Nup98 is a mobile nucleoporin that forms distinct dots in the nucleus. Our results show that Nup98 functions as a novel shuttling cofactor in Crm1-mediated nuclear export.
Abstract
Nup98 is a mobile nucleoporin that forms distinct dots in the nucleus, and, although a role for Nup98 in nuclear transport has been suggested, its precise function remains unclear. Here, we show that Nup98 plays an important role in Crm1-mediated nuclear protein export. Nuclear, but not cytoplasmic, dots of EGFP-tagged Nup98 disappeared rapidly after cell treatment with leptomycin B, a specific inhibitor of the nuclear export receptor, Crm1. Mutational analysis demonstrated that Nup98 physically and functionally interacts with Crm1 in a RanGTP-dependent manner through its N-terminal phenylalanine-glycine (FG) repeat region. Moreover, the activity of the Nup98-Crm1 complex was modulated by RanBP3, a known cofactor for Crm1-mediated nuclear export. Finally, cytoplasmic microinjection of anti-Nup98 inhibited the Crm1-dependent nuclear export of proteins, concomitant with the accumulation of anti-Nup98 in the nucleus. These results clearly demonstrate that Nup98 functions as a novel shuttling cofactor for Crm1-mediated nuclear export in conjunction with RanBP3.
INTRODUCTION
The nuclear pore complex (NPC) is a large (50–100 MDa) protein complex embedded in the nuclear envelope that mediates macromolecular traffic between the nucleus and cytoplasm (Reichelt et al., 1990; Cronshaw et al., 2002). Small molecules (e.g., ions and proteins <40 kDa) can passively diffuse through the NPC (Nigg, 1997), but the nuclear transport of larger molecules is an active process that requires energy (Hicks and Raikhel, 1995; Gorlich and Mattaj, 1996).
In yeast and higher eukaryotes, NPCs are composed of ∼30 different proteins, called nucleoporins (Nups). Although Nups comprise the NPC, they also play roles in various cellular processes, including nuclear transport, mitotic spindle assembly, and the regulation of gene expression (Suntharalingam and Wente, 2003; Tran and Wente, 2006; Dasso, 2006; Brown and Silver, 2007; Taddei, 2007).
Nups are classified according to their mobility, structural folds, and relative localization within the NPC. Mobile Nups, such as Nup50 and Nup153, reside within the NPC for a short period of time (seconds), but stable Nups including Nup107 and Nup133, can remain within the NPC for days (Rabut et al., 2004). Thus, stable Nups are thought to provide a scaffold for NPC assembly, whereas mobile Nups actively participate in nucleocytoplasmic transport (Tran and Wente, 2006).
When considered from a structural perspective, approximately one-third of Nups contain a characteristic fold consisting of tandem repeats of phenylalanine-glycine (FG) repeats, which are unfolded in the native protein (Denning et al., 2003). The FG repeats have a moderately high affinity for transport factors (Wente et al., 1992; Radu et al., 1995; Rexach and Blobel, 1995; Clarkson et al., 1996; Hu et al., 1996) and can form hydrogels in vitro that selectively allow the influx of transport factors (Frey and Gorlich, 2007). Thus, FG repeat–containing Nups (FG Nups) provide both the binding site and route for translocation of macromolecules through the NPC.
The relative localization of each Nup within the NPC differs, and Nups demonstrate either a symmetric or an asymmetric distribution. The Nup107-160 subcomplex, which consists of at least 10 proteins (Belgareh et al., 2001; Vasu et al., 2001; Harel et al., 2003; Loiodice et al., 2004; Rasala et al., 2006), is symmetrically localized to both the cytoplasmic and nuclear faces of the NPC. In contrast, asymmetrically distributed Nups include Nup214 and Nup358, which localize exclusively to the cytoplasmic aspect of the NPC (Kraemer et al., 1994; Wu et al., 1995; Yokoyama et al., 1995), and Nup153, which localizes to the nuclear face of the NPC (Sukegawa and Blobel, 1993). On the basis of their localization and differential affinity for transport factors, several groups hypothesized that asymmetric Nups may play an important role in directional nuclear transport (Floer et al., 1997; Rout et al., 2000; Ben-Efraim and Gerace, 2001; Pyhtila and Rexach, 2003).
