Abstract
The genetic code—the binding specificity of all transfer-RNAs—defines how protein primary structure is determined by DNA sequence. DNA also dictates when and where proteins are expressed, and this information is encoded in a pattern of specific sequence motifs that are recognized by transcription factors. However, the DNA-binding specificity is only known for a small fraction of the ∼1400 human transcription factors (TFs). We describe here a high-throughput method for analyzing transcription factor binding specificity that is based on systematic evolution of ligands by exponential enrichment (SELEX) and massively parallel sequencing. The method is optimized for analysis of large numbers of TFs in parallel through the use of affinity-tagged proteins, barcoded selection oligonucleotides, and multiplexed sequencing. Data are analyzed by a new bioinformatic platform that uses the hundreds of thousands of sequencing reads obtained to control the quality of the experiments and to generate binding motifs for the TFs. The described technology allows higher throughput and identification of much longer binding profiles than current microarray-based methods. In addition, as our method is based on proteins expressed in mammalian cells, it can also be used to characterize DNA-binding preferences of full-length proteins or proteins requiring post-translational modifications. We validate the method by determining binding specificities of 14 different classes of TFs and by confirming the specificities for NFATC1 and RFX3 using ChIP-seq. Our results reveal unexpected dimeric modes of binding for several factors that were thought to preferentially bind DNA as monomers.
Based on the presence of one or more of the 347 known DNA-binding domains (DBDs), it has been estimated that the human genome encodes ∼1400 potential sequence-specific transcription factors (TFs) (Vaquerizas et al. 2009). The binding specificities of these proteins are mostly unknown. The largest open-access resource for human transcription factor binding specificity models lists only 58 moderate to high quality models for humans and 93 models for all mammals (Bryne et al. 2008). By including recent profiles generated by protein-binding microarrays (Berger et al. 2008; Badis et al. 2009) and considering protein-level similarities in DBDs, these models can be extended to ∼300–400 human TFs. Lower resolution binding specificity information, such as knowledge of the strongest binding sequence (consensus sequence) or proven genomic target sites, exists for a somewhat larger proportion of TFs. However, because biologically important binding sites are often not of maximal affinity (see, for example, Jiang and Levine 1993; Tuupanen et al. 2009), this information is insufficient for most purposes, such as predicting the effect of disease-associated sequence polymorphisms on TF binding (Pomerantz et al. 2009; Tuupanen et al. 2009), and prediction of functional binding sites using bioinformatics tools such as an enhancer element locator (EEL) (Hallikas et al. 2006).
Although occupied sites for individual TFs can be accurately identified in cell lines and tissues using chromatin immunoprecipitation and the binding of TFs to genomic sequences in vivo is determined by DNA sequence (Wilson et al. 2008), we currently cannot effectively read genomic sequence to determine which sites control gene expression and/or will be occupied by a given TF. TF binding correlates with chromatin state (Robertson et al. 2008; Heintzman et al. 2009) and nucleosome occupancy (Badis et al. 2008), but comparing two types of experimental data essentially explains one set of observations with another and does not increase our understanding of the basic biochemical reactions that determine which transcription factor binding sites are occupied. To understand the biochemical basis of transcriptional regulation, we need to quantitatively measure binding affinities of transcription factors to DNA and to each other, and to use these data to develop a model of cooperative binding of TFs and transcriptional activation. The ultimate aim here is to read the genetic code of gene expression, that is, to understand the expression of genes based on DNA sequence.
Preparation of high-quality physical models of DNA–DBD interactions using existing methods is laborious and expensive. Economic and relatively simple in vitro methods, such as electrophoretic mobility shift assay (EMSA), nuclease footprinting (for review, see Lane et al. 1992; Hampshire et al. 2007), or systematic evolution of ligands by exponential enrichment (SELEX) (Kinzler and Vogelstein 1990; Tuerk and Gold 1990) using low-throughput sequencing are efficient in preparation of rough initial models, which can subsequently be refined to higher precision with competition assays, such as microwell-based binding assay (Hallikas and Taipale 2006) or competitive EMSA (Moss 2001). Currently, the only method that can produce high-quality de novo models in relatively high throughput is universal protein binding microarrays (PBMs) (Bulyk et al. 2001; Berger et al. 2006). Because the PBM method needs relatively high amounts of purified proteins, it is difficult to analyze proteins that need post-transcriptional modifications or proteins that do not express well, such as many full-length transcription factors. In addition, PBMs suffer from limitations common to microarrays, including high cost, position effects, and a limit to the number of sequences that can be placed on the array. Universal PBMs cannot practically accommodate all possible oligomers beyond 10 base pairs (bp)—therefore they cannot be used to determine preferred spacings and orientations between half-sites of dimeric TFs, or complete binding preferences for TFs that prefer longer than 10-mer DNA-motifs (e.g., RFX3) (Emery et al. 1996; Badis et al. 2009).
DNA-binding specificities can also be estimated using methods such as chromatin immunoprecipitation by sequencing (ChIP-seq) (Robertson et al. 2007). This method requires high-quality antibodies and a massive number of sequencing reads. In addition, as ChIP-seq measures occupancy of individual sites in particular cell lines or tissues, the data generated are influenced by other factors than protein–DNA-binding affinity, including protein–protein interactions between the TF analyzed and other TFs, accessibility of particular genomic sequences, and sequence biases in the genome itself.
In this work, we introduce a high-throughput protein–DNA binding specificity determination method that allows processing of hundreds of individual samples in parallel. The method requires low nanogram levels of proteins, and is thus compatible with mammalian expression systems, allowing analysis of full-length TFs and TFs that require post-transcriptional modifications. We also describe a computational pipeline designed to assess the quality of the results and to generate accurate binding profiles for the TFs analyzed. To validate the method, we use it here to generate binding specificity profiles for 19 TFs, representing 14 different structural classes.
Results
Expression of proteins and SELEX
To determine accurate DNA-binding specificities for human TFs, we cloned a set of genes representing major families of human DBDs and full-length transcription factors into a Gateway recombination cloning entry vector. We subsequently transferred the collections into a recipient vector that allows expression of the DBDs as N-terminal fusions to a streptavidin-binding–peptide-tagged luciferase enzyme from Gaussia princeps (Fig. 1A). The proteins were then expressed in transiently transfected primate cells (COS1 or 293T), and the amounts of expressed proteins were measured by a luciferase assay (data not shown). Subsequently, the different proteins were affinity-purified using streptavidin-coated 96-well plates.
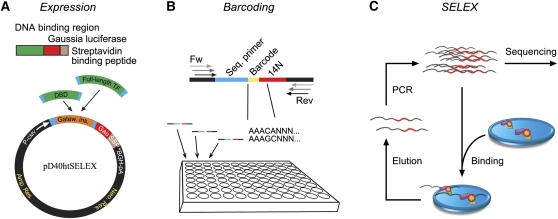
Figure 1.
Schematic description of the high-throughput SELEX process. (A) Protein expression. (Top) Proteins are expressed as fusion proteins with SBP-tagged Gaussia-luciferase. (Bottom) The GATEWAY recombination cloning system is used to transfer DNA sequences encoding DBDs or TFs from donor-vectors to the pD40htSELEX expression vector. (B) Ligand design that accommodates multiplexing of samples using barcodes. Each DNA ligand contains a 14-bp randomized region (14N), and a 5-bp barcode (Barcode) that uniquely identifies the individual SELEX sample. To increase specificity, each barcode differs from all other barcodes by at least 2 bp. These variable sequences are flanked by constant sequences that include an Illumina Genome Analyzer sequencing primer site (Seq. primer) and bridge amplification primer binding regions (Fw, Rev; arrows), which are extended in their 5′ regions to accommodate partially nested primers (used in successive SELEX rounds). (C) Basic principle of high-throughput SELEX. A double-stranded DNA mixture containing all possible 14-bp sequences (from B) is incubated with a DNA-binding protein immobilized into a well of a 96-well plate, resulting in binding of DNA to the protein. After washing and elution, the resulting population of more specific sequences is amplified by PCR and subjected to high-throughput single-molecule sequencing. The specificity of the TF can then be constructed by iterating the process and calculating the abundance of distinct sequences after different numbers of cycles. In each cycle, multiple reactions are mixed into a single sequencing lane, and the TFs are identified using the barcode sequences.
