Abstract
Pristionchus pacificus is a nematode model organism whose genome has recently been sequenced. To refine the genome annotation we performed transcriptome and proteome analysis and gathered comprehensive experimental information on gene expression. Transcriptome analysis on a 454 Life Sciences (Roche) FLX platform generated >700,000 expressed sequence tags (ESTs) from two normalized EST libraries, whereas proteome analysis on an LTQ-Orbitrap mass spectrometer detected >27,000 nonredundant peptide sequences from more than 4000 proteins at sub-parts-per-million (ppm) mass accuracy and a false discovery rate of <1%. Retraining of the SNAP gene prediction algorithm using the gene expression data led to a decrease in the number of previously predicted protein-coding genes from 29,000 to 24,000 and refinement of numerous gene models. The P. pacificus proteome contains a high proportion of small proteins with no known homologs in other species (“pioneer” proteins). Some of these proteins appear to be products of highly homologous genes, pointing to their common origin. We show that >50% of all pioneer genes are transcribed under standard culture conditions and that pioneer proteins significantly contribute to a unimodal distribution of predicted protein sizes in P. pacificus, which has an unusually low median size of 240 amino acids (26.8 kDa). In contrast, the predicted proteome of Caenorhabditis elegans follows a distinct bimodal protein size distribution, with significant functional differences between small and large protein populations. Combined, these results provide the first catalog of the expressed genome of P. pacificus, refinement of its genome annotation, and the first comparison of related nematode models at the proteome level.
Genome sequence data are useful only when genes are correctly annotated and information on their genomic localization, expression, and function is available. Comprehensive annotation of protein-coding genes is largely done in silico and is error-prone, especially when performed without experimental information on gene expression. Recent development of rapid techniques for nucleic acid sequencing has enabled comprehensive detection of transcribed genomic regions and use of this information in genome annotation. Especially, the platforms that implement high-throughput pyrosequencing, such as 454 Life Sciences (Roche) FLX, are powerful tools for genome annotation. This platform produces fewer reads (400 K–500 K) than other next-generation sequencers. However, these reads are on average longer (>200 bp vs. ∼50 bp) and are essential for an accurate reconstruction of any metazoan transcriptome. This platform has recently been used in the annotation of eukaryotic and prokaryotic genomes (Shin et al. 2008; Vera et al. 2008).
In addition to the evidence of gene expression at transcription level, mass spectrometry (MS)-based proteomics is increasingly used for experimental identification of translated genomic sequence. In a “proteogenomics” approach (Ansong et al. 2008; Gupta et al. 2008), the complete protein extract of an organism is digested into peptides, which are then mass-measured and fragmented in a mass spectrometer. Mass spectra are typically searched against a database containing a six-frame translation of the raw genome assembly and can therefore identify new, unpredicted open reading frames and refine existing gene models. Pioneered already in 1995 (Yates et al. 1995), proteogenomics has since been used to provide experimental evidence for gene expression in various model organisms, such as Arabidopsis thaliana (Baerenfaller et al. 2008), Plasmodium yoelii yoelii (Carlton et al. 2002), Toxoplasma gondii (Xia et al. 2008), and Homo sapiens (Fermin et al. 2006). A recent study of Caenorhabditis elegans identified more than 6000 gene products by mass spectrometry and refined many gene models even in this well-studied organism (Merrihew et al. 2008).
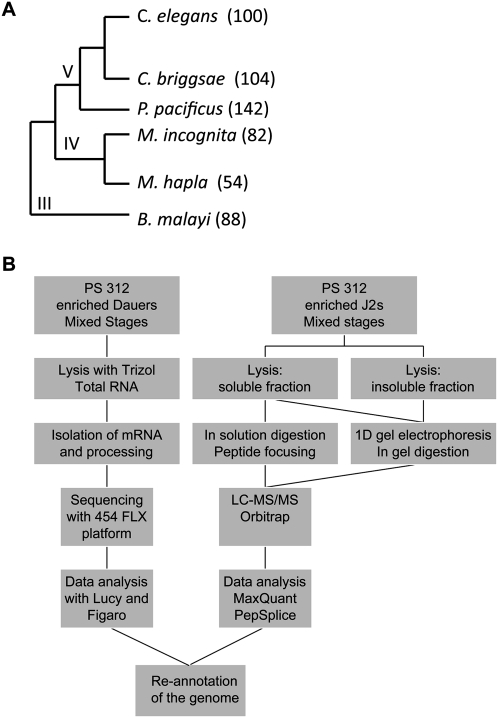
Pristionchus pacificus is a nematode that has been established as a model organism in evolutionary developmental biology (Sommer et al. 1996; Hong and Sommer 2006). It shares many advantageous features with C. elegans, in that it can be grown easily under laboratory conditions by feeding on Escherichia coli OP 50, it has a short generation time (4 d at 20°C), and it is a self-fertilizing hermaphrodite, which makes it amenable to forward and reverse genetics. The genome of P. pacificus was recently sequenced in a whole-genome shotgun approach with 10-fold coverage. The calculated genome size is 169 megabases (Mb) with a total number of 29,000 predicted protein-coding genes and a minimal gene content of 23,500 genes, as inferred from RT-PCR analyses (Dieterich et al. 2008). Many of these genes share no sequence similarity with already known genes in other nematodes and different phyla (“pioneer” genes). In comparison, the genome of C. elegans is completely assembled, consisting of a 100-Mb genome with 20,060 encoding genes (The C. elegans Sequencing Consortium 1998; Dieterich and Sommer 2009). P. pacificus and C. elegans belong to the same phylogenetic clade (Fig. 1A), which provides an ideal evolutionary distance for comparison of their proteome structures.
Figure 1.
Phylogeny of Pristionchus pacificus and proteogenomics workflow employed in this study. (A) Phylogenetic relationship of nematodes with sequenced genomes. The genome sizes (megabases) are written in brackets. The clades are depicted in the tree. (B) P. pacificus gene expression was assessed at the levels of transcription and translation and in three different developmental stages: dauer, J2, and “mixed stage” (containing all developmental stages, including eggs). In proteomics approach, several workflows for protein extraction and separation were used. ESTs detected with 454 pyrosequencing and peptide sequences detected with LTQ-Orbitrap mass spectrometry were used for genome reannotation.