Nup98 is a mobile Nup that contains an FG domain (Powers et al., 1995; Radu et al., 1995; Griffis et al., 2002). Nup98 functions in the nucleo-cytoplasmic transport of macromolecules (Powers et al., 1995, 1997; Pritchard et al., 1999; Zolotukhin and Felber, 1999; Fontoura et al., 2000; Wu et al., 2001, Iwamoto et al., 2009), but its precise role in mediating nuclear transport remains unclear. Additionally, Nup98 is predominantly asymmetrically localized to the nuclear face of the NPC (Radu et al., 1995; Frosst et al., 2002), but a fraction of Nup98 is found on the cytoplasmic side of NPC (Griffis et al., 2003). Thus, the localization pattern of Nup98 suggests that it may function in active nuclear transport. An additional, characteristic feature of Nup98 is its ability to form distinct dots in the nucleus in vivo (Powers et al., 1995; Griffis et al., 2002), and this is not seen with other FG Nups. Nevertheless, the physiological significance of nuclear Nup98 dot formation remains poorly understood.
In this report, we examined the punctate nuclear Nup98 dots and demonstrated that they disappeared rapidly upon treatment with leptomycin B (LMB), a specific inhibitor of the nuclear export factor Crm1 (Fornerod et al., 1997; Fukuda et al., 1997; Ossareh-Nazari et al., 1997; Kudo et al., 1999). Additionally, the Nup98 dots disappeared when Ran-binding protein 3 (RanBP3), a cofactor for Crm1-mediated nuclear export, was overexpressed. Further analysis revealed that Nup98 functionally and physically interacted with Crm1 through its N-terminal FG-repeat region in a RanGTP-dependent manner. Strikingly, we found that antibodies specific for the Nup98 FG repeat strongly inhibited the Crm1-mediated export of proteins when microinjected into both the nucleus and cytoplasm. Taken together, our findings indicate that the mobile nucleoporin Nup98 functions as a cofactor for nuclear protein export mediated by Crm1.
MATERIALS AND METHODS
Cell Culture and Transfections
NIH 3T3 and HeLa cells were grown in DMEM (Sigma, St. Louis, MO) supplemented with 10% fetal bovine serum. Transfections were performed using Effectene (Qiagen, Chatsworth, CA) or Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. HEK293F cells were cultured in suspension in FreeStyle 293 Expression Medium (Invitrogen), and transfected with 293fectin Transfection Reagent (Invitrogen) following the manufacturer's instructions.
Antibodies
Rat monoclonal anti-Nup98 (Fukuhara et al., 2005) and anti-Nup62 (Fukuhara et al., 2006) antibodies were prepared as described. These monoclonal antibodies were further purified in phosphate-buffered saline (PBS) before microinjection. Rabbit polyclonal anti-Crm1 and anti-RanBP3 antibodies were purchased from Novus Biologicals (Littleton, CO). Rabbit polyclonal anti-RanBP1, rabbit polyclonal anti-glutathione S-transferase (GST), goat polyclonal anti-B23, mouse monoclonal anti-fibrillarin, and mouse monoclonal anti-UBF antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Plasmids
For the generation of enhanced green fluorescent protein (EGFP)-tagged Nup98, a cDNA encoding murine Nup98 (aa 1-880) was prepared from mouse brain total RNA using the SuperScript One-Step High Fidelity system (Invitrogen) and subcloned into the pTRE2-EGFP-hyg vector containing the EGFP open reading frame upstream of the pTRE2-hyg (Clontech, Palo Alto, CA) multicloning site (pTRE2-EGFP-Nup98). pTRE2-EGFP-Nup98 was cotransfected with tTA2 expression plasmid. A series of Nup98 deletion mutants were amplified by PCR and subcloned into the pTRE2-EGFP-hyg vector. Nup98 fragments were subsequently cloned into the pEGFP-C1 vector (BD Biosciences, San Jose, CA) to generate EGFP-Nup98 expression plasmids. GST-Nup98, GST-Nup214 (aa 1864-2090), GST-Crm1, and GST-RanBP3 fusion constructs were generated by insertion of the appropriate PCR fragments into the pGEX6P vector (GE Healthcare, Piscataway, NJ). GST-Nup98 fusion protein–encoding plasmids were prepared by subcloning the Nup98 fragments in-frame with GST in pCAG-wtag, which contains GST under the control of the CAG promoter (Takeda et al., 2005). The Nup98FG-EGFP encoding plasmid (pNup98FG-EGFP) was constructed by inserting the appropriate PCR fragment of Nup98 (aa 1-480) into the pEGFP-N1 vector (Clontech). A PCR fragment of HoxA9 (aa 195-272) was inserted into pNup98FG-EGFP to generate a plasmid encoding NUP98-HoxA9. HoxA9HD-EGFP–encoding plasmid was constructed by inserting the PCR fragment of HoxA9 homeodomain (aa 195-272) with initiation codon, ATG, into the pEGFP-N1 vector.