The DNA-binding specificity of the proteins was determined using a modification of the SELEX procedure (for review, see Roulet et al. 2002; Yang et al. 2007) adapted to parallel processing of the samples through all protein production, selection, and sequencing phases. The DBDs were allowed to bind to their preferred ligands from a pool of double-stranded DNA oligonucleotides containing all possible 14-nucleotide (nt) sequences flanked by a barcode sequence indicating the identity of the sample, and two common adapter/primer sites (Fig. 1B). After washing, the bound oligonucleotides were recovered and amplified and used as a new set of ligands in the subsequent selection cycles. The process was iterated up to five times to generate a set of products for sequencing (Fig. 1C). In such samples, it is expected that the relative amount of a sequence with specific affinity to the TF will increase in each cycle until sequences with higher affinity will start to effectively compete against it. Thus, diversity of the sequence pool decreases, and the average affinity increases in each cycle, until only the absolutely highest affinity site remains.
Multiplexed sequencing and initial analysis of data
The amplified DNAs from each selection cycle were quantified, mixed together, and sequenced using massively parallel sequencing. The average output of accepted reads per lane (see Methods) was 6.6 million (varying between 4.0 and 9.6 million). Using 256 oligonucleotides with different barcode sequences allowed the analysis of binding specificities of 256 different TFs in a single sequencing run. Before SELEX analysis, all barcoded oligonucleotides were sequenced to assess the quality of the 14-mer random sequences; The barcodes had an average of 37,590 reads, with 249 out of 256 (97%) having more than 5000 reads. Three out of the 256 ligand pools that had low sequence complexity (a high number of identical 14-mer sequences) and/or strong nucleotide bias were excluded at this stage.
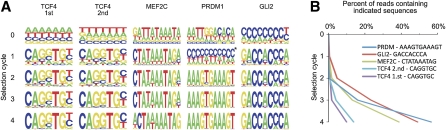
Approximately 23% of the sequences precipitated using the C2H2 zinc finger-domain TF GLI2 contained the GLI consensus sequence GACCACCCA (Kinzler and Vogelstein 1990; Hallikas et al. 2006) in either forward or reverse orientation after three rounds of SELEX (Fig. 2, 0.0046% expected by random). Analysis of other TFs that showed specific enrichment of sequences yielded similar results (Fig. 2), and very similar enrichment profiles were observed when the same TF was analyzed in two separate SELEX experiments (Fig. 2, TCF4).
Figure 2.
Enrichment of specific sequences during the SELEX process. (A) Position weight matrices built around the most enriched sequence for four different TFs (see Methods for details). The height of the letter at each position is directly proportional to the incidence of the indicated base in sequences where all other bases exactly match the most enriched sequence. Note that clear enrichment of sequences is observed after one or two SELEX rounds, and that two separate experiments for TCF4 result in a very similar enrichment pattern. In the first cycle, the algorithm used here detects incorrect binding profile for PRDM1 (asterisk) due to a low number of the relatively long consensus sequences. The enrichment of high-affinity sequences can, however, be detected by seeding the algorithm with consensus from the later cycles (see Supplemental Fig. S4A). (B) The fraction of all fragments containing the most enriched sequence from the third SELEX cycle plotted as a function of the SELEX cycle.
Bioinformatic quality control
Members from different TF families were selected for further analyses based on the following criteria: (1) robust enrichment of sequences that are related to each other without excessive loss of sequence complexity; (2) no binding to constant regions; and (3) exponential enrichment of specific sequences without detectable contamination from adjacent wells. The selection was performed using a computational pipeline “Inimotif” that allowed analysis and visualization of key aspects of the data (Fig. 3).
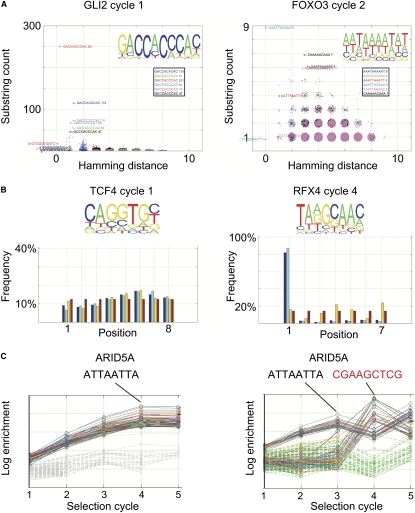
Figure 3.
Description of the bioinformatic visualization and quality-control pipeline. (A) Hamming distance plot. Incidences of all possible subsequences of length 10 (substring count) are plotted as a function of their Hamming distance (number of substitutions) from the most enriched sequence or its reverse complement. To facilitate visualization, random floating point values between −0.3 and 0.3 are added to all plotted x and y values. Note that in a successful experiment (left, GLI2), clear enrichment of sequences is observed, and many enriched sequences are found at a short Hamming distance (1 to 2). In a failed experiment (right, FOXO3), enrichment is very weak, and the enriched sequences are not clearly related to each other. (Insets) Position weight matrices from the same experiments. (B) Position plot. (Bottom) Fractional incidence (frequency) of subsequences of indicated length at each position in both strands (blue), the forward (direction indicated in top; light blue), and reverse (yellow) strand of the 14-bp random sequence. Numbers are separately normalized for each set of bars to add up to 100%, and uniform distribution (red) is shown as control. (Left) Note that in cases where flanking sequences do not interfere with binding, a very uniform distribution of sequences is observed. (Right) In cases where a part of the binding sequence for a TF is found in the constant region or barcode, a strong positional bias is observed. (C) Enrichment plot. Enrichment of a sample of the most enriched sequences and random sequences are plotted as a function of the SELEX cycle. (Left) Note that the enriched sequences show exponential enrichment (log scale), whereas the random sequences are not appreciably enriched. (Right) In cases of barcode contamination (see text), different sequences can appear to enrich in different SELEX cycles ([black sequence] correct; [red sequence] contaminating sequence). The data in C are from SELEX analysis using purified ARID5A protein-coated plates (see Methods).
For a functioning SELEX, it is expected that the most enriched sequences are related to each other. To visualize whether this is the case, we first determined the incidence of all subsequences of length 5–11. We then identified for each length the most enriched subsequence. Subsequently, we plotted the incidence of all subsequences observed as a function of the number of substitutions required (Hamming distance) to convert them to the most enriched subsequence or its reverse complement. In successful cases, the reverse complement of the most enriched sequence was also strongly enriched, together with some sequences having single-base mismatches to these sequences (Fig. 3A, left panel). In failed cases, the most enriched sequence was present in very low numbers, and the other enriched sequences were often unrelated to it (Fig. 3A, right panel).
We next assessed the contribution of the constant sequences to TF binding. If no binding to the flanking constant and/or barcode sequences is observed, the most enriched sequence should be distributed relatively evenly across all possible positions on the forward and reverse strand of the 14-bp random sequence (Fig. 3B, left panel). If, however, the factor binds to either the barcode or flanking constant sequences, a clear preference is observed in the position of the enriched sequences (Fig. 3B, right panel). In the case shown, the protein RFX4 appears to strongly prefer one position, as binding at this position allows the barcode “GTTGC” of the selection oligomer to form part of the RFX4 recognition sequence. To correct for these types of error, the experiment can be repeated with oligomers designed with different flanking regions.
Finally, to control for the quality of individual SELEX cycles, we analyzed the incidence of 100 most enriched sequences and 200 random sequences in all cycles. In functional cases, the high-affinity sequences are enriched exponentially (Fig. 3C, left panel). If a SELEX round has failed, no enrichment is observed for that round (data not shown). If DNA from one well has contaminated an adjacent well containing a different TF and DNAs from both wells are subsequently mixed to the same sequencing reaction, different sequences can appear to enrich during different SELEX rounds (Fig. 3C, right panel). This type of error can be subsequently identified and corrected by pooling the sequenced samples to smaller subpools that do not contain sequences from adjacent wells prior to sequencing (e.g., Fig. 3C, cf. left and right panels).
To assess the overall efficiency of the protein expression and SELEX methods, we analyzed DBDs for 17 of a total of 27 ETS family members, all of which are known to bind to DNA in a sequence-specific manner. Five factors (29%) expressed at low levels. Of the 12 that expressed highly, nine (75%) showed clear enrichment of specific sequences after three to five cycles in at least one experiment (see Supplemental Table S4).
