Abstract
Remediation of hazardous waste sites requires efficient and cost-effective methods to assess the extent of contamination by toxic substances including dioxin-like chemicals. Traditionally, dioxin-like contamination has been assessed by gas chromatography/high-resolution mass spectrometry (GC/MS) analysis for specific polychlorinated dibenzo-p-dioxins, dibenzofurans, and biphenyl congeners. Toxic equivalency factors for these congeners are then used to estimate the overall dioxin toxic equivalency (TEQ) of complex mixtures found in samples. The XDS-CALUX bioassay estimates contamination by dioxin-like chemicals in a sample extract by measuring expression of a sensitive reporter gene in genetically engineered cells. The output of the XDS-CALUX assay is a CALUX-TEQ value, calibrated based on TCDD standards. Soil samples taken from a variety of hazardous waste sites were measured using the XDS-CALUX bioassay and GC/MS. TEQ and CALUX-TEQ from these methods were compared, and a mathematical model was developed describing the relationship between these two data sets: log(TEQ) = 0.654 × log(CALUX-TEQ) + 0.058-(log(CALUX-TEQ))2. Applying this equation to these samples showed that predicted and GC/MS measured TEQ values strongly correlate (R2 = 0.876) and that TEQ values predicted from CALUX-TEQ were on average nearly identical to the GC/MS-TEQ. The ability of XDS-CALUX bioassay data to predict GC/MS-derived TEQ data should make this procedure useful in risk assessment and management decisions.
Introduction
Hazardous waste site remediation is a complex and expensive process, at least in part because the sites are typically contaminated with a large number of diverse chemical substances (1,2). Initial characterization of these sites requires rapid identification of the nature and amount of contaminants, and improved technologies for this purpose would significantly expedite the initial phases of remediation. Because dioxins and dioxin-like chemicals (DLCs) contaminate many hazardous waste sites under remediation, the United States Environmental Protection Agency (U.S. EPA) Superfund Innovative Technology (SITE) Program has been evaluating novel technologies and comparing them to the established high-resolution gas chromatography/mass spectrometry (GC/MS) technology for quantifying DLCs.
DLCs refer to a subset of halogenated aromatic hydrocarbons, specifically 2,3,7,8-chloro-substituted polychlorinated dibenzo-p-dioxins and dibenzofurans and 3,3′,4,4′-chloro-substituted biphenyls that produce a common spectrum of Ah receptor-dependent toxicological and biological effects (3, 4). These compounds induce similar toxicological effects in animals including weight loss, thymic atrophy, immune suppression, hepatoxicity, porphyria, chloracne and related dermal lesions, cancer, and reproductive toxicity (4–7). The spectrum of toxicological effects in humans may not include all of the effects observed in animal studies (8). The prototypical member of the dioxin family is 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). While the exact mechanism by which dioxins and related DLCs exert their toxic effects is not known, what is clear is that the alterations in gene expression induced by these compounds are mediated by their ability to bind to and activate the aryl hydrocarbon receptor (AhR), a soluble intracellular ligand-dependent transcription factor. Following binding to the AhR, the DLC/AhR complex migrates from the cytosol into the cell’s nucleus, wherein it is converted into its DNA binding form and stimulates transcription of a battery of genes (9–14). It is believed that the toxicity of TCDD and related DLCs is related to the ability of these chemicals to induce persistent expression of target genes.
The toxicity of a dioxin congener is related to the ability of the congener to bind to and activate the AhR, and the difference in relative potency between congeners can be several orders of magnitude. To assist with quantifying the overall impact of a mixture of congeners, the World Health Organization (WHO) established toxic equivalency factors (TEFs) for 17 polychlorinated dibenzo-p-dioxin (PCDD) and polychlorinated dibenzofuran (PCDF) congeners and 12 polychlorinated biphenyl (PCB) congeners. TCDD, the most potent congener, is arbitrarily assigned a TEF of 1.0, and the TEF for less toxic congeners is <1.0 (15, 16). Reevaluation of the 1998 WHO TEFs was recently reported, and minor changes were made in some of the TEF values (17). In a typical analytical study, the concentration of DLCs in a complex chemical mixture is determined by GC/MS, the amount of each congener present in the mixture is multiplied by the congener’s TEF to obtain a toxic equivalent quotient (TEQ) for that congener, and the sum of the individual TEQs represents the overall toxic potency of the chemical mixture. The TEQ estimate can be used for risk assessment studies involving different complex mixtures (15–17).
The XDS-CALUX bioassay, developed by Xenobiotic Detection Systems, is based on a mechanistic understanding of the ability of dioxin to activate gene expression, a step critical in the toxic and biological effects of dioxins and related DLCs. XDS-CALUX uses a genetically engineered murine cell line that carries a firefly luciferase reporter gene driven by an AhR-dependent promoter that is activated when the cells are exposed to dioxin or DLCs (18, 19). The magnitude of the induction response is directly proportional to the concentration of DLC in the sample and the degree of AhR receptor occupancy and activation. When specific sample processing procedures are used to eliminate the many non-dioxin-like AhR ligands/agonists (20, 21) present in a sample extract, this bioassay can detect dioxin and DLCs with a high specificity and sensitivity in extracts from various matrices (22–24). Furthermore, the XDS-CALUX bioassay is highly quantitative, and using a TCDD standard curve, the reporter gene signal can be converted to a CALUX-toxic equivalency quotient (CALUX-TEQ). The CALUX-TEQ is a surrogate for the standard TEQ generated by GC/MS. For the purpose of this paper, TEQ will refer to GC/MS estimates of toxic equivalency, and CALUX-TEQ will refer to XDS-CALUX estimates of TCDD equivalency calculated from a TCDD standard curve.
The present modeling procedure was evolved from a SITE study sponsored by the U.S. EPA. The SITE study analyzed environmental soil and sediment samples from 10 hazardous waste sites contaminated with complex mixtures of dioxin-like chemicals. Samples were analyzed both by XDS-CALUX and by GC/MS, and CALUX-TEQ and GC/MS-TEQ values were calculated and compared. Mathematical modeling was employed to determine the relationship between these two data sets. Subsequently, the equation determined by mathematical modeling of the SITE data sets was applied to a second unrelated data set of environmental samples (part of an EPA SW846 validation study) to predict GC/MS-TEQ. Predicted TEQ and GCMS TEQ values were compared to assess the validity of the modeled equation. The relevance of the current results for future dioxin risk assessment and risk management studies is discussed.
Materials and Methods
Chemicals
PCDD and PCDF standards were purchased from Wellington Laboratories (Guelph, Ontario, Canada). Solvents and chromatography matrices for sample extraction and cleanup were purchased from Fisher Scientific and were ACS grade or higher (Pittsburgh, PA).