Here, we perform a comprehensive analysis of the P. pacificus transcriptome and proteome using 454 FLX sequencing and LTQ-Orbitrap mass spectrometry, respectively. We search high-accuracy MS data against the predicted proteome and six-frame translation of the raw genomic assembly. We identify more than 700,000 expressed sequence tags (ESTs) and 27,000 nonredundant peptide sequences and use these data to refine the genome annotation and compare the predicted and detected proteome of P. pacificus with that of the other nematode models. We show that >50% of all pioneer genes are transcribed and that pioneer proteins significantly contribute to the unimodal distribution of predicted protein sizes in P. pacificus. Finally, we observe that the predicted proteome of C. elegans follows a distinct bimodal distribution, with significant functional differences between small and large protein populations.
Results
The aim of this study was to provide the first experimental catalog of the expressed genome of P. pacificus and to use this information for further refinement of the genome annotation. To obtain enhanced coverage of the expressed genome, we used two complementary approaches: transcriptome sequencing and high-accuracy MS proteomics. For the transcriptome analysis, total RNA was isolated from a mixed culture (containing all developmental stages, including eggs) and dauer stage culture of P. pacificus and sequenced on the 454 Life Sciences (Roche) FLX pyrosequencing platform.
For the proteome analysis, protein extracts were isolated from a mixed culture and second juvenile (J2) stage culture of P. pacificus. In all proteomics experiments, the protein extracts were divided into soluble and insoluble fractions, separated by 1D SDS-PAGE, and in-gel digested by trypsin. To achieve better analytical depth, soluble protein fractions were additionally digested in-solution by trypsin, and the resulting peptide mixtures were separated by isoelectric focusing. All peptide mixtures were subjected to nano-LC-MS/MS analysis on an LTQ-Orbitrap mass spectrometer. The MS data were processed and prepared for database search using the MaxQuant software suite. All MS/MS spectra were searched using the Mascot search engine against a decoy database consisting of the predicted P. pacificus proteome (based on old assembly;Dieterich et al. 2008), E. coli proteome, common laboratory contaminant proteins, and a six-frame translation of the P. pacificus raw genomic assembly. The complete workflow is summarized in Figure 1B.
Gene expression analysis of P. pacificus
In the 454 FLX transcriptome measurement, a sequencing run of the normalized mixed stage cDNA library yielded 334,441 ESTs that mapped uniquely to the genome top 965 contigs and had a median read length of 240 bp. In the normalized dauer stage library, 376,796 ESTs mapped to the top 965 contigs and had a median read length of 250 bp. In total, 711,237 ESTs were detected in both analyzed developmental stages.
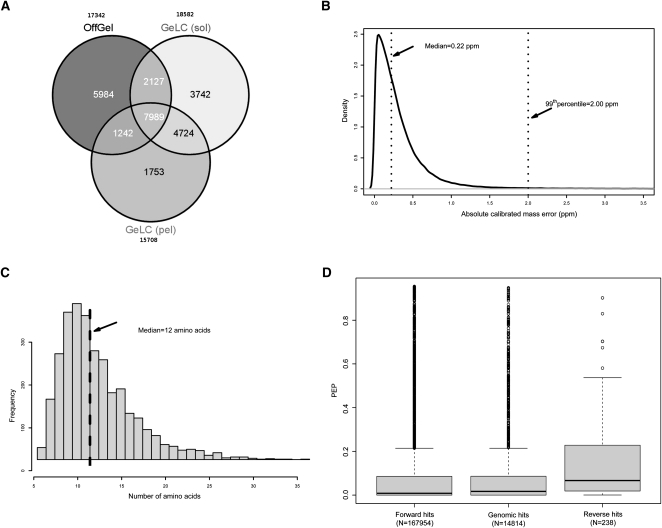
In the proteome measurement, MS data pre-processing using MaxQuant software resulted in 1,190,811 spectra that were submitted to the Mascot search engine. Database searching led to the identification of 27,561 nonredundant peptide sequences at an estimated false discovery rate (FDR) of 0.2% at the peptide level. Of these, 22,208 were detected in the mixed culture and 17,412 in the J2 stage (Supplemental Table 1). The applied biochemical workflow enabled enhanced proteome coverage, as only 7989 (29%) peptides were identified in all three approaches (Fig. 2A). Robust recalibration algorithms integrated in the MaxQuant software led to the overall average absolute peptide mass deviation of 0.345 parts per million (ppm) with a standard deviation of 0.434 ppm and enabled the use of narrow individualized precursor ion mass tolerances in the database search (Fig. 2B; Cox and Mann 2008) .
Figure 2.
Overview of the proteomics results. (A) Application of complementary biochemical workflows for protein extraction and peptide separation led to enhanced proteome coverage. (sol) Soluble fraction; (pel) pellet. (B) Peptides were detected with a mean absolute mass deviation of 0.345 ppm. (C) Peptide sequences that mapped to the genome translation (“genomic peptides”) had a median size of 12 amino acids. (D) Distribution of posterior error probabilities (PEP) was markedly different in the genomic and reversed peptide sequences.
Detected peptides were assembled into proteins and protein groups by MaxQuant software (see Methods). Of the detected protein groups, 3451 mapped to the P. pacificus predicted proteome (old assembly), 266 to the E. coli proteome, 50 to reversed sequences, 30 to contaminants included in the database, and the remainder to the raw genomic translation. The FDR at the protein group level was 1%. A list of all proteins detected by searching the six-frame translation database is available in Supplemental Table 2.
