Abstract
The regulation of gene expression is critical for organismal function and is an important source of phenotypic diversity between species. Understanding the genetic and molecular mechanisms responsible for regulatory divergence is therefore expected to provide insight into evolutionary change. Using deep sequencing, we quantified total and allele-specific mRNA expression levels genome-wide in two closely related Drosophila species (D. melanogaster and D. sechellia) and their F1 hybrids. We show that 78% of expressed genes have divergent expression between species, and that cis- and trans-regulatory divergence affects 51% and 66% of expressed genes, respectively, with 35% of genes showing evidence of both. This is a relatively larger contribution of trans-regulatory divergence than was expected based on prior studies, and may result from the unique demographic history of D. sechellia. Genes with antagonistic cis- and trans-regulatory changes were more likely to be misexpressed in hybrids, consistent with the idea that such regulatory changes contribute to hybrid incompatibilities. In addition, cis-regulatory differences contributed more to divergent expression of genes that showed additive rather than nonadditive inheritance. A correlation between sequence similarity and the conservation of cis-regulatory activity was also observed that appears to be a general feature of regulatory evolution. Finally, we examined regulatory divergence that may have contributed to the evolution of a specific trait—divergent feeding behavior in D. sechellia. Overall, this study illustrates the power of mRNA sequencing for investigating regulatory evolution, provides novel insight into the evolution of gene expression in Drosophila, and reveals general trends that are likely to extend to other species.
Phenotypic differences between species can arise from genetic changes affecting the function of gene products as well as their expression. Although there has been extensive debate over the relative importance of these two different types of changes (Carroll 2005; Hoekstra and Coyne 2007; Wray 2007; Stern and Orgogozo 2008), they both clearly contribute to phenotypic evolution. Functional divergence of a gene product has historically been much easier to detect than expression divergence; however, advances in methods for measuring gene expression during the last decade have made differences in gene expression much easier to identify. For example, microarray-based studies of gene expression in Drosophila have found that 20% of genes show expression differences between individuals of the same species (Genissel et al. 2008) and 34%–48% of genes show expression divergence between species (Ranz et al. 2003, 2004; Rifkin et al. 2003; Nuzhdin et al. 2004). Similarly, in vertebrates, expression differences have been detected for 33% of genes between individuals of the same species (Schadt et al. 2003) and 35% of genes for individuals from different species (Hsieh et al. 2003; for more complete reviews of expression differences within and between species, see Gibson and Weir 2005; Ranz and Machado 2006; Rockman and Kruglyak 2006; Whitehead and Crawford 2006).
Identifying the genetic changes underlying expression divergence, however, remains challenging. This is because gene expression is controlled by biochemical interactions between cis-regulatory DNA sequences and trans-acting RNAs and proteins that form a complex network. Every component of this network is a potential target for regulatory divergence. Indeed, most expression differences between species appear to be polygenic (Gibson and Weir 2005), indicating that multiple genetic changes have occurred that affect expression of even a single gene. Differences in the architecture of gene-specific regulatory networks (as well as differences among genes for other genomic features) complicate this matter further because they make some genes more susceptible to particular types of regulatory changes than others (Wittkopp 2005; Landry et al. 2007).
Distinguishing between cis- and trans-acting sources of expression differences is important because these molecular mechanisms can influence the way in which gene expression levels are inherited and evolve (e.g., Ronald and Akey 2007). For example, like most traits, gene expression levels can be inherited additively or nonadditively (Gibson et al. 2004); cis-regulatory changes appear to have additive effects on gene expression more often than trans-regulatory changes (Lemos et al. 2008). Such additive alleles affect the phenotype in heterozygous individuals and are thus readily “visible” to selection. Another example comes from a study of mutation accumulation lines in Caenorhabditis elegans (Denver et al. 2005), which found that selection in natural populations eliminated most trans-acting mutations that affected expression of many genes. Such a pattern of selection could cause cis-regulatory mutations to accumulate preferentially over time, and cis-regulatory divergence does indeed appear to increase with divergence time (Lemos et al. 2008; Wittkopp et al. 2008).
The net effects of cis- and trans-regulatory changes on total expression divergence can be estimated by comparing the magnitude of the expression difference between two genotypes of interest to the relative allelic expression in F1 hybrids produced by crossing these two genotypes. This is because allele-specific measures of gene expression in heterozygotes reflect the relative activity of two cis-regulatory alleles in the same trans-regulatory cellular environment (Cowles et al. 2002). The fraction of the total expression difference between the two genotypes of interest that is not explained by cis-regulatory divergence is attributed to trans-regulatory divergence (Wittkopp et al. 2004). Until recently, applications of this method for studying regulatory evolution have been limited by the paucity of techniques that could distinguish between mRNA molecules derived from different alleles of the same gene in a high-throughput manner. Advances in sequencing technology, however, are beginning to change this (e.g., Mortazavi et al. 2008; Serre et al. 2008; Wang et al. 2008; Main et al. 2009; Zhang et al. 2009). Furthermore, sequencing-based quantification of gene expression simultaneously can measure total transcript abundance and relative allele-specific mRNA abundance for each gene, can analyze an entire genome in a single experiment, and does not necessarily require genomic sequence information a priori (although the availability of reference genomes simplifies the analysis).
Here, we use next-generation sequencing to examine regulatory divergence between Drosophila melanogaster and Drosophila sechellia, which diverged ∼1.2 million years ago (Cutter 2008). After extensively validating measures of gene expression derived from our sequencing data, we examined the extent of regulatory divergence, the relative contribution of cis- and trans-regulatory changes, the inheritance of gene expression levels, and the relationships between these properties and other genomic features, such as sequence divergence. We also specifically examined regulatory evolution of genes thought to contribute to a derived phenotype in D. sechellia (Supplemental material). Taken together, these data provide a comprehensive study of regulatory divergence between species.
Results and Discussion
Using deep sequencing to measure mRNA abundance
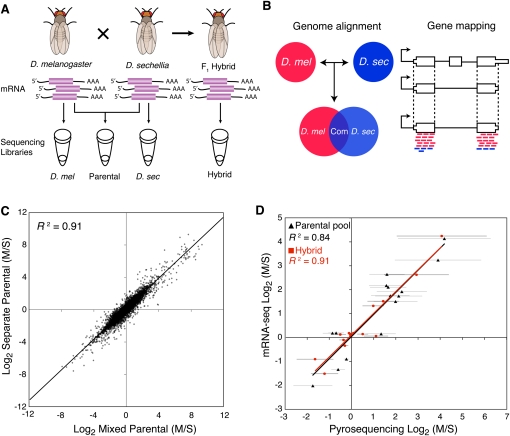
Gene expression was quantified by paired-end sequencing (mRNA-seq) of cDNA libraries prepared using RNA extracted from 2-d-old D. melanogaster, D. sechellia, and F1 hybrid females produced by crossing D. melanogaster females to D. sechellia males. (Female flies were used for this work because they allow analysis of relative allele-specific expression for genes on the X chromosome, as well as those on the autosomes.) The transcriptomes of D. melanogaster and D. sechellia were sequenced individually and as a “mixed parental” cDNA pool produced using equal amounts of RNA from each species (Fig. 1A). We obtained 26, 31, and 42 million sequence reads (13, 15.5, and 21 million mate pairs) from the D. melanogaster, D. sechellia, and mixed parental libraries, respectively, and 78 million sequence reads (39 million mate pairs) from the hybrid library (Table 1).
