Histone variant H3.3 is loaded onto chromatin in a replication-independent manner, but the epigenetic environment of H3.3 is unclear. Quantitative imaging and chromatin immunoprecipitation show that in mesenchymal stem cells H3.3 targets lineage-priming genes with a potential for activation facilitated by a permissive chromatin environment.
Abstract
In contrast to canonical histones, histone variant H3.3 is incorporated into chromatin in a replication-independent manner. Posttranslational modifications of H3.3 have been identified; however, the epigenetic environment of incorporated H3.3 is unclear. We have investigated the genomic distribution of epitope-tagged H3.3 in relation to histone modifications, DNA methylation, and transcription in mesenchymal stem cells. Quantitative imaging at the nucleus level shows that H3.3, relative to replicative H3.2 or canonical H2B, is enriched in chromatin domains marked by histone modifications of active or potentially active genes. Chromatin immunoprecipitation of epitope-tagged H3.3 and array hybridization identified 1649 H3.3-enriched promoters, a fraction of which is coenriched in H3K4me3 alone or together with H3K27me3, whereas H3K9me3 is excluded, corroborating nucleus-level imaging data. H3.3-enriched promoters are predominantly CpG-rich and preferentially DNA methylated, relative to the proportion of methylated RefSeq promoters in the genome. Most but not all H3.3-enriched promoters are transcriptionally active, and coenrichment of H3.3 with repressive H3K27me3 correlates with an enhanced proportion of expressed genes carrying this mark. H3.3-target genes are enriched in mesodermal differentiation and signaling functions. Our data suggest that in mesenchymal stem cells, H3.3 targets lineage-priming genes with a potential for activation facilitated by H3K4me3 in facultative association with H3K27me3.
INTRODUCTION
In the interphase nucleus, chromatin is organized into domains with different physical, biochemical, and functional properties influencing gene expression. These domains can be distinguished by their enrichment in various combinations of posttranslationally modified histones (Kouzarides, 2007). Notably, di- and trimethylation of H3 lysine 4 (H3K4me2/me3) together with acetylation of lysine 9 (H3K9ac) are commonly associated with transcriptionally active chromatin. In contrast, di- or trimethylated H3K9 mostly marks silent promoters and constitutive heterochromatin. Polycomb-mediated H3K27 trimethylation is a repressive modification associated with facultative heterochromatin and interestingly has been shown to mark, together with H3K4me3, repressed or weakly expressed developmentally regulated promoters in pluripotent embryonic stem cells (Bernstein et al., 2006).
A second set of factors differentially marking chromatin consists of variants of histones H1, H2, and H3 (Henikoff and Ahmad, 2005; Creyghton et al., 2008; Henikoff, 2008; Zilberman et al., 2008; Wong et al., 2009). Among these, histone H3 variant H3.3 has been shown to incorporate into nucleosomal chromatin in a replication-independent manner (i.e., both during and outside S phase), whereas the replicative H3.2 and H3.1, commonly referred to as H3, are deposited only into newly replicated DNA (Ahmad and Henikoff, 2002). H3.3 accumulates predominantly at sites of active transcription (Ahmad and Henikoff, 2002; McKittrick et al., 2004). Consistent with this finding, chromatin immunoprecipitation (ChIP) experiments have shown that sites of epitope-tagged H3.3 deposition are often associated with marks of active chromatin (Chow et al., 2005; Mito et al., 2005; Daury et al., 2006; Jin and Felsenfeld, 2007; Tamura et al., 2009). Intriguingly, posttranslational modifications of H3.3 before nucleosome assembly seem to prime its future epigenetic state once in chromatin (Loyola et al., 2006). This raises the possibility that the epigenetic fate of H3.3 may be programmed for incorporation into chromatin domains that are not necessarily transcriptionally active. Indeed, at least in hematopoietic cells, gene activity is not required to replace H3 with H3.3 (Jin and Felsenfeld, 2006). Within genes, H3.3 is found in regulatory and transcribed regions (Mito et al., 2005; Jin and Felsenfeld, 2006; Sutcliffe et al., 2009) and at sites of nucleosome displacement (Wirbelauer et al., 2005). Moreover, nucleosomes containing H3.3 are unstable, which may account for their eviction from the transcription start site (TSS) of highly expressed genes (Mito et al., 2005; Henikoff, 2008; Jin et al., 2009). This instability also has been shown to result in increased solubility of H3.3-containing nucleosomes, leading to the perception that such nucleosomes are depleted at the TSS of active genes (Jin et al., 2009).
In spite of these observations, nothing is known on the relationship between sites of H3.3 deposition and large-scale chromatin domains with a defined epigenetic composition in interphase nuclei. Fluorescence microscopy has localized epitope-tagged H3.3 at transcriptionally active regions during Cænorhabditis elegans spermatogenesis (Ooi et al., 2006) and in Drosophila melanogaster chromosomes (Schwartz and Ahmad, 2005) or interphase cells (Ahmad and Henikoff, 2002). H3.3 incorporation also occurs during mammalian meiotic sex chromosome inactivation (van der Heijden et al., 2007), and a fraction of H3.3 localizes to telomeres in undifferentiated mouse embryonic stem cells (Wong et al., 2009), providing additional evidence of H3.3 deposition not always linked to transcriptional activity. The large-scale chromatin environment of nucleosomal H3.3 is therefore likely to be intricate.
Similarly, because not all H3.3 target genes are expressed, the relationship between H3.3 enrichment and gene expression is not straightforward. This complexity may arise from sequence information (regulatory, coding, or insulator) at sites of H3.3 deposition (Jin and Felsenfeld, 2006, 2007; Jin et al., 2009), different transcriptional outputs depending on combinatorial associations of various histone modifications, and on the localization of the modified histones within a locus. Another component of this complexity is the underlying state of DNA methylation, which influences histone modifications (Mohn et al., 2008), transcription factor binding (Klose and Bird, 2006), and deposition of histone variants such as H2A.Z (Zilberman et al., 2008). The relationship between promoter DNA methylation and transcription also depends on CpG density: high CpG promoters (HCPs) are often unmethylated even when inactive, active low CpG promoters (LCPs) can be methylated or unmethylated, whereas methylation of intermediate CpG promoters (ICPs) correlates with repression (Weber et al., 2007). The CpG methylation status of sites enriched in H3.3 remains to date elusive.
Using human adipose-derived primary mesenchymal stem cells (MSCs) (Boquest et al., 2005), we integrate here nucleus-wide quantitative imaging information with ChIP-on-chip and methylated DNA immunoprecipitation (MeDIP)-chip approaches at the promoter level, to investigate the distribution of epitope-tagged H3.3 in relation to histone methylation, DNA methylation, CpG density, and transcription state.
MATERIALS AND METHODS
Constructs
pH2B-diHcRed plasmid was a gift from Jan Ellenberg (European Molecular Biology Laboratory, Heidelberg, Germany). pEGFP-H4 and pH2B-EGFP plasmids were gifts from Maïté Coppey (Institut Jacques Monod, Paris, France). D. melanogaster H3.3 and H3 (identical in amino acid sequence to mammalian H3.3 and H3.2, respectively) cDNAs were amplified by polymerase chain reaction (PCR) from plasmids pH3.3-YFP (gift from Susan Janicki, The Wistar Institute, Philadelphia, PA) and the pHS-H3-EGFP (gift from Thierry Grange, Institut Jacques Monod, Paris, France), respectively. The same primers were used for both constructs, with an EcoRI site at the 5′ end of the sense primer (5′-GCGAATTCTGATGGCACGTACCAAGCAAACA-3′) and a KpnI site at the 5′ end of the antisense primer (5′-ATGGTACCGCCCGCTCGCCACGGATGCGTCT-3′). Reaction products were digested with EcoRI and KpnI and ligated into the EcoRI and KpnI cloning sites of pEGFP-N1 (Clontech, Mountain View, CA) or pmCherry-N (gift from Maïté Coppey, Institut Jacques Monod).