Refinement of P. pacificus gene predictions
We used the transcriptomics and proteomics data to refine the gene predictions in the P. pacificus genome sequence. In the transcriptome measurement, of a total of 711,237 detected ESTs, 223,849 ESTs corresponded to genomic regions that were not predicted by the old gene model (Dieterich et al. 2008): 96,754 ESTs in the mixed culture and 127,095 ESTs in the dauer stage. In the proteomics measurement, of 27,561 detected nonredundant peptide sequences, 2783 nonredundant peptides exclusively mapped to the translated genomic sequence, providing direct expression evidence for 1537 genomic regions (contigs and their corresponding reading frames) that were previously not predicted as protein-coding. The median length of genomic peptide hits was 12 amino acids (Fig. 2C), and their posterior error probability (PEP) distribution was distinctly different than that of highest-scoring reverse database hits (Fig. 2D), confirming the high reliability of the data set. To gain additional information on the splice junctions, MS/MS peak lists derived by MaxQuant software were submitted to the PepSplice search engine, which uses raw DNA sequence information to calculate peptides with gaps corresponding to potential GT–AG introns (Roos et al. 2007). The PepSplice database search resulted in identification of 541 spliced peptide sequences (Supplemental Table 3) that enabled identification of exact exon/intron boundaries in corresponding genes.
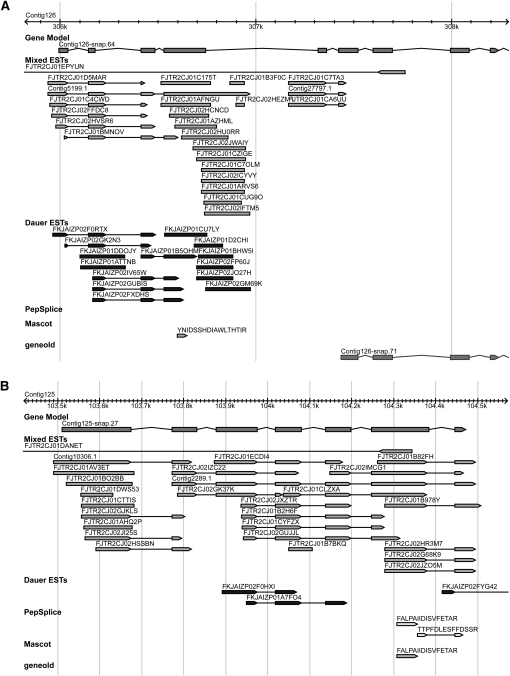
We used the information on the newly detected loci from both approaches to retrain the SNAP prediction algorithm (Dieterich and Sommer 2009), previously used for genome annotation of P. pacificus (Dieterich et al. 2008), and employed it to reannotation of gene predictions on the raw genome assembly. The genome reannotation led to a decrease in the number of predicted protein-coding genes from 29,424 (as reported at http://www.pristionchus.org), to 24,231, mainly through connection of neighboring coding regions. Consequently, 3263 old gene models were not contained in the genome reannotation, 1848 new gene models (7736 exons) have been identified, and 11,313 existing gene models were extended (Fig. 3). The new gene prediction is available at http://www.pristionchus.org.
Figure 3.
Genome reannotation resulted in new gene predictions and new gene models. (A) Example of a new gene model. New gene model “Contig126-snap.64” contains the old model “Contig126-snap.71”. (B) Example of a new gene prediction. The gene model “Contig125-snap.27” appeared only after retraining of the SNAP prediction algorithm with gene expression data.
Although our experiments were not designed to perform a direct comparison of the transcriptomics and proteomics, our study provides insights into the main contributions of the two platforms to genome reannotation. Of 1848 new gene models, only 73 were covered by peptides, demonstrating the superior genome coverage of the transcriptomics platform and pointing to the gene model refinement—rather than gene discovery—as the main contribution of the proteomics platform. Indeed, <25% of peptides that mapped to translated genomic sequence were in the intergenic regions of the old gene model, whereas the majority were in the intragenic regions (i.e., in the intron sequences), therefore exclusively affecting the existing gene models. In addition, proteomics significantly contributed to determination of exon–exon splice junctions.
For independent confirmation of expression of newly predicted genes, we chose 99 candidates and amplified them with RT-PCR on cDNA from mixed-stage animals. Of analyzed transcripts, 60 could be amplified and sequenced, confirming their expression (Supplemental Table 4).
Catalog of detected P. pacificus proteins
The refinement of the P. pacificus genome using transcriptomics and proteomics data led to its most comprehensive and accurate annotation to date. To derive a comprehensive catalog of detected P. pacificus proteins, we used the refined genome database to create the corresponding decoy protein database and search our mass spectrometry data against it. Resubmission of 1,190,811 spectra to the Mascot search engine resulted in identification of 32,126 nonredundant peptide sequences that mapped to 4029 P. pacificus protein groups at FDR 1% (Supplemental Table 5). To gain insight into the distribution of functional protein classes among detected proteins, we used the Blast2GO software to perform BLAST searches of detected protein sequences against the complete nrNCBI database and to extract the Gene Ontology (GO) terms. The GO analysis of the detected P. pacificus proteins revealed overrepresentation of cytosolic and developmental proteins, and underrepresentation of membrane proteins (Supplemental Fig. 1). The distribution of GO terms compared favorably with the recent proteomics analysis of C. elegans (Schrimpf et al. 2009) and demonstrated a sampling of similar protein classes in P. pacificus despite the more comprehensive proteome coverage in the C. elegans study.
Features of the predicted P. pacificus proteome
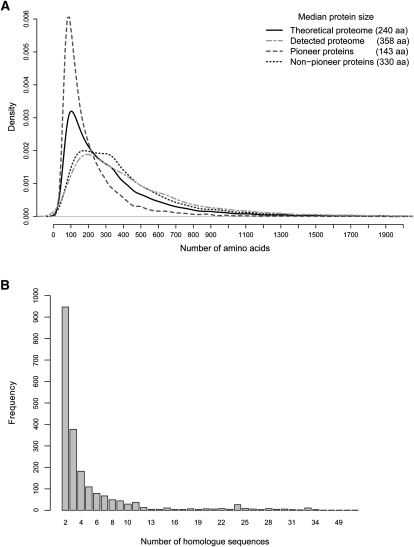
In silico translation of the predicted P. pacificus protein-coding genes showed an unusually low median predicted protein size of 240 amino acids (26.8 kDa) (Fig. 4A). BLAST analysis of the predicted proteome against the whole nrNCBI database did not retrieve any significant hits (E < 1 × 10−3) for 10,258 (42.3%) predicted proteins. We refer to them as “pioneer” proteins.