Environmental Samples
The U.S. EPA, the Battelle Corporation, and XDS conducted a U.S. EPA SITE study of 209 soil and sediment samples, including spiked samples and standard extracts, in March 2004. Samples were obtained from 10 hazardous waste sites undergoing remediation and included Warren County PCB Landfill in North Carolina; Tittabawassee River Flood Plain in Michigan; residential sites in Midland, MI; Winona Post site in Winona, MO; Solutia site in Nitro, WV; New York/New Jersey Harbors; Newark Bay in New Jersey; two sites on the Saginaw River in Michigan; and the Brunswick Wood Preserving Site in Glynn County, GA (25). Samples contained various contaminants including dioxins, PCBs, PCP, and PAHs. A complete description of the sites and samples is available in a U.S. EPA report (25). GC/MS analysis was performed by Axys Analytical Services (Sidney, British Columbia, Canada), while XDS-CALUX analysis was carried out by XDS, in a mobile laboratory stationed in Saginaw, MI (40 samples),or at XDS, in Durham, NC (169 samples). Thirty-six soil samples and eight ash samples (part of an SW846 validation study) were used to test the proposed mathematical model. Soil and ash samples collected in Japan were analyzed by GC/MS at Hiyoshi Corporation (ash and soil) (Omihachiman, Shiga, Japan), soil samples collected at a U.S. EPA remediation site in Hawaii were analyzed by GC/MS at Southwest Laboratories (Tempe, AZ), and XDS-CALUX analysis was conducted at XDS, in Durham, NC. XDS was blinded to the sample design and GC/MS data in the U.S. EPA SITE study as well as to the GC/MS data in the SW846 validation study (GC/MS data were sent to an independent statistician).
XDS-CALUX Sample Preparation
Sample extraction and cleanup were conducted as described previously (26–28). Briefly, aliquots of each sample were extracted with toluene and then purified by acid silica column chromatography and a patented carbon column chromatography procedure (29). Two fractions were subsequently collected from the carbon column. The first fraction contained chlorinated biphenyls, and the second fraction contained primarily PCDD/Fs (27, 29). Isolated fractions were exchanged into dimethylsulfoxide (DMSO), for measurement of CALUX-TEQ in the XDS-CALUX bioassay. Both fractions were analyzed for CALUX-TEQ; however, in this paper, results for only the PCDD/Fs fraction are presented and analyzed.
XDS-CALUX Bioassay Analysis
XDS-CALUX was conducted in H1L6.1c2 cells, a mouse hepatoma (hepa1c1c7) cell line stably transfected with the pGudLuc6.1 reporter plasmid that carries an AhR responsive mouse mammary tumor virus promoter and firefly luciferase gene (18, 19). Samples in DMSO were suspended in cell culture medium (RPMI 1640 supplemented with 8% fetal calf serum and 1% Pen/Strep antibiotics) and added to monolayer cultures of the H1L6.1c2 cell line in 96 well plates. Each assay was calibrated with a TCDD standard curve and included positive controls (solutions of TCDD and PCB 126) and negative controls (DMSO and solvent blanks). Plates were incubated for 20 h in a humidified 5% CO2 atmosphere at 37 °C, the medium was removed, and the luciferase activity was quantified (Promega luciferase assay kit, Promega, Madison, WI) using a Berthold Orion Microplate Luminometer (Oak Ridge, TN). All sample measurements were made in the linear range of the assay (i.e., values near the middle of the standard curve). Quality control criteria used for the analysis of XDS-CALUX bioassay results were as described in Brown et al. (26). The TCDD standard curve was modeled to a four-parameter Hill equation using the least-squares best fit method, and the resulting standard curve was used to calculate CALUX-TEQ for unknown samples (26, 27). CALUX-TEQ and TEQ data were collected and analyzed in a double-blinded manner with statistical analysis conducted by an independent statistician. The 209 EPA SITE samples were extracted and analyzed in the screening mode of analysis in the XDS-CALUX method that includes a single extraction of a sample. Upon un-blinding of the SITE study design, it became evident that there were 49 total samples within the study. A summary of the breakdown of the sample classes was 32 samples of soil and sediment analyzed in quadruplicate (124 samples), 12 performance evaluation samples analyzed in quadruplicate except for two samples that were replicated seven times (58 samples), and five extracts that were replicated from four to eight times within the 209 total samples (25). The performance evaluation samples and solutions contained reference materials with certified concentrations of dioxin, furans, and/or PCBs, spiked samples with a certified concentration of dioxin and/or other contaminants, and blank samples. A complete description of the SITE study design has been described (25). Analysis of the samples by GC/MS was not blinded, and the replicate criteria for generation of TEQ by GC/MS are not available to XDS. SW846 samples were extracted and analyzed in triplicate for CALUX-TEQ determination and presented as mean values, and GC/MS TEQ analysis was based on single instrumental determinations of DLCs by GC/MS determination of TEQ.
Mathematical Modeling
GC/MS TEQ and CALUX-TEQ values were fit to a statistical model. Initially, a generalized estimating equation (GEE) (30) that accounted for the correlation due to replicates using a base 10 logarithmic transformation of the two variables was used. However, the final model was based on the mean of the replicates. Input data for mathematical modeling initially included all samples. Subsequently, in developing a model for dioxins, seven samples were eliminated that were PCB standards, and data were limited to samples with TEQ values between 1 and 3000 pg/g with the model being generated on the means of 34 samples. Data presented are on 42 samples without elimination of the samples below 1 pg/g or greater than 3000 pg/g.
TEF/REP Conversion Factor
Calculation of conversion factors for samples based on differences between WHO TEF values and XDS-CALUX relative potency (REP) values was conducted as described previously (31). TEQ values for SW846 samples were recalculated using GC/MS congener specific concentrations and XDS-CALUX REP values (32) (see Table 1). GC/MS measurements for some congeners in SW846 samples were reported without non-detect (ND) negative control values. Therefore, for this study, all ND values were set to zero. Conversion factors (or averaged conversion factors, where n > 1) were calculated by dividing the TEQ for the sample by the TEQ calculated using CALUX REP values. The TEF/REP corrected estimate of TEQ was derived by multiplying this factor by the XDS-CALUX measured value for CALUX-TEQ.
TABLE 1.
