Id2 is a negative regulator of B cell differentiation. Its expression was found to depend on Myc–Max–Mad transcriptional complexes. Here, we show that Mad3 expression levels that are elevated in immature B cells are actively involved in inducing Id2 expression by binding to its promoter, resulting in its augmented expression.
Abstract
Immature B cells migrate to the spleen where they differentiate into mature cells. This final maturation step is crucial to enable B cells to become responsive to antigens and to participate in the immune response. Previously, we showed that Id2 acts as a negative regulator of the differentiation of immature B cells occurring in the spleen. Id2 expression has been found to depend on Myc–Max–Mad transcriptional complexes in mammary epithelial cells. Nearly all studies to date have shown that Mad proteins inhibit proliferation, presumably by antagonizing the function of Myc proteins. In the current study, we followed the Mad family members during peripheral B cell differentiation. We show that Mad3 actively regulates B cell differentiation. Our results demonstrate that high expression levels of Mad3 in immature B cells induce Id2 expression, which inhibits transcription of genes essential for B cell differentiation. During their differentiation to mature cells, B cells reduce their Mad3 expression, enabling the maturation process to occur.
INTRODUCTION
B cell development is a highly regulated process that is initiated in the bone marrow (BM). BM B-lineage precursors proliferate and progress through differentiation steps that result in the production of immature, surface immunoglobulin (Ig)-expressing B lymphocytes. These nascent, immature B lymphocytes then migrate into the spleen to complete maturation and are incorporated into the long-lived peripheral lymphocyte pool. Several groups have subdivided the immature or “transitional” B lymphocytes into two separate subpopulations termed transitional 1 (T1) and transitional 2 (T2) cells, based on surface phenotype and functional characteristics (Loder et al., 1999; Allman et al., 2001; Su and Rawlings, 2002). It is likely that the T1 → T2 → mature follicular B cell pathway is the most common (Monroe and Dorshkind, 2007). A putative, third, nonproliferating transitional subset, termed T3, also was described (Allman et al., 2001). This population resembles T2 cells; however, its assignment to the transitional compartment and its position within a stepwise maturation pathway toward the mature compartment remain to be determined. The program of B cell development and differentiation is largely controlled at the level of transcription initiation. This pathway is characterized by activation or down-regulation of stage- and lineage-specific genes that are governed by regulatory DNA elements, and combinatorial interactions of ubiquitous and cell type-specific transcription factors and cofactors. Previously, we showed that the helix-loop-helix protein (HLH), Id2, is a negative regulator of B cell differentiation. The high levels of Id2 expressed in immature B cells result in inhibition of E2A binding activity to an E2-box site. In the absence of Id2 control, the unregulated differentiation is directed toward the mature B2 population (Becker-Herman et al., 2002).
Id2 expression was found to depend on Myc–Max–Mad transcriptional complexes in mammary epithelial cells (Lasorella et al., 2000; Siegel et al., 2003). Mad family proteins are transcriptional regulators that act as antagonists of the c-Myc proto-oncogene products. Both Myc and Mad proteins form heterodimers with the cofactor Max, thereby permitting binding to specific DNA motifs known as E-box sequences (Grandori et al., 2000; Luscher, 2001). A protein–protein interaction with Max is a key element in the proper functioning of the Myc–Max–Mad transcription factor network. The DNA-bound heterodimers recruit coactivator or corepressor complexes that induce alterations in chromatin structure and thereby regulate transcriptional activity. In a complex with Max, c-Myc has been implicated in cell growth, transformation, differentiation, and apoptosis.
The Mad family of basic (b)HLH transcriptional regulators is comprised of Mad1, Mxi1, Mad3, and Mad4 (Hurlin et al., 1995). The complex between Max and proteins from the Mad family, antagonizes c-Myc transactivation and promotes differentiation (Ayer et al., 1993, 1995; Grandori et al., 2000; Luscher, 2001; Nasi et al., 2001; Zhou and Hurlin, 2001; Levens, 2003). Although Myc–Max heterodimers recognize the E-box element (CACGTG) and activate transcription by recruiting activities that mediate histone acetylation, chromatin remodeling, and transcriptional elongation, Mad–Max heterodimers recognize the same E-box sequence but function as transcriptional repressors. Mad-mediated repression occurs through the interaction between a conserved amino-terminal region (Sin3 interaction domain) of the Mad protein with the mSin3A and mSin3B corepressors. These corepressors exist as multiprotein complexes containing histone deacetylases (HDACs) and other factors (Ayer et al., 1995; Schreiberagus et al., 1995; Hassig et al., 1997; Knoepfler and Eisenman, 1999). The function of the HDACs is crucial in mediating repression.
Targeted deletion of Mad1 and Mxi1 in mice has provided evidence for their role in cell cycle exit (Foley et al., 1998; Schreiber-Agus et al., 1998; Foley and Eisenman, 1999). Mad3 deletion does not induce any defect in cell cycle exit and differentiation. However, after gamma irradiation, increased cell death of thymocytes and neural progenitor cells was observed, implicating Mad3 in the regulation of the cellular response to DNA damage (Queva et al., 2001). Mad4 transcripts are most abundant in cells that have further advanced along the differentiation pathway (Queva et al., 1998). It was recently shown that in living cells, Mad3 competes less efficiently compared with Mad4 for heterodimerization with Max. Mad3 is therefore predicted to be a less efficient repressor than Mad4, assuming that the efficiency of competition for dimerization with Max is the primary determinant of repression efficiency (Grinberg et al., 2004).
In the present study, we wanted to determine whether in B cells, members of the Mad family regulate Id2 expression and function, resulting in the control of their differentiation. We show that Mad3 inhibits B cell differentiation by regulating Id2 activity. Mad3 binds to the Id2 promoter, leading to Id2 transcription and expression, and resulting in inhibition of B cell maturation. Thus, our results suggest a novel role for Mad3 in B cell differentiation.
MATERIALS AND METHODS
Animals
C57BL/6 and CD74−/− (Shachar et al., 1995) mice (6–8 wk of age) were raised and maintained under pathogen-free conditions. All animal procedures and experiments were approved by the Animal Research Committee at the Weizmann Institute.