Figure 4.
Features of the P. pacificus predicted proteome. (A) Protein size distribution shows that the pioneer proteins are mainly responsible for the unusually low median protein size in P. pacificus. (B) BLAST results of the pioneer proteins against themselves show presence of highly homologous proteins that may have a common origin.
To gain insights into this group of proteins, we created a database consisting only of pioneer proteins and analyzed their features separately from the complete predicted proteome. Interestingly, the pioneer proteins are very short, with a median protein size of 143 amino acids (16 kDa). Their removal from the predicted proteome resulted in a significant increase in median size of the remaining proteins, from 240 amino acids (26.8 kDa) to 330 amino acids (36.9 kDa) (Fig. 4A), a value very close to the median size of proteins detected by MS (358 amino acids or 40 kDa). This leads to the conclusion that a majority of pioneer genes are not translated under tested conditions (environmental and/or developmental). Indeed, out of 4029 P. pacificus protein groups detected by MS, only 435 (10.8%) were products of pioneer genes. The search of unidentified MS spectra against a decoy database consisting only of pioneer proteins did not lead to significantly better coverage (data not shown). However, the coverage of pioneer genes was greater in the transcriptome analysis, where 5224 (51%) were detected to be expressed. To verify expression of pioneer genes we performed developmental stage-specific RT-PCR experiments. Out of 86 randomly chosen pioneer genes detected by MS, 56 could be amplified from mixed-stage cDNA and sequenced. Of this 56 transcripts, 31 showed different levels of expression in the tested second to fourth juvenile (J2–J4) stages, showing that at least some of the pioneer proteins may be functionally relevant in different developmental stages (Supplemental Table 6).
To gain further insights into primary sequence characteristics and origin of pioneer proteins, we performed a stringent BLAST analysis (E < 1 × 10−20, bit score > 90) of every entry in the pioneer protein database against the whole database. Despite the stringent criteria, 2086 entries (20%) returned multiple (1–85) BLAST hits, pointing to the existence of close structural homologs among pioneer proteins (Fig. 4B). Indeed, the first genome draft of P. pacificus has already revealed that ∼30% of the pioneer genes could be grouped into distinct protein families (Dieterich et al. 2008). We observe that some of the structurally related pioneer proteins reside on the same translated contigs, reflecting the proximity of their corresponding genes in the genome. Together, these data point to a likely common origin of part of the pioneer genes.
Comparison of the predicted P. pacificus proteome with proteomes of nematode models
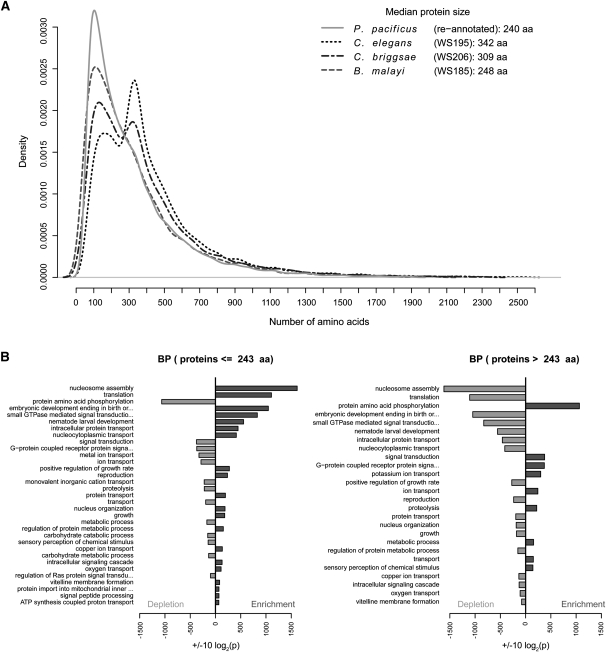
We used the reannotated genome as a starting point for comparison of the predicted proteome of P. pacificus with three published nematode proteomes: C. elegans (The C. elegans Sequencing Consortium 1998), C. briggsae (Stein et al. 2003), and Brugia malayi (Ghedin et al. 2007). Surprisingly, the predicted protein sizes followed a unimodal distribution in P. pacificus and B. malayi and a distinct bimodal distribution in C. elegans and C. briggsae (Fig. 5A). To test whether this was a consequence of different qualities of gene annotations, we extended this comparison to three additional members of the Caenorhabditis genus: C. remanei, C. brenneri, and C. japonica, whose genome assemblies are publicly available (http://www.wormbase.org/), but are not yet peer-reviewed. These three organisms also showed distinct distributions of protein sizes, with C. japonica matching the unimodal protein size distribution and C. remanei and C. brenneri matching the bimodal distribution (Supplemental Fig. 2A,B). To assess the functional relevance of this observation, we performed GO analysis of the predicted P. pacificus and C. elegans proteomes. Whereas the GO analysis of the two proteomes showed a very similar overall distribution of GO classes (Supplemental Fig. 3), the GO analysis applied separately to the small and large protein populations in C. elegans pointed to a significant functional relevance (Fig. 5B). The short protein population was enriched in functions related to nucleosome assembly, translation, and development, while the long protein population was enriched in functions related to protein phosphorylation, signal transduction, and ion transport. Notably, protein functions (GO terms) enriched in one tested data set were depleted in the other, pointing to the functional complementarity of the two protein populations.
Figure 5.
Comparison and functional analysis of protein size distributions in nematode models. (A) Predicted protein sizes in P. pacificus and B. malayi have a unimodal distribution, whereas C. elegans and C. briggsae have distinct bimodal distributions. (B) Gene Ontology enrichment analysis for short and long proteins in C. elegans shows distinct functional differences between the two classes of proteins.