Expression and Purification of Recombinant Proteins
GST-fusion proteins were expressed in Escherichia coli and purified. Briefly, BL21 (DE3) cells transformed with GST-fusion vector were grown to an OD600 of 0.5–0.8 at 37°C, and the expression of GST-fusion protein was induced with 0.2 mM IPTG at 18°C for 20 h. GST-fusion proteins were then purified using glutathione Sepharose 4B (GE Healthcare). The recombinant proteins were either eluted with 20 mM glutathione or, if necessary, digested with Pre-scission protease (GE Healthcare) according to the manufacturer's protocol.
GST Pulldown Experiments
Recombinant GST-fusion proteins bound to glutathione-Sepharose beads were incubated with HeLa cell lysates (300 μg of lysate was used for each pulldown experiment) freshly prepared with CHAPS lysis buffer (50 mM Tris-Cl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.2% CHAPS, 1 mM DTT, 1 mM PMSF, 1 μg/ml leupeptin, and 1 μg/ml aprotinin) for 3 h at 4°C in the presence or absence of 2 μM RanQ69L-GTP. The beads were then washed four times with lysis buffer, and GST-fusion proteins were eluted with glutathione elution buffer (100 mM Tris-Cl, pH 8.3, 100 mM NaCl, 1 mM EDTA, 2 mM DTT, 20 mM glutathione, 1 mM PMSF, 1 μg/ml pepstatin, 1 μg/ml leupeptin, and 1 μg/ml aprotinin). The purified protein complexes were analyzed by SDS-PAGE and immunoblotting.
For the in vivo GST pulldown assay, HEK293F cells (Invitrogen) were transfected with GST-fusion protein–expressing plasmids using 293fectin (Invitrogen) according to the manufacturer's instruction. Forty-eight hours after transfection, the cells were washed with PBS and resuspended in 10 volumes of CHAPS lysis buffer (50 mM Tris-Cl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.2% CHAPS, 1 mM DTT, 1 mM PMSF, 1 μg/ml pepstatin, 1 μg/ml leupeptin, and 1 μg/ml aprotinin) and sonicated. After centrifugation, the supernatants was transferred into a fresh tube and incubated with glutathione beads for 3 h at 4°C. The beads were washed four times with lysis buffer, and GST-fusion proteins were eluted with glutathione elution buffer. The purified fusion protein complexes were analyzed by SDS-PAGE followed by immunoblotting.
Binding Assay
Recombinant GST-fusion proteins bound to glutathione-Sepharose beads were incubated with various amounts of recombinant proteins in transport buffer (20 mM HEPES, pH 7.3, 110 mM potassium acetate, 2 mM magnesium acetate, 5 mM sodium acetate, 0.5 mM ethylene glycol bis-P-aminoethylether)-N,N,N′,N′-tetraacetic acid (EGTA), 2 mM DTT, and 1 mM PMSF, 1 μg/ml pepstatin, 1 μg/ml leupeptin, and 1 μg/ml aprotinin) for 2 h at 4°C. Beads were then washed four times with transport buffer, and GST-fusion proteins were eluted with glutathione elution buffer (100 mM Tris-Cl, pH 8.3, 100 mM NaCl, 1 mM EDTA, 2 mM DTT, 20 mM glutathione, 1 mM PMSF, 1 μg/ml pepstatin, 1 μg/ml leupeptin, and 1 μg/ml aprotinin) for 20 min on ice. The purified protein complexes were analyzed by SDS-PAGE and immunoblotting.