Cells and B Cell Separation
Spleen cells were obtained from C57BL/6 mice or CD74−/− mice at 6–8 wk of age, as described previously (Shachar et al., 1995). B cells were then purified from each mouse strain, by using CD45R (B220) beads (BD Biosciences, San Jose, CA).
Separation of T1, T2, and Mature B Cell Populations
Splenocytes were stained with anti-CD45R/B220, anti-CD21 monoclonal antibody (mAb), anti-CD24 mAb, and anti-CD23 mAb (eBioscience, San Diego, CA). After gating on the CD45R-positive population, which represents total B cells, the different subpopulations were sorted by FACSAria cell sorter (BD Biosciences) as described previously (Loder et al., 1999; Petro et al., 2002). The following markers were used to define each population: T1 B cells: CD21− CD23− CD24+; T2 B cells: CD21+ CD23+ CD24+; and mature B cells: CD21+ CD23+ CD24−.
Constructs and Molecular Cloning
The pcB6-mad3 and pcB6-mad4 plasmids were a kind gift from Dr. Bernhard Lüscher (Institut fur Biochemie, Aachen, Germany) (Sommer et al., 1998).
For the luciferase construct, an Id2 promoter fragment was prepared by polymerase chain reaction (PCR) with Pfu DNA polymerase (Promega, Madison, WI), using genomic mouse DNA as a template.
For PGL2-Id2, the cloning primer sequences were as follows: 5′ primer, 5′-TCACGCGTCTGCAGTCTCCAAACTGAGA-3′ and 3′ primer, 5′-ATCAAGCTTAGGCTTTCATGCTGCTCGTA-3′.
For all the constructs, the PCR products were separated by agarose electrophoresis to ensure the correct size and purified from the gel using Gel Extraction kit (QIAGEN, Valencia, CA). The product and the cloning plasmid (PGL2) were digested by MluI (New England Biolabs, Ipswich, MA) followed by BglII (BioLab), and the product was ligated into the PGL2 vector (Promega). Clones were subjected to automated DNA sequencing by standard protocols using an ABI377 machine (Applied Biosystems, Foster City, CA). The sequence of Id2 was verified, with no errors in the sequence.
Preparation of Cell Extracts
Purified B cells were lysed in lysis buffer containing 25 mM Tris, pH 7.4, 2 mM vanadate, 75 mM β-glycophosphate, pH 7.2, 2 mM EDTA, 2 mM EGTA, 10 mM NaPPi, and 0.5% NP-40 in the presence of the following protease inhibitors: 10 μg/ml leupeptin, 10 μg/ml aprotinin, 10 μg/ml pepstatin, 10 μg/ml chymostatin (Roche Diagnostics, Mannheim, Germany), 1 mM phenylmethylsulfonyl fluoride (Sigma-Aldrich, St. Louis, MO), and 20 mM N-ethylmaleimide (Sigma-Aldrich).
RNA Isolation and Reverse Transcription
Total RNA was isolated from cells using the TRI Reagent kit (Molecular Research Center, Cincinnati, OH). Reverse transcription was carried out using Moloney murine leukemia virus RT (Invitrogen, Carlsbad, CA). Primers that were used included Mad3, 5′-CCCGATACACTACACTAAGC-3′:5′-GCCCAGGAACAAGAAGT-3′; Mad4, 5′-CCAAACTCCATGCCCTCTAT-3′:5′-CCCGAACAACAGGTCTTCAC-3′; Max, 5′-ATGACCTCAAGCGGCAGAA-3′:5′-CTGGTAGAGGCGTCCTC-3′; and HPRT, 5′-GTTGGATACAGGCCAGACTTTGTTG-3′:5′-GAGGGTAGGCTGGCCTATGGCT-3′.
Transfection into 293 Cells
Human embryonic kidney 293 cells (human embryonic kidney fibroblasts) were maintained in F-12/DMEM supplemented with 10% fetal calf serum. Transfections were performed using the standard CaCl2 method. In transfections to be used for total protein extraction or cell fractionation, the cells were seeded in a 10 cm2/dish.
For luciferase reporter assays, cells were seeded in a 24-well multidish and transfected with mad3 or mad4 vectors by using a total amount of 1 μg of DNA. For Id2 reporter assay, 2 ng of the reporter plasmid was cotransfected.
Electrophoretic Mobility Shift Assay (EMSA)
Nuclear extracts were prepared as described previously (Becker-Herman et al., 2002) from CD74−/− and control B cells. A double-stranded oligonucleotide (oligo), containing the Oct or the Id2 promoter site, was 32P labeled using Klenow fragment of DNA polymerase I for use as a probe. The oligonucleotide sequences were as follows: AGGAACACTACAAATGTGACTTC-3′:5′-GTGATGTTTACACTGAAGGCAG-3′, TGTCACGTGGAGGTCTT-3′:5′-TTGGGACAGTGTACCT-3′, and TTGGGGCACATGGCTGT-3′:5′-GTGTACCGACAGGGTTT-3′.
The antibodies (Abs) were preincubated with the extract for 30 min at 4°C and then with the probes for 20 min at 4°C. The Abs used included anti-Mad3, anti-Max, anti-Mad4, and isotype control Ab (Santa Cruz Biotechnology, Santa Cruz, CA). The probed lysates were separated on an 8% acrylamide gel.
Chromatin Immunoprecipitation (ChIP)
CD74−/− (1.5 × 108) cells were fixed by adding 1/10 volume of 11% formaldehyde in phosphate-buffered saline (PBS) to the cell suspension for 10 min at room temperature (RT). Fixation was stopped by adding glycine to final concentration 0.125 M. Cells were washed once with PBS. Nuclei were isolated, lysed, and DNA was sonicated to get fragments of 500-2000 base pairs, as described previously (Lantner et al., 2007). For immunoprecipitation, 2 μg of anti-Mad3 (sc770; Santa Cruz Biotechnology), anti-Mad4 (sc-771; Santa Cruz Biotechnology), anti-acetylated H3 (H3A; 06-559, Millipore, Billerica, MA), or 2 μg of rabbit IgG (H+L) (Jackson ImmunoResearch Laboratories, West Grove, PA) as a control antibody were used. Washing, reversal of cross-linking, and purification were performed as described previously (Lantner et al., 2007).