Generation of binding models
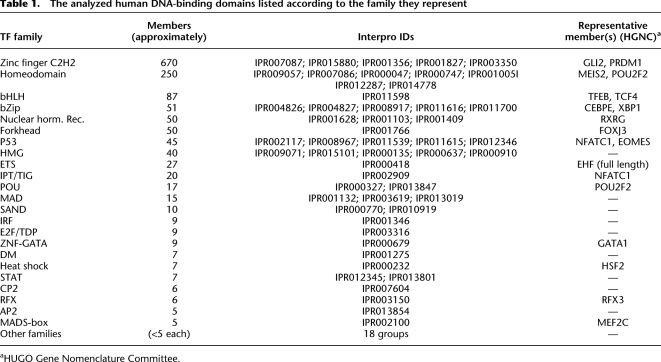
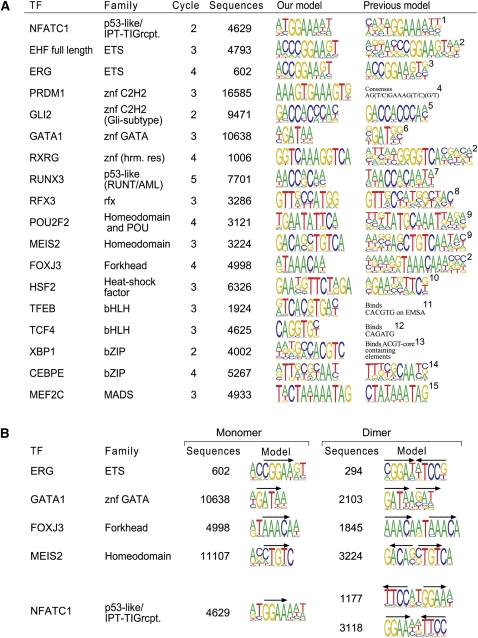
Using the tools described above, we could identify a successful SELEX enrichment for at least one member for 14 of the 23 major DBD classes (Table 1; Supplemental Table S2; Supplemental Archive S1). We next identified the most enriched subsequence for all lengths between 4 and 13 for all the factors after all of the SELEX cycles. Subsequently, we generated 5–12 base-long position weight matrices for all samples by counting the number of occurrences of all subsequences that differed from the most enriched subsequences by 1 base. This analysis was done for two consecutive cycles, with the first of the cycles used as background to correct for nonspecific carryover of DNA from the starting material (for details, see Methods). The following criteria were then used to select the optimal matrix for each protein: To limit statistical error, profiles were selected that were of minimum length allowing incorporation of all highly specific positions and that were derived from at least 500 but preferably more than 3000 subsequences. To minimize the distortion caused by the exponential enrichment of sequences, such profiles were generally selected from the earliest possible SELEX cycle. The resulting binding models were based on between 602 and 16,585 sequences. In cases where the same factor appeared to bind in monomeric and dimeric configurations, optimal lengths were selected for all such binding modes.
Table 1.
The analyzed human DNA-binding domains listed according to the family they represent
aHUGO Gene Nomenclature Committee.
Validation of results
To validate the SELEX binding method, we analyzed whether our method gives results that are similar to those obtained using PBMs, using the same protein purified from Escherichia coli that was used in an earlier PBM experiment (mouse EOMES DBD fused to GST). The profile generated for EOMES using our method was very similar to the PBM-derived profile (Supplemental Fig. S1).
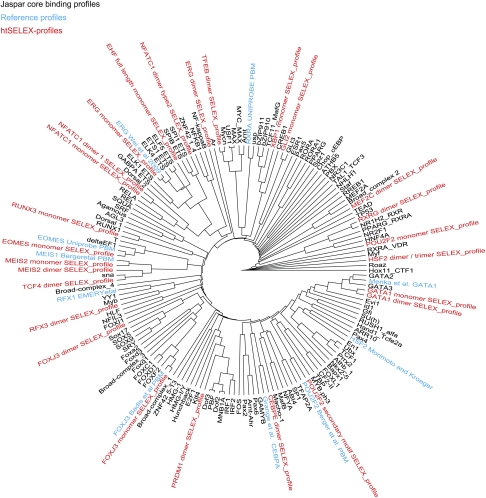
To further evaluate the quality of the obtained matrices, we first analyzed them by comparing them computationally with existing profiles from the Jaspar2 database (Bryne et al. 2008) and from the literature (Fisher et al. 1991; Grange et al. 1991; Pollock and Treisman 1991; Verrijzer et al. 1992; Mader et al. 1993; Merika and Orkin 1993; Meyers et al. 1993; Kroeger and Morimoto 1994; Clauss et al. 1996; Emery et al. 1996; Kel et al. 1999; Hallikas et al. 2006; Berger et al. 2008; Badis et al. 2009; Lord et al. 2009). For the comparison, we used the method based on minimal Kullback–Leibler divergence described in Wei et al. (2010). Most of the binding profiles clustered near the existing profiles for the same or related factors where such profiles were available (Fig. 4). Consistent with the presence of one or two factors from most DBD families in our analysis, the binding profiles distributed relatively evenly across the binding specificity space (Fig. 4).
Figure 4.
Distance dendogram based on the minimum Kullback-Leibler divergences between TF position weight matrices from the Jaspar database and reference matrices of Figure5. Note that the binding profiles generated using the SELEX method are in general similar to existing matrices for the same or related factors in cases where they are available. Note also that the profiles generated using SELEX for 14 of the 23 major DNA-binding domain families occupy most major branches of TF binding specificities, highlighting the broad utility of the method.
More detailed analysis of the generated profiles revealed that they were generally in very good agreement with existing position weight matrices where such data were available for the same protein or for a related protein from human or other species (Fig. 5A). In many cases, the same factor was found to bind both in monomeric and dimeric configurations. If a factor had a dimeric mode(s) of binding, strongly preferred orientations and spacings between sequences that resembled the corresponding monomeric sequences were observed for each factor (Fig. 5B).
Figure 5.
Binding profiles. (A) Comparison of determined binding-specificity models with previously known data. The left columns indicate the transcription factor analyzed and its DNA-binding domain family. The SELEX cycle from where the model is derived and the number of independent sequences included in it are also indicated. The previous model for the same protein or for the closest related ortholog (o) or paralog (p) is shown, including reference. These are RUNX1 for RUNX3, mouse RXRA for RXRG, RFX1 for RFX3, mouse MEIS1 for MEIS2, mouse EHF for EHF, mouse POU2F2 for POU2F2, mouse MEIS1 for MEIS2, CEBPA for CEBPE, and MEF2A for MEF2C. References: (1) Kel et al. (1999); (2) Badis et al. (2009); (3) Wei et al. (2010); (4) Lord et al. (2009); (5) Hallikas et al. (2006); (6) Merika and Orkin (1993); (7) Meyers et al. (1993); (8) Emery et al. (1996); (9) Berger et al. (2008); (10) Kroeger and Morimoto (1994); (11) Fisher et al. (1991); (12) Pscherer et al. (1996); (13) Clauss et al. (1996); (14) Grange et al. (1991); (15) Pollock and Treisman (1991). (B) Monomeric and dimeric binding modes. Factors that can bind DNA either as monomers or as dimers are shown; arrows indicate orientations of the monomeric sites. Two dimeric motifs, dimer1 (top) and dimer2 (bottom), were found for NFATC1; a profile similar to the dimer2 has been previously reported by Falvo et al. (2008) for the paralog NFATC2.