TEF REP Conversion for Representative Soil Samplea
| congener | GC/MS (pg/g) |
WHO-TEF | CALUX-REP | WHO-TEQ (pg of TCDD/g) |
CALUX-TEQ (pg of TCDD/g) |
|---|---|---|---|---|---|
| 2378-TCDD | ND | 1 | 1 | ||
| 12378-PeCDD | 4.6 | 1 | 0.73 | 4.6 | 3.358 |
| 123478-HxCDD | 7.2 | 0.1 | 0.075 | 0.72 | 0.54 |
| 123678-HxCDD | 17 | 0.1 | 0.098 | 1.7 | 1.666 |
| 123789-HxCDD | 13 | 0.1 | 0.061 | 1.3 | 0.793 |
| 1234678-HpCDD | 320 | 0.01 | 0.031 | 3.2 | 9.92 |
| OCDD | 7100 | 0.0001 | 0.00034 | 0.71 | 2.414 |
| 2378-TCDF | 2.5 | 0.1 | 0.067 | 0.25 | 0.1675 |
| 12378-PeCDF | 10 | 0.05 | 0.14 | 0.5 | 1.4 |
| 23478-PeCDF | 13 | 0.5 | 0.58 | 6.5 | 7.54 |
| 123478-HxCDF | 30 | 0.1 | 0.13 | 3.0 | 3.9 |
| 123678-HxCDF | 34 | 0.1 | 0.14 | 3.4 | 4.76 |
| 123789-HxCDF | 4.3 | 0.1 | 0.11 | 0.43 | 0.473 |
| 234678-HxCDF | 82 | 0.1 | 0.31 | 8.2 | 25.42 |
| 1234678-HpCDF | 270 | 0.01 | 0.024 | 2.7 | 6.48 |
| 1234789-HpCDF | 44 | 0.01 | 0.044 | 0.44 | 1.936 |
| OCDF | 380 | 0.0001 | 0.0016 | 0.038 | 0.608 |
| 37.688 | 71.376 | ||||
| TEQ/CALUX-TEQ = 37.688/71.376 = 0.528 | |||||
TEF and REP values are shown for PCDD and PCDF congeners in a representative SW846 soil sample. Conversion factor was calculated as described previously (31).
Results
Comparison of GC/MS-TEQ and CALUX-TEQ
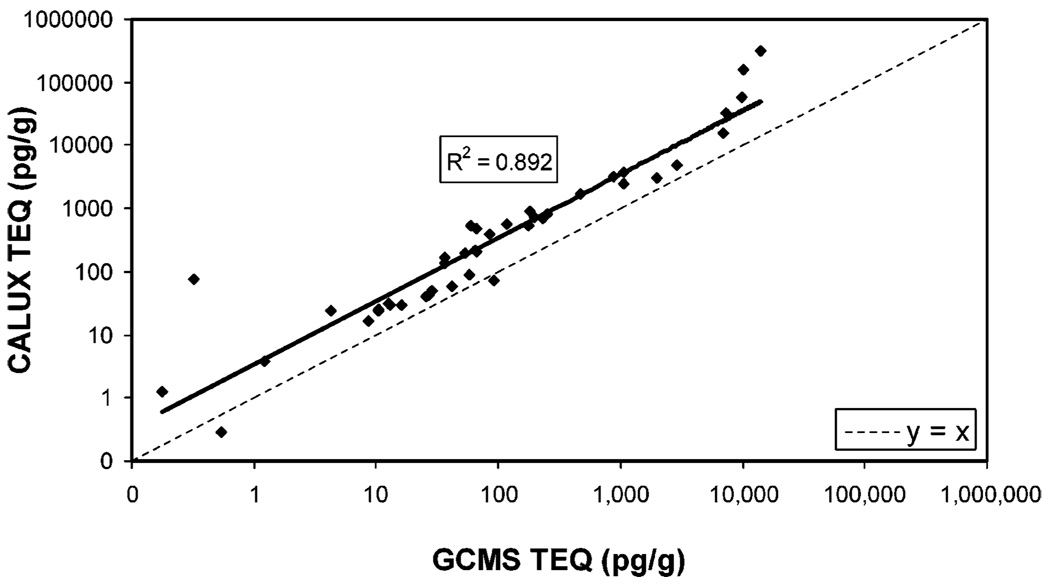
GC/MS and XDS-CALUX assays were performed on 209 SITE samples as described in the Materials and Methods. During data analysis, sample replicates within the data set were identified, the data presented in the graphs represent the average for each of 42 samples analyzed in quadruplicate, and the mean was calculated for the samples after un-blinding of the data. GC/MS-TEQ and CALUX-TEQ values calculated from GC/MS and XDS-CALUX, respectively, are compared graphically in Figure 1. The results show that GC/MS-TEQ and CALUX-TEQ values are highly correlated (R2 = 0.892). Variance is observed between the two methods at very low or high GC/ MS-TEQs or CALUX-TEQs. This may indicate that GC/MS and XDS-CALUX assays demonstrate increased variance at or near the limits of detection of both analytical procedures. The variance is also greater at extremely high concentrations of dioxin-like chemicals. This is probably due to the XDS-CALUX bioassay detecting additional compounds that activate AhR but are not detected or measured by GC/MS methods. The extreme dilution of the sample to allow detection in the linear range of the standard curve also contributes to the variance in estimating the CALUX-TEQ for the sample. CALUX-TEQ values are consistently higher than TEQ values (Figure 1), with CALUX-TEQs averaging 9.4 times higher than TEQ values. This is consistent with what has been observed previously for environmental samples (22, 33).
FIGURE 1.
Comparison of GC/MS-TEQ and CALUX-TEQ for SITE samples. TEQ was determined by GC/MS, CALUX-TEQ was determined by XDS-CALUX as described, and the average results for each quadruplicate analysis of 42 soil and sediment samples are graphed. Solid line is the data trend line; for comparison, the dashed line represents y = x(slope = 1 and intercept = 0). The regression line equation for the data is y = 3.356x1.0067.