Eluted DNA was analyzed by PCR. PCR was performed on 10 ng of extracted DNA using oligomers for Id2 promoter and HPRT. The following oligomers were used: Id2 promoter (582 CAAATG), 5′-GTTGCAAAGCTTCACGCT and 3′-GCTAGCGTTAGGGAAGACAG; and HPRT, 5′-TCACTGATAATTGGGAATACTGT and 3′-CCCGACTTACCTTTATTACCAT.
Antisense Oligonucleotide-mediated Knockdown
To inhibit gene expression, 1.6 × 107 (4 × 106/ml) primary B cells derived from control or CD74−/− mice were incubated in the presence of 10 μM of Mad3 antisense, Mad3 sense, Mad4 antisense, or Mad4 sense oligonucleotides (with phosphorothioate modification at both ends). Cells were incubated in six-well plates in RPMI 1640 medium without fetal calf serum (FCS) at RT for 1 h. Next, 20% (vol/vol) of FCS was added to the medium, and cells were incubated at 37°C for 24 or 48 h.
The oligonucleotide sequences were as follows: Mad3 sense, 5′-TCGCCGACATGGAACCCG-3′; Mad3 antisense, 5′-CGGGTTCCATGTCGGCGA-3′; Mad4 sense, 5′-GGTGGATATAGAGGGCATGGA-3′; and Mad4 antisense, 5′-TCCATGCCCTCTATATCCACC-3′.
Real-Time Reverse Transcription (RT)-PCR Analysis
Levels of mRNA of actin, Mad3, and Id2 were analyzed by quantitative real-time RT-PCR by using a LightCycler instrument (Roche Diagnostics). Total RNA was isolated from cells using the TRI Reagent kit (Molecular Research Center). Reverse transcription was carried out using SuperScript II RT (Invitrogen). The reaction volume (10 μl) contained 3 mM MgCl2, LightCycler HotStart DNA SYBR Green I mix (Roche Diagnostics), specific primer pairs, and 2.5 ml of cDNA. Conditions for PCR were as follows: 10 min at 95°C followed by 40–60 cycles of 15 s at 95°C, 15 s at 60°C, and 15 s at 72°C. Primer sequences were as follows: Mad3, 5-AGAGCACGGTTATGCG-3 and 5-AGTGTATCGGGTACAGTC-3; Id2, 5-ATATTGAGTGAACCTTGTG-3 and 5-AGCACTGGTTGTCTGA-3; and Actin, 5-CAGTAACAGTCCGCCT-3 and 5- GTGACGTTGACATCCG-3. β-Actin levels were used to normalize samples for calculation of the relative expression levels of the Id2 gene.
Western Blot Analysis
To detect changes in protein expression, lysates were separated by 15%(wt/vol) SDS-polyacrylamide gel electrophoresis (PAGE). The proteins were transferred onto a nitrocellulose membrane and probed with anti-Mad3 Ab (Santa Cruz Biotechnology), anti-Mad4 Ab (Santa Cruz Biotechnology), or anti-Max Ab (Santa Cruz Biotechnology) followed by horseradish peroxidase-conjugated anti-rabbit Ab (The Jackson Laboratory, Bar Harbor, ME). The membrane was then stripped and reprobed with anti-tubulin antibody (Sigma-Aldrich) followed by peroxidase-conjugated anti-mouse Ab (The Jackson Laboratory).
Coimmunoprecipitation and Western Blot
Protein G-Sepharose beads (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) were conjugated to Mad3 (H-206; sc-770, Santa Cruz Biotechnology), p300 (C-20; sc-585, Santa Cruz Biotechnology), or rabbit anti-mouse IgG (Jackson ImmunoResearch Laboratories) mAb for 2 h at 4°C, followed by three washes in PBS. Beads were added to the cell lysates, and proteins were immunoprecipitated overnight. The protein G-bound material was washed three times with PBS containing 0.1% SDS and 0.5% NP-40. Immunoprecipitates were separated by 10% (wt/vol) SDS-PAGE. The protein bands were transferred onto a nitrocellulose membrane and probed with anti-Mad3 or anti-p300 respectively, followed by horseradish peroxidase-conjugated anti rabbit.
Analysis of B Cell Populations
Characterization of B cell populations was performed using the following antibodies: RA3-6B2 anti-CD45R/B220, anti-CD21 (CR2/CR), and anti-CD24 (HSA) or anti-AA4.1 (all from eBioscience). Staining was analyzed by fluorescence-activated cell sorting (FACS).
RESULTS
Mad3 Expression Level Is Reduced during B Cell Maturation
To identify genes that are differentially expressed during B cell maturation, we compared the expression pattern of genes in the T1, T2, and mature B cell populations of C57BL/6 mice, and in CD74−/− mice, whose peripheral B cells are mainly T1 (Shachar and Flavell, 1996), by using the GeneChip expression analysis system (Affymetrix, Santa Clara, CA). The expression of Mad3 was found to be markedly elevated in the immature population, whereas it was down-regulated in the T2 and mature populations.
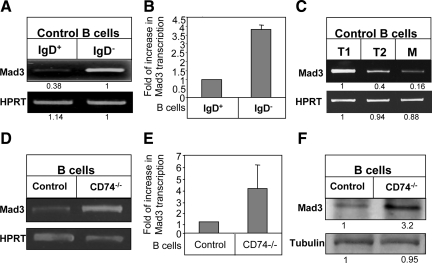
To verify that Mad3 expression is indeed differently expressed during B cell development in the spleen, we first compared its mRNA levels in the immature and mature B cell populations. To this end, we analyzed the immature (IgD−) and mature (IgD+) B populations purified from control (C57BL/6) mice for, Mad3 transcription. As shown in Figure 1, A and B, high levels of Mad3 mRNA were detected in IgD− immature B cells, whereas these levels were dramatically reduced in the mature (IgD+) population. To further dissect the stages of B cell development at which Mad3 levels are regulated, B cells derived from control mice were sorted by FACS for T1, T2, and mature populations based on the CD21, CD23, and CD24 markers (as described in Materials and Methods). As shown in Figure 1C, Mad3 message was most highly expressed in the immature population (T1), and its transcription was down-regulated during B cell development to mature cells.