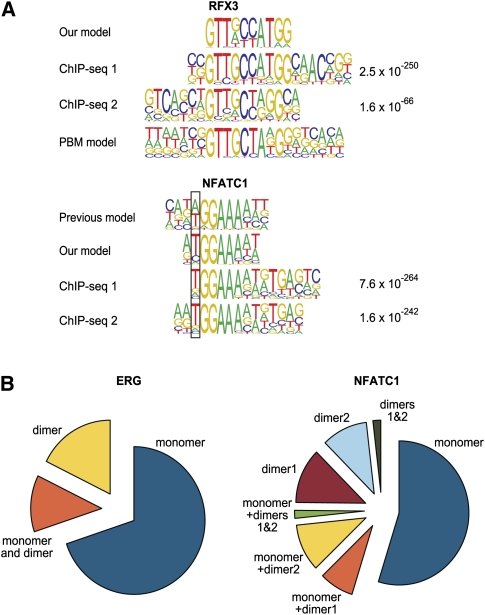
To analyze whether the binding profiles obtained were relevant for the in vivo situation, we performed chromatin immunoprecipitation by sequencing (ChIP-seq) experiments for RFX3 and NFATC1 in K562 and Jurkat cells, respectively. In both cases, identification of the enriched sequence motifs from the ChIP-seq peaks using the MEME algorithm (Bailey and Elkan 1995) revealed a profile that was very similar to that obtained using SELEX (Fig. 6A). Furthermore, both dimeric NFATC1 sites were also enriched in the ChIP-seq peaks. Similarly, the dimeric ERG profile we identify here was enriched in ERG ChIP-seq peaks identified by Wei et al. (2010) (Fig. 6B). The ERG dimer profile and both NFATC1 dimer profiles were also enriched in peaks that did not contain matches for the respective monomer sites (in all cases, the P-value was lower than 2.2E-16, the limit imposed by computational precision for binomial distribution).
Figure 6.
Validation of binding models by ChIP-seq. (A) Comparison of in vitro derived binding models for RFX3 and NFATC1 and previously described models (Kel et al. 1999; Badis et al. 2009) with motifs that are enriched in peaks from a ChIP-seq experiment for the same factors. The MEME algorithm was used to identify enriched motifs in the ChIP-seq peaks. For both factors, two different antibodies were used in the experiments shown; the expectation value for the motifs is indicated on the right. (Top) Note that both our model and the model of Badis et al. (2009) (PBM model) are supported by ChIP-seq. For NFATC1, the position where our model matches the ChIP-seq-derived model better than the previous model is boxed. Note also that the ChIP-seq-enriched motif for NFATC1 (bottom) appears to contain also signal (g/a)TGA(g/c) that is located right of the NFATC1 monomer profile tGGAAAa(t/a). This signal is likely derived from a dimerization partner, as it is well known that NFAT proteins dimerize with many other TFs (Macian 2005). (B) Relative fractions of peaks containing monomer and dimer sites and combinations thereof for NFATC1 and ERG (ChIP-seq data from Wei et al. 2010). For all matrices, the cut-off score was set to yield 1 site per 10 kb of human genome. Note that a significant fraction of peaks contain motifs that match the dimer model but not the monomer model. The total fraction of peaks with sites was 24% and 14% for NFATC1 and ERG, respectively.
In all cases, the binding sites were preferentially located near the summits of the ChIP-seq peaks (Supplemental Fig. S6). These results suggest that the obtained profiles and the identified dimeric binding modes are biologically relevant.
Discussion
High-throughput SELEX method
We report here a SELEX-based method that allows high-throughput analysis of transcription factor binding specificity. The method utilizes massively parallel single-molecule sequencing technology, which eliminates all cloning steps and results in generation of a very large number of individual sequencing reads. The number of samples that can be analyzed in parallel is increased by the use of selection oligonucleotides containing barcoded flanking sequences and constant regions that contain binding sites for bridge-amplification and sequencing primers. The selected fragments can thus be directly sequenced without a ligation or template-switching step, decreasing the risk of sequence bias and DNA contamination. The design of the selection oligomer is similar to that recently described by Zykovich et al. (2009).
These changes in aggregate result in both dramatically increased sequence yield and throughput over previously described SELEX methods. The method was used here to study DNA-binding proteins, but can easily be adapted also to the analysis of protein–RNA binding and for generation of oligonucleotide-based affinity reagents (aptamers).
We also miniaturized the protein production and purification steps, making the method compatible with mammalian expression systems. The method can thus be applied to the analysis of full-length transcription factors (Fig. 5), and also proteins that require native expression, dimerization partners, and/or post-transcriptional modifications for activity.
Compared to earlier SELEX-based TF DNA-binding profiles, the profiles we describe here are based on 100–1000-fold higher numbers of sequences. For each of the factors we describe, the number of sequences is on the same order of magnitude as that attainable using SELEX-SAGE (Roulet et al. 2002), a much more costly and labor-intensive protocol. The large number of sequences also improves effectiveness of quality control (Fig. 3) and decreases statistical error in the profiles.
High number of reads allows effective quality control of results
SELEX can produce errors by multiple mechanisms. First, having too few molecules in the reactions can result in bottleneck effects, which reduce the complexity of the selectable oligonucleotide pool and result in amplification of erroneous sequences. Second, PCR amplification can introduce bias to the pool of sequences analyzed. Third, constant linker sequences included in the ligand DNA can contain regions with high affinity to the DNA-binding protein inducing either a total failure of the selection process, or positional biases into the location of the DNA-binding elements. To correct for these error sources, we developed a computational pipeline that allows quality control of the data and identification of common problems, including lack of enrichment, binding of TFs to constant sequences, cross-contamination of samples, and failure of individual SELEX rounds. All of these quality-control steps critically depend on the fact that enough data can be generated to assess the enrichment of a large number of sequences in each cycle.
Our method also allows generation of profiles from early SELEX cycles, decreasing the distortion caused by the exponential enrichment of the ligands. Furthermore, the large amount of data allows analysis of composite/dimeric sites (Fig. 5B) and pairwise correlation between sequence positions (data not shown).
A particular feature of SELEX is the exponential enrichment of high-affinity binding sequences. Each round enriches sequences in a manner related to their affinity toward the ligands, that is, if all DNA binding is specific and each sequence is initially present at equal concentration and in large excess compared to the TF, a sequence that has 10-fold lower affinity to a TF than the sequence with highest affinity will be present at 10-fold lower concentration than the highest affinity sequence after the first cycle, and 100-fold lower concentration after the second cycle. In practice, when random sequence libraries are used, the high-affinity sites will be approaching saturation by the TF, resulting in lower than expected discrimination between medium- and high-affinity sites. Thus, although we consistently observe exponential enrichment of bound sequences, the enrichment ratios do not exactly follow biochemical affinity—the enrichment for lower-affinity sequences is consistently higher in each cycle than what is expected from their affinity relative to the maximum affinity sequence. Thus, in effect, the exponential enrichment and saturation counteract each other, and the models generated using our multinomial model (see Methods) after two to four SELEX cycles are much closer to biochemical affinity than what is expected from a purely relative-affinity-based exponential enrichment. For example, the models for ERG and GLI2 that are closest to biochemical affinity measured by a competition-ELISA-like approach (Hallikas et al. 2006; Wei et al. 2010) are from selection cycle 3 (Supplemental Fig. S3).
The downside of the saturation of the sites is that although the model correctly describes the rank of affinities of the included sequences and gives a surprisingly good estimate of affinity after two to four cycles, the derivation of exact biochemical affinity from the sequencing data is not trivial. Two methods have recently been developed to correct for the saturation effect, QPMEME (Djordjevic et al. 2003) and BEEML (Zhao et al. 2009). Of these, we were able to analyze our cycle 1 GLI2 data with BEEML, which resulted in a very similar profile to that we obtained using our data analysis method (Supplemental Fig. S4). Theoretical considerations suggest that a method such as QPMEME that generates binding models from enrichment ratios of individual sequences rather than from the absolute number of occurrences of the sequences should be able to generate a model that more accurately represents biochemical affinity. Unfortunately, QPMEME could not be used to analyze our data as it requires an internal control for each factor and cannot account for nonspecific DNA carryover. In addition to dealing with carryover and saturation, future method development in such enrichment ratio-based models should also concentrate on correcting for sequencing errors. Although the ∼0.5% error rate due to PCR and sequencing does not materially affect absolute count-based models such as ours, such errors badly confound purely ratio-based approaches. This is because sequencing errors cause sequences that have no affinity and high affinity to enrich at the same rate if they are within 1 bp of each other (absolute numbers are more than 100 times different, but the rate of enrichment is the same).
Comparison with existing methods
Compared to the previous high-throughput microwell-based method that we developed (Hallikas et al. 2006), the method described here allows a larger number of parallel samples (only a maximum of five 96-well plate wells are needed per TF), and can reveal complex binding specificities without previous knowledge of a high-affinity site. Compared to existing state-of-the-art methods that can identify binding profiles without prior information—SELEX-SAGE (Roulet et al. 2002) and universal protein-binding microarrays (Berger et al. 2006)—our method has 100–1000-fold higher throughput. The application of massively parallel sequencing eliminates complicated and expensive cloning steps and makes the method easy to set up in any modern laboratory. In addition, compared to PBMs, our method requires much less protein and is thus more compatible with mammalian expression. In addition, it can also identify >10-bp-long binding sequences, which are commonly observed when TFs bind in dimeric or multimeric configurations. In theory, even in the miniaturized format used here, all possible 14-bp-long sequences are expected to be present more than 3000 times in the reactions at the start of cycle 1. Thus, with real TFs that do not have absolute specificity for all bases, we expect that a similar method using a somewhat longer random region could be used to identify binding preferences that are at least 20 bp long.