Modeling the Relationship between GC/MS-TEQ and CALUX-TEQ
The data in Figure 1 (excluding samples with TEQ values ≤1 or ≥3000 pg of TCDD/g) were used to derive a statistical model for the relationship between GC/MS-TEQ and CALUX-TEQ as follows:
| (1) |
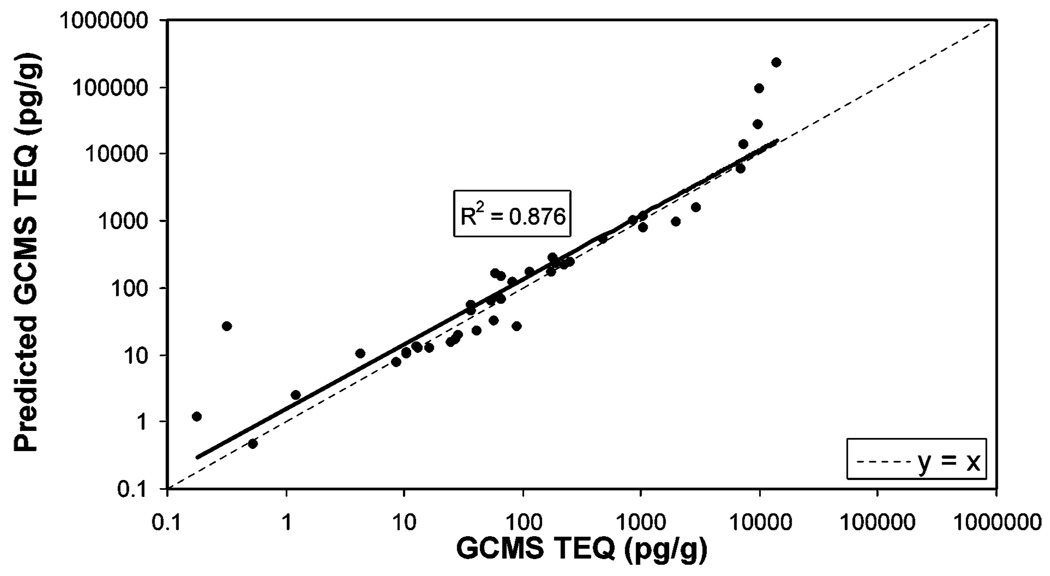
This model was evaluated by predicting TEQ from CALUX-TEQ using input CALUX-TEQ data from the SITE study (Figure 1). The results showed a linear relationship with a correlation of 0.876 (Figure 2), which is slightly lower than the direct correlation between GC/MS-TEQ and CALUX-TEQ. However, since the data trend line almost coincides with the dashed diagonal line through the origin, the predicted TEQ values are very close to the TEQ values measured by GC/MS. For these data, CALUX predicted TEQ was on average 0.997 times the TEQ measured by GC/MS. As observed previously, the discrepancy between GC/MS measured TEQ and CALUX predicted TEQ was greater for samples with very low or high TEQ values.
FIGURE 2.
Modeling the relationship between predicted GC/MS-TEQ and GC/MS-TEQ. The TEQ was predicted from CALUX-TEQ values for SITE samples using eq 1. Solid line is the data trend line; for comparison, the dashed line represents y = x(slope = 1 and intercept = 0). The regression line equation for the data is y = 1.7789x0.9384.
Testing the Mathematical Model
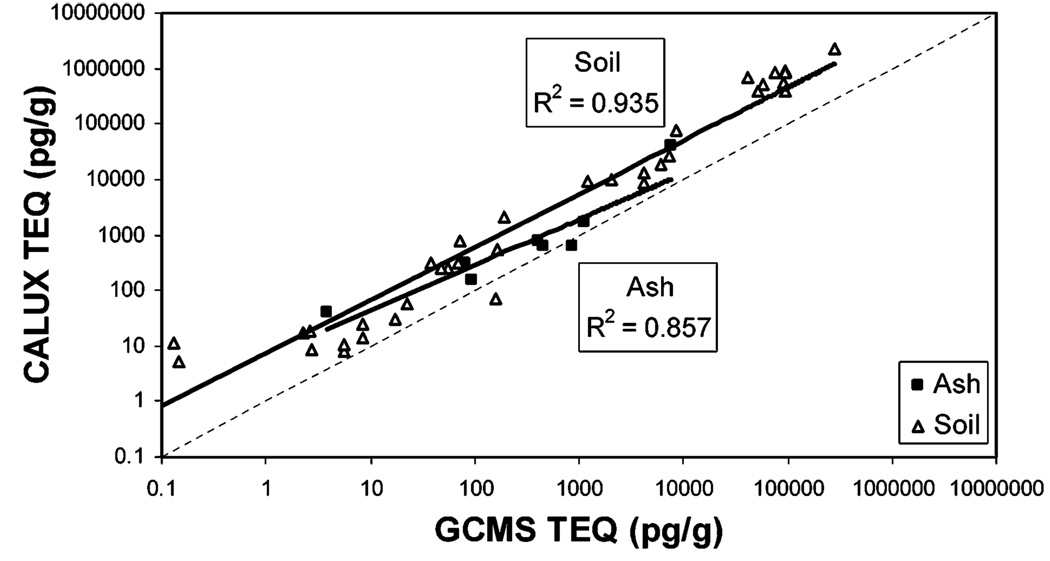
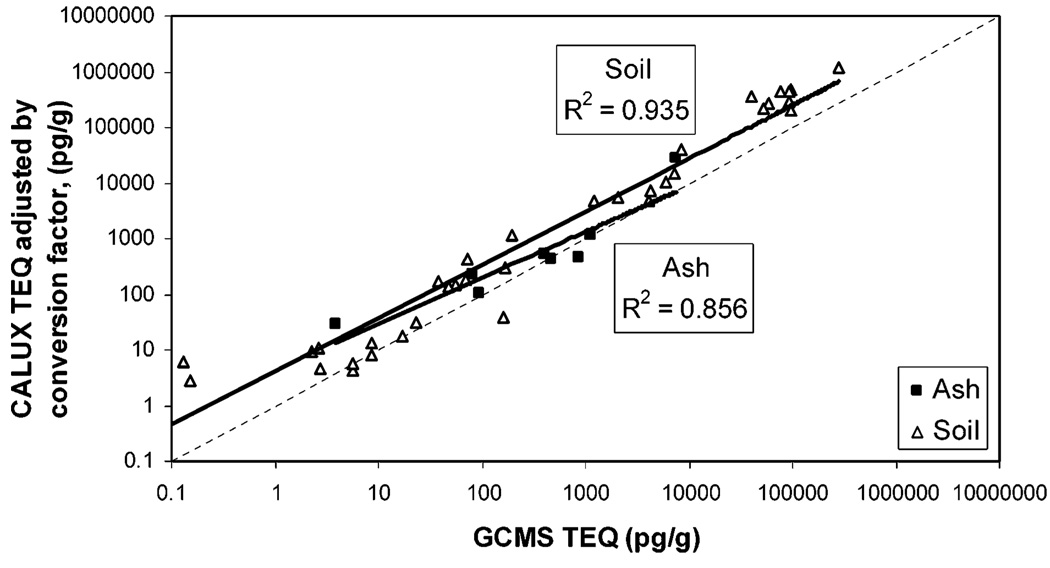
The mathematical model was tested with XDS-CALUX data from an independent data set, SW846, that included 36 dioxin contaminated soil samples and eight ash samples. CALUX-TEQ and GC/MS-TEQ values for these samples had correlation values of 0.857 and 0.935 for ash and soil samples, respectively (Figure 3). As observed previously, CALUX-TEQ values for the SW846 samples were consistently higher than TEQ values. For these data, CALUX-TEQ was on average 12.6-fold higher than TEQ (3.4-fold for ash samples and 14.6–fold for soil samples).
FIGURE 3.
Comparison of GC/MS-TEQ and CALUX-TEQ for SW846 samples. TEQ was determined by GC/MS, and CALUX-TEQ was determined by XDS-CALUX as described. Solid line is the data trend line; for comparison, the dashed line represents y = x(slope = 1 and intercept = 0). Sample matrix was ash (■) or soil (△).