Discussion
In this study, we performed a comprehensive analysis of gene expression in P. pacificus with the goals of (1) genome refinement, (2) in-depth analysis of the detectable proteome, and (3) comparative predicted proteome analysis of the nematode model organisms. To achieve optimal gene expression coverage, we performed transcriptome and proteome analyses of P. pacificus cultures from different developmental stages, covering the mixed population, dauer, and J2 stages. By sequencing ESTs we achieved comprehensive coverage of the transcribed genomic regions and complemented it with information on the translated genomic regions from the proteomics experiment. Both technological platforms used in this study—454 FLX nucleic acid pyrosequencing and LTQ-Orbitrap mass spectrometry—represent the state of the art in the fields of transcriptomics and proteomics, respectively. The use of the 454 platform in this study enabled acquisition of one of the most comprehensive collection of ESTs so far used for genome refinement. In addition, the use of LTQ-Orbitrap MS resulted in one of the most accurate proteogenomics data sets to date, both in terms of mass accuracy and FDR (0.2%). We note that in this study the MS/MS spectra were recorded in the low-resolution linear ion trap analyzer, whereas the MS spectra were recorded in the high-resolution Orbitrap mass analyzer. This was needed to achieve a high speed of MS/MS acquisition at high precursor ion mass accuracy, both of which are crucial in proteogenomics. One of the challenges in the use of mass spectrometry in proteogenomics applications is the use of six-frame translation protein databases that result in the increase of search space and decrease in search specificity. Although high mass accuracy has an obvious potential to resolve this problem, the proteome complexity and high dynamic range of gene expression have so far made fast-scanning and low-accuracy mass spectrometers almost exclusively used in proteogenomics applications. While such instrumentation ensures in-depth proteome coverage, it requires the use of wide mass tolerance windows (up to 4 Da) during database search. In this study, the sub-ppm measurement mass accuracy of precursor ions enabled the use of narrow initial mass tolerance window during database search (7 ppm or 0.007 Da at m/z = 1000), and even higher tolerances for peptide acceptance (Cox and Mann 2008), thereby significantly increasing search specificity and reducing the FDR. This is especially important when searching the peptide mass spectra against the translation of the complete genome assembly as >80% of entries present nonsense protein sequences (e.g., wrong reading frames).
The refined P. pacificus gene predictions provided a unique opportunity to study its proteome features and compare them with C. elegans, a related and well-studied nematode model. C. elegans was recently a subject of a comprehensive proteogenomics study, in which 245 novel genes were identified and 151 existing gene models were modified (Merrihew et al. 2008). The drastically higher number of newly predicted and modified P. pacificus gene models in our study is caused in part by using a transcriptomics platform for genome reannotation, but also reflects more comprehensive existing genome annotation of C. elegans.
A distinct feature of the P. pacificus proteome is the presence of short proteins with no apparent homologs in current protein databases such as nrNCBI (“pioneer” proteins). A high proportion of genes without any known homologs was previously reported in the P. pacificus genome (Dieterich et al. 2008), but their expression was never demonstrated. Here, we show that at least a portion of these genes is expressed under tested conditions. Although only ∼10% of predicted pioneer proteins were detected by MS, their coverage was higher in the transcriptome data set, where >50% were detected. At present, it is not clear whether this discrepancy is due to better transcriptome coverage, impaired translation, or low abundance (and therefore undersampling) of this class of proteins. The RT-PCR data showed that at least part of the pioneer genes show stage-specific expression, pointing to their potential roles in development. Interestingly, >20% of the pioneer proteins show similarity in primary structure and may therefore have a common origin. Although these data show that a fraction of pioneer proteins are synthesized and may be functional, their exact function and origin remain to be elucidated.
An interesting aspect that arose from the comparison of P. pacificus and C. elegans proteomes is the bimodality of protein size distribution in the C. elegans proteome. To our knowledge, this is the first reported example of a bimodal distribution of protein sizes in any proteome, with pronounced functional differences between the two protein populations. At present it is not clear whether this distinct proteome feature is of biological relevance; however, it seems to represent a phylogenetic trait, as only species of the Caenorhabditis crown group show the bimodal distribution, whereas C. japonica follows a unimodal distribution. Also, we note that the enrichment of GO terms related to protein phosphorylation among the larger protein population may reflect the unusually high number of protein kinases reported in C. elegans (Manning et al. 2002). Since the recently published phosphoproteome of C. elegans showed an unusual functional distribution of phosphoproteins (Zielinska et al. 2009), a quantitative comparison of P. pacificus and C. elegans organisms at the phosphoproteome level is likely to give valuable insights into evolution of phosphorylation networks in Metazoa.
Methods
Culturing of worms and preparation of protein extracts
P. pacificus strain PS312 was grown on 10-cm NGM agar plates spotted with 2 mL of E. coli OP50 solution. Plates were inoculated with 50–100 worms and incubated at 25°C. The mixed-stage population was harvested shortly after the bacterial lawn was consumed, avoiding starvation of the animals. After thoroughly washing with distilled water and 0.9% sodium chloride, the animals were incubated with ampicillin (100 μL/mL) and chloramphenicol (34 μL/mL) in 0.9% sodium chloride for 48 h to remove residual bacteria. The worms were then pelleted and prepared for proteomics measurements. The animals in the J2 developmental stage were harvested as follows: Plates full of eggs were washed with distilled water and the animals were bleached with hydrogen peroxide and 5 M sodium hydroxide, leaving just the eggs alive. Animals were then spotted on 10-cm NGM agar plates for hatching for 24 h, and collected by washing after removing debris and corpses. Animals were pelleted and stored frozen until further analysis. For protein isolation, 100 μL of animals was solubilized in 300 μL of denaturation buffer (6 M urea, 2 M thiourea, 10 mM Tris at pH 8.0). After three cycles of freeze (liquid nitrogen) and thaw (37°C), 100 μL of glass beads were added and the solution was vortexed for 20 min. After centrifugation (20 min, 20.800g, 4°C), the protein concentration of the supernatant was determined using the Bradford assay. The pellet was solubilized in sample buffer for gel electrophoresis and further analysis.
