpine et al. 2010). Therefore, it is possible that a loss of HP1 results in chromatin decompaction, which might expose origins of replication to the replication machinery, which are normally suppressed.
Replication timing of pericentromeric heterochromatin and the fourth chromosome is affected by HP1 knockdown
Interestingly, our results show that cells with reduced HP1 protein levels display differences in replication timing also at genes embedded in dispersed repeats. These differences mostly occur in HP1-bound heterochromatin and on the fourth chromosome, suggesting that replication timing changes are directly dependent on HP1. In contrast to centromeric repeats, however, these unique sequences show a delay in replication timing after HP1 knockdown. HP1 is highly enriched on chromosome 4, where we observe a global replication delay. Chromosome 4 is entirely heterochromatic; however, in the distal 1.2 Mbp, the gene density is typical of euchromatin (Riddle and Elgin 2006; Riddle et al. 2009). In Kc cells, most genes within this distal region are active, and it replicates in early S phase (Fig. 4). Thus we conclude that HP1 is required for early replication of genes embedded in repetitive DNA.
This questions a simple model where chromatin opening in the absence of HP1 advances replication per se. Furthermore, it argues that the influence of heterochromatin on DNA replication is very different for regions that replicate early in S phase versus those that replicate late in S phase. There are several potential models to explain this result. It is conceivable that some of the observed delays in replication timing after HP1 knockdown are a result of down-regulated transcription and concomitant changes in histone modifications. Some regions of delayed replication timing indeed show slightly down-regulated transcription. Importantly, however, many do not show any changes in gene expression levels (Fig. 5), making this explanation unlikely.
Alternatively, up-regulation of repeats and the resulting transcriptional activity could interfere with normal origin activity and fork movement. Such an interference model has been postulated as a potential explanation of transcriptional misregulation of unique genes upon activation of neighboring repeats such as in the case of heterochromatic genes in Drosophila (Yasuhara and Wakimoto 2006). It seems plausible that transcription could locally interfere with proper origin activation and delay replication timing as a consequence. Indeed, transcription can have a negative influence on origin selection in mammalian cells (Mesner and Hamlin 2005; Gregoire et al. 2006; Sasaki et al. 2006). It has been suggested that eukaryotic replication origins preferentially locate in intergenic, nontranscribed regions, yet in proximity to actively transcribed genes (MacAlpine and Bell 2005; Macalpine et al. 2010). This would be consistent with a model where aberrant transcription of intergenic, repetitive regions interferes with origin selection as well as gene expression. While the zones of replication initiation seem to be similar in control and HP1 knockdown cells (Fig. 5A; Supplemental Fig. 7C,D), only a detailed characterization of the location of origins of replication after HP1 knockdown would allow one to distinguish between a delay in origin firing and the use of different origins. An additional explanation for the observed replication timing changes could be that the HP1 knockdown affects the speed of the replication fork or leads to its stalling.
Repeat up-regulation might account for the reduction of transcription of heterochromatic genes after loss of HP1 or translocation to euchromatin (Wakimoto and Hearn 1990; Clegg et al. 1998; Lu et al. 2000; Schulze et al. 2005) due to transcriptional interference. It seems feasible that this also affects origin activity or replication fork movement negatively even though the overall level of transcription within a region might have increased in the absence of HP1.
An alternative scenario for altered replication in the absence of HP1 originated from a described interaction of HP1 with ORC2 (Pak et al. 1997). Recently, it was shown that Swi6, the S .pombe HP1 homolog, activates origins of replication within early replicating heterochromatin by interacting with an essential kinase (Hayashi et al. 2009). It is tempting to speculate that Drosophila HP1 activates replication origins within certain repeat-rich regions in the same way. A reduction of HP1 levels in the cell would then result in delayed or less efficient origin firing. This might then lead to the observed replication timing delay in HP1 RNAi cells. Interestingly, mutations in both Swi6 and Clr4, the S. pombe H3K9 histone methyltransferase, result in early replication of heterochromatin (Hayashi et al. 2009). This suggests that H3K9 methylation-mediated chromatin compaction suppresses origin firing, while HP1 directly activates it. Our observations in Drosophila are consistent with the results in S. pombe, since the fourth chromosome, which has high HP1, but low H3K9me2 levels, replicates generally earlier than the pericentric heterochromatin of chromosome 2, which shows high HP1 binding and H3K9 methylation (Supplemental Figs. 4, 6A). Most importantly, the three types of heterochromatin in S. pombe show three completely different types of replication timing regulation. We show that also in Drosophila, the replication of different types of heterochromatin responds differently to a reduction of HP1 protein levels.