Cells and Antibodies
MSCs were purified from liposuction material from three donors and cultured as described previously (Boquest et al., 2005). HeLa cells were cultured in DMEM, 1% glutamine, 1% Na-pyruvate, 1% nonessential amino acids, and 10% fetal calf serum. C2C12 cells were cultured in DMEM, 1% glutamine, 1% Na-pyruvate, and 15% fetal calf serum. MSCs were transfected using AMAXA Human MSC Nucleofector (Lonza, Allendale, NJ). HeLa and C2C12 cells grown to 50% confluence on coverslips were transfected using FuGENE6 (Roche Diagnostics, Basel, Switzerland). Antibodies to H3K9ac (06-942), H3K9me2 (07-441), H3K9me3 (07-442), and H3K27me3 (05-851) were from Upstate Biotechnologies (now Millipore, Billerica, MA). Antibodies to H3K4me2 (Ab7766) and H3K4me3 (Ab8580) were from Abcam (Cambridge, United Kingdom). Anti-H3 antibodies (1326-1) were from Epitomics (Burlingame, CA). Anti-H3K9me3 antibodies (pAb-056-050) used for ChIP were from Diagenode (Liège, Belgium). Living Colors anti-enhanced green fluorescent protein (EGFP) antibodies were from Clontech. Anti-5-methylcytosine antibodies (MAb-5MCYT) were from Diagenode. Cy2- and Cy3- and horseradish peroxidase-conjugated antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA).
Immunofluorescence
Cells were plated onto coverslips; fixed with 3% paraformaldehyde for 15 min; and permeabilized with 0.1% Triton X-100, 0.01% Tween 20, and 2% bovine serum albumin for 30 min. Primary and secondary antibodies were diluted 1:100 and 1:200, respectively. DNA was labeled with 0.25 mg/ml 4,6-diamidino-2-phenylindole (DAPI). Cells were observed on an IX71 microscope (Olympus, Tokyo, Japan) fitted with a piezo-driven 100× objective (numerical aperture 1.4) and a CellR̂ wide-field Imaging Station (Olympus). For analysis of correlation between H3.3-mCherry and immunolabeled histones (Håkelien et al., 2008), the middle plane of the nucleus was acquired in the two channels, and images were treated with ImageJ 1.37v (National Institutes of Health, Bethesda, MD). For each cell, the same area of the nucleus was selected in the mCherry and Cy2 images. The two fluorescent profiles were compared and r-based analysis based on Pearson's coefficient (Bolte and Cordelieres, 2006), calculated using Image CorrelationJ. Intensities of mCherry fluorescence were obtained by measuring mean fluorescence intensity over the whole nucleus in middle plane images.
Western Blotting
SDS-polyacrylamide gel electrophoresis and Western blotting were performed as described previously (Håkelien et al., 2008). Membranes were incubated with either of the following primary antibodies: anti-H3K4me2, anti-H3K4me3, anti-H3K9me2, anti-H3K9me3, anti-H3K27me3 (all at a 1:250 dilution), and anti-H3 (1:3000). Secondary antibodies were horseradish peroxidase-conjugated anti-mouse and anti-rabbit immunoglobulin Gs (1:7500).
Fluorescence-activated Cell Sorting (FACS)
For expression microarray, ChIP-PCR, and ChIP-on-chip analyses, MSCs were harvested 168 h posttransfection and sorted using a FACS DiVa cell sorter (BD Biosciences, Franklin Lakes, NJ).
Expression Microarrays
RNA was isolated from the samples using the RNeasy Mini kit (QIAGEN, Hilden, Germany). Biotin-labeled cRNA (1.5 μg) was hybridized onto Illumina Human-6 v2 Expression BeadChips (Illumina, San Diego, CA). Data were analyzed with GeneSpring GX (Agilent Technologies, Santa Clara, CA). Expression array data are deposited under National Center for Biotechnology Information (NCBI) GEO GSE17053.
Chromatin Immunoprecipitation
Histone ChIPs were done essentially as described previously (Dahl and Collas, 2007) by using 0.5 A260 units chromatin per ChIP. In short, cells were cross-linked with formaldehyde, lysis buffer was added to ∼120 μl, and samples were incubated for 5 min on ice. Cells were sonicated for 14 × 30 s on ice with 30-s pauses by using a probe sonicator to produce fragments of ∼400–500 base pairs. The lysate was centrifuged, the supernatant collected, chromatin was diluted to 0.5 A260 units, and 100 μl was incubated with 2.4 μg of antibody coupled to Dynabeads Protein A (Invitrogen, Oslo, Norway) for 2 h at 4°C. ChIP material was washed, and DNA was eluted with 1% SDS and 50 μg/ml proteinase K for 2 h at 68°C. ChIP DNA was purified and dissolved in 50 μl of Tris-EDTA, pH 8.0, buffer. H3.3-EGFP and H3.2-EGFP were immunoprecipitated from transfected and sorted cells as described above, by using 50 μl of Dynabeads Protein G (Invitrogen), 5 μl of Living Color anti-EGFP antibody, and 0.5 A260 units of chromatin (∼50,000 cells) per ChIP. For ChIP-on-chip, 5 μg of RNAse A was added before ChIP DNA elution, and ChIP DNA samples were dissolved in 10 μl of MilliQ water (Millipore). ChIP DNA was analyzed either by duplicate quantitative (q)PCR (Dahl and Collas, 2007) by using primers listed in Supplemental Table S1, or by microarray hybridization.
Microarray Hybridization and Data Analysis
ChIP and input DNA were amplified using the WGA4 kit (Sigma-Aldrich, St. Louis, MO), cleaned up (QIAquick PCR Purification kit; QIAGEN), and eluted in 30 μl of MilliQ water to 250–500 ng/μl. ChIP and input DNA fragments were labeled with Cy5 and Cy3, respectively, and hybridized on human HG18 RefSeq Promoter arrays (Nimblegen, Madison, WI) with tiled regions covering −2.2 to +0.5 kb relative to the TSS. Data were analyzed using NimbleScan (Johnson et al., 2008) and deposited under NCBI GEO GSE17053. Peaks were detected by searching for at least four probes with a signal above a cut-off value by using a 500-base pair sliding window. Ratio data were randomized 20 times to evaluate probability of false positives, and each peak was assigned a false discovery rate (FDR) of ≤0.1. Gene ontology (GO) term enrichments within a target gene set were calculated using Bioconductor GOstats (Falcon and Gentleman, 2007).
Methyl-DNA Immunoprecipitation and Data Analysis
MeDIP was performed and data were analyzed as described previously (Sørensen and Collas, 2009). In short, genomic DNA was purified and fragmented by sonication to 300- to 500-base pair fragments. Methylated fragments were immunoprecipitated using anti-5-methylcytosine antibodies. Immunoprecipitated (MeDIP) and input DNA was amplified by 14 PCR cycles using the WGA2 kit (Sigma-Aldrich) and cleaned up as described above. Input and MeDIP DNA were labeled with Cy3 and Cy5, respectively, and hybridized on similar promoter arrays as those used for ChIP-on-chip. Scaled log2 ratios of MeDIP/input signals were centered on zero by subtracting the biweight mean for the log2 ratio values for all features on the array from each log2 ratio value. To determine p values from scaled log2 ratios, a 750-base pair window was placed around each consecutive probe, and a one-sided Kolmogorov–Smirnov test was applied to determine whether the probes were drawn from a significantly more positive distribution of intensity log2 ratios than those in the rest of the array. Resulting score for each probe was converted into a −log10 p value. Peak data were generated from p values by searching for at least two probes above a −log10 p value minimum cut-off of 2 (i.e., p < 0.01). Peaks within 500 base pairs of each other were merged. DNA methylation was defined by the detection of at least one methylation peak in the promoter regions (Weber et al., 2007). MeDIP-chip data are deposited under NCBI GEO GSE17053.
RESULTS
H3.3 Preferentially Incorporates in Chromatin Domains Marked by Histone Modifications of Active Genes
Previous investigations of the epigenetic environment of H3.3 have relied on biochemical approaches, and no information exists on the relationship between H3.3 deposition and posttranslational histone modifications at the nucleus level. We have used a quantitative imaging approach to measure, in normal primary human MSCs, the extent of overlap between transcriptionally activating or repressing histone modifications and epitope-tagged H3.3, and we compared the results with the extent of colocalization between these modifications and epitope-tagged canonical H2B, a core histone used as a bona-fide chromatin marker, or H3.2, a replicative H3 variant.