Transcription factor binding models
The utility of the SELEX method and the associated informatics pipeline was analyzed by generation of binding profiles for 14 different transcription factor DNA-binding domain subfamilies. In cases where detailed binding information was available, our data were in broad agreement with the existing data. However, in many cases, our data revealed much more information about the binding preferences of the analyzed TFs than what was previously available.
Out of the four cases where to our knowledge, only consensus binding data or relatively limited numbers of bound sequences were publicly available, our data were consistent with the earlier results for three of the factors—TFEB (Fisher et al. 1991), XBP1 (Clauss et al. 1996), and PRDM1 (Lord et al. 2009). In the case of TCF4, our optimal binding sequence CAGGTG(C/T) differed by one position from the previously reported bound sequence CAGATGT identified from the somatostatin receptor II gene by Pscherer et al. (1996). However, significant enrichment was also observed for the CAGATGT sequence (Fig. 5A; Supplemental Fig. S2a), suggesting that CAGATGT does bind TCF4 albeit at a lower affinity than the CAGGTG(C/T) motif. Taken together, where existing profiles were derived from a limited number of sequences, our data significantly improved on the existing binding models.
In cases where the existing profiles were derived from more sequences or were generated using protein-binding microarrays (Berger et al. 2008; Badis et al. 2009), our profiles were in good agreement with the existing data. However, some notable differences were seen, including POU2F2 and RFX3. In the case of the POU2F2 protein, whose DBD is composed of two subdomains—a POU-homeodomain and a POU-specific domain—the sequences that enrich most efficiently in our SELEX are consistent with the earlier described consensus gAATAT(g/t)CA (Verrijzer et al. 1992) for the POU-specific DBD, whereas the motif found using PBM is the classical TATGCAAAT motif, which is thought to be a composite of the POU-specific and homeodomain DBDs (Verrijzer et al. 1992). This classical motif is also enriching during the SELEX rounds, but not as efficiently (Supplemental Fig. S2b). These differences could be due to the presence of two binding modes for POU2F2, of which the composite mode likely displays slower dissociation kinetics (see Verrijzer et al. 1992), and could thus be preferentially identified using PBMs due to the long washes used in that protocol.
In the case of RFX3, the most enriched sequences in our data are closer to the model described by Emery et al. (1996) than the profiles derived using PBMs. The most enriched sequences are consistent with a model of dimeric binding, wherein the ideal substrate for the RFX3 protein is a 14-mer palindromic sequence that is composed of two GTT(G/A)CC sequences in head-to-tail orientation with an “AT” spacer. In contrast, all PBM-derived profiles are clearly different from the earlier data or the profile generated from our most enriched sequences. However, we do observe weaker enrichment of a 10-base sequence that corresponds to the PBM-derived primary consensus sequence, GTTGCTANGG (Supplemental Fig. S2c). Both of these models are supported by our ChIP-seq experiments as well (Fig. 6A). Thus, our results are consistent with the presence of multiple alternative DNA-recognition modes for RFX family of proteins as suggested by Badis et al. (2009). However, the preferred mode of binding appears to be the dimeric mode reported by Emery et al. (1996) that apparently cannot be identified using PBMs that are optimally designed to identify short binding sites (10 bp or shorter).
Dimeric modes of binding
In addition to analysis of monomeric binding, the large amount of sequence data generated allows analysis of cases where a protein binding to DNA as a homodimer can accommodate multiple different orientations and/or spacings of the monomers. This is illustrated in the case of Retinoid X receptor (RXR) proteins, which are known to form hetero- and homodimeric complexes with themselves and other members of the nuclear hormone receptor protein family, and to prefer these kinds of interactions over monomeric binding. In our model, RXRG shows a highly similar, apparently homodimeric binding pattern to that first described by Mader et al. (1993), enriching head-to-tail repeats of two GGTCA-motifs with an almost invariable 2-bp “AA”-spacer. Thus, while our prediction appears to have large differences to the PBM-predicted model for the RXRG homolog RXRA (Badis et al. 2009), the difference is probably the result of the two methods identifying the dimeric and monomeric sites, respectively.
Dimeric modes of binding were also identified for many factors that also bound to DNA as monomers (Fig. 5B). In all cases, we observed a preferential spacing and orientation of the “half-sites” of the dimer, and in some cases (e.g., MEIS2, ERG) the half-sites appeared to be of relatively low affinity. Whereas it is theoretically possible that the observed multimeric binding is due to the multivalency of streptavidin, and the spacing and orientation preferences are caused by steric effects, this is very unlikely, as the DBDs are connected to the streptavidin via two flexible linkers and a globular protein (Gaussia luciferase). In addition, members of the same family of TFs showed different types of dimeric interactions (e.g., EHF shows no dimer and ERG has a strong dimeric component) (Fig. 5B; data not shown). Furthermore, dimeric sites identified for ERG and NFATC1 were also found to be enriched within in vivo occupied sites identified for the same factors using ChIP-seq (Fig. 6B), suggesting that the dimeric profiles identified are, indeed, biologically relevant.
We have shown previously that in vitro generated binding profiles such as those described here can be used together with computational models to identify target genes of human TFs (Hallikas et al. 2006), and polymorphisms that affect TF binding and disease predisposition (Tuupanen et al. 2009). However, in cases in which biologically relevant cell or tissue models exist, direct measurements such as ChIP-seq and RNAi followed by expression profiling are generally more efficient at identifying TF sites and target genes, respectively. The in vitro binding profiles and computational models are thus best used for generating hypotheses on which factors may bind to a region of interest identified by genetics, and for global identification of TF targets in all tissues. In addition, they are required for systems biology models of regulatory element activity.
ChIP analyses have revealed that many regions occupied in vivo by a given TF contain only relatively weak affinity sites for the same TF, suggesting that cooperative reactions play a critical role in determining which genomic sites are occupied by TFs. Understanding the “second genetic code” that explains how DNA sequence controls gene expression thus requires both determination of binding specificities of transcription factors and identification of their preferred orientations and spacings with regard to each other. The method developed here has the potential to greatly improve the number and quality of DNA-binding profiles, and also to reveal preferential orientations and spacings between TFs. Incorporation of such information into a model of transcription would ultimately allow movement beyond observations (i.e., ChIP, expression profiling) and toward understanding of transcriptional regulation based on biochemical principles.
Methods
DNA binding domain assignment
For each protein sequence (obtained from the IPI database) (Apweiler et al. 2001) encoding a human transcription factor (Vaquerizas et al. 2009), we mapped all matching DBD-containing Interpro entries. As each Interpro entry contains sequence models from multiple sources (e.g., Pfam, superfamily, etc.), we defined the DBD from the most N-terminal to the most C-terminal amino acid. Some Interpro entries have parent–child relationships when two or more different entries model the same DBD but with different levels of specificity. For example, IPR007087 and IPRO007086 have a parent–child relationship, where both are zinc finger C2H2-type DBDs; but the parent, IPR007087, models a sequence of about 28 amino acids containing both the C2 and the H2 ends, whereas the child, IPR007086, models a subset of that sequence containing either the C2 end (around 14 amino acids) or the H2 end (around 10 amino acids). These parent–child entries were treated independently. We also defined tags for the N- and C-terminal ends of DBDs beginning with a 4-amino-acid sequence at either end and extending them toward each other until they are unique within the protein sequence for each TF. If a TF had multiple DBDs, all of them and sequences between them were included in the DBD clones.