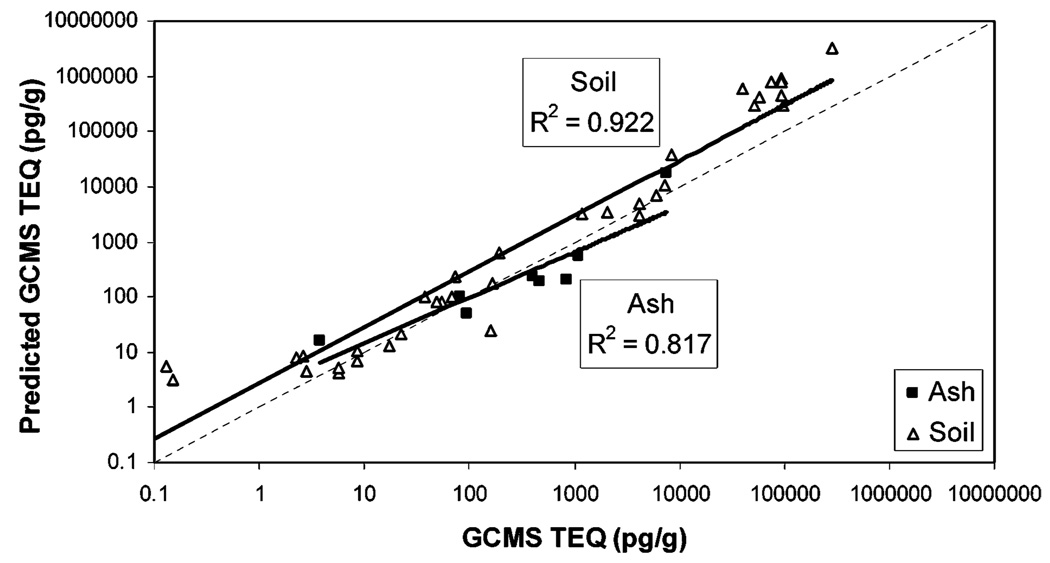
On the basis of our mathematical model and eq 1, CALUX-TEQ values were used to predict TEQ values, and the predicted and measured GC/MS-TEQ values for the SW846 data set were compared (Figure 4). Correlation coefficients were 0.817 for ash samples and 0.922 for soil samples. Predicted TEQ was on average 6.5-fold higher than measured GC/MS-TEQ (1.3-fold for ash and 7.7-fold for soil). Note that these values change when samples with extreme GC/MS-TEQ values (i.e., ≤1 or ≥3000 pg/g) were excluded from the analysis (see Table 2 and Discussion).
FIGURE 4.
Predicting GC/MS-TEQ from CALUX-TEQ for SW846 samples. TEQ was predicted from CALUX-TEQ determinations using eq 1. Solid line is the data trend line; for comparison, the dashed line represents y = x(slope = 1 and intercept = 0). Sample matrix was ash (■) or soil (△). The regression line equation for ash is y = 6.49x0.82 and for soil is y = 5.86x0.97.
TABLE 2.
Summary of Overall Discrepancy between CALUX-TEQ and TEQ Values for SW846 Samplesa
| transformation method | all samples | all samples in range (1–3000 pg/g) |
ash samples | ash samples in range (1–3000 pg/g) |
soil samples | soil samples in range (1–3000 pg/g) |
|---|---|---|---|---|---|---|
| none | 12.6 | 4.2 | 3.4 | 1.9 | 14.6 | 5.4 |
| mathematical model | 6.5 (52%) |
1.6 (82%) |
1.3 (89%) |
1.1 (88%) |
7.7 (51%) |
1.7 (83%) |
| TEF REP conversion factor | 7.0 (48%) |
2.3 (57%) |
2.3 (45%) |
1.3 (68%) |
8.0 (48%) |
3.0 (55%) |
Average fold difference between CALUX-TEQ and TEQ is shown for all samples or sample subsets as indicated. TEQ values were derived using no transformation, mathematical modeling (eq 1), or TEF REP conversion, as indicated. Values in parentheses show what fraction of the discrepancy between CALUX-TEQ and TEQ is accounted for by mathematical modeling or TEF REP conversion, respectively.
Prediction of SW846 Data using TEF REP Conversion Factors
The XDS-CALUX bioassay has been used to define congener specific toxic equivalencies called REP values (32) based on the method described in refs 34 and 35. REP is an XDS-CALUX specific equivalent of TEF, as determined by the WHO using GC/MS. TEF REP conversion factors were calculated for each sample in the SW846 data set; the result for a representative sample is presented in Table 1, and average results for all ash and soil samples are summarized in Table 3. Results for individual samples are summarized graphically in Figure 5. By using TEF REP conversion factors, the overall discrepancy between CALUX-TEQ and TEQ is reduced but not eliminated (also see Table 2), with predicted TEQ on average 7.0–fold higher than TEQ (2.3-fold for ash samples and 8.0-fold for soil samples). Thus, applying a TEF REP conversion eliminates approximately half of the difference between CALUX-TEQ and GC/MS-TEQ.
TABLE 3.
Average TEF/REP Conversion Factors for Soil and Ash Samples in SW846 Data Set
| matrix | N | min | max | median | av | SD | rel SD (%) |
|---|---|---|---|---|---|---|---|
| ash | 8 | 0.52 | 0.80 | 0.69 | 0.69 | 0.11 | 16 |
| soil | 36 | 0.35 | 0.75 | 0.53 | 0.55 | 0.10 | 18 |
FIGURE 5.
Predicted CALUX-TEQ by TEF/REP conversion comparison with GC/MS TEQ. TEQ was calculated from XDS-CALUX data using TEF REP conversion factors as described previously (ref 32; also see Tables 1 and 3). Solid line is the data trend line; for comparison, the dashed line represents y = x(slope = 1 and intercept = 0). Sample matrix was ash (■) or soil (△). The regression line for ash is y = 4.49x0.82 and for soil is y = 3.88x0.97.
Discussion
The XDS-CALUX bioassay is a sensitive method for measuring the bioactivity of dioxin and dioxin-like chemicals. Using a recombinant cell line containing a stably transfected AhR responsive promoter and firefly luciferase reporter gene, the XDS-CALUX bioassay estimates the concentration of DLCs by measuring the ability of a sample to activate AhR-dependent luciferase gene expression. While the XDS-CALUX bioassay generates a CALUX-TEQ value, which reflects the relative bioactivity of DLCs in the sample, the traditional GC/MS analytical method for measuring dioxin-like compounds generates a TEQ value by physical measurement of the concentrations of individual chlorinated dioxin congeners and multiplication by a TEF and summation to generate a TEQ in the mixture. While both methods are efficient and accurate, they are based on different methods with differences in the underlying mechanisms. CALUX-TEQ and GC/MS-TEQ values are not expected to be identical due to differences in the TEF and REP values, measurement of other DLCs, or non-additive interactions that may occur in the XDS-CALUX method. The goal of this study was to define the relationship between CALUX-TEQ and GC/MS-TEQ. To this end, CALUX-TEQ and GC/MS-TEQ values were measured and compared for two data sets, a mathematical model was derived using the first data set, and the model was tested with a second data set. The results suggest that XDS-CALUX can be a useful and relatively accurate method for estimating the concentration of DLCs in complex environmental samples and that CALUX-TEQ and GC/MS-TEQ data can be compared.