OffGel electrophoresis and in-solution digestion
For OffGel fractionation the proteins were reduced by incubation in 1 mM dithiotreitol (DTT) for 1 h at room temperature. Alkylation was performed in 5.5 mM iodoacetamide (IAA) in 50 mM ABC for 1 h at room temperature in the dark. Proteins were digested using LysC (1:100 w/w) for 3 h at room temperature and trypsin (1:100 w/w) overnight at room temperature after diluting the sample with four volumes of 20 mM ammonium bicarbonate (ABC). The resulting peptides were separated using the 3100 OffGel fractionator (Agilent) according to manufacturer's instructions with a 12- or 24-well setup. Focusing was done with 13-cm (12-well) or 24-cm (24-well) Immobiline DryStrips pH 3–10 (GE Healthcare) at a maximum current of 50 μA for 50 kVh. Peptide fractions were harvested and desalted using C18 StageTips as previously described (Ishihama et al. 2006).
GeLC-MS and in-gel digestion
For GeLC-MS analysis 100 μg of the supernatant and the solubilized pellet were loaded on a NuPAGE Bis-Tris 4%–12% gradient gel (Invitrogen). After brief Coomassie staining, each lane was cut into 10 slices that were further cut into small pieces. Destaining was performed by washing three times with 10 mM ABC and acetonitrile (ACN) (1:1, v/v) and was followed by protein reduction with 10 mM DTT in 20 mM ABC for 45 min at 56°C, and alkylation with 55 mM iodoacetamide in 20 mM ABC for 30 min at room temperature in the dark. The gel pieces were then washed twice for 20 min in destaining solution followed by dehydration with ACN. The liquid was removed and gel pieces were swollen at room temperature by adding 13 ng/μL sequencing-grade trypsin (Promega) in 20 mM ABC. Digestion of proteins was performed at 37°C overnight. The resulting peptides were extracted in three subsequent incubation steps with 30% ACN/3% TFA; with 80% ACN/0.5% acetic acid; and with 100% ACN. Supernatants were combined, ACN was evaporated in a vacuum centrifuge, and peptides were desalted using C18 StageTips.
NanoLC-MS/MS analysis
All digested peptide mixtures were separated on a nanoLC-2D HPLC (Eksigent) coupled to a LTQ-Orbitrap-XL (Thermo Fisher Scientific) through a nano-LC-MS interface (Proxeon Biosystems). Binding and chromatographic separation of the peptides was performed on a 15-cm fused silica emitter of 75-μm inner diameter (Proxeon Biosystems), in-house packed with reversed-phase ReproSil-Pur C18-AQ 3-μm resin (Dr. Maisch GmbH). The peptide mixtures were injected onto the column in HPLC solvent A (0.5% acetic acid) at a flow rate of 500 nL/min and subsequently eluted with a 107-min segmented gradient of 2%–80% HPLC solvent B (80% ACN in 0.5% acetic acid) at a flow rate of 200 nL/min.
The mass spectrometer was operated in the data-dependent mode to automatically switch between MS and MS/MS acquisition. Survey full-scan MS spectra were acquired in the mass range of m/z 300–2000 in the orbitrap mass analyzer at a resolution of 60,000. An accumulation target value of 106 charges was set and the lock mass option was used for internal calibration (Olsen et al. 2005). The 10 most intense ions were sequentially isolated and fragmented in the linear ion trap using collision-induced dissociation (CID) at the ion accumulation target value of 5000 and default CID settings. The ions already selected for MS/MS were dynamically excluded for 90 sec. The resulting peptide fragment ions were recorded in the linear ion trap. In total, 101 LC-MS measurements were performed, corresponding to 10 d of measurement time.
The mass spectrometry data associated with this manuscript may be downloaded from the Proteome Commons Tranche repository (https://proteomecommons.org/tranche/) using the following hash: QgF9ukyrC8Y74IIE8L/y2ccmTd02ElO8UnFcVLVFwvy1C+/41QDGWVzZIR96f33MKIui57iuS6x8 8KNT2v4RiIuHRN4AAAAAAAALhA==.
Data processing and analysis
MaxQuant data processing and Mascot database search
All raw files were processed together using the MaxQuant software suite (v. 1.12.35) (Cox and Mann 2008; Cox et al. 2009). Raw MS spectra were first processed by the Quant module to generate peak lists. This module performs a nonlinear mass recalibration for each individual precursor ion and calculates precise masses as well as individual mass errors. To retrieve peptide sequences from the processed spectra, we used the Mascot search engine v.2.2 (Matrix Science). The processed MS spectra were searched against an in-house assembled target-decoy database that consisted of the in silico–predicted proteome of P. pacificus (SNAPNG2.aa.annot, 27,103 sequences); a complete six-frame translation of its genome (sctg_plus_2000.fas, 14,654 contigs; 87,924 sequences after six-frame translation); E. coli proteome (4256 sequences); and 262 commonly observed contaminants. All protein sequences in the database were reversed and appended to the database. This enabled the estimation of false discovery rate (FDR) in the data set by a target-decoy search strategy (Elias and Gygi 2007).
In the database search, carbamidomethylation (Cys) was set as fixed modification, whereas oxidation (Met) and acetylation (protein N termini) were set as variable modifications. The mass tolerances for precursor and fragment ions were set to 7 ppm and 0.5 Da, respectively.
The retrieved peptide sequences were further processed with the Identify module of the MaxQuant software. This module considers all 10 peptide candidates suggested by Mascot for each fragmentation spectrum and filters them according to consistency with a priori information, e.g., the individual precursor mass errors. Furthermore, the probability that an individual peptide is a false hit given its score and length is estimated by a Bayesian probability (posterior error probability [PEP]). All filtered peptide sequences are sorted according to their PEP values, starting with the best PEP. To control the FDR the peptides are accepted until 1% of reversed peptides have accumulated within the list. The identified peptides are then assembled back into proteins. If a set or subset of identified peptides can be assigned to more than one protein, these proteins are joined into a protein group (Nesvizhskii and Aebersold 2005; Cox and Mann 2008). Finally, the FDR on protein group level was also controlled to be at 1%.
PepSplice database search
The PepSplice search engine (Roos et al. 2007) was used to complement Mascot-based searches. PepSplice uses a cache-optimized peptide database search algorithm for aligning spectra to genome-wide spliced six-frame translations. MaxQuant-processed MS/MS spectra (J2 + Mixed Stage) were searched against a target database, which contained all spliced six-frame translations of the 965 largest supercontigs (≥2 kb). All possible splicing events up to an intron size of 2 kb were considered and the maximal FDR was set to 1% on the peptide level. PepSplice also employs a target-decoy search strategy to estimate the FDR.