Cloning
Amplification of the TF- and DBD-encoding sequences was performed using a two-step PCR-procedure; in the first phase, DBD-specific primers that contained the following 5′ linker sequences coding for partial AttB sites were used: Forward, AAAAAAGTTGGCATG; Reverse, AGAAAGTTGGGTA. The secondary reactions used a generic primer pair (GGGGACAACTTTGTACAAAAAAGTTGGC and GGGGACAACTTTGTACAAGAAAGTTGGG) to complete the AttB recombination regions. A Megaman cDNA library (Stratagene) and the mammalian gene collection (Strausberg et al. 2002) clones where available were used as templates. The linker sequences used here were the modified versions described by Rual et al. (2004). The amplified products were recombined directly into Gateway pDONR223-vector, and all clones were confirmed by sequencing (amino acid sequences in Supplemental Table S1).
Expression vector construction
Our expression vector design combines small and highly efficient Gaussia luciferase to a streptavidin-binding peptide (SBP) tag, which allows effective and economic fusion protein capture and quantification. Luciferase and SBP-coding regions were PCR-amplified separately from pGLuc-Basic (NEB) and pCeMM-CTAP(SG)-GW (a kind gift from Giulio Superti-Furga) (Burckstummer et al. 2006), respectively, using the following forward and reverse primers: attaatactagtatgggagtcaaagttctgtttgcc and ctcgtccatgtcaccaccggcccccttg, ggtggtgacatggacgagaagaccaccg and attaatgtttaaacttaatcgataggctcgcgttgcccctg. The fragments were concatenated using fusion-PCR, after which the insert was cloned into XbaI and PmeI sites of the Gateway recipient-vector pDEST40 (Invitrogen). The final recipient vector pDEST40_Gau-SBP was generated by shifting the frame between the Gateway-cloned inserts and the Gaussia-SBP coding sequence using QuickChange mutagenesis (Stratagene) with the primers aactttgactcccattcgagcaaccactttgtacaa and tacaaagtggttgctcgaatgggagtcaaagttctg.
Cell culture, expression and purification of the fusion proteins
Jurkat and K562 cells were grown in RPMI1640, supplemented with penicillin/streptomycin and fetal bovine serum (FBS, 10%). COS1 and 293T cells were grown in DMEM supplemented with penicillin/streptomycin and 10% FBS. Transfection of 70%–80% confluent cells growth in 6-well plates was performed using FugeneHD (Roche) according to the manufacturer's instructions or by using polyethyleneimine (PEI, 25 kDa average molecular mass; Sigma cat. nr. 408727).
For PEI transfection, 2 μL of 0.45% (w/v) PEI in distilled water was diluted to 50 μL of DMEM, and incubated for ∼5 min. Subsequently, 3 μg of plasmid DNA dissolved in 50 μL of 150 mM NaCl was added, and the incubation continued for 15 min, after which the mixture was added to cells in full medium. Cells were grown for 48 h with one medium exchange 6–12 h after transfection.
Cells were then washed three times with PBS at room temperature and lysed by addition of 200 μL of ice-cold lysis buffer (50 mM Tris-Cl at pH 7.4 containing 150 mM NaCl, 1% Triton X-100, and EDTA-free protease inhibitor cocktail [Roche 04693159001; according to the manufacturer's instructions]) followed by incubation for 30 min on ice with gentle rotation. Lysates were cleared by centrifugation (5 min at 3600g) to remove debris and chromatin/genomic DNA, and aliquoted into 50-μL proportions that were either used directly in the binding assay or stored at −75°C. Protein amounts in lysates were measured by a luciferase assay (Promega Renilla Luciferase Assay System E2820). Briefly, 1 μL of lysate was added to 20 μL of Promega lysis buffer, after which 20 μL of substrate buffer was added, and flash-luminescence was measured with the Perkin-Elmer TopCount luminometer (40,000,000 cps corresponds approximately to a 1-ng amount of average-sized fusion protein [70 kDa]).
Before binding of fusion proteins into streptavidin binding plates (Thermo Fisher; AB-1226/W), the NaCl concentration of the lysates was raised to 1 M to inhibit interactions between DBDs and residual genomic DNA from the lysates. The plates were first washed twice with 300 μL of lysis buffer, after which the fusion-protein-containing lysates were added to the wells and incubated for 30 min on ice. The wells were then washed three times with 300 μL of lysis buffer. Wells were then blocked by 10 min of incubation with 0.5% BSA (w/v) in binding buffer (20 mM HEPES-Cl at pH 7 containing 140 mM KCl, 5 mM NaCl, 1 mM K2HPO4, 2 mM MgSO4, 100 μM EGTA, and 1 μM ZnSO4). Based on the estimation of luciferase counts from duplicate wells, the amount of actual affinity-immobilized and purified protein was estimated to be between 1 and 10 ng (data not shown).
For analysis of mouse ARID5A and EOMES, E. coli expression and protein purification was performed as described in Badis et al. (2009). About 100 ng of protein in 50 μL of PBS was used to coat Nunc Maxisorp plates for a minimum of 16 h at 4°C, and the plates were subsequently washed with PBS and blocked as described above. Plates directly coated with purified proteins and streptavidin-coated plates containing fusion proteins were then used in SELEX as described below.
SELEX and massively parallel sequencing
Sequence of the DNA ligand is described in Supplemental Table S3. The ligands contain all the sequence features necessary for direct sequencing using an Illumina Genome Analyzer. The ligands were synthesized from two single-stranded primers (Supplemental Table S3) using Taq polymerase. The 256 barcodes used consist of all possible 4-bp identifier sequences and a 1-bp “checksum” nucleotide, which allows identification of most mutated sequences. The products bearing different barcodes can be mixed and later identified based on the unique sequence barcodes.
For SELEX, 50–100 ng of barcoded DNA fragments was added to the TF or DBD-containing wells in 50 μL of binding buffer containing 150–500 ng of poly(dI/dC)-oligonucleotide (Amersham 27-7875-01[discontinued] or Sigma P4929-25UN) competitor. The resulting molar protein-to-DNA and protein-to-binding site ratios are on the order of 1:25 and 1:15,000, respectively. The plate was sealed and mixtures were left to compete for 2 h in gentle shaking at room temperature. Unbound oligomers were cleared away from the plates by five rapid washes with 100–300 μL of ice-cold binding buffer. After the last washing step, the residual moisture was cleared by centrifuging the plate inverted on top of paper towels at 500g for 30 sec. The bound DNA was eluted into 50 μL of TE buffer (10 mM Tris-Cl at pH 8.0 containing 1 mM EDTA) by heating for 25 min to 85°C, and the TE buffer was aspirated directly from the hot plate into a fresh 96-well storage plate.
The efficiency of the SELEX was initially evaluated by real-time quantitative PCR (qPCR) on a Roche light cycler using the SYBR-green-based system and calculating the differences in eluted oligomer amount by crossing-point analysis. Seven microliters of eluate was amplified using PCR (19–25 cycles), and the products were used in subsequent cycles of SELEX. Nesting primers (Supplemental Table S3) moving at least 2 bp inward in each cycle were used to prevent amplification of contaminating products. For sequencing, approximately similar amounts of DNA from each sample were mixed to generate a multiplexed sample for sequencing.
Sequencing sample preparation and sequencing
PCR-product concentrations were approximated by visual comparison of the products and DNA marker run on EtBr-stained samples on 2% agarose gels. Oligonucleotides were pooled from samples in roughly equivalent amounts, and the pool was either purified directly (QIAGEN min-elute PCR-purification kit) or, if EtBr gels showed other bands than the expected 105-bp product, by gel-extraction of the 105-bp band (QIAGEN min-elute gel-extraction kit). DNA concentration was determined (Nanodrop-spectrophotometer) and a 3 pM amount was sequenced (Illumina Genome Analyzer mk.2) with a SELEX-sequencing-primer that is identical to the standard Illumina Genomic DNA sequencing primer except that it lacks the 3′ terminal thymine base (Supplemental Table S3).
Data analysis
The sequences containing different barcodes were separated from each other using a Perl script. Only sequences containing (1) no bases annotated as N and (2) a valid barcode assessed by the presence of a correct 1-bp checksum nucleotide, and (3) a 5-bp exact match to the common sequence after the 14-bp random region were analyzed further.
These sequences (Supplemental Archive S1) were fed into the initial quality control program IniMotif, which takes the DNA-read-containing files as its input, finds the most enriched 5–11-bp-long sequence substrings, and uses these as consensus sequences. It then counts the occurrence of all subsequences of a given length and plots them into a Hamming distance plot (see Fig. 3A). The Hamming distance (the number of positions for which the corresponding bases are different) for each sequence is calculated from both the consensus sequence and its reverse complement, and the lower value is used. Inimotif also generates a position plot (Fig. 3B) that describes the distribution of subsequences within the 14-bp random sequence, using all sequences within a Hamming distance of 2 from the consensus or its reverse complement. An enrichment plot is also generated that describes the enrichment of subsequences in the SELEX cycles (see Fig. 3C).