CALUX-TEQ and GC/MS-TEQ were calculated and compared for 209 SITE samples that represented 32 soil and sediment samples with quadruplicate analysis by GC/MS after un-coding of the blinded data, allowing comparison to XDS-CALUX determinations. During data analysis, replicates of samples were identified, and some samples (blanks, PCB standards, TCDD standards, and other DLCs to evaluate limits of detection and specificity) were eliminated from the final data used for modeling. On average, CALUX-TEQ values were 9.4-fold higher than TEQ values, and these values demonstrated greater variance at extreme ends of the CALUX-TEQ/ TEQ distribution. Mathematical modeling of these data (excluding samples with TEQ values ≤1 and ≥3000 pg of TCDD/g) produced a model for the relationship between CALUX-TEQ and TEQ (eq 1). Applying eq 1 to SITE samples showed that predicted and GC/MS measured TEQ values are strongly correlated (R2 = 0.876) and that TEQ values predicted from CALUX-TEQ values were on average nearly identical to the GC/MS-TEQ (multiple of 0.997). The model was further tested using soil and ash samples from an independent data set, SW846. When eq 1 was applied to this data set, a strong correlation was again observed between predicted and TEQ measured values by GC/MS (R2 = 0.922 for soil samples and R2 = 0.817 for ash samples). Prior to transformation, CALUX-TEQ was on average 12.6-fold higher than TEQ, but after transformation using eq 1, the difference was reduced by 52% (Table 2). If the analysis was limited to samples with TEQ values in the range of 1–3000 pg of TCDD/g, predicted TEQ was 1.6-fold higher than TEQ. Thus, the model accounted for approximately 82% of the systematic difference between CALUX-TEQ and GC/MS-TEQ for this data set. Table 2 also shows that the difference between CALUX-TEQ and GC/MS-TEQ is generally higher in soil than in ash samples. While the reason for this difference is not entirely clear, it suggests that CALUX-TEQ and GC/MS-TEQ may be more similar for some sample types/matrices than others. This also suggests that the use of mathematical modeling should be conducted on a matrix-by-matrix basis. Analysis comparing XDS-CALUX and GC/MS on a matrix basis should aid in estimating TEQ since it would provide an estimate of the contribution of other halogenated dioxins/ furans and biphenyls and other active compounds such as chlorinated napthalenes that may contribute to CALUX-TEQ but not to GC/MS estimates of TEQ.
A previous study proposed a way of comparing GC/MS-TEQ and CALUX-TEQ results based solely on the difference between WHO TEF values and bioassay REP values (31). While this method takes into account only one source of variation between GC/MS-TEQ and CALUX-TEQ results, in some sample matrices, it appears to provide acceptable results. XDS-CALUX REP values, which are a measure of the relative potency of specific dioxin congeners, are similar but non-identical to WHO estimates of TEF values (32). To determine how much of the observed variation between GC/MS-TEQ and CALUX-TEQ values resulted from differences in the relative potency for specific PCDD and PCDF congeners, this method of conversion was also conducted on the SW846 data set. Table 3 and Figure 5 show that this conversion reduced the average fold difference between CALUX-TEQ and GC/MS-TEQ from 12.6 to 7.0 (all samples) or 4.2 to 2.3 (all samples in the range of 1–3000 pg of TCDD/g). Thus, TEF REP conversion accounts for approximately half of the difference between CALUX-TEQ and TEQ. Given that the TEF values themselves may possess an order of magnitude in variance in establishing these values, the point estimates between CALUX-TEQ and TEQ correlate remarkably well (15–17).
On the basis of the previous analysis and results from prior studies, we propose that at least three factors account for the observed differences between CALUX-TEQ and GC/ MS-TEQ: (i) XDS-CALUX may detect bioactivity of AhR agonists that are not normally measured by GC/MS such as polychlorinated naphthalenes (34, 35) or brominated dioxin-like compounds (36, 37); (ii) XDS-CALUX, but not GC/MS, can detect non-additive interactions between AhR ligands (refs 38–40 and reviewed in ref 41); and (iii) specific dioxin-like chemicals have different relative potency values using GC/MS and XDS-CALUX (i.e., TEF and REP, respectively). The use of conversion factors based on the difference between TEF and REP values can correct for the third factor, but for the ash and soil samples in the SW846 study, this only accounted for about half of the observed differences. In contrast, mathematical modeling as conducted in this study can take into account all three factors. The model provides an average basis for estimation of GC/MS-derived TEQ from CALUX-TEQ based upon sampling of 10 sites that are under remediation for these contaminates.
Studies are ongoing with the U.S. EPA and the Battelle Corporation to identify the hypothesized DLCs and other chemical entities in these sample extracts that are acting as AhR agonists. The patented sample processing method (U.S. Patent 6,720,431 (29)) has been demonstrated to remove greater than 99.8% of 14 different non-halogenated aromatic hydrocarbons that are AhR agonists (28). Two aromatic hydrocarbons (beno (b) fluoranthene and indeno (1,2,3 cd) pyrene) were removed from sample extracts to a lesser extent (94.3 and 97.5%, respectively). This suggests that non-halogenated aromatic hydrocarbons that are AhR agonists are probably not responsible for the higher estimates of CALUX-TEQ observed in the samples from the SITE study. However, this possibility cannot be completely discounted if extremely high concentrations of aromatic hydrocarbon AhR agonists are present in the samples. Brominated and/or mixed bromo/chloro dioxins and furans and halogenated naphthalenes are two classes of chemicals that could contribute to the higher XDS-CALUX activity.
The previous analysis also suggests that XDS-CALUX and GC/MS agree more closely for samples falling in the midrange of detection limits for these analytical methods (i.e., samples with TEQ >1 and <3000 pg of TCDD/g in this study). The variance observed with very dilute or concentrated samples in this study was likely due to threshold and saturation effects that are typical for most analytical methods. It is also possible that for samples with low levels of dioxin contamination, the bioactivity of other AhR agonists that are not measured by GC/MS could be relatively more important contributors to the overall biological response. Because of its broader specificity, XDS-CALUX may provide a better assessment of relative potency in extremely complex samples.