Figure 1.
Overview of allele-specific expression profiling by mRNA-seq. (A) mRNA prepared from D. melanogaster, D. sechellia, and their F1 hybrids were used to create four different sequencing libraries: F1 hybrid (Hybrid), mixed parental (Parental), and two species-specific libraries (D. mel and D. sec). (B) Allele-specific sequences were chosen by comparative genomic alignments and mapped to constitutive regions of genes. (C) Comparison of relative gene expression levels (M/S) measured from mixed and separate parental samples with at least 20 mapped reads per gene. (D) Comparison of allele-specific expression ratios determined by mRNA-seq and pyrosequencing. Error bars indicate 95% confidence intervals for pyrosequencing data.
Table 1.
Overview of transcriptome analysis via Illumina sequencing
aHybrid and parental results are total number of sequences obtained, regardless of allele specificity.
bExcluding mitochondrial genes.
NA, Not available.
Sequence reads from the individual D. melanogaster and D. sechellia samples were aligned to the respective D. melanogaster (Adams et al. 2000; Celniker et al. 2002) and D. sechellia (Drosophila 12 Genomes Consortium 2007) reference genomes to identify reads that mapped uniquely and without mismatches. Sequence reads from the mixed parental and hybrid samples were independently aligned to both the D. melanogaster and D. sechellia genomes (Fig. 1B), and reads that mapped to only one genome were assigned to that species (Table 1). In some cases, reads that matched both species (“common” in Table 1) could be assigned to one species or the other based on the sequence of the mate-pair. Reads that could not be unambiguously mapped to either D. melanogaster or D. sechellia were excluded from further analysis. Each read was then assigned to a specific gene based on annotations of the D. melanogaster genome (FlyBase 5.11) and their corresponding sequences in the D. sechellia genome (see Methods). Over 90% of all aligned reads mapped to annotated exons, and 93%–94% of these mapped to constitutive exons that appear in all known isoforms of a gene (Table 1). To eliminate complications from alternative isoforms, only the reads mapping to constitutive regions of exons were used for our analyses. In all, 81% and 80% of genes in D. melanogaster and D. sechellia, respectively, were represented by at least one sequence read in the mixed parental sample.
To examine the accuracy of computational methods used to assign reads to D. melanogaster or D. sechellia, as well as to evaluate the reproducibility of expression levels measured in replicate mRNA-seq experiments, we compared the gene expression levels observed in individual D. melanogaster and D. sechellia libraries to the species-specific expression inferred from the mixed parental pool. Measurements of relative gene expression (M/S) between D. melanogaster (M) and D. sechellia (S) were similar in the mixed parental and single species libraries when comparing all orthologous genes (R2 > 0.81). Excluding the 7% of genes with fewer than 20 mapped reads in both species combined (i.e., M + S < 20) produced an even greater correlation (Fig. 1C; R2 > 0.91). Only genes that exceeded this minimum read number threshold (i.e., M + S ≥ 20) were used for subsequent analyses.
The accuracy of our computational allele assignments was also investigated by examining sequences derived from mitochondrial genes in the F1 hybrid sample. Because D. melanogaster females were used to produce the F1 hybrids, all mitochondrial mRNAs should originate from this species. Indeed, 139,295 of the 140,467 (99.2%) sequence reads that mapped to mitochondrial genes were assigned to D. melanogaster, indicating a combined error rate of 0.8% for sequencing and species-specific allele assignment.
Finally, to determine the consistency of gene expression levels measured using different techniques, we compared estimates of relative allelic expression derived from our mRNA-seq data to estimates of relative allelic expression determined using pyrosequencing (Ahmadian et al. 2000; Wittkopp et al. 2004). Fifteen genes, with sequence coverage ranging from 212 to 6860 mapped reads per gene (Supplemental Table S1), were analyzed using pyrosequencing in the hybrid and mixed parental cDNA samples. In both cases, we found that pyrosequencing measurements of relative gene expression were strongly correlated with those derived from mRNA-seq data (mixed parental: R2 = 0.84; F1 hybrids: R2 = 0.91; Fig. 1D).
Wide-spread expression divergence between Drosophila species
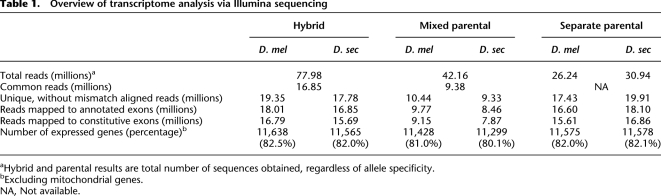
To characterize expression divergence between D. melanogaster and D. sechellia, we compared total levels of gene expression for the 9966 genes that had at least 20 total mapped reads in the mixed parental pool (i.e., M + S ≥ 20). Binomial exact tests with an experiment-wide false discovery rate (FDR) of 0.5%, which corresponded to a significance threshold of P = 0.024 (Storey and Tibshirani 2003), were used to identify genes that were differentially expressed between species. 7739 (78%) of the genes studied were detected as significantly differentially expressed between species (39 false-positives expected), including genes with previously identified mutant alleles in one or both of the parental strains (Supplemental Fig. S1).
Over half (64%) of the 7739 differentially expressed orthologous genes had lower expression in D. sechellia (Fig. 2; M/S > 1). This is a significant deviation from a neutral model assuming that increases and decreases in gene expression are equally likely to have occurred in both species since they diverged from their last common ancestor (binomial exact tests, P < 2 × 10−16). Interestingly, a similar pattern was also reported for Drosophila simulans and D. sechellia based on a microarray analysis of mRNA from adult bodies: 90% of the 130 genes shown to have significant expression differences between species had lower expression in D. sechellia than D. simulans (Dworkin and Jones 2009). Of these 130 genes, 107 were also detected as differentially expressed in our study, 80% of which showed an expression difference between D. sechellia and D. melanogaster in the same direction as that observed between D. sechellia and D. simulans (Supplemental Fig. S2; Kendall's tau (τ) = 0.44, P = 2.6 × 10−11). D. simulans and D. sechellia last shared a common ancestor ∼0.4 million years ago (Kliman et al. 2000; Cutter 2008). Given the phylogenetic relationships among these three species, the divergence of D. sechellia expression relative to D. melanogaster and D. simulans suggests that many of the mutations contributing to these interspecific expression differences are unique to the D. sechellia lineage.