Immunofluorescence Staining and Confocal Microscopy
Cells were grown on coverslips and fixed with 3.7% formaldehyde in PBS for 10 min at room temperature. After permeabilization with 0.5% Triton X-100 in PBS for 5 min, the cells were incubated in blocking buffer (PBS containing 3% skim milk) for 30 min and then incubated with primary antibodies overnight at 4°C. The cells were washed five times with PBS and incubated with secondary antibodies for 45 min at room temperature. The cells were washed as described above five times in PBS; the third wash included DAPI (0.1 μg/ml). Coverslips were mounted with Vectashield (Vector Laboratories, Burlingame, CA). After fixation of the cells with 3.7% formaldehyde in PBS for 10 min at room temperature, the cells were processed for immunofluorescence staining.
Images were acquired using a confocal microscope (LSM 510 META, Carl Zeiss, Thornwood, NY) equipped with 63× 1.4 NA oil objective lens (Carl Zeiss).
Antibody Microinjection
Purified monoclonal antibodies (0.3 mg/ml) were microinjected into the cytoplasm or the nucleus of HeLa cells grown on coverslips, together with GST-GFP-NES (nuclear export signal; 1 mg/ml) and/or Alexa568-conjugated donkey anti-sheep IgG as an injection marker. Fifteen minutes after microinjection, the cells were fixed with 3.7% formaldehyde in PBS for 10 min at room temperature and processed for microscopic observation.
RESULTS
Nuclear Dots of EGFP-Nup98 Are Sensitive to LMB Treatment
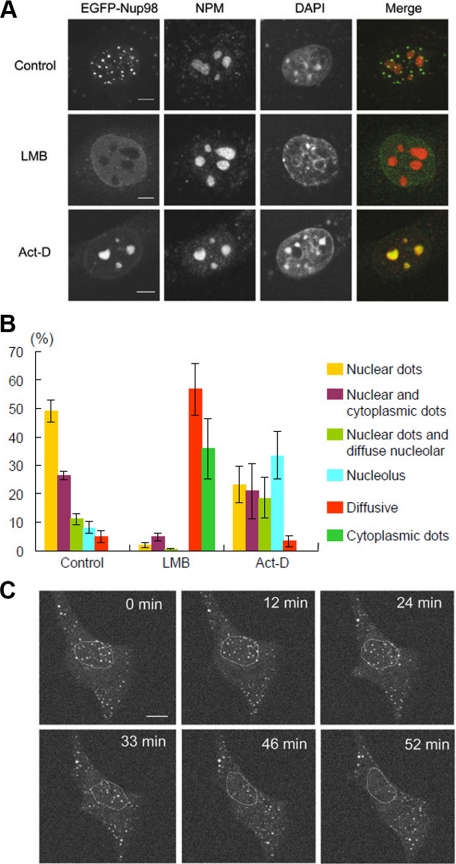
To better understand the role of Nup98 in nucleocytoplasmic transport, we first wanted to monitor the dynamic behavior of Nup98, and we expressed N-terminally EGFP-tagged murine Nup98 (EGFP-Nup98) in NIH 3T3 cells. As reported previously (Griffis et al., 2002), EGFP-Nup98 predominantly localized in the nucleus as distinct dots, and some nuclear envelope localization was also seen (Figure 1A). A similar pattern of localization was observed for N-terminally hemagglutinin (HA)-tagged Nup98 (data not shown), consistent with published results (Zolotukhin and Felber, 1999). The EGFP-Nup98 signal at the nuclear envelope increased when soluble nuclear materials were extracted before fixation (data not shown), suggesting that a fraction of EGFP-Nup98 was soluble and diffusely spread throughout the nucleoplasm.