The position weight matrix (PWM) models were generated as follows: First, we adopted the standard assumption that the binding affinities of individual sites of a PWM are independent of each other (Benos et al. 2002). Then the PWM represents a multinomial distribution. To estimate this distribution at site j, we take the consensus and the three other sequences obtained from the consensus by replacing the j-th base in turn with each of the other three bases. The occurrence counts of these four sequences within the DNA-reads give an unbiased estimate of the PWM at site j. For nonpalindromic sequences, only those reads were used that contained exactly one sequence that was within Hamming distance 1 of the consensus or its reverse complement.
For the matrices shown in Figure 5 and Supplemental Figures S1–S3, the PWMs were corrected to decrease the distortion caused by nonspecific carryover of DNA from the previous cycle. First, the fraction of DNA that was carried over nonspecifically (λ) was estimated. In one SELEX cycle, the fraction f of any set of nonspecific sequences out of total sequences is expected to decrease according to the equation fk+1 = λfk, where fk denotes the fraction of the set of nonspecific sequences in cycle k. Using defined sets of nonspecific sequences can lead to sampling error. However, reasonable estimates for fk+1 and fk can be derived from the sum of the number of occurrences of all 8-mers that rank between 25% and 75% in relative abundance divided by the total number of sequences Nk+1 and Nk in cycles k + 1 and k, respectively. This is because it is likely that <25% of all possible 8-mer sequences bind specifically to any TF. The observed values for λ calculated from our data ranged from 0.326 to 1.018. The PWM for the later cycle (Mcorrected, final result shown) was then corrected for background caused by the nonspecific DNA carryover by the equation
where both of the PWMs Mk and Mk+1 were generated using the same consensus. Biologically implausible values that can arise due to the discrete nature of sequence counts were corrected as follows: Values for λ that were >1 were replaced by 1, and negative values in the final PWM were replaced by 0. In the worst cases, such correction affected λ by 1.8% and a PWM position by 3.1% (of total reads in the model).
We also experimented with using an EM algorithm to derive the PWMs. For this purpose, we reimplemented the MEME EM algorithm described in Bailey and Elkan (1995). This method was more sensitive and could derive PWMs from a lower number of sequences. However, using all of our data, the EM algorithm generally resulted in a very similar profile to that of our multinomial method (Supplemental Fig. S4B). In the end, we chose to use the multinomial models instead of the EM models as it is not clear how nonspecific DNA carryover can be accounted for in the EM algorithm.
We also tested a variant of the multinomial method in which we had in the role of the consensus all the sequences at Hamming distance 1 from it. The four counts for each such sequence were formed analogously and added together. As these counts are larger than in the basic variant, they should lead to a better estimate. However, while the resulting PWMs turned out to be very similar to the basic ones, they also were systematically somewhat less restrictive, obviously because the added distance from the consensus means decreased affinity, and hence the relative effect of background sequences becomes larger in the larger counts. All profiles shown were therefore generated using the multinomial model with Hamming distance 1. Use of a smaller Hamming distance also allowed for better separation between monomeric and dimeric binding modes.
Chromatin immunoprecipitation by sequencing
Antibodies against NFATC1 (mouse monoclonal sc-7294x for ChIP-seq1 and rabbit polyclonal sc-13033x for ChIP-seq2 in Fig. 6), RFX3 (sc-10662x for ChIP-seq1 and sc-10663x for ChIP-seq2), and normal mouse and goat IgG were purchased from Santa Cruz Biotechnology. K562 and Jurkat cell lines were used for RFX3 and NFATC1 ChIP-seq analyses. To induce nuclear localization of NFATC1, Jurkat cells were exposed to 2 μM ionomycin and 100 ng/mL PMA for 24 h (Jin et al. 2003).
ChIP-seq and data analysis was performed essentially as described in Tuupanen et al. (2009). Briefly, proteins were cross-linked to DNA by incubation of cells for 10 min in medium containing 1% formaldehyde at room temperature, after which the cross-linking was quenched, nuclei were extracted, and DNA was fragmented by sonication. Precleared samples were incubated with 4 μg of specific antibody overnight at 4°C, and the antibodies were collected by incubation with 30 μL of protein G–Sepharose beads for 2–3 h at +4°C followed by centrifugation at 800g. Subsequently, the beads were washed, precipitated chromatin complexes were eluted, and the cross-links were reversed by incubating overnight at 65°C. DNA was extracted with a QiaQuick PCR purification kit (QIAGEN).
ChIP DNA was quantitated by PicoGreen dsDNA quantitation reagent (Molecular Probes). A ChIP library was prepared for sequencing as described in Tuupanen et al. (2009), and 120–350-bp fragments were size-selected on a 2% agarose gel. The fragments were enriched by 18 cycles of PCR amplification and size-selected again (to 150–300 bp). Purified DNA (QIAGEN gel purification kit) was quantified (Nanodrop 1000 spectrophotometer) and used for massively parallel sequencing (Illumina Genome Analyzer) according to the manufacturer's instructions. Sequencing reads were mapped to the human genome (NCBI36) using MAQ software by Heng Li, version 0.6.5 (Li et al. 2008). Only high-quality reads that could be reliably mapped (mapping quality score at least 30) were accepted, resulting in total of 9.35 million reads from NFATC1 ChIP samples and 9.85 million reads from IgG control in Jurkat cells, 9.1 million reads for RFX3 ChIP samples, and 9.2 million reads from IgG control in K562 cells. The data were then analyzed essentially as described in Tuupanen et al. (2009). Peaks (NCBI36 coordinates, P < 0.05) are given in tab-delimited text format in Supplemental Data Files S1–S4. Primary sequence reads are available in the NCBI Sequence Read Archive under accession number SRA012198. ChIP-seq data were validated using two different antibodies for both factors, and by confirming a random set of significant peaks using qPCR (antibodies sc-7294x and sc-10663x) (Supplemental Fig. S5; Supplemental Table S5).
The sequences of the top 150 or 500 peaks of length <400 bp according to P-value were selected for the motif analysis from each sample. Motifs between widths 6 and 50 bases were searched from both strands assuming zero or one binding site per sequence (mod=zoops) using MEME version 4.1.0 and a third-order background Markov model estimated from the human genome (NCBI36).
Acknowledgments
We thank Ritva Nurmi and Sini Miettinen for technical assistance; Drs. Sampsa Hautaniemi, Anna Saramäki, and Minna Taipale for critical review of the manuscript; and the EMBL gene core, Uppsala SNP platform and Dr. Outi Monni for massively parallel sequencing. This work was supported by the EU FP6 STREP projects NET2DRUG and REGULATORY GENOMICS, Academy of Finland Center of Excellence in Translational Genome-Scale Biology, and the NEURO program of the Academy of Finland.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession no. SRA012198.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.100552.109.