Figure 2.
Gene expression differences between D. melanogaster and D. sechellia. The distribution of log2-transformed expression ratios between species is shown. Negative values indicate higher expression in D. sechellia (D. sec > D. mel), whereas positive values indicate higher expression in D. melanogaster (D. mel > D. sec).
Cis- and trans-regulatory differences underlying expression divergence between D. melanogaster and D. sechellia
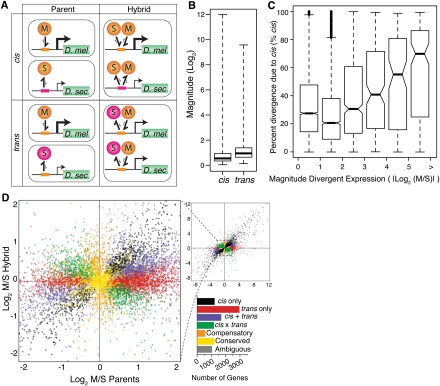
As described in the introduction, expression differences between species can arise from changes in either cis- or trans-regulation. cis-regulatory sequences have allele-specific effects on gene expression, whereas trans-regulatory factors impact expression of both alleles in a diploid cell. Comparing relative allelic expression in hybrids to relative expression between the parental strains allows these two types of changes to be distinguished (Fig. 3A; Wittkopp et al. 2004). In hybrids, the maternal and paternal cis-regulatory alleles are exposed to the same set of trans-acting factors; thus, the relative allelic expression in hybrids provides a direct readout of relative cis-regulatory activity (Cowles et al. 2002). (Note that this assumes that expression of each allele is independent of the other allele; that is, there is no transvection.) We calculated cis-regulatory divergence as the log2-transformed ratio of reads mapping to D. melanogaster and D. sechellia in the hybrid sample: log2(MF1/SF1). Expression differences observed between species that were not attributable to cis-regulatory divergence were assumed to be caused by trans-regulatory divergence (Wittkopp et al. 2004), and trans-regulatory divergence was calculated as the difference between log2-transformed ratios of species-specific reads in the mixed parental and hybrid samples: log2(MMP/SMP) − log2(MF1/SF1). Binomial exact tests and Fisher's exact tests, with an FDR of 0.5%, were used to identify genes with significant cis- and/or trans-regulatory divergence (Supplemental Table S2). Using alternative FDR cutoffs of 1% and 5% had minimal effect on the patterns observed (Supplemental Table S3).
Figure 3.
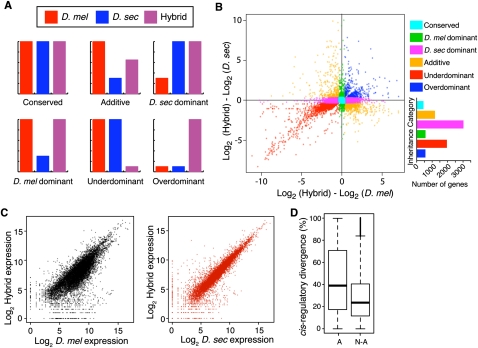
Dissecting cis and trans regulatory differences. (A) Two hypothetical regulatory divergence scenarios are shown. In the top section, a cis-regulatory change in D. sechellia (pink boxes) causes a reduction in affinity for both the conserved D. melanogaster and D. sechellia transcription factors (orange circles). In the bottom section, a trans-regulatory change in a D. sechellia transcription factor (pink circles) reduces its binding affinity for the conserved D. melanogaster and D. sechellia cis-regulatory sequences (orange boxes). The relative expression level is indicated by the promoter arrow thickness. The size of the one-sided arrows show the relative “on” and “off” rates for the transcription factor binding to the cis-regulatory DNA sequence. Note that the cis-regulatory change affects expression of only the D. sechellia allele in F1 hybrids, whereas the trans-regulatory change affects expression of both the D. melanogaster and D. sechellia alleles. (B) Box plots summarize the fold expression differences resulting from cis and trans regulatory divergence. (C) The distribution of percent cis is shown for six groups of genes that differ in the magnitude of total expression divergence between species. Genes were binned based on the absolute value of log2-transformed expression divergence (|log2(M/S)|), and 4086, 2590, 625, 171, 104, and 163 genes were contained within bins 0–1, 1–2, 2–3, 3–4, 4–5, and >5, respectively. Nonoverlapping notches depict statistically significant differences in the median percent cis between pairs of bins. (D) Plot summarizes the relative, allele-specific expression levels in parental and hybrid data sets. Each point represents a single gene and is color-coded according to the mechanism of regulatory evolution inferred from a hierarchical series of statistical tests (FDR = 0.5%; Supplemental Fig. S4). The bar graph depicts the number of genes in each category.
All 9966 genes with at least 20 mapped reads in the mixed parental pool (M + S ≥ 20) were tested for evidence of cis- and trans-regulatory divergence by comparing allele-specific expression in the mixed parental and hybrid samples. This analysis was also performed using minimum read thresholds of 50, 100, and 200, and all showed similar results (Supplemental Table S4). Of the 9966 genes with at least 20 mapped reads, 6546 (66%) showed evidence of significant trans-regulatory divergence and 5042 (51%) genes showed significant evidence of cis-regulatory divergence, with 3473 (35%) of these genes showing significant evidence of both (Supplemental Table S2). The median significant trans-regulatory difference between species (1.89-fold) was larger than the median cis-regulatory difference between species (1.43-fold; Wilcoxon's rank-sum test, P < 2.2 × 10−16; Fig. 3B), and expression differences between species correlated more strongly with trans-regulatory divergence (τ = 0.65, P < 2.2 × 10−16) than cis-regulatory divergence (τ = 0.29, P < 2.2 × 10−16). However, the amount of total regulatory divergence explained by cis-regulatory differences (% cis) increased with the magnitude of expression divergence (Fig. 3C). Over one thousand (1222) genes showed no evidence of significant expression differences between species nor any significant evidence of cis- or trans-regulatory divergence and were classified as “conserved.” An “ambiguous” pattern of significance tests was shown in 1051 genes (e.g., significant expression divergence between species, but no significant evidence of cis- or trans-regulatory differences), and were excluded from further analysis.