Figure 1.
Mad3 expression is down-regulated as part of the transition from immature to mature B cells. (A–E) A and B, control immature (IgD−) and mature (IgD+) cells; C, sorted T1 B cell (CD21-CD23− CD24+), T2 B cell (CD21+, CD23+ CD24+), and mature B cell (CD21+, CD23+, and CD24−) populations derived from control mice were purified. (D and E) B cells from CD74−/− or control mice. (A, C, and D) RT-PCR for Mad3 and HPRT was performed. (B and E) Quantitative RT-PCR results are expressed as fold change in Mad3 expression in stimulated cells compared with nonstimulated cells, which was defined as 1 (N = 3). (F) Western blot showing steady-state levels of Mad3 in total cell lysates of B cells from control and CD74−/− mice. Results presented are representative of at least three separate experiments.
Furthermore, we analyzed Mad3 mRNA levels in immature B cells derived from CD74−/− mice (Shachar and Flavell, 1996; Matza et al., 2001, 2002, 2003) and mature B cells (IgD+) derived from control mice. As shown in Figure 1, D and E, elevated levels of Mad3 mRNA were detected in the immature CD74−/− population, compared with the mature cells. Thus, immature B cells derived from control or CD74−/− mice express higher levels of Mad3. Control and CD74−/− immature B cells behave similarly in our various studies (Becker-Herman et al., 2002). Moreover, because the CD74−/− immature B cell population is enriched and more available compared with the immature population derived from control mice, we continued our analysis using this population.
We next compared Mad3 protein expression in immature and mature B populations by Western blot analysis. As shown in Figure 1F, reduced expression of Mad3 protein was observed in the mature compared with the immature cells. Thus, the bHLH transcription factor Mad3 is specifically down-regulated during differentiation of immature cells to mature B cells.
There were no significant differences in the mRNA levels of either Mad2 or Mad4, and the expression levels of Mad4 and Max proteins were similar in both populations (Supplemental Figure 1). These results suggest that Mad3 rather than Max, Mad4, and Mad2 might play a role during B cell differentiation.
Mad3 Binds to the Id2 Promoter
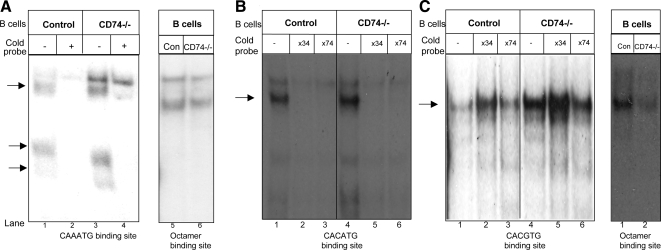
Because the Myc–Max–Mad transcription complex regulates Id2 expression in epithelial cells (Lasorella et al., 2000; Siegel et al., 2003), we next wanted to determine whether Mad3 similarly regulates Id2 expression in B cells. We investigated this potential relationship by analyzing Mad3 binding activity to the Id2 promoter in the transition between immature and mature B cell populations. The promoter region of the Id2 gene contains several E-box consensus binding sites (CANNTG). To determine whether proteins in the lysates of immature and mature cells bind to the Id2 promoter, we analyzed binding to five potential E-box sites that were synthesized as DNA probes: CAAATG, CACGTG, and CACATG (Figure 2) and CAATTG and CAGCTG (data not shown). Whole-cell extracts from the immature and mature B populations were prepared and analyzed using the EMSA, with these oligonucleotide probes or the octamer binding site serving as a probe control. Several complexes of proteins from lysates of both mature and immature cells were formed with all the probes. To determine the specificity of the binding, cold Id2 promoter oligonucleotide probes were added to cell extracts from CD74−/− and control B cells. Each unlabeled probe had the identical sequence to the labeled probe used in the particular experiment. As demonstrated in Figure 2A, extracts from immature (CD74−/−) B cells bound more strongly to the CAAATG probe compared with mature B cell extract. Addition of the cold probe resulted in a reduction in the intensity of the specific bands that were generated after incubation with the labeled probes in both immature and mature populations (Figure 2A). These results suggest existence of greater levels of heterodimeric complexes in the immature population compared with the mature population, whereas Oct-1 binding remained unchanged. In addition, we analyzed the specificity of the binding of the CACGTG and CACATG probes. Addition of the cold CACATG probe resulted in a reduction in the intensity of the specific bands that were generated after incubation with the CACATG-labeled probes in both immature and mature populations (Figure 2B), whereas no change was detected after the addition of the CACGTG probe (Figure 2C).
Figure 2.
Binding activity to the Id2 promoter in immature and mature B cells. Control or CD74−/− B cells extracts were subjected to EMSA using labeled probes representing the Id2 or Oct-1 promoters in the presence or absence of unlabeled Id2 probes (34-fold excess [B and C] or 74-fold excess [A–C]). The probes used were CAAATG (A), CACATG (B), and CACGTG (C). Arrows indicate complexes of proteins and probes. Results presented are representative of at least three separate experiments.