In summary, this study demonstrates that mathematical modeling can expand our ability to quantify the toxic potential of complex environmental samples containing dioxin and dioxin-like chemicals and compares the results obtained through different analytical methods. While GC/MS is used in current U.S. EPA regulatory requirements for quantifying dioxin contamination, XDS-CALUX is an important alternative method that can provide data for risk assessment and remediation decisions. Because of its high capacity and broad specificity, XDS-CALUX may be the preferred method (due to lower cost and time) for high-throughput screening or to accommodate samples with high or unknown complexity. The results presented here make it possible to compare analytical studies that use XDS-CALUX and GC/MS. Thus, the XDS-CALUX bioassay, coupled with the model presented here, may have important future applications in risk assessment and risk management associated with dioxin contamination and/or hazardous waste site remediation.
Acknowledgments
This work was supported by a grant from the National Institutes of Environmental Health Sciences, Small Business Initiated Research (SBIR) Grant ES08372 and an NIEHS Superfund Basic Research Grant (M.S.D., ES04699). The authors acknowledge the valuable contributions of Dr. Miriam Sanders of Page One editorial services in revising the manuscript.
Literature Cited
- 1.Safe SH. Polychlorinated biphenyls (PCBs): Environmental impact, biochemical and toxic responses, and implications for risk assessment. Crit. Rev. Toxicol. 1994;24:87–149. doi: 10.3109/10408449409049308. [DOI] [PubMed] [Google Scholar]
- 2.Birnbaum LS. TEFs: A practical approach to a real world problem. Hum. Ecol. Risk Assess. 1999;5:13–24. [Google Scholar]
- 3.Goldstein JA, Safe S. Mechanism of action and structure–activity relationships for chlorinated dibenzo-p-dioxins and related compounds. In: Jenson KA, editor. Halogenated Biphenyls, Terphenyls, Napthalenes, Dibenzofurans, and Related Products. New York: Elsevier Science Publishers; 1989. pp. 239–293. [Google Scholar]
- 4.Safe S. Polychlorinated biphenyls (PCBs), dibenzo-p-dioxins (PCDDs), dibenzofurans (PCDFs), and related compounds: Environmental and mechanistic considerations which support the development of toxic equivalency factors (TEFs) Crit. Rev. Toxicol. 1990;21:51–88. doi: 10.3109/10408449009089873. [DOI] [PubMed] [Google Scholar]
- 5.McConnell EE, Moore JA, Haseman JK, Harris MW. The comparative toxicity of chlorinated dibenzo-p-dioxins in mice and guinea pigs. Toxicol. Appl. Pharmacol. 1978;44:335–356. doi: 10.1016/0041-008x(78)90195-3. [DOI] [PubMed] [Google Scholar]
- 6.Birnbaum LS. The mechanism of dioxin toxicity: Relationship to risk assessment. Environ. Health Perspect. 1994;102 Suppl. 9:157–167. doi: 10.1289/ehp.94102s9157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Vito MJ, Birnbaum LS. Toxicology of the dioxins and related chemicals. In: Schecter A, editor. Dioxins and Health. New York: Elsevier Science Publishers; 1994. [Google Scholar]
- 8.Committee on EPA’s Exposure and Human Health Reassessment of TCDD and Related Compounds National Research Council. Washington, DC: National Academy Press; 2006. http://www.nap.edu/catalog/11688.html. [Google Scholar]
- 9.Sogawa K, Fujisawa-Sehara A, Yamane M, Fujii-Kuriyama Y. Location of regulatory elements responsible for drug induction in the rat cytochrome P-450c gene. Proc. Natl. Acad. Sci. U.S.A. 1986;83:8044–8048. doi: 10.1073/pnas.83.21.8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denison MS, Fisher JM, Whitlock JP., Jr The DNA recognition site for the dioxin–Ah receptor complex. Nucleotide sequence and functional analysis. J. Biol. Chem. 1988;263:17221–17224. [PubMed] [Google Scholar]
- 11.Okey AB, Riddick DS, Harper PA. The Ah receptor: Mediator of the toxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) and related compounds. Toxicol. Lett. 1994;70:1–22. doi: 10.1016/0378-4274(94)90139-2. [DOI] [PubMed] [Google Scholar]
- 12.Hankinson O. The aryl hydrocarbon receptor complex. Annu. Rev. Pharmacol. Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- 13.Sewall CH, Lucier GW. Receptor-mediated events and the evaluation of the Environmental Protection Agency (EPA) of dioxin. Mutat. Res. 1995;333:111–122. doi: 10.1016/0027-5107(95)00137-9. [DOI] [PubMed] [Google Scholar]
- 14.Denison MS, Phelen D, Elferink CJ. The Ah receptor signal transduction pathway. In: Denison MS, Helferich WG, editors. Xenobiotics, Receptors, and Gene Expression. Philadelphia: Taylor and Francis; 1998. pp. 3–33. [Google Scholar]
- 15.Van den Berg M, Birnbaum L, Bosveld AT, Brunstrom B, Cook P, Feeley M, Giesy JP, Hanberg A, Hasegawa R, Kennedy SW, Kubiak T, Larsen JC, van Leeuwen FX, Liem AK, Nolt C, Peterson RE, Poellinger L, Safe S, Schrenk D, Tillitt D, Tysklind M, Younes M, Waern F, Zacharewski T. Toxic equivalency factors (TEFs) for PCBs, PCDDs, and PCDFs for humans and wildlife. Environ. Health Perspect. 1998;106:775–792. doi: 10.1289/ehp.98106775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haws LC, Su SH, Harris M, Devito MJ, Walker NJ, Farland WH, Finley B, Birnbaum LS. Development of a refined database of mammalian relative potency estimates for dioxin-like compounds. Toxicol. Sci. 2006;89:4–30. doi: 10.1093/toxsci/kfi294. [DOI] [PubMed] [Google Scholar]
- 17.Van den Berg M, Birnbaum LS, Denison M, De Vito M, Farland W, Feeley M, Fiedler H, Hakansson H, Hanberg A, Haws L, Rose M, Safe S, Schrenk D, Tohyama C, Tritscher A, Tuomisto J, Tysklind M, Walker N, Peterson RE. The 2005 World Health Organization reevaluation of human and mammalian toxic equivalency factors for dioxins and dioxin-like compounds. Toxicol. Sci. 2006;93:223–241. doi: 10.1093/toxsci/kfl055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garrison PM, Tullis K, Aarts JM, Brouwer A, Giesy JP, Denison MS. Species specific recombinant cell lines as bioassay systems for the detection of 2,3,7,8-tetrachlorodibenzo-p-dioxin-like chemicals. Fundam. Appl. Toxicol. 1996;30:194–203. doi: 10.1006/faat.1996.0056. [DOI] [PubMed] [Google Scholar]