The 3473 genes with significant evidence of both cis- and trans-regulatory differences were subdivided into three groups: “cis + trans,” differentially expressed genes for which both cis- and trans-regulatory divergence favored expression of the same allele; “cis × trans,” differentially expressed genes for which cis- and trans-regulatory divergence favored expression of opposite alleles; and “compensatory,” genes with no significant expression differences between species, despite evidence for both cis- and trans-regulatory divergence. These classes are identical to those used previously (Landry et al. 2005; see Methods for classification procedures). Genes in the amounts of 1703, 1187, and 583 fell into the cis + trans, cis × trans, and compensatory categories, respectively (Fig. 3D; Supplemental Table S3). Genes classified as cis + trans are more likely to have divergent expression resulting from directional selection than genes classified as cis × trans or compensatory (Tirosh et al. 2009), whereas genes that fall into the latter classes may contribute to hybrid incompatibilities (Landry et al. 2005).
The frequency and magnitude of cis- and trans-regulatory changes we observed between D. melanogaster and D. sechellia show a larger relative contribution of trans-regulatory changes to expression divergence than was reported for D. melanogaster and D. simulans (Wittkopp et al. 2004; Wittkopp et al. 2008; Graze et al. 2009). While this pattern may reflect differences in the sensitivity of methods used in different studies, it may also reflect the unique evolutionary history of D. sechellia. This species is an island endemic that exhibits little intraspecific genetic variation and appears to have maintained a much smaller population size than D. melanogaster or D. simulans, since it was established (Kliman et al. 2000; Legrand et al. 2009). Consequently, natural selection is expected to have been less efficient in D. sechellia than in these other species, suggesting that many divergent sites in D. sechellia may have been fixed by genetic drift rather than selection.
Mechanisms of regulatory divergence influence the inheritance of gene expression
One of the main advantages of mRNA-seq over other techniques used to measure allelic expression is that it simultaneously measures total levels of gene expression and relative allelic expression. By comparing total expression levels among D. melanogaster, D. sechellia, and their hybrids, we determined the mode of inheritance for expression of 7739 divergently expressed genes. As shown in Figure 4A, genes for which expression in the hybrid was less than D. melanogaster and greater than D. sechellia (or vice versa) were classified as additive; genes for which expression in the hybrid was similar to one of the parents were classified as dominant; and genes for which expression in the hybrid was either greater than or less than both D. melanogaster and D. sechellia were considered misexpressed and classified as over-dominant and under-dominant, respectively. Regardless of statistical significance, any pair of genotypes with expression that differed less than 1.25-fold was considered to have similar expression for this analysis (Gibson et al. 2004). Consequently, despite showing a statistically significant difference in expression between species, 440 genes (6%) were classified having conserved expression in this analysis.
Figure 4.
Inheritance of gene expression levels in F1 hybrids between D. melanogaster and D. sechellia. (A) Hypothetical patterns of gene expression in D. melanogaster (red), D. sechellia (blue), and F1 hybrids (purple) illustrating conserved, additive, dominant, under-dominant, and over-dominant modes of inheritance are shown. (B) The scatterplot compares the differences in expression level between the F1 hybrid and each of the parental species (D. melanogaster on x-axis; D. sechellia on y-axis) for each differentially expressed gene. The bar graph (right) shows the frequency of genes in each inheritance category. (C) Plots compare total expression levels (i.e., log2 normalized read-hits) in F1 hybrids to total expression levels in D. melanogaster (black) and D. sechellia (red). Expression of genes with divergent expression between species was more similar in F1 hybrids to D. sechellia (τ = 0.80, P < 2 × 10−16) than D. melanogaster (τ = 0.57, P < 2 × 10−16). This similarity may result from dominance of D. sechellia alleles within homologous cells or a greater morphological similarity of F1 hybrids to D. sechellia than to D. melanogaster. Over half of the genes that showed dominance of the D. sechellia allele (59%, N = 3026; binomial exact test, P < 2 × 10−16) had lower expression in F1 hybrids than in D. melanogaster, while over half of the genes showing dominance of the D. melanogaster expression level had higher expression in F1 hybrids than in D. sechellia (62%, N = 572; binomial exact test, P < 3 × 10−8). (D) Distributions of percent cis for genes showing additive (A) and nonadditive (N-A) inheritance.
Of the remaining 7299 genes, 1183 (16%) were classified as additive, 3598 (49%) were classified as dominant, and 2518 (35%) were misexpressed in hybrids with 564 (8%) classified as over-dominant and 1954 (27%) classified as under-dominant (Fig. 4B). Interestingly, 3026 (84%) of the 3598 dominant genes showed D. sechellia-like expression, and overall gene expression levels in hybrids were more similar to D. sechellia than D. melanogaster (Fig. 4C). These inheritance patterns are expected to primarily reflect dominance relationships between orthologous alleles; however, because mRNA was extracted from whole flies for this study, expression levels are also affected by differences in the abundance of cell types expressing a particular gene. For example, D. sechellia produces fewer ovarioles than D. melanogaster (Jones 2004), suggesting that mRNA from genes expressed primarily in these cells will be less abundant in D. sechellia than D. melanogaster, even if the number of mRNA molecules produced per cell is conserved. Similarly, hybrids with D. melanogaster often lack ovary tissue (Santamaria 1977) and this would cause a gene to show under-dominant inheritance in our analysis (Ranz et al. 2004).
To examine the relationship between dominance and the molecular mechanisms of regulatory divergence, we tested whether genes with cis-regulatory divergence were more likely to show additive inheritance than genes with trans-regulatory divergence. Such a pattern was reported for variation within D. melanogaster (Lemos et al. 2008) and is expected because transcripts from the maternal and paternal alleles are assumed to contribute independently to total levels of hybrid gene expression. Comparing the amount of total regulatory divergence attributable to cis-regulatory differences (% cis) between sets of genes with additive and nonadditive inheritance showed that the median percent cis was indeed significantly higher for genes with additive (39%) than those with nonadditive (24%) inheritance (Wilcoxon's rank-sum test, P < 2 × 10−16; Fig. 4D). The magnitude of the cis-regulatory difference between species-specific alleles was also greater for genes with additive (median = 1.67-fold) than those with nonadditive (median = 1.20-fold) expression (Fisher's exact test, P < 2.2 × 10−16).
Finally, we used our mRNA-seq data to explore the possibility that regulatory divergence contributes to hybrid incompatibilities. Over time, the interaction of cis-regulatory sequences with trans-regulatory factors can co-evolve, causing divergence at the biochemical level between species. When such co-evolution occurs, cis- and trans-regulatory elements may interact incorrectly in hybrids, causing misexpression. Landry et al. (2005) found that cis- and trans-regulatory changes with opposite effects on gene expression (cis × trans) were enriched in a set of 23 genes misexpressed in the hybrids of D. melanogaster and D. simulans. Consistent with this much smaller survey, we found that 21% (n = 521) of the 2518 misexpressed genes in hybrids between D. melanogaster and D. sechellia showed cis × trans regulatory divergence, compared with only 12% (n = 591) of the 4781 genes with additive or dominant inheritance (Fisher's exact test, P = 3.6 × 10−15). Thus, our data support a model in which cis- and trans-regulatory divergence with opposite effects on gene expression contributes to misexpression in hybrids, and may therefore also contribute to hybrid incompatibilities.