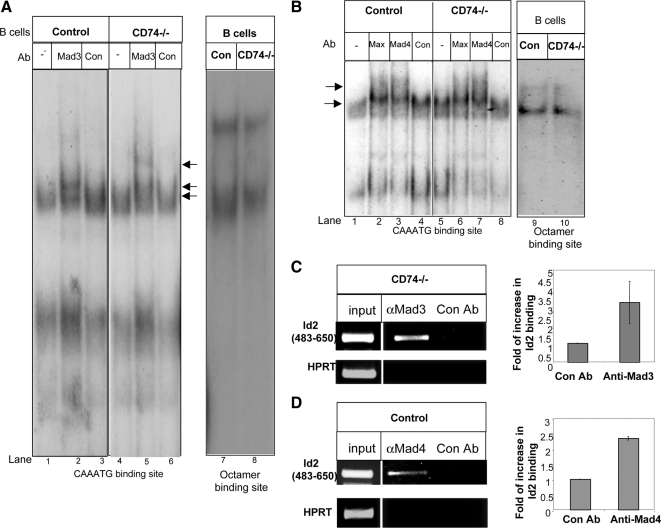
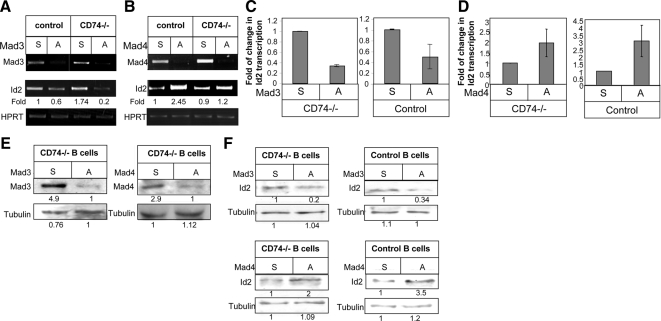
Next, to determine whether the observed DNA binding activity is Mad3 specific, the protein extracts were incubated with anti-Mad3 Ab (Figure 3A, lanes 2 and 5) or an isotype control antibody (Figure 3A, lanes 3 and 6). Incubation of extract from immature cells with Abs against this protein resulted in a supershift complex in EMSA using the CAAATG oligonucleotide (Figure 3A, lane 5), whereas incubation with an extract of mature cells (Figure 3A, lane 2) induced significantly lower levels of complex formation. No supershift complex could be observed using the CACATG DNA sequences (Supplemental Figure 2B). Moreover, we compared the binding activity of Mad4 and Max to the CAAATG oligonucleotide in the immature and mature populations. As shown in Figure 3B, similar levels of supershift complexes were detected in the immature and mature populations. These results show that Mad3 binds to Id2 promoter, primarily in immature B cells.
Figure 3.
Mad3 binds to the Id2 promoter. (A) Cell extracts from CD74−/− or control B cells were incubated in with or without an anti-Mad3 Ab (Mad3) or an isotype control antibody (Con) and analyzed by EMSA with the oligonucleotide CAAATG. (B) Cell extracts from CD74−/− or control B cells were incubated with or without anti-Mad4 (Mad4), anti-Max (Max), or isotype control (Con) Abs and analyzed by EMSA with the CAAATG oligonucleotide probe. (C and D) ChIP analysis of Mad3 (C) or Mad 4 (D) binding to the Id2 promoter. Chromatin prepared from CD74−/− (C) or control (D) B cells was immunoprecipitated with control or anti-Mad3 (C) or anti-Mad4 (D) antibodies. Presence of the promoter sequence was then quantified by quantitative real-time PCR. Results presented are representative of at least three separate experiments.
To gain further insight into the regulation of Id2 expression, we analyzed whether Mad3 bind directly to the CAAATG site in the Id2 promoter. We therefore designed oligonucleotides spanning this potential site, and analyzed Mad3 binding by ChIP. As seen in Figure 3C, CD74−/− derived Mad3 was found to be associated with the CAAATG site in the Id2 promoter. We next analyzed the ability of Mad4 to bind directly to the CAATG site in the Id2 promoter. As shown in Figure 3D, Mad4 derived from control extract was able to bind to this site as well.
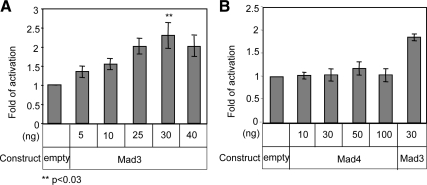
To directly determine the role of Mad3 and Mad4 in regulation of Id2 transcription activity, we analyzed Id2 promoter activity using a luciferase assay. Id2 reporter plasmid was generated as described in Materials and Methods. 293 cells were cotransfected with Mad3 (Figure 4A) or Mad4 (Figure 4B) expression plasmids along with a luciferase reporter containing the Id2 promoter and the Rous sarcoma virus promoter, which was used as a reference; luciferase activity was measured after 24 h. As demonstrated in Figure 4A, Mad3 enhanced the Id2 activity in a dose dependent manner, peaking at 30 ng of Mad3 plasmid, whereas Mad4 expression did not affect Id2 promoter activity (Figure 4B), confirming the specificity of the effect of Mad3 on Id2.
Figure 4.
Mad3 activates Id2 expression. (A and B) 293 cells were transfected with Mad3 (A) or Mad4 (B), and an Id2 promoter/luciferase construct, or with an empty vector. After 24 h, the cells were lysed and luciferase expression from the Id2 promoter was determined. Results presented are representative of at least three separate experiments.
We next sought to understand the mechanism through which Mad3 contributes to Id2-dependent transcription. We reasoned that Mad3 might regulate transcription by inducing alterations at the level of chromatin, which could control accessibility of DNA to initiation and elongation factors. It was shown previously that retinoid-induced Id2 gene suppression was associated with decreased levels of histone H3 and H4 acetylation (Memezawa et al., 2007). Therefore, we proposed that Mad3 might elevate Id2 transcription by inducing histone H3 acetylation. To test this hypothesis, we used ChiP to probe for H3 status at the Id2 promoter in immature and mature B cells. This analysis (Figure 5, A and B) revealed higher levels of Id2 promoter acetylation in immature B cells compared with mature cells.
Figure 5.
Mad3 binds to HAT p300, which induces histone H3 acetylation. (A and B) ChIP analysis of histone H3 acetylation in the Id2 promoter. Chromatin prepared from CD74−/− or control B cells was immunoprecipitated with control or anti-acetylated H3 antibodies. Presence of the promoter sequence was then quantified by RT-PCR (A) and quantitative real-time PCR (B). Results presented are representative of two separate experiments. (C and D) Immunoprecipitations: CD74−/− B220+ B cells were lysed. (C) After anti-p300 immunoprecipitation, proteins were separated on SDS-PAGE and transferred onto nitrocellulose. Mad3 was detected by Western blot analysis. (D) After anti-Mad3 immunoprecipitation, proteins were separated on SDS-PAGE and transferred onto nitrocellulose. p300 was detected by Western blot analysis. The results presented are representative of at least three separate experiments.