In summary, we report a dual role of HP1 in controlling replication timing of repetitive and unique heterochromatin sequences in Drosophila cells. Centromeric repeats replicate earlier after HP1 knockdown, while many regions of pericentric heterochromatin and the fourth chromosome display a replication delay independent of gene expression differences. While the exact mechanism of this surprising function of HP1 for the replication timing program remains to be determined, it provides a further link between the control of chromatin structure and the temporal control of replication.
Methods
RNAi in Kc cells
dsRNA was prepared from a PCR product spanning the entire HP1 coding region, generated with primers containing a T7 RNA polymerase binding site using the MEGASCRIPT T7 in vitro transcription kit (Ambion).
The following primers were used: forward primer, 5′-TTAATACGACTCACTATAGGGAGAatgggcaagaaaatcgacaac-3′; reverse primer, 5′-TTAATACGACTCACTATAGGGAGAatcttcattatcagagtaccag-3′. Capital letter sequences represent the T7 binding site. We tested the specificity of our dsRNA using dsCheck (Naito et al. 2005). This highly sensitive program uses 19-bp sequences for homology search. We found more than 600 siRNAs targeting SuVAR205 and only one siRNA each for three off target genes. However, none of these genes was down-regulated after HP1 knockdown according to Affymetrix expression array data (GEO accession no. GSE18092). Other HP1 variants did not receive any predicted siRNAs and were also not affected by HP1 knockdown (Supplemental Fig. 1D).
The in vitro transcribed RNA was purified and heated for 10 min to 70°C and slowly cooled down to room temperature for about 30 min to enhance annealing. Fifty micrograms of dsRNA was added to 106 cells every second day and 8 d after initial addition of dsRNA cells were harvested, and the efficiency of HP1 reduction was estimated by Western blot analysis using a monoclonal α-HP1 antibody (HP1–CIA9), which was kindly provided by Fang-Lin Sun (Tsinghua University, Beijing, China) and has been previously described (James and Elgin 1986).
Immunofluorescence analysis
Immunofluorescence staining was carried out as described (Wirbelauer et al. 2005), using a polyclonal rabbit α-HP1 (HP1a) antibody, which was provided by Sarah Elgin (Pal-Bhadra et al. 2004). For replication timing analysis by immunofluorescence, cells were labeled with BrdU for 1 h, and BrdU was detected using the 5′-bromo-2′-deoxy-uridine labeling and detection kit I (Roche). DNA was counterstained with 0.04 μg/mL DAPI. Stainings were analyzed using a laser scanning confocal microscope LSM510 META (Zeiss) and LSM510 software.
Cell cycle analysis
Control and HP1 knockdown cells were incubated with BrdU for 1 h and subjected to BrdU and DNA staining using the BrdU flow kit (BD PharMingen). Fluorescence was measured using a FACSCalibur (Becton Dickinson). Data collection and analysis were performed using CellQuest software.
Transcription analysis
Total RNA was isolated from cells using TRIzol (Invitrogen) and subsequently purified using an RNeasy kit (QIAGEN). For hybridization to Affymetrix tiling arrays, we made double-stranded cDNA by performing two rounds of cDNA synthesis using random primers and addition of 2 mM dUTPs using the GeneChip WT Double-Stranded cDNA Synthesis Kit (Affymetrix). This cDNA was fragmented and end-labeled using the GeneChip WT double-stranded DNA terminal labeling kit (Affymetrix) and hybridized to GeneChip Drosophila Tiling 1.0R arrays (Affymetrix) according to the manufacturer's instructions. For hybridization to expression arrays, cDNA synthesis and hybridizations were carried out according to standard Affymetrix procedures.
Chromatin immunoprecipitation
ChIP was carried out as described (Bell et al. 2007), using antibodies against H3K9me2 (LPBio) and H3K27me3 (Upstate).
Replication timing analysis
Replication timing was measured by sorting BrdU-labeled cells into different S-phase fractions followed by Brdu-IPs, as described by Schwaiger et al. (2009). Detailed descriptions of replication timing, transcription, and ChIP data analysis can be found in the Supplemental Methods.