Downstream bioninformatics analysis
All downstream bionformatics was done in R (v. 2.9.0; http://www.r-project.org).
Protein size distributions were determined from the most recent versions of publicly available protein databases (http://www.wormbase.org). Distributions were determined by the “density” function from the base R package using default bandwidth. For robust estimation of protein size distribution, 99% of all proteins within the particular databases were considered. All BLAST searches in this study were performed by BLASTP v.2.2.21.
Gene Ontology analysis
GO annotation for the predicted P. pacificus and C. elegans (WS140, WS195) proteomes was derived using Blast2GO software (Conesa et al. 2005). For each query sequence the software first detects up to 20 homolog sequences in the nrNCBI database (nrNCBI version was from August, 2009; E-value <1 × 10−3) by a BLAST search. Based on the GO terms associated with these candidate sequences the software applies an annotation rule that filters and reports the most specific annotations.
To test for enrichment or depletion of specific GO terms among the identified proteome, the topGO R package was used (Alexa et al. 2006). This package implements two scoring methods that take care of the underlying GO graph topology. We used the “elim” algorithm that starts at the leaves of the induced GO graph and subsequently removes all proteins from the corresponding parent nodes that have been already used for testing the children nodes. Therefore, only the most specific GO terms for each protein were considered.
Fisher's exact test served as test statistic assuming the hypergeometric distribution as null-distribution. The derived P-values were further adjusted for multiple hypothesis testing by the method of Benjamini and Hochberg (Benjamini and Hochberg 1995) to control the FDR.
PCR analysis
To validate expression of proteins that were identified by MS, we chose 184 genes for RT-PCR. The primers were designed with the online tool Primer3 (Rozen and Skaletsky 2000) with an average amplicon size of 100 bp and were purchased from Eurofins MWG. For stage-specific cDNA, J2 stages were collected as described above and grown to J3 and J4, respectively. Total RNA was isolated using TRIzol (Invitrogen) according to the manufacturer's instructions. cDNA was produced using the Superscript III cDNA synthesis kit (Invitrogen) for 2 h at 42°C for the reverse transcription. PCR reactions were performed for 35 cycles of 20 sec at 95°C, 30 sec at 55°C, and 30 sec at 72°C. The reactions were subsequently separated on a 2% TBE agarose gel, stained with ethidium bromide, and visualized under UV light.
Transcriptome sequencing on the 454 Life Sciences (Roche) FLX platform
Total RNA was isolated from a mixed and dauer stage culture of P. pacificus (Ppa 312, California) using TRIzol (Invitrogen) according to the manufacturer's instructions. The RNA was sequenced at the Genome Sequencing Center at Washington University, St. Louis, MO using the 454 FLX for 454 sequencing.
Transcriptome assembly
The 454 reads were processed prior to assembly. Low-quality base calls were removed from read ends by Lucy (Chou and Holmes 2001) using default settings. Highly repetitive sequence segments were removed by Figaro (White et al. 2008) using default settings. We assembled 28,599/26,092 contigs from the 350,839/394,453 remaining sequences with the EST version of PCAP.REP. These contigs encompass >10 Mb of transcribed sequence.
Transcriptome mapping
The assembled contigs and all trimmed reads were aligned to the genome using Exonerate (Slater and Birney 2005) with a maximal intron size of 20 kb. In summary, we could identify 98,254 unique acceptor and 95,210 unique donor sites. This data set was subsequently used to improve the Pristionchus genome annotation.
Gene prediction
We took the 11 largest supercontigs from the Hybrid Genome Assembly (Sanger + 454). We predicted a new set of genes with the current hidden Markov model (HMM) gene model plus external evidence as given by the 454 transcriptome data (98254 Acceptor and 95210 Donor sites). This new gene set was subsequently used to retrain our HMM gene model (SNAPNG2).
All protein database searches for peptide identification were carried out on this reference data set. We used the genomic hits from Mascot and PepSplice as additional external evidence in the next gene model training step (4431 data points/coding segments). We updated our gene model to its final version and reran the gene predictions including all available external evidence (MS/MS + 454).
Acknowledgments
This work is supported by grants of Ministerium fuer Wissenschaft und Kunst and Landesstiftung Baden-Wuerttemberg to the Proteome Center Tübingen and by the Innovation Fund of the Max-Planck Society grant to R.J.S.
Footnotes
[Supplemental material is available online at http://www.genome.org. The 454 Life Sciences (Roche) sequencing data from this study have been submitted to the NCBI Short Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession no. SRA010772. Sequences from targeted RT-PCR reactions have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/Genbank/) (accession numbers provided in Supplemental Tables 4 and 6). Mass spectrometry data have been uploaded to the Proteome Commons Tranche repository (https://proteomecommons.org/tranche/).]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.103119.109.