The correlation we observed between Mad3-mediated repression and H3 acetylation suggested to us that histone acetyltransferases (HATs) may be recruited to the Id2 promoter. We therefore determined whether there is a physical interaction between Mad3 and the HAT p300. As shown in Figure 5C, Mad3 coimmunoprecipitated with p300, and immunoprecipitation of Mad3 pulled down p300 (Figure 5D). Thus, Mad3 induces Id2 expression by recruiting p300, which elevates histone H3 acetylation at the Id2 promoter.
Mad3 Negatively Regulates B Cell Differentiation
Next, we artificially down-regulated the levels of intracellular Mad3, to directly demonstrate the role of Mad3 in Id2 expression. Mad3 and Mad4 expression levels were down-regulated using phosphorothioate-modified oligonucleotides complementary to Mad3 or Mad4 mRNA. Purified B cells derived from CD74−/− mice were incubated in the presence of antisense or sense Mad3 or Mad4 oligonucleotides (10 mM) for 24 h at 37°C, and Id2 transcription levels were then analyzed. As shown in Figure 6, A and C, in immature B cells, knockdown of Mad3 expression by using the antisense oligomers led to a significant decrease in Id2 mRNA levels compared with control (sense) oligos. However, silencing Mad4 expression resulted in an increase in Id2 mRNA levels (Figure 6, B and D). These results were further confirmed on the protein level (Figure 6, E and F). Thus, in immature B cells, Mad3 binds to the Id2 promoter and activates its transcription; when Mad3 expression is down-regulated, Id2 mRNA levels are similarly reduced. The elevated Id2 mRNA levels in cells in which Mad4 expression is reduced suggests that Mad4 has an inhibitory effect on Id2 expression levels in immature B cells.
Figure 6.
Knockdown of Mad3/Mad4 expression in immature and mature cells using antisense oligos. Purified B cells derived from CD74−/− or control mice were incubated in the presence of 10 nM Mad3 (A) or Mad4 (B) antisense (A) or sense (S) oligonucleotides for 24 h at 37°C. Id2 transcription levels were analyzed using RT-PCR. Results presented are representative of at least seven independent experiments. (C and D) Quantitative RT-PCR results are expressed as a fold change in Id2 expression after treatment with antisense Mad3 (C) or Mad4 (D). Results presented are representative of at least three independent experiments. (E and F) Western blot showing levels of Mad3 or Mad4 (E) or Id2 (F) in S- or A-treated B cells from control and CD74−/− mice.
We next followed the role Mad3 and Mad4 in mature B cells. B cells derived from control mice were incubated with antisense or sense Mad3 or Mad4 oligonucleotides, and their effects on Id2 mRNA levels were analyzed. Silencing of Mad3 expression reduced, although to a lesser extent, Id2 mRNA and protein levels in mature cells (Figure 6, A and C). However, reduced Mad4 expression resulted in augmented Id2 mRNA levels (Figure 6, B and D). These results were further confirmed on the protein level (Figure 6, E and F). Thus, in mature cells, where Mad3 expression levels are low, Mad4 plays an important role in inhibiting Id2 expression. In its absence, the low levels of Mad3 can induce Id2 expression.
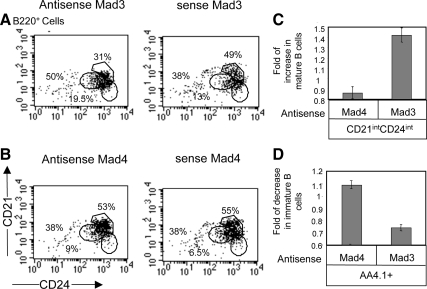
Finally, to directly show the effect of Mad3 on B cell differentiation, total splenocytes derived from CD74−/− mice were incubated in the presence of Mad3 antisense or sense oligonucleotides, and B cell subpopulations were analyzed using FACS analysis. As demonstrated in Figure 7, down-regulation of Mad3 shifted the cells toward the mature population (CD21intCD24int; AA4.1−; Figure 7, A, C, and D), whereas cells in which Mad4 expression was silenced showed a small elevation in the immature compartment compared with the sense-treated (control) cells (Figure 7, B–D). These results demonstrate that high expression levels of Mad3 in immature B cells induce Id2 expression. Id2 itself was shown previously to inhibit transcription of genes essential for B cell differentiation by inhibiting E2A binding activity (Becker-Herman et al., 2002). After differentiation to mature cells, B cells reduce their Mad3 expression; the low Mad3 levels allow Mad4 binding to the Id2 promoter and inhibition of Id2 expression, enabling the maturation process to occur.
Figure 7.
Mad3 negatively regulates B cell differentiation. (A–C) Total splenocytes derived from CD74−/− mice were incubated in the presence of 10 nM Mad3 (A) or Mad4 (B) antisense or sense oligonucleotides at 37°C. After 48 h, B cell subpopulations were analyzed for the mature B population by using anti-B220, anti-CD21, and anti-CD24 antibodies. (C) Graph summarizes the results of four independent experiments. (D) Total splenocytes derived from CD74−/− mice were incubated in the presence of 10 nM Mad3 or Mad4 antisense or sense oligonucleotides at 37°C. After 48 h, B cell subpopulations were analyzed for the mature B population by using anti-AA4.1 antibody. Graph summarizes the results of four independent experiments.
DISCUSSION
We have shown previously that the HLH protein Id2 is a negative regulator of immature B cell differentiation in the spleen (Becker-Herman et al., 2002). Id2 expression has been found to depend on Myc–Max–Mad transcriptional complexes in mammary epithelial cells (Lasorella et al., 2000; Siegel et al., 2003). Thus, in the current study, we wanted to determine the role of Mad proteins during B cell differentiation. Our gene expression screening revealed that Mad3 mRNA levels are elevated in cells of the immature stage; therefore, we hypothesized that this factor may be relevant to peripheral B cell differentiation.