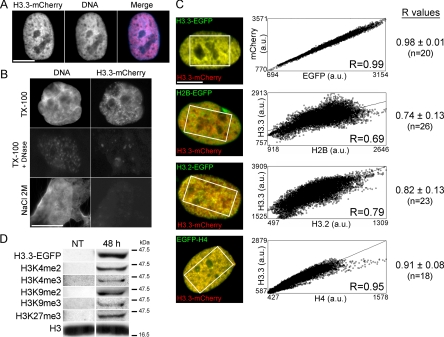
To visualize chromatin regions incorporating H3.3, we transiently expressed H3.3 tagged in its C terminus with mCherry or EGFP, and then we imaged cells 168 h posttransfection, the time at which epitope-tagged H3.3 assumed a chromatin distribution (Figure 1A). Tagged histones have been used as general chromatin markers because they behave as endogenous histones (Kimura and Cook, 2001; Gerlich et al., 2003; Tagami et al., 2004) and can be posttranslationally modified (Loyola et al., 2006; Tamura et al., 2009; also see below). Tagged H3.3 colocalized with DNA (Figure 1, A and B) independently of expression level (data not shown) and of the tag used (Figure 1C). In situ extraction of cells expressing H3.3-mCherry with 0.1% of the nonionic detergent Triton X-100 preserved H3.3-mCherry fluorescence, whereas 1 mg/ml DNase I or 2 M (but not 1 M) NaCl, a concentration that removes core histones from DNA (Stokes and Perry, 1995), eliminated H3.3-mCherry labeling (Figure 1B). Thus, in MSCs, exogenous H3.3 is bound to DNA with a strength comparable with endogenous core histones. Furthermore, Western blot analysis of HeLa cells showed that EGFP-tagged H3.3 is posttranslationally modified by di- and trimethylation of K4, as well as di- or trimethylation of K9, and trimethylation of K27 (Figure 1D and Supplemental Figure S1).
Figure 1.
Incorporation of epitope-tagged H3.3 into chromatin in human primary cells. (A) Intranuclear distribution of H3.3-mCherry 168 h after transient transfection. DNA was labeled with DAPI. (B) Cells expressing H3.3-EGFP were extracted with 0.1% Triton X-100, 0.1% Triton X-100 and 1 mg/ml DNase I, or 2 M NaCl before fixation and DAPI staining. (C) Correlation analysis of colocalization of H3.3-mCherry with H3.3-EGFP, H2B-EGFP, H3.2-EGFP, and EGFP-H4. Scatter plots show for each cell illustrated on the left, pixel distribution whereby the intensity of a given pixel in the green image corresponds to the x-coordinate, and the intensity of the corresponding pixel in the red image is shown as the y-coordinate. Correlation was done in the area delineated on the images. Correlation coefficients (R) of overlaps between H3.3-mCherry and H3.3-EGFP, H2B-EGFP, H3.2-EGFP or EGFP-H4 are also shown for all cells (n, number of cells analyzed). (D) Western blot analysis of indicated posttranslational modifications on overexpressed H3.3-EGFP in HeLa cells. NT, nontransfected cells; 48 h, H3.3-EGFP-expressing cells 48 h posttransfection. Endogenous H3 was probed as a loading control (see Supplemental Figure S1).
In cotransfected cells, H2B-EGFP-labeled chromatin regions only partially overlapped with H3.3-mCherry-containing regions, unlike H3.3-EGFP, which perfectly colocalized with H3.3-mCherry (Figure 1C). In particular, we observed areas of H2B-EGFP deprived of H3.3-mCherry, whereas essentially all H3.3-mCherry regions were colabeled with H2B-EGFP (Figure 1C, second panels from top). Moreover, the replicative H3 variant H3.2 tagged in its C terminus with EGFP also showed a distribution similar, but not identical to, H3.3-mCherry, with areas of H3.3 without H3.2 and vice versa (Figure 1C, third panels from top). Thus, a pool of H3.3 localizes to chromatin domains other than those occupied by H3.2. In addition, H3.3-mCherry colocalized better with EGFP-tagged histone H4 than with H3.2-EGFP (p = 0.003) or H2B-EGFP (p = 10−5). These data suggest that a given genomic site can be defined with respect to its content in the various histones (H3.2, H3.3, H4, and H2B) examined here by fluorescence microscopy.
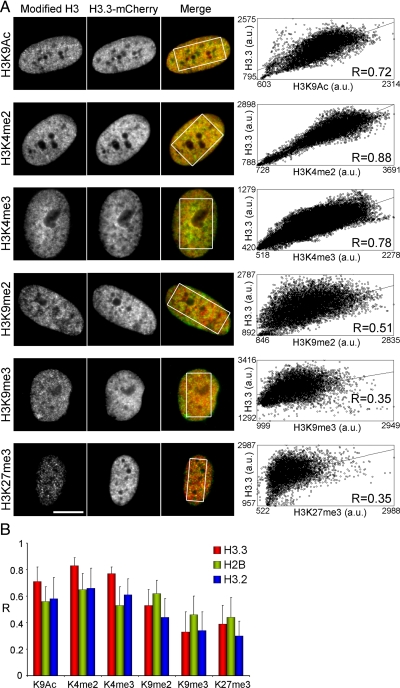
We next investigated the epigenetic environment of H3.3 by quantitative imaging. Cells expressing H3.3-mCherry were analyzed by immunofluorescence using antibodies to three H3 modifications associated with transcriptionally active chromatin (H3K9ac, H3K4me2, H3K4me3), and three modifications associated with repressed chromatin (H3K9me2, H3K9me3, and H3K27me3). The extent of overlap between mCherry fluorescence and modified histone immunolabeling was measured in >40 individual cells using intensity r-based analysis to generate Pearson's correlation coefficients (Bolte and Cordelieres, 2006). We found a greater overlap between H3.3-mCherry and H3K9ac, H3K4me2 and H3K4me3 than between H3.3-mCherry and H3K9me2, H3K9me3 or H3K27me3 (Figure 2, A and B). Relative enrichment of H3.3-mCherry with marks of active chromatin also was detected in HeLa cells (Supplemental Figure S2, A and B) and in mouse C2C12 myoblasts (Supplemental Figure S2, C and D). Low correlation values with di- and trimethylated H3K9 were striking in C2C12 cells due to the exclusion of H3.3-mCherry from heterochromatic foci (Supplemental Figure S2E). This shows that a given genomic site also can be defined with respect to its content in the various histone modifications (H3K9ac, H3K4me2, H3K4me3 and H3K9me2, H3K9me3, H3K27me3) examined here by fluorescence microscopy. The data further suggest that tagged H3.3 is preferentially found in chromatin regions labeled with posttranslational histone modifications of active or potentially active genes.
Figure 2.
H3.3 preferentially incorporates into chromatin domains marked by histone modifications of active genes. (A) Colocalization analysis of overlap between H3.3-mCherry and immunolabeled modified histones in MSCs. Correlation coefficients (R) of overlaps were determined in individual cells as in Figure 1C, as shown on the graphs (each graph is for a single cell). (B) Average ± SD of correlation coefficients (R values) of H3.3-mCherry and H3 modification immunolabeling overlaps for each H3 modification in MSCs (n = 40–56 cells). R values for modified H3 and either H2B-diHcRed (21–35 cells) or H3.2-mCherry (n = 24–56 cells) overlaps are also shown. Bars, 10 μm.