- 19.Han D, Nagy SR, Denison MS. Comparison of recombinant cell bioassays for the detection of Ah receptor agonists. Biofactors. 2004;20:11–22. doi: 10.1002/biof.5520200102. [DOI] [PubMed] [Google Scholar]
- 20.Denison MS, Seidel SD, Rogers WJ, Ziccardi M, Winter GM, Heath-Pagliuso S. Natural and synthetic ligands for the Ah receptor. In: Puga A, Wallace KB, editors. Molecular Biology Approaches to Toxicology. Philadelphia: Taylor and Francis; 1999. pp. 393–410. [Google Scholar]
- 21.Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu. Rev. Pharmacol. Toxicol. 2003;43:309–334. doi: 10.1146/annurev.pharmtox.43.100901.135828. [DOI] [PubMed] [Google Scholar]
- 22.Brown DJ, Kishimoto Y, Ikeno O, Chu M, Nomura J, Murakami T, Murata H. Validation study for the use of the dioxin responsive CALUX assay for analysis of Japanese ash and soil samples. Organohalogen Compd. 2000;45:200–203. [Google Scholar]
- 23.Van Overmeir I, Goeyens L, Beernaert H, Srebrnik S, De Poorter G, Baeyens W, Clark GC, Chu M, Chu A, Chu D, Morris R, Brown DJ. A comparative study of GC-HRMS and CALUX-TEQ determinations in food samples by the Belgian federal ministries of public health and agriculture. Organohalogen Compd. 2000;45:196–199. [Google Scholar]
- 24.Tsutsumi T, Amakura Y, Nakamura M, Brown DJ, Clark GC, Sasaki K, Toyoda M, Maitani T. Validation of the CALUX bioassay for the screening of PCDD/Fs and dioxin-like PCBs in retail fish. Analyst. 2003;128:486–492. doi: 10.1039/b300339f. [DOI] [PubMed] [Google Scholar]
- 25.United States Environmental Protection Agency. Washington, DC: Technologies for Monitoring and Measurement of Dioxin and Dioxin-like Compounds in Soil and Sediment (Xenobiotic Detection Systems, Inc. CALUX by XDS) 2005 http://www.epa.gov/ord/SITE/reports/540r05001/540r05001.htm.
- 26.Brown DJ, Goeyens L, Van Overmeir I, Chu M, Murata H, Clark GC. Quality control criteria implemented for monitoring the use of the CALUX bioassay. Organohalogen Compd. 2001;54:32–35. [Google Scholar]
- 27.Brown DJ, Nakamura M, Chu MD, Denison MS, Murata H, Clark GC. Recovery determinations for bioassay analysis: Considerations and results. Organohalogen Compd. 2002;58:357–360. [Google Scholar]
- 28.Brown DJ, Van Overmeir I, Goeyens L, Chu MD, Denison MS, Clark GC. Elimination of interfering compounds in preparation for analysis by an Ah receptor-based bioassay. Organohalogen Compd. 2002;58:401–404. [Google Scholar]
- 29.Chu MD, Clark GC, inventors. Methods and Apparatus for Separating and Detecting Specific Polyhalogenated Diaromatic Hydrocarbons. 6,720,431. U.S. Patent. 2004
- 30.Zeger SL, Liang KY. Longitudinal data analysis for discrete and continuous outcomes. Biometrics. 1986;42:121–130. [PubMed] [Google Scholar]
- 31.Besselink H, Jones A, Pijnappels M, Swinkels A, Fjellanger K, Brouwer A. Comparison of DR CALUX and HRGCMS-derived TEQs. Organohalogen Compd. 2003;60:203–206. [Google Scholar]
- 32.Brown DJ, Chu M, Van Overmeir I, Chu A, Clark GC. Determination of REP values for the CALUX bioassay and comparison to the WHO TEF values. Organohalogen Compd. 2001;53:211–214. [Google Scholar]
- 33.Denison MS, Zhao B, Baston DS, Clark GC, Murata H, Han DH. Recombinant cell bioassay systems for the detection and relative quantification of halogenated dioxins and related chemicals. Talanta. 2004;63:1123–1133. doi: 10.1016/j.talanta.2004.05.032. [DOI] [PubMed] [Google Scholar]
- 34.Villeneuve DL, Kannan K, Khim JS, Falandysz J, Nikiforov VA, Blankenship AL, Giesy JP. Relative potencies of individual polychlorinated naphthalenes to induce dioxin-like responses in fish and mammalian in vitro bioassays. Arch. Environ. Contam. Toxicol. 2000;39:273–281. doi: 10.1007/s002440010105. [DOI] [PubMed] [Google Scholar]
- 35.Villeneuve DL, Khim JS, Kannan K, Giesy JP. Relative potencies of individual polycyclic aromatic hydrocarbons to induce dioxin-like and estrogenic responses in three cell lines. Environ. Toxicol. 2002;17:128–137. doi: 10.1002/tox.10041. [DOI] [PubMed] [Google Scholar]
- 36.Brown DJ, Van Overmeire I, Goeyens L, Denison MS, De Vito MJ, Clark GC. Analysis of Ah receptor pathway activation by brominated flame retardants. Chemosphere. 2004;55:1509–1518. doi: 10.1016/j.chemosphere.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 37.Behnisch PA, Hosoe K, Sakai S. Brominated dioxin-like compounds: In vitro assessment in comparison to classical dioxin-like compounds and other polyaromatic compounds. Environ. Int. 2003;29:861–877. doi: 10.1016/s0160-4120(03)00105-3. [DOI] [PubMed] [Google Scholar]
- 38.Windal I, Schroijen C, Van Wouwe N, Carbonnelle S, Van Overmeir I, Brown DJ, Clark GC, Baeyens W, Goeyens L. Non-additive interactions in CALUX. Organohalogen Compd. 2003;60:215–218. [Google Scholar]
- 39.Windal I, Denison MS, Birnbaum LS, Van Wouwe N, Baeyens W, Goeyens L. Chemically activated luciferase gene expression (CALUX) cell bioassay analysis for the estimation of dioxin-like activity: Critical parameters of the CALUX procedure that impact assay results. Environ. Sci. Technol. 2005;39:7357–7364. doi: 10.1021/es0504993. [DOI] [PubMed] [Google Scholar]
- 40.Windal I, Van Wouwe N, Eppe G, Xhrouet C, Debacker V, Baeyens W, De Pauw E, Goeyens L. Validation and interpretation of CALUX as a tool for the estimation of dioxin-like activity in marine biological matrixes. Environ. Sci. Technol. 2005;39:1741–1748. doi: 10.1021/es049182d. [DOI] [PubMed] [Google Scholar]
- 41.Safe S. Limitations of the toxic equivalency factor approach for risk assessment of TCDD and related compounds. Teratog., Carcinog., Mutagen. 1997;17:285–304. [PubMed] [Google Scholar]