Here, we demonstrate that Mad3 is most highly expressed in the immature population (T1), and its expression is down-regulated during B cell development to mature cells. Mad3 binds to the Id2 promoter primarily in immature B cells, resulting in induction of Id2 transcription. Antisense oligonucleotides complementary to Mad3 mRNA resulted in reduced Id2 mRNA levels and an increase in the mature population, whereas reduced Mad4 mRNA levels resulted in elevated levels of Id2 message. These results suggest that Mad3 and Mad4 have opposing effects in regulating Id2 expression. Higher levels of Mad3 at the immature stage compete with Mad4 in binding to the Id2 promoter, allowing expression of Id2. In the absence of Mad3, Mad4 binding to the Id2 promoter inhibits its transcription. In the mature stage, silencing of Mad3 expression slightly inhibited Id2 mRNA levels, whereas reduced Mad4 expression resulted in augmented Id2 mRNA levels. Thus, Mad4, which is expressed at similar levels in both immature and mature populations, can mediate its inhibitory role resulting in reduced Id2 expression when Mad3 expression levels are low.
Nearly all studies to date have shown that Mad proteins inhibit proliferation, presumably by antagonizing the function of Myc family members. Our studies show a novel function of Mad3 in regulating B cell differentiation by directly controlling Id2 expression, rather than as an antagonist of Myc function. We show that Mad3 induces transcription of Id2 by a process that involves histone H3 acetylation. It is generally accepted that Myc acts as a transcription activator by recruiting HATs, whereas Mad recruits HDAC-containing corepressors to inhibit target gene transcription. However, Id2 may be an exception, and HDACs are apparently recruited to the Id2 promoter by Myc to repress its transcription (Kurland and Tansey, 2008), whereas HAT is recruited by Mad3 in immature B cells. Together, our studies show that Mad3 is actively involved in differentiation. A recent study supports the conclusion that Mad3 has an active rather that inhibitory role, showing that Mad3 promotes cell proliferation in cultured granule neuron precursors and seems to have overlapping activity with Nmyc (Yun et al., 2007).
It was shown previously that during acute signaling, transforming growth factor (TGF)-β inhibits myc transcription and the subsequent formation of Myc–Max heterodimers on the Id2 promoter, leading to decreased Id2 synthesis. During chronic TGF-β signaling, mxi1/mad2 and mad4 expression is induced, resulting in increased association of Mad–Max heterodimers on the Id2 promoter, which contributes to the inhibition of Id2 transcription (Siegel et al., 2003). Our study adds an important player to the pathways regulating Id2 expression. We show that in immature B cells, Mad3 is actively involved in inducing Id2 expression, by directly binding to its promoter.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge members of the Shachar laboratory team for helpful discussions. This research was supported by The Israel Science Foundation and The Israel Science Foundation (Morasha), the Israel Cancer Association, and the Minerva foundation. I. S. is the incumbent of the Dr. Morton and Ann Kleiman Professorial Chair.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-09-0813) on April 7, 2010.
REFERENCES
- Allman D., Lindsley R. C., DeMuth W., Rudd K., Shinton S. A., Hardy R. R. Resolution of three nonproliferative immature splenic B cell subsets reveals multiple selection points during peripheral B cell maturation. J. Immunol. 2001;167:6834–6840. doi: 10.4049/jimmunol.167.12.6834. [DOI] [PubMed] [Google Scholar]
- Ayer D. E., Kretzner L., Eisenman R. N. Mad—a heterodimeric partner for Max that antagonizes myc transcriptional activity. Cell. 1993;72:211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- Ayer D. E., Lawrence Q. A., Eisenman R. N. Mad-Max transcriptional repression is mediated by ternary complex-formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- Becker-Herman S., Lantner F., Shachar I. Id2 negatively regulates B cell differentiation in the spleen. J. Immunol. 2002;168:5507–5513. doi: 10.4049/jimmunol.168.11.5507. [DOI] [PubMed] [Google Scholar]
- Foley K. P., Eisenman R. N. Two MAD tails: what the recent knockouts of Madl and Mxil tell us about the MYC/MAX/MAD network. Biochim. Biophys. Acta Rev. Cancer. 1999;1423:M37–M47. doi: 10.1016/s0304-419x(99)00012-8. [DOI] [PubMed] [Google Scholar]
- Foley K. P., McArthur G. A., Queva C., Hurlin P. J., Soriano P., Eisenman R. N. Targeted disruption of the MYC antagonist MAD1 inhibits cell cycle exit during granulocyte differentiation. EMBO J. 1998;17:774–785. doi: 10.1093/emboj/17.3.774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C., Cowley S. M., James L. P., Eisenman R. N. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- Grinberg A. V., Hu C. D., Kerppola T. K. Visualization of Myc/Max/Mad family dimers and the competition for dimerization in living cells. Mol. Cell. Biol. 2004;24:4294–4308. doi: 10.1128/MCB.24.10.4294-4308.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassig C. A., Fleischer T. C., Billin A. N., Schreiber S. L., Ayer D. E. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- Hurlin P. J., Queva C., Koskinen P. J., Steingrimsson E., Ayer D. E., Copeland N. G., Jenkins N. a., Eisenman R. N. Mad3 and Mad4—novel Max-interacting transcriptional repressors that suppress C-myc dependent transformation and are expressed during neural and epidermal differentiation. EMBO J. 1995;14:5646–5659. doi: 10.1002/j.1460-2075.1995.tb00252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler P. S., Eisenman R. N. Sin meets NuRD and other tails of repression. Cell. 1999;99:447–450. doi: 10.1016/s0092-8674(00)81531-7. [DOI] [PubMed] [Google Scholar]