Figure 1.
EGFP-Nup98 dots redistribute after LMB or Act-D treatment. (A) NIH 3T3 cells were transfected with EGFP-Nup98 expression vector. After 48 h, the cells were treated with DMSO (Control), 5 nM LMB, or 40 ng/ml Act-D for 2 h. Cells were then fixed, immunostained, and examined by confocal microscopy. Anti-nucleophosmin/B23 (NPM) antibody was used to stain the nucleolus. DAPI staining was used to visualize nuclei. Bar, 10 μm. (B) Summary of the results obtained in A. At least 150 EGFP-Nup98–expressing cells were observed for each condition. The localization patterns of EGFP-Nup98 were categorized into six groups as follows: nuclear dots, nuclear and cytoplasmic dots, cytoplasmic dots and diffuse nuclear staining, nuclear dots and diffuse nucleolar staining, diffuse nucleolar staining, and diffuse nuclear staining. Data are represented as mean ± SD of three independent experiments. (C) Time-lapse analysis of EGFP-Nup98 dots after LMB treatment. EGFP-Nup98–expressing NIH3T3 cells were treated with 5 nM LMB and analyzed by time-lapse microscopy. The position of the nucleus is indicated by a dashed circle. Bar, 10 μm.
To determine whether the Nup98-containing nuclear dots function in nucleocytoplasmic transport, we treated cells with LMB, a specific inhibitor of the nuclear export receptor Crm1. After LMB treatment, the Nup98 nuclear dots rapidly disappeared within 2 h (Figure 1, A and B). Time-lapse analysis demonstrated that the effects of LMB treatment were apparent as early as 10 min after the addition of LMB, and by 40 min, most of the dots either disappeared or became substantially reduced in size (data not shown). Interestingly, more than 20% of cells treated with LMB exhibited punctate cytoplasmic dots of EGFP-Nup98 with few or no nuclear dots, and this localization pattern was not observed in the absence of drug treatment (Figure 1B). Furthermore, when LMB-treated cells were examined by time-lapse microscopy the appearance and disappearance of cytoplasmic and nuclear dots, respectively, occurred in concert (Figure 1C). These results suggest that the behavior of the nuclear Nup98 dots is closely related to the function of Crm1.
Consistent with a previous report (Fornerod et al., 1997), Crm1 accumulated in the nucleolus after cell treatment with a low-dose of actinomycin D (Act-D; see Figure 7C), an RNA polymerase I inhibitor (Perry and Kelley, 1970). Therefore we treated EGFP-Nup98–expressing cells with low-dose Act-D to monitor changes in EGFP-Nup98 localization. Similar to what was seen for Crm1, EGFP-Nup98 accumulated at the nucleolus after Act-D treatment (Figure 1, A and B), and this phenomenon was only rarely seen in cells growing under normal culture conditions. Taken together, these results suggest that exogenously expressed EGFP-Nup98 associates with Crm1.
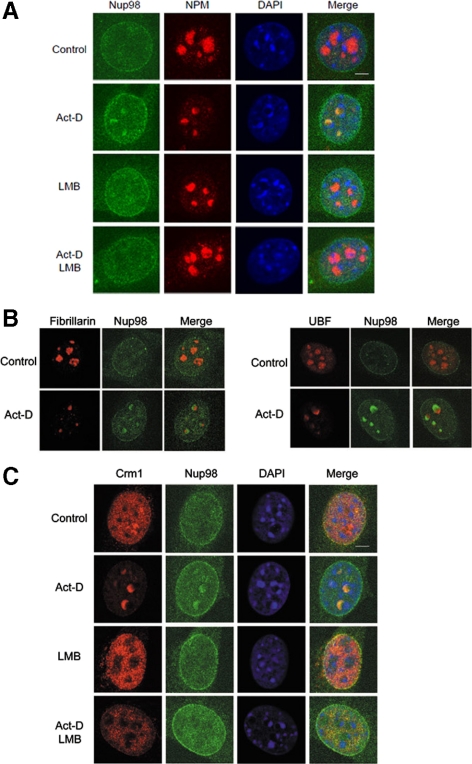
Figure 7.