Genomic features that correlate with cis-regulatory divergence
Unlike trans-regulatory divergence, cis-regulatory divergence is expected to be caused by changes in DNA sequence near the affected gene. To test this hypothesis, we examined the frequency of nucleotide substitutions, insertions, and deletions in the 1 kb of DNA sequence located immediately 5′ of each gene's transcription start site. As expected, genes with significant cis-regulatory divergence (n = 1056) had more sequence changes (substitutions and indels) than genes without significant cis-regulatory divergence (n = 891), with medians of 14.8% vs. 13.9% sequence divergence, respectively (Fisher's exact test, P < 0.006). This difference was observed throughout the 1-kb region surveyed (Supplemental Fig. S5). In addition, the magnitude of cis-regulatory divergence (|MF1/SF1|) showed a small, but significant, correlation with the extent of sequence divergence (τ = 0.05, P < 0.009, N = 1056), as well as with the frequency of indels (τ = 0.06, P < 0.006), suggesting that more divergent promoter sequences have greater cis-regulatory divergence. Similar patterns have previously been reported for expression variation and sequence divergence in other organisms (e.g., Tirosh et al. 2009; Zhang and Borevitz 2009), suggesting that sequence divergence generally correlates with cis-regulatory divergence. We also observed a small, but significant, positive correlation between the size of the upstream intergenic region (measured as the distance in kilobases between adjacent genes) and the magnitude of cis-regulatory divergence (τ = 0.03, P = 0.002, N = 4481): The median size of the upstream intergenic region was larger for genes with significant cis-regulatory divergence than for genes without (597 and 506 bp, respectively; Wilcoxon's rank-sum test, P < 0.0002). Genes with larger upstream intergenic regions may tend to show greater cis-regulatory divergence because these regions contain more cis-regulatory elements (Nelson et al. 2004).
Unexpected patterns of regulatory divergence between D. melanogaster and D. sechellia
In addition to illustrating the power of mRNA-seq as a tool for investigating regulatory divergence, this study provides novel insight into the evolution of gene expression between D. melanogaster and D. sechellia. For example, we observed an excess of regulatory changes that decrease expression in D. sechellia, as well as many more dominant regulatory alleles in D. sechellia than D. melanogaster. These findings appear to contradict a simple null model in which regulatory mutations with different types of effects are fixed at similar rates in the two lineages. We also observed fewer genes affected by cis-regulatory divergence than were reported for D. melanogaster and D. simulans (Wittkopp et al. 2008), which have approximately the same divergence time. As discussed above, this difference may reflect the fixation of many (presumably trans-acting) deleterious alleles in D. sechellia facilitated by a dramatic reduction in population size, and we predict that other species with similar demographic histories may also exhibit less cis-regulatory divergence than species that have maintained large population sizes throughout their history. Of course, positive selection may also have contributed to the higher than expected proportion of genes with trans-regulatory divergence. D. sechellia has undergone extensive environmental specialization (Jones 2005), and work in yeast suggests that divergent expression of genes responding to the environment may be preferentially caused by trans-regulatory changes (Tirosh et al. 2009). To explore this idea further, a detailed discussion of regulatory divergence putatively affecting the evolution of divergent feeding behavior in D. sechelllia is provided in the Supplemental material.
Emerging trends in Drosophila regulatory evolution
In addition to revealing features of regulatory evolution unique to D. melanogaster and D. sechellia, the comprehensive nature of this study allowed us to explore the potential generality of trends reported in prior (often smaller scale) studies. First, we found that 84% of differentially expressed genes showed nonadditive (i.e., dominant, over-dominant, or under-dominant) inheritance of expression levels between D. melanogaster and D. sechellia, which is consistent with the extensive nonadditive inheritance of gene expression reported within D. melanogaster (Gibson et al. 2004) and between D. melanogaster and D. simulans (Ranz et al. 2004). However, only 35% of differentially expressed genes between D. melanogaster and D. sechellia were classified as misexpressed in F1 hybrids, compared with 69% of genes that were differentially expressed in D. melanogaster and D. simulans (Ranz et al. 2004). In both analyses of interspecific hybrids, underexpression was more common than overexpression. Second, antagonistic interactions between cis- and trans-regulatory elements were found to be more common among misexpressed than nonmisexpressed genes in our analysis of 7355 differentially expressed genes, just as they were in a comparison of 31 genes between D. melanogaster and D. simulans (Landry et al. 2005). Third, our data indicate that cis-regulatory divergence contributes more to expression differences between species for genes that show additive (as opposed to nonadditive) inheritance, consistent with a previous study of genes on the second chromosome of D. melanogaster (Lemos et al. 2008). Finally, we found that cis-regulatory divergence correlates with sequence divergence of proximal promoters and the size of upstream intergenic sequences, consistent with prior studies of species from different biological kingdoms (e.g., Tirosh et al. 2009; Zhang and Borevitz 2009).
This study provides a significant advance in understanding regulatory evolution on a genomic scale, yet it reveals evolutionary changes that affect only a single developmental time point, under a single set of conditions, and for a single species pair. We expect many of the general trends reported here to be robust to these variables; however, we also anticipate that a meta-analysis of similar studies conducted at different developmental stages, under different environmental conditions, and in different species will reveal insights into regulatory evolution that are not apparent when studying only a single condition. With the decreasing cost and increasing availability of high-throughput sequencing technologies, such a meta-analysis of regulatory polymorphism and divergence among different stages, species, and environmental conditions should soon be possible.