To exclude the possibility that these results might arise from differences in antibody staining patterns and occupancy rather than from specific H3.3 targeting, we performed a similar intensity correlation analysis using mCherry-tagged H3.2 and diHcRed-tagged H2B as internal controls. t test analysis of correlation coefficients of overlap with each histone H3 modification showed that H3.3 correlated significantly better with active chromatin marks than H3.2 (p = 10−6 to 10−11 for H3K4me2, H3K4me3, and H3K9ac) or H2B (p = 10−7–10−10 for the same modifications) in MSCs (Figure 2B and Supplemental Table S2). Similar results were found for H3.3 and H2B in HeLa and C2C12 cells (Supplemental Figure S2, B and D). H2B-diHcRed showed no marked preference for active or inactive chromatin regions (Figure 2B and Supplemental Figure S2, B and D), as would be expected from a bona fide chromatin marker. However, both H3.2 and H3.3 were less correlated than H2B with repressive marks (p = 10−4–10−10 for H3K9me2, H3K9me3, and H3K27me3; Supplemental Table S2). These results indicate, relative to replicative H3.2 or canonical H2B, a quantitative enrichment of epitope-tagged H3.3 in regions of transcriptionally active or potentially active chromatin and an impoverishment of H3.3 in regions marked by histone modifications of silent genes.
H3.3-containing Promoters Are Enriched in H3K4me3 Relative to All RefSeq Promoters and Promoters Enriched in Replicative H3.2
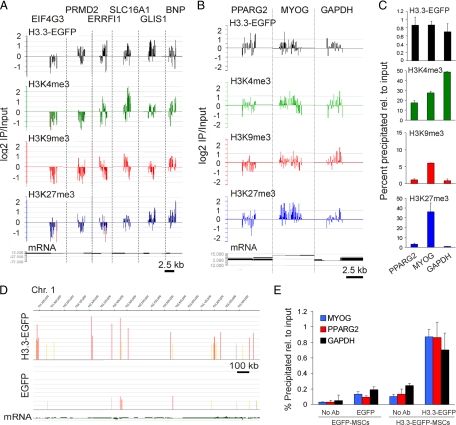
Imaging data provide rapid and quantitative information to define large chromatin domains by their respective contents in various histones, histone variants, and histone modifications, at the global nucleus level. However, imaging resolution is currently insufficient to investigate the targeting of H3.3 to specific genomic regions. In particular, H3.3 targeting to promoters in human primary cells has not been specifically addressed, and the chromatin environment associated with the deposition of H3.3 in these regulatory regions has not been characterized. To address these issues, we mapped H3.3-EGFP enrichment on promoters by ChIP-on-chip using anti-EGFP antibodies for immunoprecipitation after sorting cells for H3.3-EGFP expression (Supplemental Figure S3), in combination with hybridization to microarrays including 18,028 annotated human RefSeq promoters. To provide information on the epigenetic environment of H3.3 on promoters, trimethylated H3K4, H3K9, and H3K27 were also immunoprecipitated from the same sorted cells. ChIP-on-chip profiles for H3.3-EGFP and modified histones (Figure 3A) were confirmed by ChIP-qPCR at the single gene level (Figure 3, B and C). Specificity of H3.3-EGFP targeting was supported by the absence of consistent enrichment of control EGFP expressed alone in ChIP-on-chip and ChIP-qPCR experiments (Figure 3, D and E) and by enrichment profiles for replicative H3.2 (see below).
Figure 3.
H3.3-EGFP ChIP-on-chip validation on target promoters. (A) Combinatorial enrichment profiles detected by ChIP-on-chip for H3.3-EGFP, H3K4me3, H3K9me3, and H3K27me3 on the promoters of indicated genes. (B and C) H3.3-EGFP, H3K4me3, H3K9me3, and H3K27me3 profiles on the PPARG2, MYOG, and GAPDH promoters determined by ChIP-on-chip (B) and ChIP-qPCR 9C). (D) EGFP ChIP-on-chip peak detection (FDR ≤ 0.1) in a 1.7-Mb segment of chromosome 1 in sorted H3.3-EGFP-expressing cells (top track) and control EGFP-expressing cells (bottom track). (E) ChIP-qPCR analysis of control EGFP (left) and H3.3-EGFP (right) enrichment on indicated promoters (data from 4 independent ChIPs each analyzed by duplicate qPCRs).
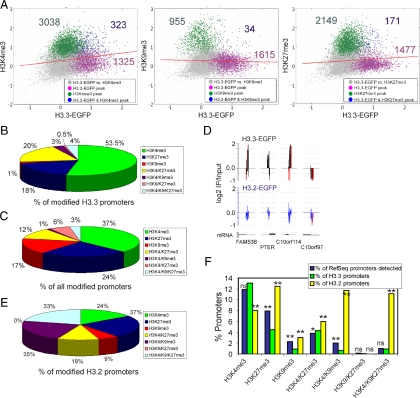
We identified 1649 promoters enriched in H3.3-EGFP (referred to as “H3.3 promoters” hereafter) based on peak calling with FDR ≤ 0.1 (Figure 4A and Supplemental Figure S4). Plotting MaxTen values computed from log2 IP/Input ratios for H3.3-EGFP versus each histone modification highlighted promoters coenriched in H3.3 and either of these modifications (Figure 4A). Among H3.3 promoters coenriched in modified histones, 80% harbored H3K4me3, for the most part alone (53.5% of the total), or together with H3K27me3 (20%; Figure 4B). Furthermore, the proportion of H3K4me3-enriched promoters, alone or in combination with other histone modifications, was significantly higher among H3.3 promoters harboring modifications than among RefSeq promoters harboring modifications in the genome (p < 10−4, chi-square test with Yates' correction; Figure 4, B and C, green sections). Conversely, H3.3-EGFP targeted fewer promoters enriched in H3K27me3 alone or in combination with H3K9me3; and strikingly, H3K9me3 alone was absent from H3.3 promoters (Figure 4, B and C). There is therefore a preferred association of H3.3-EGFP with H3K4me3-containing promoters over any of the other marks examined. Nonetheless, a minor fraction of H3.3-EGFP also can be found in a facultative heterochromatic environment (Figure 4B; also see Figure 2A).
Figure 4.
H3.3-EGFP associates with promoters enriched in H3K4me3 but not H3K9me3. (A) Two-dimensional scatter plots of MaxTen values for log2 IP/Input ratios for H3.3-EGFP versus H3K4me3 (left), H3K9me3 (middle), and H3K27me3 (right). MaxTen values are the average of both ChIP-on-chip replicates for each condition. Data points were colored to indicate classification according to peak calling, to show promoters enriched in H3.3-EGFP (purple); H3K4me3, H3K9me3, or H3K27me3 (green); or H3.3-EGFP together with either modification (blue). Red bars show regression lines for all points. (B) Proportions of modified H3.3-EGFP promoters enriched in indicated histone modifications. (C) Proportions of all modified RefSeq promoters enriched in indicated histone modifications. (D) Representative profiles of H3.3-EGFP and H3.2-EGFP occupancy on selected promoters on chromosome 10; red bars reflect out-of-scale signals. (E) Proportions of modified H3.2-EGFP promoters enriched in indicated histone modifications. (F) Percentages of promoters enriched in indicated histone modification (x-axis) expressed as a percentage of all RefSeq promoters on the array (blue bars), as a percentage of H3.3 promoters (green bars), and as a percentage of H3.2 promoters (yellow bars). **p < 10−4 and *p < 0.05 relative to the percentage of RefSeq promoters; chi-square test with Yates' correction; ns, nonsignificant.
Specificity of H3.3-EGFP enrichment on H3K4me3 promoters was corroborated upon examination of the histone modification environment on promoters targeted by the replicative H3 variant H3.2 tagged in its C terminus with EGFP. H3.2-EGFP immunoprecipitated using anti-EGFP antibodies was detected at some level, albeit below peak detection threshold, in a large number of promoters, independently of H3.3 enrichment (Figure 4D). Enrichment of H3.2-EGFP above genome-average level was significant only in a small number of promoters (n = 297), corroborating the view of a widespread distribution of H3.2 in the genome. Importantly, modifications associated with H3.2-EGFP-enriched promoters were strongly shifted toward enrichment in heterochromatic marks, compared with those associated with H3.3-EGFP or among all RefSeq promoters harboring modifications (compare Figure 4E with 4, B and C). Similar conclusions were drawn with statistical significance for H3.3 and H3.2 when we considered all RefSeq promoters identified on the array as opposed to modified promoters only (Figure 4F).