Acknowledgments
We thank Ichiro Hiratani, David M. Gilbert, Oliver Bell, and members of our laboratory for helpful comments on the manuscript; Herbert Angliker for Affymetrix microarray processing; and Sarah C. Elgin and Fang-Lin Sun for providing HP1 antibodies. M.S. acknowledges support by an EMBO long-term fellowship and by a predoctoral fellowship of the Boehringer Ingelheim Fonds. Research in the laboratory of D.S. is supported by the Novartis Research Foundation, by the European Union (NoE “The Epigenome” LSHG-CT-2004-503433, LSHG-CT-2006-037415), the European Research Council (EpiGePlas 204264), and the EMBO Young Investigator program.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequence data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://ncbi/nlm.nih.gov/geo) under accession no. GSE18092.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.101790.109.
References
- Abad JP, Carmena M, Baars S, Saunders RD, Glover DM, Ludena P, Sentis C, Tyler-Smith C, Villasante A 1992. Dodeca satellite: A conserved G+C-rich satellite from the centromeric heterochromatin of Drosophila melanogaster. Proc Natl Acad Sci 89: 4663–4667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal BD, Calvi BR 2004. Chromatin regulates origin activity in Drosophila follicle cells. Nature 430: 372–376 [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124 [DOI] [PubMed] [Google Scholar]
- Bell SP, Dutta A 2002. DNA replication in eukaryotic cells. Annu Rev Biochem 71: 333–374 [DOI] [PubMed] [Google Scholar]
- Bell O, Wirbelauer C, Hild M, Scharf AN, Schwaiger M, MacAlpine DM, Zilbermann F, van Leeuwen F, Bell SP, Imhof A, et al. 2007. Localized H3K36 methylation states define histone H4K16 acetylation during transcriptional elongation in Drosophila. EMBO J 26: 4974–4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaeva ES, Zhimulev IF, Volkova EI, Alekseyenko AA, Moshkin YM, Koryakov DE 1998. Su(UR)ES: A gene suppressing DNA underreplication in intercalary and pericentric heterochromatin of Drosophila melanogaster polytene chromosomes. Proc Natl Acad Sci 95: 7532–7537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caizzi R, Caggese C, Pimpinelli S 1993. Bari-1, a new transposon-like family in Drosophila melanogaster with a unique heterochromatic organization. Genetics 133: 335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvi BR, Byrnes BA, Kolpakas AJ 2007. Conservation of epigenetic regulation, ORC binding and developmental timing of DNA replication origins in the genus Drosophila. Genetics 177: 1291–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg NJ, Honda BM, Whitehead IP, Grigliatti TA, Wakimoto B, Brock HW, Lloyd VK, Sinclair DA 1998. Suppressors of position-effect variegation in Drosophila melanogaster affect expression of the heterochromatic gene light in the absence of a chromosome rearrangement. Genome 41: 495–503 [PubMed] [Google Scholar]
- Devlin RH, Bingham B, Wakimoto BT 1990. The organization and expression of the light gene, a heterochromatic gene of Drosophila melanogaster. Genetics 125: 129–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Greil F, van Steensel B 2005. Genome-wide HP1 binding in Drosophila: Developmental plasticity and genomic targeting signals. Genome Res 15: 1265–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Greil F, van Steensel B 2007. High-resolution mapping reveals links of HP1 with active and inactive chromatin components. PLoS Genet 3: e38 doi: 10.1371/journal.pgen.0030038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC, James TC, Foster-Hartnett DM, Hartnett T, Ngan V, Elgin SC 1990. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc Natl Acad Sci 87: 9923–9927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM 2002. Replication timing and transcriptional control: Beyond cause and effect. Curr Opin Cell Biol 14: 377–383 [DOI] [PubMed] [Google Scholar]