Methods
Sample preparation and sequencing
D. melanogaster strain 14021-0231.36 (y[1]; Gr22b[1] Gr22d[1] cn[1] CG33964[R4.2] bw[1] sp[1]; LysC[1] MstProx[1] GstD5[1] Rh6[1]) and D. sechellia strain 14021-0248.25 (wild-type) were used for this study. All flies were reared on cornmeal/molasses medium. F1 hybrids were produced by crossing seven D. melanogaster females to ∼30 D. sechellia males. Reciprocal crosses were not performed because the F1 offspring of this cross die as embryos (Sawamura et al. 1993). Virgin females were collected from each parental species and their F1 hybrids shortly after eclosion and aged 2 d in isolation. For each genotype, total RNA was extracted from seven flies using TRIzol (Invitrogen). Total RNA in the amount of 4.5 μg from each species was mixed to create the mixed parental pool. Total RNA samples were treated with DNase I (Invitrogen) to remove any contaminating genomic DNA. Poly(A)+ transcripts were isolated from each sample using Dynal magnetic beads (Invitrogen) and fragmented for 2 min using the “RNA Fragmentation Reagent” (Ambion). Double-stranded cDNA was prepared from fragmented mRNA using random hexamer primers and SuperScriptII reverse transcriptase (Invitrogen). Following the isolation of ∼370-bp fragments from a 2% agarose gel, cDNA libraries were prepared for sequencing by following the manufacturer's protocol for the Paired-end Genomic DNA Library Preparation Kit (Illumina). The D. melanogaster and D. sechellia parental libraries were each subjected to paired-end sequencing in three lanes on an Illumina Genome Analyzer II (GAII) using 27 cycles per read. The mixed parental and F1 hybrid libraries were subjected to four and six lanes, respectively, of paired-end sequencing on a GAII using 37 cycles per read. Images were analyzed using the Firecrest and Bustard software modules to generate sequence and quality scores for each read.
Assigning reads to genes and species
Sequence reads that passed purity filtering were aligned individually to the D. melanogaster (dm3 assembly) (Adams et al. 2000; Celniker et al. 2002) and D. sechellia (droSec1 assembly) (Drosophila 12 Genomes Consortium 2007) genomes with Bowtie (Langmead et al. 2009) allowing for zero mismatches. Alignment results for the mixed parental and hybrid libraries were used to classify each read as either species-specific (i.e., clearly derived from D. melanogaster or D. sechellia) or common to both species. Mate-pair information was used to assign some common reads to one species or the other by virtue of their linkage to species-specific reads. These assignments were performed using a combination of publicly available (http://sysbio.harvard.edu/csb/resources/computational/scriptome/) and custom Perl scripts (see Supplemental material). Because the D. sechellia genome assembly consists of many unconnected contigs, as well as contigs containing gaps and errors, we limited our analysis to regions of the genome present in both assemblies. Gapped regions were identified from the UCSC pairwise alignment file (http://hgdownload.cse.ucsc.edu/goldenPath/dm3/vsDroSec1/) and removed using Galaxy (http://main.g2.bx.psu.edu/; Taylor et al. 2007). The D. sechellia genome assembly also contains multiple overlapping contigs, complicating the ability to accurately identify uniquely mapping sequence reads. To compensate for this, reads were initially allowed to map to as many as three genomic locations. The genomic coordinates of the D. sechellia specific reads were converted to D. melanogaster syntenic genomic coordinates using the UCSC lift-over tool (http://genome.ucsc.edu). Reads that mapped to multiple contigs in the D. sechellia genome, but corresponded to a single location in the D. melanogaster genome were retained for further analysis and counted once. In contrast, reads that mapped to multiple contigs in the D. sechellia genome that corresponded to multiple locations in the D. melanogaster genome were discarded (Supplemental Fig. S3). In many cases, the number of reads mapped to a gene exceeded the number of mappable positions (i.e., gene length − read length). Therefore, identical sequencing reads were expected and were included in the final analysis to avoid imposing an artificial cap on gene expression levels based on gene length. The aligned species-specific reads were then mapped to constitutive regions of all genes (i.e., exons common to all annotated isoforms) to mask species-specific differences in isoform levels (alternative transcription start sites, splicing, and termination) that might bias expression level comparisons (Fig. 1B; Supplemental Fig. S3). Gene mapping was performed using the program exonhitter.pl (see Supplemental material).
Gene expression levels are reported as the number of reads that mapped to each gene, with one exception: The number of reads reported for a gene in the mixed parental sample was adjusted such that the percentage of all species-specific reads in the mixed parental sample was equal to that observed in the F1 hybrid sample (i.e., 51.7% D. melanogaster). This adjustment was performed to account for any imbalance caused by mixing the two parental samples to create the mixed pool. No correction for gene size was used because all comparisons were between identically sized regions of orthologous genes. In the mixed parental library, 9332 and 9256 genes had at least 20 mapped reads from D. melanogaster and D. sechellia, respectively, with 8881 genes having more than 20 reads in both species.
Eighty-five genes with at least 20 reads from both species combined (i.e., M + S ≥ 20) had zero read-hits from one of the two species (M = 0 or S = 0). These genes could truly not be expressed in one species, but this could also be an artifact of the bioinformatic procedures used for allele assignment. For example, if one species contained multiple paralogs of a gene, requiring unique alignments would cause all of the reads to be excluded, resulting in 0 reads. To deal with this issue, we manually curated these genes and removed 34 that were likely to be bioinformatic artifacts. For the remaining 51 genes where one species had a read count of 0 (and the other had a read count of at least 20), we changed the number of mapped reads from 0 to 1 in order to calculate ratios and perform statistical analyses on these genes (Marioni et al. 2008).
Pyrosequencing
New hybrid and mixed parental cDNA pools were synthesized from the same RNA samples used for Illumina sequencing. Genomic DNA was also extracted from F1 hybrids and used to normalize cDNA measurements (Landry et al. 2005). Pyrosequencing was performed as described previously (Wittkopp et al. 2004). Briefly, cDNA prepared from pooled parental flies or hybrids was used for three replicate PCR amplifications and subsequent pyrosequencing reactions to determine relative expression. Pyrosequencing assays were developed for 15 genes (Supplemental Table S1) that had various levels of expression and cis- and trans-regulatory divergence in the mRNA-seq analysis. Hybrid genomic DNA was analyzed in duplicate PCR and pyrosequencing reactions and used to account for any amplification bias. After normalization (Landry et al. 2005), ratios of allelic expression for genes measured using Illumina and pyrosequencing were compared using linear regressions.
Cis- and trans-regulatory divergence assignment
We tested for evidence of cis- and trans-regulatory divergence using hierarchical statistical analyses (Supplemental Fig. S4). Three different significance thresholds were used for this analysis (FDR = 5%, 1%, and 0.5%). Only the results of the most conservative analysis (i.e., FDR = 0.5%) are discussed in the main text (but see Supplemental Table S3). Both the parental (P) and hybrid (H) data sets were first analyzed for evidence of differential expression using the binomial exact test, followed by FDR analysis (Storey and Tibshirani 2003). Any significant difference in the abundance of D. melanogaster and D. sechellia alleles in the P data set was considered evidence of expression divergence, and any significant difference in the abundance of D. melanogaster and D. sechellia alleles in the H data set was considered evidence of cis-regulatory divergence. Genes found to be differentially expressed in either the P or H data sets were further analyzed for trans effects (T) by comparing species-specific mRNA abundance ratios between the P and H samples using Fisher's exact tests followed by FDR analysis. These data were also analyzed using a χ2 test with a Yates correction and >99% of the significant trans-regulatory differences identified using the χ2 test were also identified by Fisher's exact test (data not shown). We used a custom Perl script to sort genes into the following seven categories:
Cis only: Significant differential expression in P and H. No significant T.