These results illustrate the association of a transcriptionally permissive chromatin environment enriched in H3K4me3 with H3.3 deposition, even on promoters coharboring repressive modifications. One possibility is that the presence of H3K4me3 enables the deposition of H3.3; alternatively, H3.3 may promote the modification of H3K4 into a trimethylated state. These possibilities remain to be investigated. The correlation between H3K4me3 enrichment and deposition of H3.3 contrasts with H3K9me3, which seems to be antagonistic to the deposition of H3.3 but not of H3.2. Although we examined a small fraction of the genome on microarrays (∼1.8%; 18,028 promoters with 2.8 kb tiled), the ChIP-on-chip data corroborate the large-scale enrichment of epitope-tagged H3.3 with H3K4me3 versus H3K9me3 or H3K27me3 observed by imaging at the nucleus level.
H3.3-EGFP Incorporation Correlates with Promoter CpG Methylation
Our results show thus far that among the histone modifications examined, H3.3 deposition correlates with H3K4me3 enrichment on promoters. The reported preference of H3K4me3 for unmethylated DNA (Ooi et al., 2007; Mohn et al., 2008) predicts, then, that promoters co-occupied by H3.3 and H3K4me3 would be predominantly unmethylated. To address this issue, we profiled promoter DNA methylation by MeDIP-chip by using the same promoter arrays as for ChIP-on-chip experiments.
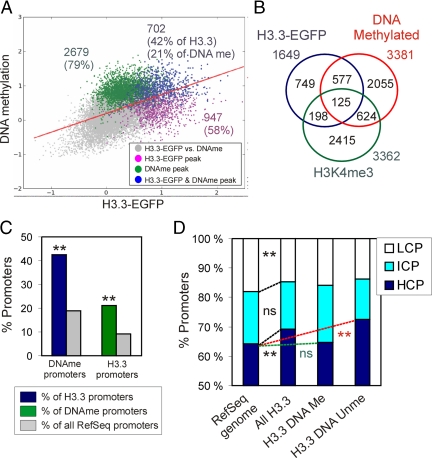
Two-dimensional scatter plot analysis and peak intersect analysis identified 702 promoters enriched in both H3.3 and methylated DNA (Figure 5A, blue data points; and B). Remarkably, DNA methylated promoters represented 42% of H3.3 promoters versus 19% of the entire set of RefSeq promoters on the array (p < 10−4; chi-square test with Yates' correction; Figure 5C, left bars). Conversely, H3.3 promoters made up 21% of DNA methylated promoters, a 2.2-fold enrichment over the proportion of H3.3 promoters among RefSeq promoters (p < 10−4; Figure 5C, right bars). Thus, although a large number of H3.3 promoters are not DNA methylated and vice versa (Figure 5A), the data indicate a significant enrichment (p < 10−4) of DNA methylation among H3.3 promoters relative to the proportion of DNA methylated RefSeq promoters.
Figure 5.
H3.3-EGFP promoters are enriched in methylated CpGs relative to all RefSeq promoters. (A) Two-dimensional scatter plot of MaxTen values for log2 IP/Input ratios for H3.3-EGFP versus DNA methylation. Average values for both MeDIP and ChIP replicates are shown. Data points were colored to indicate classification according to peak calling, to show promoters enriched in H3.3-EGFP (purple), methylated DNA (green), or in both (blue). Red bar shows the regression line for all data points. (B) Venn diagram analysis of DNA methylation, H3.3-EGFP enrichment, and H3K4me3 enrichment. (C) Proportions of DNA methylated promoters and H3.3 promoters relative to indicated reference (see legend). **p < 10−4 relative to the percentage of all RefSeq promoters; chi-square test with Yates' correction. (D) CpG content distribution among all H3.3-EGFP promoters and as reference among all human RefSeq promoters. **p < 0.001; ns, nonsignificant relative to proportions among all RefSeq promoters; chi-square test with Yates' correction.
H3.3-EGFP Preferentially Targets CpG-rich Promoters
We next asked whether H3.3-EGFP deposition was influenced by CpG density in the target promoters. Previous analysis of CpG content more than ∼3.5 kb in all human RefSeq promoters revealed a trimodal distribution of promoters as a function of CpG content based on observed/expected CpG ratios, identifying HCPs, ICPs, and LCPs (Weber et al., 2007). We applied the algorithm of Weber et al. (2007) to the 1649 H3.3-EGFP promoters identified in our study. Among those, 81 contained two or more sequences localizing to distinct genomic positions with a different CpG content and thus were removed from the analysis. Figure 5D shows that 69% of H3.3 promoters were HCPs, 16% were ICPs, and 15% were LCPs. H3.3 promoters were in fact significantly enriched (p < 10−4; chi-square test with Yates' correction) in HCPs relative to the proportion of HCPs among all RefSeq promoters, at the expense of LCPs (p < 10−3). This enrichment was significantly enabled (p < 10−3) by unmethylated promoters rather than methylated promoters (Figure 5D), in agreement with the overall unmethylated state of HCPs in the genome (Weber et al., 2007).
Promoter Targeting of H3.3 Correlates with Enhanced Proportion of Expressed Genes
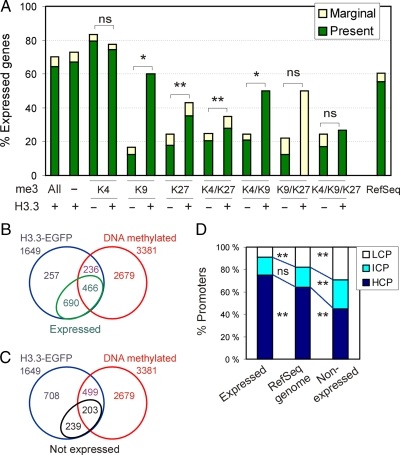
The absence of significant enrichment of H3 modifications linked to active or repressed genes on 75% of H3.3-EGFP promoters makes prediction of the transcriptional state of these promoters challenging. To address this question, we assessed expression of H3.3 target genes in H3.3-EGFP-transfected MSCs by using Illumina microarrays by defining present (expressed), marginal (weakly expressed), and absent (not expressed) cells. We showed that overexpression of H3.3-EGFP altered expression of 2.1% (n = 232) of the total number of genes represented on the array (p < 0.05; Fisher's exact test) relative to untransfected cells and control EGFP-expressing cells. Only 10% of these genes were enriched in H3.3-EGFP. Thus, and in line with previous observations (Jin and Felsenfeld, 2006), overexpression of H3.3-EGFP did not significantly alter the gene expression profile of MSCs. Figure 6A first shows that ∼70% of all genes with a promoter containing H3.3 were expressed, representing an enrichment (p < 0.001) compared with the proportion of expressed RefSeq genes detected in MSCs; thus, H3.3-EGFP targets predominantly, albeit not exclusively, active promoters. Second, 75–80% of genes enriched in H3K4me3 were expressed regardless of H3.3 detection (Figure 6A), indicating that H3.3 does not confer additional transcriptional activity potential to H3K4me3-only promoters. In contrast, H3K9me3 and H3K27me3 alone or in facultative combination with H3K4me3, mainly occupied promoters of repressed genes (Figure 6A, H3.3 minus). Interestingly, however, the proportions of expressed genes marked by H3K9me3, H3K27me3, H3K4/K27me3, or H3K4/K9me3 were enhanced when these were in addition enriched in H3.3 (Figure 6A). The proportions of expressed genes with a promoter enriched in H3K9/K27me3 or H3K4/K9/K27me3 combinations also tended to increase when H3.3 was codetected, although not significantly due to the low number of genes in these categories (Figure 6A and Supplemental Figure S4). These results are consistent with reports that not all genes harboring trimethylated H3K9 or H3K27 are repressed (Bernstein et al., 2006; Dahl et al., 2009), an outcome likely to depend on positioning of the repressive mark in the gene (Martin and Zhang, 2005). These results indicate that H3.3 enrichment correlates with an enhanced proportion of expressed genes, in particular when those are co-occupied by otherwise repressive marks. Nonetheless, not all H3.3 promoters are active.