- Goren A, Tabib A, Hecht M, Cedar H 2008. DNA replication timing of the human β-globin domain is controlled by histone modification at the origin. Genes & Dev 22: 1319–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoire D, Brodolin K, Mechali M 2006. HoxB domain induction silences DNA replication origins in the locus and specifies a single origin at its boundary. EMBO Rep 7: 812–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greil F, van der Kraan I, Delrow J, Smothers JF, de Wit E, Bussemaker HJ, van Driel R, Henikoff S, van Steensel B 2003. Distinct HP1 and Su(var)3-9 complexes bind to sets of developmentally coexpressed genes depending on chromosomal location. Genes & Dev 17: 2825–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen RS, Thomas S, Sandstrom R, Canfield TK, Thurman RE, Weaver M, Dorschner MO, Gartler SM, Stamatoyannopoulos JA 2010. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc Natl Acad Sci 107: 139–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi MT, Takahashi TS, Nakagawa T, Nakayama J, Masukata H 2009. The heterochromatin protein Swi6/HP1 activates replication origins at the pericentromeric region and silent mating-type locus. Nat Cell Biol 11: 357–362 [DOI] [PubMed] [Google Scholar]
- Heitz E 1934. Über α und β heterochromatin sowie konstanz und bau der chromomeren bei Drosophila. Biol Zentralbl 45: 588–609 [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang CW, Lyou Y, Townes TM, Schubeler D, Gilbert DM 2008. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol 6: e245 doi: 10.1371/journal.pbio.0060245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Rathjen J, Kulik M, Papp B, Fussner E, Bazett-Jones DP, Plath K, Dalton S, et al. 2010. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res 20: 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James TC, Elgin SC 1986. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol Cell Biol 6: 3862–3872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen HF, Azuara V, Amoils S, Spivakov M, Terry A, Nesterova T, Cobb BS, Ramsahoye B, Merkenschlager M, Fisher AG 2007. The impact of chromatin modifiers on the timing of locus replication in mouse embryonic stem cells. Genome Biol 8: R169 doi: 10.1186/gb-2007-8-8-r169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SM, Dubey DD, Huberman JA 2003. Early-replicating heterochromatin. Genes & Dev 17: 330–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koryakov DE, Reuter G, Dimitri P, Zhimulev IF 2006. The SuUR gene influences the distribution of heterochromatic proteins HP1 and SU(VAR)3-9 on nurse cell polytene chromosomes of Drosophila melanogaster. Chromosoma 115: 296–310 [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116–120 [DOI] [PubMed] [Google Scholar]
- Locke J, Kotarski MA, Tartof KD 1988. Dosage-dependent modifiers of position effect variegation in Drosophila and a mass action model that explains their effect. Genetics 120: 181–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu BY, Emtage PC, Duyf BJ, Hilliker AJ, Eissenberg JC 2000. Heterochromatin protein 1 is required for the normal expression of two heterochromatin genes in Drosophila. Genetics 155: 699–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas I, Palakodeti A, Jiang Y, Young DJ, Jiang N, Fernald AA, Le Beau MM 2007. High-throughput mapping of origins of replication in human cells. EMBO Rep 8: 770–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine DM, Bell SP 2005. A genomic view of eukaryotic DNA replication. Chromosome Res 13: 309–326 [DOI] [PubMed] [Google Scholar]
- MacAlpine DM, Rodriguez HK, Bell SP 2004. Coordination of replication and transcription along a Drosophila chromosome. Genes & Dev 18: 3094–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macalpine HK, Gordan R, Powell SK, Hartemink AJ, Macalpine DM 2010. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res 20: 201–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makunin IV, Volkova EI, Belyaeva ES, Nabirochkina EN, Pirrotta V, Zhimulev IF 2002. The Drosophila suppressor of underreplication protein binds to late-replicating regions of polytene chromosomes. Genetics 160: 1023–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesner LD, Hamlin JL 2005. Specific signals at the 3′ end of the DHFR gene define one boundary of the downstream origin of replication. Genes & Dev 19: 1053–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ, Altenburg E 1930. The frequency of translocations produced by X-rays in Drosophila. Genetics 15: 283–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito Y, Yamada T, Matsumiya T, Ui-Tei K, Saigo K, Morishita S 2005. dsCheck: Highly sensitive off-target search software for double-stranded RNA-mediated RNA interference. Nucleic Acids Res 33: W589–W591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieduszynski CA, Knox Y, Donaldson AD 2006. Genome-wide identification of replication origins in yeast by comparative genomics. Genes & Dev 20: 1874–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak DT, Pflumm M, Chesnokov I, Huang DW, Kellum R, Marr J, Romanowski P, Botchan MR 1997. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell 91: 311–323 [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA, Elgin SC 2004. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303: 669–672 [DOI] [PubMed] [Google Scholar]