Trans only: Significant differential expression in P, but not H. Significant T.
Cis + trans: Significant differential expression in P and H. Significant T. The log2-transformed allele specific ratios of these genes in parental and hybrid data sets have the same sign (i.e., the species with higher expression also had the higher expressing allele in F1 hybrids). Regulation of these genes has diverged such that cis- and trans-regulatory differences favor expression of the same allele.
Cis × trans: Significant differential expression in P and H. Significant T. The log2-transformed allele specific ratios of these genes in parental and hybrid data sets have opposite signs (i.e., the species with higher expression contributed the lower expressing allele in F1 hybrids). Regulation of these genes has diverged such that cis- and trans-regulatory differences favor expression of opposite alleles.
Compensatory: Significant differential expression in H, but not P. Significant T. Regulation of these genes has diverged such that cis- and trans-regulatory differences perfectly compensate each other, resulting in no expression difference between species.
Conserved: No significant differential expression in H or P. No significant T. These genes are expressed at a similar level in each species, as well as in the hybrid, indicating conserved regulation.
Ambiguous: All other patterns of significance tests, which have no clear biological interpretation.
Inheritance classifications
The mode of inheritance for differentially expressed genes was determined using R (v 2.9.0, CRAN). Total expression was normalized by dividing the number of mapped reads at each gene by the total number of mapped reads for the entire genome, and multiplying by 100 to give a percent expression value. Log-transformed percent expression values of each parent were subtracted from those of the hybrid to examine changes in expression. Genes whose total expression in hybrids deviated more than 1.25-fold from that of either parent were considered to have nonconserved inheritance, and were classified as having additive, dominant, under-dominant, or over-dominant inheritance, based on the magnitude of the difference between total expression in the hybrid and in each parental species, as described in the main text. Parent-of-origin effects on gene expression (e.g., Gibson et al. 2004) are not described in this study because only one direction of cross generates viable F1 hybrid females.
Comparison of genomic features and regulatory divergence
Pairwise alignments of the D. melanogaster and D. sechellia sequences 1 kb upstream of annotated transcription start sites were extracted from the 12 species multialignment file available from UCSC Genome Browser (http://hgdownload.cse.ucsc.edu/goldenPath/dm3/multiz15way/maf/upstream1000.maf.gz). These regions were filtered to remove genes that have more than one annotated start site and genes whose 1-kb upstream region overlaps other annotated genes. The remaining pairs of sequences were aligned using the EMBOSS needle program. The summary results of this program (% match and % gap) were extracted for statistical comparisons of sequence divergence. Sequence divergence was also compared between the upstream regions of genes with and without cis-regulatory divergence using a 75-bp sliding window. The magnitude of cis-regulatory divergence was compared to the EMBOSS needle summary results using Wilcoxon's rank sum test.
The sizes of upstream intergenic regions were calculated from the D. melanogaster annotation dm3 (April 2006) by first determining the minimum sequence coordinate of all annotated start sites and maximum sequence coordinate of all annotated end sites for each gene. Intergenic distances were calculated as the difference between the minimum base pair coordinates of a gene and the maximum base pair coordinates of the nearest upstream neighboring gene encoded on the same DNA strand. Intergenic sizes were compared to the magnitude of cis-regulatory divergence using Wilcoxon's rank sum test. All test statistics were calculated in R (v 2.9.0, CRAN).
Acknowledgments
We thank J. Long and the University of Michigan's Department of Human Genetics for use of the pyrosequencing machine; the UCHC Translational Genomics Core for use of the Illumina Genome Analyzer; J. Gruber and T. Theara for technical assistance, and I. Dworkin, D. Presgraves, and S. Celniker for discussions and comments on the manuscript. This work was supported by a Basil O'Connor Starter Scholar Research Award from the March of Dimes (P.J.W.) (5-FY07-181) and the National Institutes of Health (B.R.G.) (5R01GM062516-09). P.J.W. is also a research fellow supported by the Alfred P. Sloan foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequencing data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE20421.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.102491.109.
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195 [DOI] [PubMed] [Google Scholar]
- Ahmadian A, Gharizadeh B, Gustafsson AC, Sterky F, Nyren P, Uhlen M, Lundeberg J 2000. Single-nucleotide polymorphism analysis by pyrosequencing. Anal Biochem 280: 103–110 [DOI] [PubMed] [Google Scholar]
- Carroll SB 2005. Evolution at two levels: On genes and form. PLoS Biol 3: e245 doi: 10.1371/journal.pbio.0030245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker SE, Wheeler DA, Kronmiller B, Carlson JW, Halpern A, Patel S, Adams M, Champe M, Dugan SP, Frise E, et al. 2002. Finishing a whole-genome shotgun: Release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol 3: research0079 doi: 10.1186/gb-2002-3-12-research0079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles CR, Hirschhorn JN, Altshuler D, Lander ES 2002. Detection of regulatory variation in mouse genes. Nat Genet 32: 432–437 [DOI] [PubMed] [Google Scholar]
- Cutter AD 2008. Divergence times in Caenorhabditis and Drosophila inferred from direct estimates of the neutral mutation rate. Mol Biol Evol 25: 778–786 [DOI] [PubMed] [Google Scholar]
- Denver DR, Morris K, Streelman JT, Kim SK, Lynch M, Thomas WK 2005. The transcriptional consequences of mutation and natural selection in Caenorhabditis elegans. Nat Genet 37: 544–548 [DOI] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450: 203–218 [DOI] [PubMed] [Google Scholar]
- Dworkin I, Jones CD 2009. Genetic changes accompanying the evolution of host specialization in Drosophila sechellia. Genetics 181: 721–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genissel A, McIntyre LM, Wayne ML, Nuzhdin SV 2008. Cis and trans regulatory effects contribute to natural variation in transcriptome of Drosophila melanogaster. Mol Biol Evol 25: 101–110 [DOI] [PubMed] [Google Scholar]
- Gibson G, Weir B 2005. The quantitative genetics of transcription. Trends Genet 21: 616–623 [DOI] [PubMed] [Google Scholar]