Figure 6.
H3.3-EGFP preferentially targets transcriptionally active promoters. (A) Percentage of expressed genes with a promoter occupied by indicated combinations of trimethylated (me3) H3K4, H3K9, and H3K27, with or without H3.3. *p < 0.05; **p < 0.001; ns, nonsignificant (Fisher's t test). Percentage of expressed genes among all RefSeq genes included in both Nimblegen and Illumina arrays also is shown (right). (B) Venn diagram analysis of H3.3, DNA methylated, and expressed genes. (C) Venn diagram analysis of H3.3, DNA methylated, and nonexpressed genes. (D) Percentage of H3.3 target HCPs, ICPs, and LCPs as a function of gene expression. **p < 0.001; ns, not significant; chi-square test with Yates' correction.
DNA-methylated promoters made up 40% of active H3.3 promoters (466/1156; Figure 6B) and 46% of inactive H3.3 promoters (203/442; Figure 6C). These proportions were not different from those of all DNA-methylated H3.3 promoters; thus, expression status does not affect enrichment of H3.3 for methylated or unmethylated promoters. Finally, relative to the proportion of HCPs, ICPs, and LCPs among all RefSeq promoters, we found that active H3.3 promoters were enriched in HCPs to the detriment of LCPs (p < 0.001), in contrast to inactive H3.3 promoters which were enriched in LCPs (p < 0.001; Figure 6D).
H3.3 Target Genes Are Enriched in Developmental and Signaling Functions
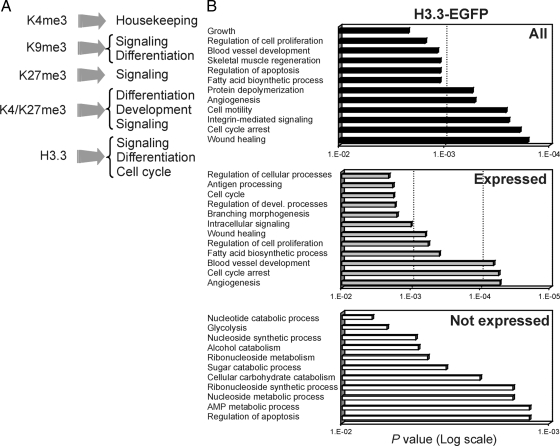
GO analysis revealed distinct functional categories enriched (relative to what would be expected from the total number of genes fitting these GO terms in the genome) among genes marked by the histone modifications or variants (Figure 7A and Supplemental Figure S5). In particular, H3K4me3 genes were enriched in housekeeping functions, including metabolic, synthetic, RNA processing and DNA repair processes. H3K27me3 genes were linked to cell signaling, cell communication, developmental and differentiation functions, and H3K9me3 genes were associated with sensory perception, signal transduction, cell communication, and metabolic processes. H3K4/K27m3 genes were enriched in differentiation, developmental, and transcriptional processes. These functional categories are consistent with those identified in embryonic stem cells (Bernstein et al., 2006; Mikkelsen et al., 2007; Pan et al., 2007; Mohn et al., 2008) and in hematopoietic progenitors (Cui et al., 2009), albeit on distinct sets of genes.
Figure 7.
H3.3-EGFP targets genes are enriched in signaling and developmental functions involved in mesodermal differentiation. (A) Summary of GO term analysis of genes with promoters enriched in indicated H3 modifications or H3.3-only (see Supplemental Figure S5). (B) Top 12 most significant GO terms enriched among all H3.3-EGFP target genes and among expressed (present and marginal calls on Illumina arrays) and nonexpressed (absent calls) genes.
Enriched GO terms of genes marked by H3.3 encompassed cell cycle functions; and interestingly, signaling and developmental processes focused on tissue repair, angiogenesis, muscle regeneration, and adipogenesis (Figure 7, A and B). Connecting GO terms to gene expression, we found that these functions were associated with expressed or weakly expressed H3.3 target genes, whereas unexpressed H3.3 target genes, representing a minor proportion of all H3.3 targets (Figure 6A) were enriched in metabolic or catabolic functions (Figure 7B). In addition, enriched GO terms could be clearly distinguished on the basis of co-occupancy of H3.3 with various combinations of repressive histone modifications (Table 1). Indeed, most H3.3/H3K27me3 genes were linked to cell signaling and differentiation functions; H3.3/H3K4/K27me3 genes were predominantly associated with transcription regulation and developmental functions; H3.3/H3K4/K9me3 genes were linked to DNA metabolic processes, whereas most H3.3/H3K4/K9/K27me3 genes were associated with transcription regulation and metabolic processes. Thus, in MSCs, H3.3-EGFP preferentially associates with promoters of genes encoding various functions with a strong propensity toward mesodermal differentiation pathways, as well as transcription regulation and signaling functions particularly when it co-occupies promoters with transcriptionally repressive histone modifications.
Table 1.
Functional GO terms associated with genes with a promoter enriched in H3.3-EGFP and indicated combinations of repressive histone modifications
| Combination on promoter | % of genes making up GO terms |
|---|---|
| H3.3/H3K27me3 (n = 74) | |
| Regulation of Rab GTPase activity | 21 |
| Regulation of Ras signal transduction | 8.5 |
| Signal transduction | 15 |
| Muscle differentiation and function | 11 |
| Transport | 11 |
| Other | 33.5 |
| H3.3/H3K4me3/H3K27me3 (n = 80) | |
| Regulation of transcription | 41 |
| Development and differentiation | 38 |
| Macromolecule biosynthesis | 8 |
| Regulation of metabolism | 8 |
| Other | 5 |
| H3.3/H3K9me3 (n = 5) | |
| Response to stimulus | 100 |
| H3.3/H3K9me3/H3K27me3 (n = 2) | |
| No GO terms identified | |
| H3.3/H3K4me3/H3K9me3 (n = 12) | |
| RNA metabolic process | 100 |
| H3.3/H3K4me3/H3K9me3/H3K27me3 (n = 15) | |
| Regulation of transcription | 57 |
| Macromolecule biosynthesis | 11 |
| Metabolic/biological process | 26 |
| Other | 6 |
DISCUSSION
We have combined imaging with chromatin and methylated DNA immunoprecipitation approaches to refine the chromatin environment of H3.3 deposition in the human genome. Cell imaging and immunoprecipitation approaches operate at different resolution levels: one pixel covers 4.2 × 10−3 μm2, with the number of nucleosomes within this area varying with the degree of chromatin compaction, whereas in our study ChIP-chip and MeDIP-chip tiled sequences at 100-base pair resolution. This enables linking global nucleus-wide representations to high-resolution genomic data to define chromatin domains at these different scales by their respective contents in various histones, histone variants, and histone modifications.
Quantitative imaging of human MSCs, HeLa cells and mouse myoblasts reveals that, relative to canonical H2B, H3.3 is enriched in chromatin domains marked by histone modifications of active genes and impoverished in regions marked by repressive modifications. In contrast, canonical H3.2 is not enriched in active chromatin marks, but as H3.3, it is less correlated with repressive marks than H2B. This observation is somewhat counterintuitive, considering the widespread genomic distribution of H3.2 reflected by the majority of promoters containing some H3.2 (see NCBI GEO GSE17053) and by the low number of promoters showing H3.2 enrichment over genome-average. Human somatic cells contain two replicative H3 (H3.1 and H3.2) in addition to H3.3 and the centromeric CENP-A. Thus, the absence of strong correlation of H3.3 or H3.2 with negative marks at the nucleus level could be explained by the fact that one or both of the other two H3 variants, H3.1 or CENP-A, correlates better with repressive marks. Indeed, epitope-tagged H3.1 localizes in heterochromatic foci (Tamura et al., 2009) and is preferentially modified by repressive marks (Loyola et al., 2006; Tamura et al., 2009). CENP-A localizes to centromeres (Black et al., 2004), strongly enriched in heterochromatin.