- Pindyurin AV, Moorman C, de Wit E, Belyakin SN, Belyaeva ES, Christophides GK, Kafatos FC, van Steensel B, Zhimulev IF 2007. SUUR joins separate subsets of PcG, HP1 and B-type lamin targets in Drosophila. J Cell Sci 120: 2344–2351 [DOI] [PubMed] [Google Scholar]
- Pindyurin AV, Boldyreva LV, Shloma VV, Kolesnikova TD, Pokholkova GV, Andreyeva EN, Kozhevnikova EN, Ivanoschuk IG, Zarutskaya EA, Demakov SA, et al. 2008. Interaction between the Drosophila heterochromatin proteins SUUR and HP1. J Cell Sci 121: 1693–1703 [DOI] [PubMed] [Google Scholar]
- Pope BD, Hiratani I, Gilbert DM 2009. Domain-wide regulation of DNA replication timing during mammalian development. Chromosome Res 18: 127–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivy JP, Gerard A, Cook AJ, Roche D, Almouzni G 2008. The HP1-p150/CAF-1 interaction is required for pericentric heterochromatin replication and S-phase progression in mouse cells. Nat Struct Mol Biol 15: 972–979 [DOI] [PubMed] [Google Scholar]
- Riddle NC, Elgin SC 2006. The dot chromosome of Drosophila: Insights into chromatin states and their change over evolutionary time. Chromosome Res 14: 405–416 [DOI] [PubMed] [Google Scholar]
- Riddle NC, Shaffer CD, Elgin SC 2009. A lot about a little dot: Lessons learned from Drosophila melanogaster chromosome 4. Biochem Cell Biol 87: 229–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Ramanathan S, Okuno Y, Kumagai C, Shaikh SS, Gilbert DM 2006. The Chinese hamster dihydrofolate reductase replication origin decision point follows activation of transcription and suppresses initiation of replication within transcription units. Mol Cell Biol 26: 1051–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, Rea S, Jenuwein T, Dorn R, Reuter G 2002. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J 21: 1121–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze SR, Sinclair DA, Fitzpatrick KA, Honda BM 2005. A genetic and molecular characterization of two proximal heterochromatic genes on chromosome 3 of Drosophila melanogaster. Genetics 169: 2165–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaiger M, Stadler MB, Bell O, Kohler H, Oakeley EJ, Schubeler D 2009. Chromatin state marks cell-type- and gender-specific replication of the Drosophila genome. Genes & Dev 23: 589–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seum C, Reo E, Peng H, Rauscher FJ 3rd, Spierer P, Bontron S 2007. Drosophila SETDB1 is required for chromosome 4 silencing. PLoS Genet 3: e76 doi: 10.1371/journal.pgen.0030076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng TY, Lee CH, Chan LW, Shen CK 2007. Epigenetic regulation of the Drosophila chromosome 4 by the histone H3K9 methyltransferase dSETDB1. Proc Natl Acad Sci 104: 12691–12696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakimoto BT, Hearn MG 1990. The effects of chromosome rearrangements on the expression of heterochromatic genes in chromosome 2L of Drosophila melanogaster. Genetics 125: 141–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrath LL, Elgin SC 1995. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes & Dev 9: 1263–1277 [DOI] [PubMed] [Google Scholar]
- Wirbelauer C, Bell O, Schubeler D 2005. Variant histone H3.3 is deposited at sites of nucleosomal displacement throughout transcribed genes while active histone modifications show a promoter-proximal bias. Genes & Dev 19: 1761–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Singh PB, Gilbert DM 2006. Uncoupling global and fine-tuning replication timing determinants for mouse pericentric heterochromatin. J Cell Biol 174: 185–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wustmann G, Szidonya J, Taubert H, Reuter G 1989. The genetics of position-effect variegation modifying loci in Drosophila melanogaster. Mol Gen Genet 217: 520–527 [DOI] [PubMed] [Google Scholar]
- Yasuhara JC, Wakimoto BT 2006. Oxymoron no more: The expanding world of heterochromatic genes. Trends Genet 22: 330–338 [DOI] [PubMed] [Google Scholar]
- Yasuhara JC, Wakimoto BT 2008. Molecular landscape of modified histones in Drosophila heterochromatic genes and euchromatin-heterochromatin transition zones. PLoS Genet 4: e16 doi: 10.1371/journal.pgen.0040016 [DOI] [PMC free article] [PubMed] [Google Scholar]