Endogenous Nup98 relocalizes to the granular compartment of the nucleolus after Act-D treatment in a Crm1-dependent manner. (A) NIH 3T3 cells were treated with either Act-D or LMB, or both for 2 h. The cells were fixed, permeabilized, and immunostained with anti-nucleophosmin/B23 (NPM) and anti-Nup98. The nuclei were visualized by DAPI staining. Bar, 5 μm. (B) NIH 3T3 cells were treated with Act-D or DMSO (Control) for 2 h. The cells were fixed, permeabilized, and immunostained with anti-Nup98 together with the indicated antibody to stain the components of the nucleolus. Merged images are shown. Bar, 5 μm. (C) NIH 3T3 cells were treated with either Act-D or LMB, or both for 2 h. The cells were fixed, permeabilized, and immunostained with anti-Crm1 and anti-Nup98. The nuclei were visualized by DAPI staining. Bar, 5 μm.
The N-Terminal FG-Repeat Region of Nup98 Forms Dots That Colocalize with Crm1
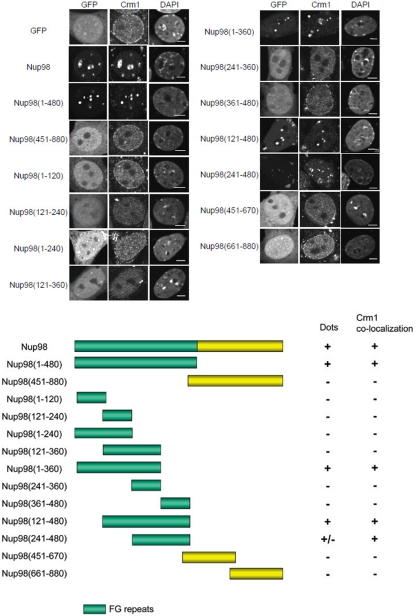
We next wanted to determine the domain(s) of Nup98 responsible for mediating its possible interaction with Crm1. As such, we transiently transfected NIH 3T3 cells with plasmids encoding full-length EGFP-Nup98 or deletion mutants. As shown in Figure 2, in control EGFP-expressing cells, endogenous Crm1 was localized diffusely throughout the nucleoplasm and at the nuclear envelope. In contrast, when EGFP-Nup98 was expressed, endogenous Crm1 colocalized with distinct Nup98 dots in the nucleus. In addition, the staining intensity of Crm1 both in the nucleoplasm and at the nuclear envelope was diminished in the EGFP-Nup98–expressing cells.
Figure 2.
The N-terminal FG-repeat region of Nup98 is required for the formation of nuclear dots that colocalize with Crm1. NIH 3T3 cells were transfected with EGFP-Nup98 or the indicated EGFP-Nup98 deletion mutants. After 48 h, the cells were fixed, permeabilized, and immunostained with anti-Crm1 antibody. The formation of nuclear dots and the localization of Crm1 were determined. DAPI staining was used to visualize nuclei. Bar, 5 μm.
Nup98 can be divided into two regions, an N-terminal half containing FG repeats and a C-terminal half lacking FG repeats (Radu et al., 1995), and we constructed plasmids encoding the Nup98 N-terminus (aa 1-480) or C-terminus (aa 451-880) N-terminally tagged with EGFP. As shown in Figure 2, the N-terminal half of Nup98 formed nuclear dots that colocalized with endogenous Crm1, but the C-terminal Nup98 construct was diffusely expressed throughout the cells, consistent with a previous report (Griffis et al., 2002).
To better define the residues of Nup98 responsible for mediating the colocalization with Crm1, we prepared an additional series of deletion mutants tagged to EGFP. We found that aa 121-360 of Nup98 were necessary but not sufficient for the formation of Nup98 dots in the nucleus, and additional residues from the N-terminus (1-120) or C-terminus (361-480) were required for dot formation. The constructs colocalized with endogenous Crm1 when cells were examined by coimmunostaining with anti-Crm1. These results indicate that the FG-repeat domain of Nup98 is required for the formation of nuclear dots that colocalize with endogenous Crm1.