The greater extent of colocalization of H3.3 with epitope-tagged H4 than with H3.2 or H2B is consistent with the formation of an H3/H4 heterodimer as an initial step of nucleosome assembly (Kornberg and Thomas, 1974). Moreover, because H4 has no known variant, only H4 itself is available for dimerization with any H3 variant; this accounts for the highest correlation (R = 0.91), determined by imaging, of H3.3 with any of the histones examined other than itself (Figure 1C).
Our results indicate that H3.2 and H3.3 are distinguished by their enrichment in trimethylated H3K4 and acetylated H3K9 domains. The differential epigenetic fate of H3.3 and H3.2 is further highlighted at the promoter level, where H3.2 is found on a larger proportion of promoters occupied by H3K9me3 and/or H3K27me3. Furthermore, unlike H3.3-EGFP promoters, all H3.2-EGFP promoters are co-occupied with any of the modifications examined. This again agrees with a wider genomic distribution of H3.2 than H3.3, and with a local affinity of H3.2 for a repressive chromatin environment or a propensity of H3.2 to be modified by repressive marks (Hake et al., 2006). The latter observation remains compatible with the propensity of H3.2 to be less correlated with repressive marks than H2B, because local repressive chromatin configurations are likely to exist to modulate gene expression within large-scale transcriptionally permissive domains.
Distinct assembly pathways for H3 variants may contribute to specifying the epigenetic fate of H3.1, H3.3, and perhaps H3.2. Replication-coupled integration of the H3/H4 dimer into nucleosomes is mediated by chromatin assembly factor CAF-1, whereas incorporation of H3.3/H4 occurs through the histone repression A (HIRA) protein (Ray-Gallet et al., 2002; Tagami et al., 2004). Upstream of these chaperones is anti-silencing factor 1 (Asf1), found in a complex with H3.3 and H3.1 and involved in both replication-coupled and replication-independent nucleosome assembly (Tagami et al., 2004; De Koning L. et al., 2007). Asf1 interacts with single H3.1/H4 or H3.3/H4 dimers (English et al., 2005) and has been proposed to distribute these dimers to, respectively, CAF-1 and HIRA (Orsi et al., 2009). Whether Asf1 is also involved in H3.2 deposition is currently unknown. Additional specificity of H3.3 toward active chromatin would be subsequently provided by HIRA, targeting H3.3 to sites of transcription in a replication-independent manner. Intriguingly, the chromatin environment of H3.2 and H3.3 may also be influenced by their differential posttranslational modification before assembly into nucleosomes, as suggested for H3.1 and H3.3 (Loyola et al., 2006). These modifications could occur at transit sites associated with chaperones. Our preliminary data suggest an intermediate assembly step of epitope-tagged H3.3 (Delbarre, Küntziger, and Collas, unpublished data), but whether posttranslational modifications occur at this stage remains to be investigated.
Interrogating a fraction of the genome by ChIP-on-chip first reveals that most H3.3-EGFP promoters are not enriched in any of the modified histones examined here, extending recent findings from a few genes in avian erythroid cells (Jin and Felsenfeld, 2006). Moreover, only a minor fraction (<10–20%) of endogenous H3.3 has been shown in tissues to be trimethylated on K4, K9 or K27 (Garcia et al., 2008), consistent with the low proportion of trimethylated forms of oligonucleosomal exogenous H3.3 in HeLa cells (Loyola et al., 2006). Our results therefore concur with the idea that not all H3.3 may be subject to the posttranslational modifications examined here or in other studies.
H3.3-EGFP promoters are preferentially CpG-methylated, when analyzed against all Refseq promoters, an observation in line with findings that H3K4me3 is antagonistic to DNA methylation (Ooi et al., 2007). Active genes (trimethylated on H3K4) show little H3.3 turnover at promoters that incorporate H3.3 constitutively, in contrast to coding regions that are more dynamic (Jin and Felsenfeld, 2006; Tamura et al., 2009). Thus, one would predict a low incorporation frequency of H3.3 in H3K4me3 promoters. However, among H3.3-EGFP promoters coenriched in any of the trimethylated marks examined here, H3K4me3 largely prevails, whereas H3K9me3 is essentially nonexistent. Mutual exclusion of H3.3 and H3K9me3 on promoters is in accordance with the segregation of these marks by quantitative imaging at the nucleus level in human and mouse cells. Our imaging data together with earlier studies argue that H3.3 is also incorporated in genomic regions extending beyond or other than promoters (Mito et al., 2005; Wirbelauer et al., 2005; Jin and Felsenfeld, 2006, 2007; Mito et al., 2007), which massively parallel sequencing has recently shown to include gene bodies, transcription termination sites, and insulator elements (Jin et al., 2009).
Several studies including ours show that exogenous H3.3 incorporates in promoters even in the absence of detectable transcripts for these genes, suggesting that promoter activity is not required to replace H3 with H3.3 (Chow et al., 2005; Jin and Felsenfeld, 2006). In hematopoietic progenitors, some of these inactive genes are expressed at a later developmental stage (Jin and Felsenfeld, 2006), suggesting that H3.3 marks a subset of genes primed for activation. Notably, these genes are marked by a so-called “bivalent” H3K4me3 and H3K27me3 combination (Barski et al., 2007; Cui et al., 2009). We show here that in MSCs, H3.3 can be found on differentiation-regulated H3K4me3 and H3K27me3 coenriched promoters, notably on adipogenic promoters that we have previously shown are demethylated on H3K27 after adipogenic stimulation (Noer et al., 2009). These observations collectively raise the possibility that H3.3 may be an additional component of lineage priming.
Interestingly, in homeotic gene clusters of Drosophila S2 cells, H3.3 marks genes for activation by histone turnover through nucleosome eviction (Mito et al., 2007), suggesting that H3.3 marks H3K4me3 nucleosomes for turnover, rather than H3K27me3 nucleosomes that would be stable (Henikoff, 2008). We identified in MSCs a narrow region apparently depleted of H3K4me3, H3.3, and H3K27me3 over the TSS in metapromoter analyses (Jacobsen, Reiner, and Collas, unpublished data). This suggests that on these promoters in progenitor cells, H3K27me3 nucleosomes may also be of lower density at the TSS as a result of their instability. Consistent with the view that H3.3 may mark promoters for transcriptional activation are the parallel and in-phase occupancy profiles of H3.3, H3K4me3, and H3K27me3 on promoters enriched in all marks, and the concept that H3.3 may serve as a memory mark of active chromatin states during mitosis (Chow et al., 2005) and development (Ng et al., 2008) in the absence of transcription.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Marie Rogne for in situ extractions. This work was funded by the Association Française pour la lutte contre les Myopaties (to E. D.), the University of Oslo (to B.M.J.), the Research Council of Norway, and South-East Health Norway.
Abbreviations used:
- Asf1
anti-silencing factor 1
- ChIP
chromatin immunoprecipitation
- FDR
false discovery rate
- GO
gene ontology
- HCP
high CpG promoter
- HIRA
histone repression A
- ICP
intermediate CpG promoter
- LCP
low CpG promoter
- me
methylated
- MeDIP
methylated DNA immunoprecipitation
- TSS
transcription start site.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-09-0839) on April 7, 2010.