- Kurland J. F., Tansey W. P. Myc-mediated transcriptional repression by recruitment of histone deacetylase. Cancer Res. 2008;68:3624–3629. doi: 10.1158/0008-5472.CAN-07-6552. [DOI] [PubMed] [Google Scholar]
- Lantner F., et al. CD74 induces TAp63 expression leading to B cell survival. Blood. 2007;110:4303–4311. doi: 10.1182/blood-2007-04-087486. [DOI] [PubMed] [Google Scholar]
- Lasorella A., Noseda M., Beyna M., Iavarone A. Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature. 2000;407:592–598. doi: 10.1038/35036504. [DOI] [PubMed] [Google Scholar]
- Levens D. L. Reconstructing MYC. Genes Dev. 2003;17:1071–1077. doi: 10.1101/gad.1095203. [DOI] [PubMed] [Google Scholar]
- Loder F., Mutschler B., Ray R. J., Paige C. J., Sideras P., Torres, Lamers M. C., Carsetti R. B cell development in the spleen takes place in discrete steps and is determined by the quality of B cell receptor-derived signals. J. Exp. Med. 1999;190:75–90. doi: 10.1084/jem.190.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscher B. Function and regulation of the transcription factors of the Mye/Max/Mad network. Gene. 2001;277:1–14. doi: 10.1016/s0378-1119(01)00697-7. [DOI] [PubMed] [Google Scholar]
- Matza D., Kerem A., Shachar I. Invariant chain, a chain of command. Trends Immunol. 2003;24:246–248. doi: 10.1016/s1471-4906(03)00073-5. [DOI] [PubMed] [Google Scholar]
- Matza D., Lantner D., Bogoch Y., Flaishon L., Hershkoviz R., Shachar I. Invariant chain induces B cell maturation in a process which is independent of its chaperonic activity. Proc. Natl. Acad. Sci. USA. 2002;99:3018–3023. doi: 10.1073/pnas.052703299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matza D., Wolstein O., Dikstein R., Shachar I. Invariant chain induces B cell maturation by activating TAFII105-NF-kB dependent transcription program. J. Biol. Chem. 2001;276:27203–27206. doi: 10.1074/jbc.M104684200. [DOI] [PubMed] [Google Scholar]
- Memezawa A., Takada I., Takeyama K., Igarashi M., Ito S., Aiba S., Kato S., Kouzmenko A. P. Id2 gene-targeted crosstalk between Wnt and retinoid signaling regulates proliferation in human keratinocytes. Oncogene. 2007;26:5038–5045. doi: 10.1038/sj.onc.1210320. [DOI] [PubMed] [Google Scholar]
- Monroe J. G., Dorshkind K. Fate decisions regulating bone marrow and peripheral B lymphocyte development. Adv. Immunol. 2007;95:1–50. doi: 10.1016/S0065-2776(07)95001-4. [DOI] [PubMed] [Google Scholar]
- Nasi S., Ciarapica R., Jucker R., Rosati J., Soucek L. Making decisions through Myc. FEBS Lett. 2001;490:153–162. doi: 10.1016/s0014-5793(01)02118-4. [DOI] [PubMed] [Google Scholar]
- Petro J. B., Gerstein R. M., Lowe J., Carter R. S., Shinners N., Khan W. N. Transitional type 1 and 2 B lymphocyte subsets are differentially responsive to antigen receptor signaling. J. Biol. Chem. 2002;277:48009–48019. doi: 10.1074/jbc.M200305200. [DOI] [PubMed] [Google Scholar]
- Queva C., Hurlin P. J., Foley K. P., Eisenman R. N. Sequential expression of the MAD family of transcriptional repressors during differentiation and development. Oncogene. 1998;16:967–977. doi: 10.1038/sj.onc.1201611. [DOI] [PubMed] [Google Scholar]
- Queva C., McArthur G. A., Iritani B. M., Eisenman R. N. Targeted deletion of the S-phase-specific Myc antagonist Mad3 sensitizes neuronal and lymphoid cells to radiation-induced apoptosis. Mol. Cell. Biol. 2001;21:703–712. doi: 10.1128/MCB.21.3.703-712.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber-Agus N., Meng Y., Hoang T., Hou H., Chen K., Greenberg R., Cordon-Cardo C., Lee H. W., DePinho R. A. Role of Mxi1 in ageing organ systems and the regulation of normal and neoplastic growth. Nature. 1998;393:483–487. doi: 10.1038/31008. [DOI] [PubMed] [Google Scholar]
- Schreiberagus N., Chin L., Chen K., Torres R., Rao G., Guida P., Skoultchi a. I., Depinho R. A. An amino-terminal domain of Mxi1 mediates anti-myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor Sin3. Cell. 1995;80:777–786. doi: 10.1016/0092-8674(95)90356-9. [DOI] [PubMed] [Google Scholar]
- Shachar I., Elliot E. A., Chasnoff B., Grewal I. S., Flavell R. A. Reconstitution of invariant chain function in transgenic mice in vivo by individual p31 and p41 isoforms. Immunity. 1995;3:373–383. doi: 10.1016/1074-7613(95)90121-3. [DOI] [PubMed] [Google Scholar]
- Shachar I., Flavell R. A. Requirement for invariant chain in B cell maturation and function. Science. 1996;274:106–108. doi: 10.1126/science.274.5284.106. [DOI] [PubMed] [Google Scholar]
- Siegel P. M., Shu W. P., Massague J. Mad upregulation and Id2 repression accompany transforming growth factor (TGF)-beta-mediated epithelial cell growth suppression. J. Biol. Chem. 2003;278:35444–35450. doi: 10.1074/jbc.M301413200. [DOI] [PubMed] [Google Scholar]
- Sommer A., Bousset K., Kremmer E., Austen M., Luscher B. Identification and characterization of specific DNA-binding complexes containing members of the Myc/Max/Mad network of transcriptional regulators. J. Biol. Chem. 1998;273:6632–6642. doi: 10.1074/jbc.273.12.6632. [DOI] [PubMed] [Google Scholar]
- Su T. T., Rawlings D. J. Transitional B lymphocyte subsets operate as distinct checkpoints in murine splenic B cell development. J. Immunol. 2002;168:2101–2110. doi: 10.4049/jimmunol.168.5.2101. [DOI] [PubMed] [Google Scholar]
- Yun J. S., Rust J. M., Ishimaru T., Diaz E. A novel role of the Mad family member Mad3 in cerebellar granule neuron precursor proliferation. Mol. Cell. Biol. 2007;27:8178–8189. doi: 10.1128/MCB.00656-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z. Q., Hurlin P. J. The interplay between Mad and Myc in proliferation and differentiation. Trends Cell Biol. 2001;11:S10–S14. doi: 10.1016/s0962-8924(01)02121-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.