Overexpression of the FG-Repeat Region of Nup98 Inhibits Crm1-mediated Nuclear Export
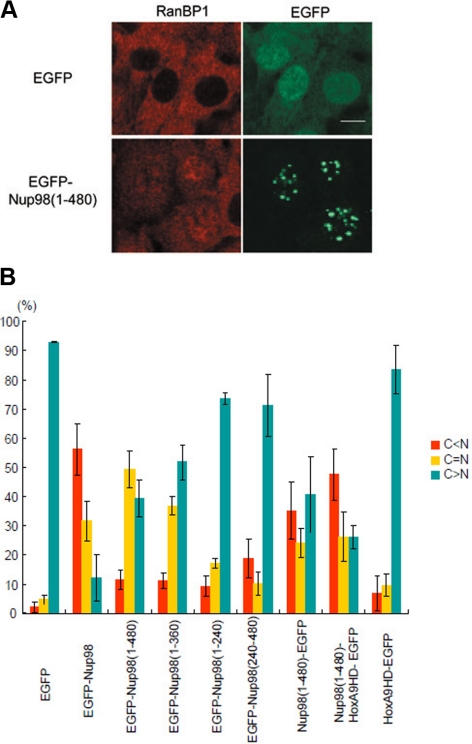
To examine whether the FG repeats of Nup98 are functionally involved in Crm1-mediated nuclear export in vivo, we overexpressed the Nup98 FG-repeat region in NIH 3T3 cells to determine its effect on the localization of endogenous RanBP1, a protein shuttles between the nucleus and cytoplasm in a Crm1-dependent manner (Richards et al., 1996; Zolotukhin and Felber, 1997). We found that the expression of the EGFP-Nup98 (1-480) (Figure 3, A and B) or Nup98 (1-360) as well as EGFP-Nup98 effectively inhibited the nuclear export of RanBP1 (Figure 3B). We also expressed two Nup98 deletion mutants that did not efficiently form nuclear dots (i.e., 1-240 and 240-480 aa), and their ability to inhibit RanBP1 export was decreased (Figure 3B). Thus, there appears to be a relationship between Nup98 nuclear dot formation and the inhibition of nuclear export, suggesting that sequestration of endogenous Crm1 to the Nup98 dots (Figure 2) inhibits nuclear export. In contrast, Nup98 expression showed no obvious inhibitory effect on the nuclear import of mRFP-NLS, which is imported into the nucleus in an importin α/β–dependent manner (Figure S1).
Figure 3.
The FG-repeat region of Nup98 affects the function of Crm1. (A) NIH 3T3 cells were transfected with EGFP or EGFP-Nup98 (aa 1-480) expression vectors. After 48 h, the cells were immunostained with anti-RanBP1 antibody. Bar, 10 μm. (B) NIH 3T3 cells were transfected with the indicated expression vectors. After 48 h, the cells were immunostained with anti-RanBP1, and the cellular localization of RanBP1 in EGFP-positive cells was categorized as C > N, C = N, or C < N. Data are represented as mean ± SD of three independent experiments.
In some forms of acute myeloid leukemia, the N-terminal FG-repeat region of Nup98 forms oncogenic fusion genes with several different partner genes. Therefore, we next examined whether one of these oncogenic Nup98-fusion genes could affect the function of Crm1. We overexpressed a fusion protein consisting of the Nup98 FG-repeat region combined with the homeodomain (HD) of HoxA9 (Borrow et al., 1996; Nakamura et al., 1996), which is C-terminally tagged with EGFP (Nup98-HoxA9-EGFP). When Nup98-HoxA9-EGFP was expressed, it localized as fine nuclear dots (Figure S2), as reported previously (Kasper et al., 1999). Confocal microscopic analysis revealed that these dots were not targeted to the nuclear pore complex (Figure S3), but they partially colocalized with Crm1 (Figure S4). Furthermore, expression of Nup98-HoxA9-EGFP (Figure 3B) as well as untagged Nup98-HoxA9 (data not shown) inhibited the export of RanBP1. Collectively, these results demonstrate that the FG-repeat domain of Nup98 is involved in nuclear protein export in vivo.