REFERENCES
- Ahmad K., Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bernstein B. E., et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Black B. E., Foltz D. R., Chakravarthy S., Luger K., Woods V. L., Jr, Cleveland D. W. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- Bolte S., Cordelieres F. P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- Boquest A. C., Shahdadfar A., Fronsdal K., Sigurjonsson O., Tunheim S. H., Collas P., Brinchmann J. E. Isolation and transcription profiling of purified uncultured human stromal stem cells: alteration of gene expression after in vitro cell culture. Mol. Biol. Cell. 2005;16:1131–1141. doi: 10.1091/mbc.E04-10-0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow C. M., Georgiou A., Szutorisz H., Maia e Silva, Pombo A., Barahona I., Dargelos E., Canzonetta C., Dillon N. Variant histone H3.3 marks promoters of transcriptionally active genes during mammalian cell division. EMBO Rep. 2005;6:354–360. doi: 10.1038/sj.embor.7400366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton M. P., Markoulaki S., Levine S. S., Hanna J., Lodato M. A., Sha K., Young R. A., Jaenisch R., Boyer L. A. H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell. 2008;135:649–661. doi: 10.1016/j.cell.2008.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui K., Zang C., Roh T. Y., Schones D. E., Childs R. W., Peng W., Zhao K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl J. A., Collas P. Q2ChIP, a quick and quantitative chromatin immunoprecipitation assay unravels epigenetic dynamics of developmentally regulated genes in human carcinoma cells. Stem Cells. 2007;25:1037–1046. doi: 10.1634/stemcells.2006-0430. [DOI] [PubMed] [Google Scholar]
- Dahl J. A., Reiner A. H., Collas P. Fast genomic ChIP-chip from 1,000 cells. Genome Biol. 2009;10:R13. doi: 10.1186/gb-2009-10-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daury L., Chailleux C., Bonvallet J., Trouche D. Histone H3.3 deposition at E2F-regulated genes is linked to transcription. EMBO Rep. 2006;7:66–71. doi: 10.1038/sj.embor.7400561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Koning L., Corpet A., Haber J. E., Almouzni G. Histone chaperones: an escort network regulating histone traffic. Nat. Struct. Mol. Biol. 2007;14:997–1007. doi: 10.1038/nsmb1318. [DOI] [PubMed] [Google Scholar]
- English C. M., Maluf N. K., Tripet B., Churchill M. E., Tyler J. K. ASF1 binds to a heterodimer of histones H3 and H4: a two-step mechanism for the assembly of the H3–H4 heterotetramer on DNA. Biochemistry. 2005;44:13673–13682. doi: 10.1021/bi051333h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon S., Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- Garcia B. A., Thomas C. E., Kelleher N. L., Mizzen C. A. Tissue-specific expression and post-translational modification of histone H3 variants. J. Proteome. Res. 2008;7:4225–4236. doi: 10.1021/pr800044q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlich D., Beaudouin J., Kalbfuss B., Daigle N., Eils R., Ellenberg J. Global chromosome positions are transmitted through mitosis in mammalian cells. Cell. 2003;112:751–764. doi: 10.1016/s0092-8674(03)00189-2. [DOI] [PubMed] [Google Scholar]
- Hake S. B., Garcia B. A., Duncan E. M., Kauer M., Dellaire G., Shabanowitz J., Bazett-Jones D. P., Allis C. D., Hunt D. F. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J. Biol. Chem. 2006;281:559–568. doi: 10.1074/jbc.M509266200. [DOI] [PubMed] [Google Scholar]
- Håkelien A. M., Delbarre E., Gaustad K. G., Buendia B., Collas P. Expression of the myodystrophic R453W mutation of lamin A in C2C12 myoblasts causes promoter-specific and global epigenetic defects. Exp. Cell Res. 2008;314:1869–1880. doi: 10.1016/j.yexcr.2008.02.018. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat. Rev. Genet. 2008;9:15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Ahmad K. Assembly of variant histones into chromatin. Annu. Rev. Cell Dev. Biol. 2005;21:133–153. doi: 10.1146/annurev.cellbio.21.012704.133518. [DOI] [PubMed] [Google Scholar]
- Jin C., Felsenfeld G. Distribution of histone H3.3 in hematopoietic cell lineages. Proc. Natl. Acad. Sci. USA. 2006;103:574–579. doi: 10.1073/pnas.0509974103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C., Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C., Zang C., Wei G., Cui K., Peng W., Zhao K., Felsenfeld G. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat. Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. S., et al. Systematic evaluation of variability in ChIP-chip experiments using predefined DNA targets. Genome Res. 2008;18:393–403. doi: 10.1101/gr.7080508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H., Cook P. R. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 2001;153:1341–1353. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose R. J., Bird A. P. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Kornberg K. D., Thomas J. O. Chromatin structure: oligomers of the histones. Science. 1974;184:865–868. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Loyola A., Bonaldi T., Roche D., Imhof A., Almouzni G. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol. Cell. 2006;24:309–316. doi: 10.1016/j.molcel.2006.08.019. [DOI] [PubMed] [Google Scholar]
- Martin C., Zhang Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- McKittrick E., Gafken P. R., Ahmad K., Henikoff S. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl. Acad. Sci. USA. 2004;101:1525–1530. doi: 10.1073/pnas.0308092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T. S., et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y., Henikoff J. G., Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- Mito Y., Henikoff J. G., Henikoff S. Histone replacement marks the boundaries of cis-regulatory domains. Science. 2007;315:1408–1411. doi: 10.1126/science.1134004. [DOI] [PubMed] [Google Scholar]
- Mohn F., Weber M., Rebhan M., Roloff T. C., Richter J., Stadler M. B., Bibel M., Schübeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol. Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Ng R. K., Dean W., Dawson C., Lucifero D., Madeja Z., Reik W., Hemberger M. Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat. Cell Biol. 2008;10:1280–1290. doi: 10.1038/ncb1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noer A., Lindeman L. C., Collas P. Histone H3 modifications associated with differentiation and long-term culture of mesenchymal adipose stem cells. Stem Cells Dev. 2009;18:725–735. doi: 10.1089/scd.2008.0189. [DOI] [PubMed] [Google Scholar]
- Ooi S. K., et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi S. L., Priess J. R., Henikoff S. Histone H3.3 variant dynamics in the germline of Caenorhabditis elegans. PLoS. Genet. 2006;2:e97. doi: 10.1371/journal.pgen.0020097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi G. A., Couble P., Loppin B. Epigenetic and replacement roles of histone variant H3.3 in reproduction and development. Int. J. Dev. Biol. 2009;53:231–243. doi: 10.1387/ijdb.082653go. [DOI] [PubMed] [Google Scholar]
- Pan G., Tian S., Nie J., Yang C., Ruotti V., Wei H., Jonsdottir G. A., Stewart R., Thomson J. A. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Ray-Gallet D., Quivy J. P., Scamps C., Martini E. M., Lipinski M., Almouzni G. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell. 2002;9:1091–1100. doi: 10.1016/s1097-2765(02)00526-9. [DOI] [PubMed] [Google Scholar]
- Schwartz B. E., Ahmad K. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev. 2005;19:804–814. doi: 10.1101/gad.1259805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen A. L., Collas P. Immunoprecipitation of methylated DNA. Methods Mol. Biol. 2009;567:249–261. doi: 10.1007/978-1-60327-414-2_16. [DOI] [PubMed] [Google Scholar]
- Stokes D. G., Perry R. P. DNA-binding and chromatin localization properties of CHD1. Mol. Cell Biol. 1995;15:2745–2753. doi: 10.1128/mcb.15.5.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe E. L., et al. Dynamic histone variant exchange accompanies gene induction in T cells. Mol. Cell Biol. 2009;29:1972–1986. doi: 10.1128/MCB.01590-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H., Ray-Gallet D., Almouzni G., Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- Tamura T., Smith M., Kanno T., Dasenbrock H., Nishiyama A., Ozato K. Inducible deposition of the histone variant H3.3 in interferon-stimulated genes. J. Biol. Chem. 2009;284:12217–12225. doi: 10.1074/jbc.M805651200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heijden G. W., et al. Chromosome-wide nucleosome replacement and H3.3 incorporation during mammalian meiotic sex chromosome inactivation. Nat. Genet. 2007;39:251–258. doi: 10.1038/ng1949. [DOI] [PubMed] [Google Scholar]
- Weber M., Hellmann I., Stadler M. B., Ramos L., Paabo S., Rebhan M., Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- Wirbelauer C., Bell O., Schubeler D. Variant histone H3.3 is deposited at sites of nucleosomal displacement throughout transcribed genes while active histone modifications show a promoter-proximal bias. Genes Dev. 2005;19:1761–1766. doi: 10.1101/gad.347705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L. H., et al. Histone H3.3 incorporation provides a unique and functionally essential telomeric chromatin in embryonic stem cells. Genome Res. 2009;19:404–414. doi: 10.1101/gr.084947.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D., Coleman-Derr D., Ballinger T., Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–129. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.