References
- Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MD, et al. 2001. The InterPro database, an integrated documentation resource for protein families, domains, and functional sites. Nucleic Acids Res 29: 37–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, et al. 2008. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell 32: 878–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, et al. 2009. Diversity and complexity in DNA recognition by transcription factors. Science 324: 1720–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C 1995. The value of prior knowledge in discovering motifs with MEME. Technical Report CS95-413. Department of Computer Science, University of California, San Diego: [PubMed] [Google Scholar]
- Benos PV, Bulyk ML, Stormo GD 2002. Additivity in protein–DNA interactions: How good an approximation is it? Nucleic Acids Res 30: 4442–4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Philippakis AA, Qureshi AM, He FS, Estep PW III, Bulyk ML 2006. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat Biotechnol 24: 1429–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, et al. 2008. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell 133: 1266–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryne JC, Valen E, Tang MH, Marstrand T, Winther O, da Piedade I, Krogh A, Lenhard B, Sandelin A 2008. JASPAR, the open access database of transcription factor-binding profiles: New content and tools in the 2008 update. Nucleic Acids Res 36: D102–D106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulyk ML, Huang X, Choo Y, Church GM 2001. Exploring the DNA-binding specificities of zinc fingers with DNA microarrays. Proc Natl Acad Sci 98: 7158–7163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckstummer T, Bennett KL, Preradovic A, Schutze G, Hantschel O, Superti-Furga G, Bauch A 2006. An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat Methods 3: 1013–1019 [DOI] [PubMed] [Google Scholar]
- Clauss IM, Chu M, Zhao JL, Glimcher LH 1996. The basic domain/leucine zipper protein hXBP-1 preferentially binds to and transactivates CRE-like sequences containing an ACGT core. Nucleic Acids Res 24: 1855–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djordjevic M, Sengupta AM, Shraiman BI 2003. A biophysical approach to transcription factor binding site discovery. Genome Res 13: 2381–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery P, Strubin M, Hofmann K, Bucher P, Mach B, Reith W 1996. A consensus motif in the RFX DNA binding domain and binding domain mutants with altered specificity. Mol Cell Biol 16: 4486–4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falvo JV, Lin CH, Tsytsykova AV, Hwang PK, Thanos D, Goldfeld AE, Maniatis T 2008. A dimer-specific function of the transcription factor NFATp. Proc Natl Acad Sci 105: 19637–19642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher DE, Carr CS, Parent LA, Sharp PA 1991. TFEB has DNA-binding and oligomerization properties of a unique helix–loop–helix/leucine-zipper family. Genes & Dev 5: 2342–2352 [DOI] [PubMed] [Google Scholar]
- Grange T, Roux J, Rigaud G, Pictet R 1991. Cell-type specific activity of two glucocorticoid responsive units of rat tyrosine aminotransferase gene is associated with multiple binding sites for C/EBP and a novel liver-specific nuclear factor. Nucleic Acids Res 19: 131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallikas O, Taipale J 2006. High-throughput assay for determining specificity and affinity of protein–DNA binding interactions. Nat Protoc 1: 215–222 [DOI] [PubMed] [Google Scholar]
- Hallikas O, Palin K, Sinjushina N, Rautiainen R, Partanen J, Ukkonen E, Taipale J 2006. Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell 124: 47–59 [DOI] [PubMed] [Google Scholar]
- Hampshire AJ, Rusling DA, Broughton-Head VJ, Fox KR 2007. Footprinting: A method for determining the sequence selectivity, affinity and kinetics of DNA-binding ligands. Methods 42: 128–140 [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Levine M 1993. Binding affinities and cooperative interactions with bHLH activators delimit threshold responses to the dorsal gradient morphogen. Cell 72: 741–752 [DOI] [PubMed] [Google Scholar]
- Jin L, Sliz P, Chen L, Macian F, Rao A, Hogan PG, Harrison SC 2003. An asymmetric NFAT1 dimer on a pseudo-palindromic kappa B-like DNA site. Nat Struct Biol 10: 807–811 [DOI] [PubMed] [Google Scholar]
- Kel A, Kel-Margoulis O, Babenko V, Wingender E 1999. Recognition of NFATp/AP-1 composite elements within genes induced upon the activation of immune cells. J Mol Biol 288: 353–376 [DOI] [PubMed] [Google Scholar]
- Kinzler KW, Vogelstein B 1990. The GLI gene encodes a nuclear protein which binds specific sequences in the human genome. Mol Cell Biol 10: 634–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeger PE, Morimoto RI 1994. Selection of new HSF1 and HSF2 DNA-binding sites reveals difference in trimer cooperativity. Mol Cell Biol 14: 7592–7603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D, Prentki P, Chandler M 1992. Use of gel retardation to analyze protein–nucleic acid interactions. Microbiol Rev 56: 509–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord CA, Savitsky D, Sitcheran R, Calame K, Wright JR, Ting JP, Williams KL 2009. Blimp-1/PRDM1 mediates transcriptional suppression of the NLR gene NLRP12/Monarch-1. J Immunol 182: 2948–2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macian F 2005. NFAT proteins: Key regulators of T-cell development and function. Nat Rev Immunol 5: 472–484 [DOI] [PubMed] [Google Scholar]
- Mader S, Leroy P, Chen JY, Chambon P 1993. Multiple parameters control the selectivity of nuclear receptors for their response elements. Selectivity and promiscuity in response element recognition by retinoic acid receptors and retinoid X receptors. J Biol Chem 268: 591–600 [PubMed] [Google Scholar]
- Merika M, Orkin SH 1993. DNA-binding specificity of GATA family transcription factors. Mol Cell Biol 13: 3999–4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers S, Downing JR, Hiebert SW 1993. Identification of AML-1 and the (8;21) translocation protein (AML-1/ETO) as sequence-specific DNA-binding proteins: The runt homology domain is required for DNA binding and protein–protein interactions. Mol Cell Biol 13: 6336–6345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T 2001. DNA–protein interactions: Principles and protocols Humana Press, Totowa, NJ [Google Scholar]
- Pollock R, Treisman R 1991. Human SRF-related proteins: DNA-binding properties and potential regulatory targets. Genes & Dev 5: 2327–2341 [DOI] [PubMed] [Google Scholar]
- Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, Beckwith CA, Chan JA, Hills A, Davis M, et al. 2009. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet 41: 882–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pscherer A, Dorflinger U, Kirfel J, Gawlas K, Ruschoff J, Buettner R, Schule R 1996. The helix–loop–helix transcription factor SEF-2 regulates the activity of a novel initiator element in the promoter of the human somatostatin receptor II gene. EMBO J 15: 6680–6690 [PMC free article] [PubMed] [Google Scholar]
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. 2007. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods 4: 651–657 [DOI] [PubMed] [Google Scholar]
- Robertson AG, Bilenky M, Tam A, Zhao Y, Zeng T, Thiessen N, Cezard T, Fejes AP, Wederell ED, Cullum R, et al. 2008. Genome-wide relationship between histone H3 lysine 4 mono- and tri-methylation and transcription factor binding. Genome Res 18: 1906–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulet E, Busso S, Camargo AA, Simpson AJ, Mermod N, Bucher P 2002. High-throughput SELEX SAGE method for quantitative modeling of transcription-factor binding sites. Nat Biotechnol 20: 831–835 [DOI] [PubMed] [Google Scholar]
- Rual JF, Hirozane-Kishikawa T, Hao T, Bertin N, Li S, Dricot A, Li N, Rosenberg J, Lamesch P, Vidalain PO, et al. 2004. Human ORFeome version 1.1: A platform for reverse proteomics. Genome Res 14: 2128–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF, et al. 2002. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci 99: 16899–16903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C, Gold L 1990. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249: 505–510 [DOI] [PubMed] [Google Scholar]
- Tuupanen S, Turunen M, Lehtonen R, Hallikas O, Vanharanta S, Kivioja T, Bjorklund M, Wei G, Yan J, Niittymaki I, et al. 2009. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet 8: 885–890 [DOI] [PubMed] [Google Scholar]
- Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM 2009. A census of human transcription factors: Function, expression, and evolution. Nat Rev Genet 10: 252–263 [DOI] [PubMed] [Google Scholar]
- Verrijzer CP, Alkema MJ, van Weperen WW, Van Leeuwen HC, Strating MJ, van der Vliet PC 1992. The DNA binding specificity of the bipartite POU domain and its subdomains. EMBO J 11: 4993–5003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G-H, Badis G, Berger MF, Kivioja T, Palin K, Enge M, Bonke M, Jolma M, Varjosalo M, Gehrke AR, et al. 2010. Genome-wide analysis of ETS family DNA-binding in vitro and in vivo. EMBO J (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MD, Barbosa-Morais NL, Schmidt D, Conboy CM, Vanes L, Tybulewicz VL, Fisher EM, Tavare S, Odom DT 2008. Species-specific transcription in mice carrying human chromosome 21. Science 322: 434–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Yang D, Schluesener HJ, Zhang Z 2007. Advances in SELEX and application of aptamers in the central nervous system. Biomol Eng 24: 583–592 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Granas D, Stormo GD 2009. Inferring binding energies from selected binding sites. PLoS Comput Biol 5: e1000590 doi: 10.1371/journal.pcbi.1000590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zykovich A, Korf I, Segal DJ 2009. Bind-n-Seq: High-throughput analysis of in vitro protein–DNA interactions using massively parallel sequencing. Nucleic Acids Res 37: e151 doi: 10.1093/nar/gkp802 [DOI] [PMC free article] [PubMed] [Google Scholar]