References
- Alexa A, Rahnenfuhrer J, Lengauer T 2006. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22: 1600–1607 [DOI] [PubMed] [Google Scholar]
- Ansong C, Purvine SO, Adkins JN, Lipton MS, Smith RD 2008. Proteogenomics: Needs and roles to be filled by proteomics in genome annotation. Brief Funct Genomics Proteomics 7: 50–62 [DOI] [PubMed] [Google Scholar]
- Baerenfaller K, Grossmann J, Grobei MA, Hull R, Hirsch-Hoffmann M, Yalovsky S, Zimmermann P, Grossniklaus U, Gruissem W, Baginsky S 2008. Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320: 938–941 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y 1995. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57: 289–300 [Google Scholar]
- The C. elegans Sequencing Consortium 1998. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science 282: 2012–2018 [DOI] [PubMed] [Google Scholar]
- Carlton JM, Angiuoli SV, Suh BB, Kooij TW, Pertea M, Silva JC, Ermolaeva MD, Allen JE, Selengut JD, Koo HL, et al. 2002. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature 419: 512–519 [DOI] [PubMed] [Google Scholar]
- Chou HH, Holmes MH 2001. DNA sequence quality trimming and vector removal. Bioinformatics 17: 1093–1104 [DOI] [PubMed] [Google Scholar]
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M 2005. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676 [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Cox J, Matic I, Hilger M, Nagaraj N, Selbach M, Olsen JV, Mann M 2009. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat Protoc 4: 698–705 [DOI] [PubMed] [Google Scholar]
- Dieterich C, Sommer RJ 2009. How to become a parasite—lessons from the genomes of nematodes. Trends Genet 25: 203–209 [DOI] [PubMed] [Google Scholar]
- Dieterich C, Clifton SW, Schuster LN, Chinwalla A, Delehaunty K, Dinkelacker I, Fulton L, Fulton R, Godfrey J, Minx P, et al. 2008. The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. Nat Genet 40: 1193–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias JE, Gygi SP 2007. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 4: 207–214 [DOI] [PubMed] [Google Scholar]
- Fermin D, Allen BB, Blackwell TW, Menon R, Adamski M, Xu Y, Ulintz P, Omenn GS, States DJ 2006. Novel gene and gene model detection using a whole genome open reading frame analysis in proteomics. Genome Biol 7: R35 doi: 10.1186/gb-2006-7-4-r35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghedin E, Wang S, Spiro D, Caler E, Zhao Q, Crabtree J, Allen JE, Delcher AL, Guiliano DB, Miranda-Saavedra D, et al. 2007. Draft genome of the filarial nematode parasite Brugia malayi. Science 317: 1756–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N, Benhamida J, Bhargava V, Goodman D, Kain E, Kerman I, Nguyen N, Ollikainen N, Rodriguez J, Wang J, et al. 2008. Comparative proteogenomics: Combining mass spectrometry and comparative genomics to analyze multiple genomes. Genome Res 18: 1133–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong RL, Sommer RJ 2006. Pristionchus pacificus: A well-rounded nematode. Bioessays 28: 651–659 [DOI] [PubMed] [Google Scholar]
- Ishihama Y, Rappsilber J, Mann M 2006. Modular stop and go extraction tips with stacked disks for parallel and multidimensional peptide fractionation in proteomics. J Proteome Res 5: 988–994 [DOI] [PubMed] [Google Scholar]
- Manning G, Plowman GD, Hunter T, Sudarsanam S 2002. Evolution of protein kinase signaling from yeast to man. Trends Biochem Sci 27: 514–520 [DOI] [PubMed] [Google Scholar]
- Merrihew GE, Davis C, Ewing B, Williams G, Kall L, Frewen BE, Noble WS, Green P, Thomas JH, MacCoss MJ 2008. Use of shotgun proteomics for the identification, confirmation, and correction of C. elegans gene annotations. Genome Res 18: 1660–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Aebersold R 2005. Interpretation of shotgun proteomic data: The protein inference problem. Mol Cell Proteomics 4: 1419–1440 [DOI] [PubMed] [Google Scholar]
- Olsen JV, de Godoy LM, Li G, Macek B, Mortensen P, Pesch R, Makarov A, Lange O, Horning S, Mann M 2005. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics 4: 2010–2021 [DOI] [PubMed] [Google Scholar]
- Roos FF, Jacob R, Grossmann J, Fischer B, Buhmann JM, Gruissem W, Baginsky S, Widmayer P 2007. PepSplice: Cache-efficient search algorithms for comprehensive identification of tandem mass spectra. Bioinformatics 23: 3016–3023 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386 [DOI] [PubMed] [Google Scholar]
- Schrimpf SP, Weiss M, Reiter L, Ahrens CH, Jovanovic M, Malmstrom J, Brunner E, Mohanty S, Lercher MJ, Hunziker PE, et al. 2009. Comparative functional analysis of the Caenorhabditis elegans and Drosophila melanogaster proteomes. PLoS Biol 7: e48 doi: 10.1371/journal.pbio.1000048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Hirst M, Bainbridge MN, Magrini V, Mardis E, Moerman DG, Marra MA, Baillie DL, Jones SJ 2008. Transcriptome analysis for Caenorhabditis elegans based on novel expressed sequence tags. BMC Biol 6: 30 doi: 10.1186/1741-7007-6-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater GS, Birney E 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6: 31 doi: 10.1186/1471-2105-6-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer RJ, Carta LK, Kim SY, Sternberg PW 1996. Morphological, genetic and molecular description of Pristionchus pacificus sp n (Nematoda: Neodiplogastridae). Fundam Appl Nematol 19: 511–521 [Google Scholar]
- Stein LD, Bao Z, Blasiar D, Blumenthal T, Brent MR, Chen N, Chinwalla A, Clarke L, Clee C, Coghlan A, et al. 2003. The genome sequence of Caenorhabditis briggsae: A platform for comparative genomics. PLoS Biol 1: e45 doi: 10.1371/journal.pbio.0000045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera JC, Wheat CW, Fescemyer HW, Frilander MJ, Crawford DL, Hanski I, Marden JH 2008. Rapid transcriptome characterization for a nonmodel organism using 454 pyrosequencing. Mol Ecol 17: 1636–1647 [DOI] [PubMed] [Google Scholar]
- White JR, Roberts M, Yorke JA, Pop M 2008. Figaro: A novel statistical method for vector sequence removal. Bioinformatics 24: 462–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia D, Sanderson SJ, Jones AR, Prieto JH, Yates JR, Bromley E, Tomley FM, Lal K, Sinden RE, Brunk BP, et al. 2008. The proteome of Toxoplasma gondii: Integration with the genome provides novel insights into gene expression and annotation. Genome Biol 9: R116 doi: 10.1186/gb-2008-9-7-r116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates JR III Eng JK, McCormack AL 1995. Mining genomes: Correlating tandem mass spectra of modified and unmodified peptides to sequences in nucleotide databases. Anal Chem 67: 3202–3210 [DOI] [PubMed] [Google Scholar]
- Zielinska DF, Gnad F, Jedrusik-Bode M, Wisniewski JR, Mann M 2009. Caenorhabditis elegans has a phosphoproteome atypical for metazoans that is enriched in developmental and sex determination proteins. J Proteome Res 8: 4039–4049 [DOI] [PubMed] [Google Scholar]