- Gibson G, Riley-Berger R, Harshman L, Kopp A, Vacha S, Nuzhdin S, Wayne M 2004. Extensive sex-specific nonadditivity of gene expression in Drosophila melanogaster. Genetics 167: 1791–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graze RM, McIntyre LM, Main BJ, Wayne ML, Nuzhdin SV 2009. Regulatory divergence in Drosophila melanogaster and D. simulans, a genomewide analysis of allele-specific expression. Genetics 183: 547–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh W-P, Chu T-M, Wolfinger RD, Gibson G 2003. Mixed-model reanalysis of primate data suggests tissue and species biases in oligonucleotide-based gene expression profiles. Genetics 165: 747–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra HE, Coyne JA 2007. The locus of evolution: Evo devo and the genetics of adaptation. Evolution 61: 995–1016 [DOI] [PubMed] [Google Scholar]
- Jones CD 2004. Genetics of egg production in Drosophila sechellia. Heredity 92: 235–241 [DOI] [PubMed] [Google Scholar]
- Jones CD 2005. The genetics of adaptation in Drosophila sechellia. Genetica 123: 137–145 [DOI] [PubMed] [Google Scholar]
- Kliman RM, Andolfatto P, Coyne JA, Depaulis F, Kreitman M, Berry AJ, McCarter J, Wakeley J, Hey J 2000. The population genetics of the origin and divergence of the Drosophila simulans complex species. Genetics 156: 1913–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry CR, Wittkopp PJ, Taubes CH, Ranz JM, Clark AG, Hartl DL 2005. Compensatory cis-trans evolution and the dysregulation of gene expression in interspecific hybrids of Drosophila. Genetics 171: 1813–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry CR, Lemos B, Rifkin SA, Dickinson WJ, Hartl DL 2007. Genetic properties influencing the evolvability of gene expression. Science 317: 18–21 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand D, Tenaillon MI, Matyot P, Gerlach J, Lachaise D, Cariou ML 2009. Species-wide genetic variation and demographic history of Drosophila sechellia, a species lacking population structure. Genetics 182: 1197–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos B, Araripe LO, Fontanillas P, Hartl DL 2008. Dominance and the evolutionary accumulation of cis- and trans-effects on gene expression. Proc Natl Acad Sci 105: 14471–14476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Main BJ, Bickel RD, McIntyre LM, Graze RM, Calabrese PP, Nuzhdin SV 2009. Allele-specific expression assays using Solexa. BMC Genomics 10: 422 doi: 10.1186/1471-2164-10-422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y 2008. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18: 1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Nelson CE, Hersh BM, Carroll SB 2004. The regulatory content of intergenic DNA shapes genome architecture. Genome Biol 5: R25 http://genomebiology.com/2004/5/4/R25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuzhdin SV, Wayne ML, Harmon KL, McIntyre LM 2004. Common pattern of evolution of gene expression level and protein sequence in Drosophila. Mol Biol Evol 21: 1308–1317 [DOI] [PubMed] [Google Scholar]
- Ranz JM, Machado CA 2006. Uncovering evolutionary patterns of gene expression using microarrays. Trends Ecol Evol 21: 29–37 [DOI] [PubMed] [Google Scholar]
- Ranz JM, Castillo-Davis CI, Meiklejohn CD, Hartl DL 2003. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science 300: 1742–1745 [DOI] [PubMed] [Google Scholar]
- Ranz JM, Namgyal K, Gibson G, Hartl DL 2004. Anomalies in the expression profile of interspecific hybrids of Drosophila melanogaster and Drosophila simulans. Genome Res 14: 373–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifkin SA, Kim J, White KP 2003. Evolution of gene expression in the Drosophila melanogaster subgroup. Nat Genet 33: 138–144 [DOI] [PubMed] [Google Scholar]
- Rockman MV, Kruglyak L 2006. Genetics of global gene expression. Nat Rev Genet 7: 862–872 [DOI] [PubMed] [Google Scholar]
- Ronald J, Akey JM 2007. The evolution of gene expression QTL in Saccharomyces cerevisiae. PLoS One 2: e678 doi: 10.1371/journal.pone.0000678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria P 1977. On the causes of sterility in some interspecific hybrids from the melanogaster subgroup of Drosophila. Wilhelm Roux's Archives 82: 305–310 [DOI] [PubMed] [Google Scholar]
- Sawamura K, Watanabe TK, Yamamoto M-T 1993. Hybrid lethal systems in the Drosophila melanogaster species complex. Genetica 88: 175–185 [DOI] [PubMed] [Google Scholar]
- Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, Ruff TG, Milligan SB, Lamb JR, Cavet G, et al. 2003. Genetics of gene expression surveyed in maize, mouse and man. Nature 422: 297–302 [DOI] [PubMed] [Google Scholar]
- Serre D, Gurd S, Ge B, Sladek R, Sinnett D, Harmsen E, Bibikova M, Chudin E, Barker DL, Dickinson T, et al. 2008. Differential allelic expression in the human genome: A robust approach to identify genetic and epigenetic cis-acting mechanisms regulating gene expression. PLoS Genet 4: e1000006 doi: 10.1371/journal.pgen.1000006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL, Orgogozo V 2008. The loci of evolution: How predictable is genetic evolution? Evolution 62: 2155–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J, Schenck I, Blankenberg D, Nekrutenko A 2007. Using galaxy to perform large-scale interactive data analyses. Curr Protoc Bioinformatics 19: 10.5 doi: 10.1002/0471250953.bi1005s19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Reikhav S, Levy AA, Barkai N 2009. A yeast hybrid provides insight into the evolution of gene expression regulation. Science 324: 659–662 [DOI] [PubMed] [Google Scholar]
- Wang X, Sun Q, McGrath SD, Mardis ER, Soloway PD, Clark AG 2008. Transcriptome-wide identification of novel imprinted genes in neonatal mouse brain. PLoS One 3: e3839 doi: 10.1371/journal.pone.0003839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead A, Crawford DL 2006. Variation within and among species in gene expression: Raw material for evolution. Mol Ecol 15: 1197–1211 [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ 2005. Genomic sources of regulatory variation in cis and in trans. Cell Mol Life Sci 62: 1779–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG 2004. Evolutionary changes in cis and trans gene regulation. Nature 430: 85–88 [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG 2008. Regulatory changes underlying expression differences within and between Drosophila species. Nat Genet 40: 346–350 [DOI] [PubMed] [Google Scholar]
- Wray GA 2007. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet 8: 206–216 [DOI] [PubMed] [Google Scholar]
- Zhang X, Borevitz JO 2009. Global analysis of allele-specific expression in Arabidopsis thaliana. Genetics 182: 943–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Li JB, Gao Y, Egli D, Xie B, Deng J, Li Z, Lee JH, Aach J, Leproust EM, et al. 2009. Digital RNA allelotyping reveals tissue-specific and allele-specific gene expression in human. Nat Methods 6: 613–618 [DOI] [PMC free article] [PubMed] [Google Scholar]