Cytoplasmic Nup98 Dots Are Not Colocalized with Crm1
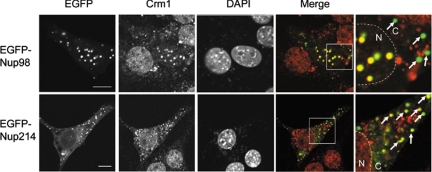
In addition to its nuclear localization, some Nup98 dots were seen in the cytoplasm when overexpressed. Thus, we attempted to determine whether these cytoplasmic Nup98 dots also colocalized with Crm1. However, there was no Crm1 colocalized to these structures (Figure 4). Crm1 is predominantly found in the nucleoplasm and the nuclear envelope, and it is possible that the preferential colocalization of Crm1 with the nuclear Nup98 dots may be secondary to the increased amounts of Crm1 in the nucleus. To test this possibility, we expressed EGFP fusion to Nup214, a Nup that forms cytoplasmic dots (Bastos et al., 1997), and stained the cells with anti-Crm1. As shown in Figure 4, cytoplasmic Nup214 dots colocalized with endogenous Crm1, consistent with a previous report (Boer et al., 1998), and this demonstrates that cytoplasmic concentrations of Crm1 are sufficient to mediate dot formation. Thus, nuclear, but not cytoplasmic, Nup98 preferentially associates with Crm1. Additionally, because there is a concentration gradient of RanGTP across the nuclear envelope, Ran GTPase, a key regulator of directed nucleocytoplasmic transport, may play a role in the association of Nup98 and Crm1 (Dasso, 2001; Kalab et al., 2002).
Figure 4.
Nuclear, but not cytoplasmic, Nup98 dots colocalize with Crm1. NIH 3T3 cells were transfected with EGFP-Nup98– or EGFP-Nup214– encoding plasmids. After 48 h, the cells were fixed, permeabilized, and immunostained with anti-Crm1. DAPI staining was used to visualize nuclei. Arrows indicate the EGFP-Nup dots in the cytoplasm. Merged images of EGFP-Nup98 or EGFP-Nup214 (green) and Crm1 (red) are shown. N, nucleus; C, cytoplasm. Bar, 10 μm.
Nup98 Directly Interacts with Crm1 in a RanGTP-dependent Manner
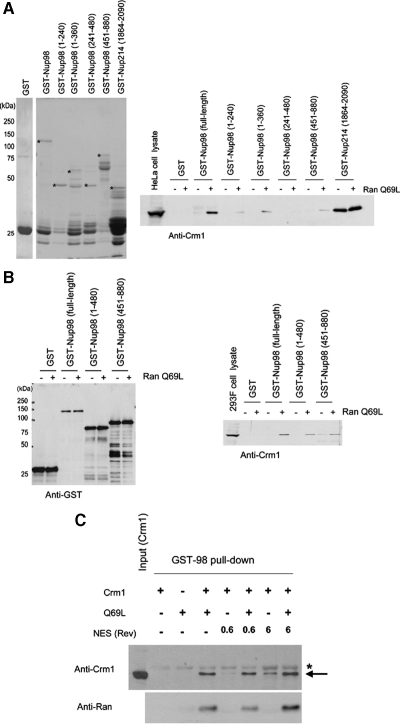
We have shown that Nup98 colocalizes with Crm1, and our data suggest a possible role for RanGTP in this process. However, it is unknown whether these proteins directly interact. Therefore, we performed a pulldown assay using recombinant Nup98 N-terminally tagged with GST (GST-Nup98) using HeLa cell lysates in the presence or absence of GTP-loaded RanQ69L, a GTPase-deficient Ran mutant that remains GTP-bound in cell extracts (Bischoff et al., 1994). As shown in Figure 5A, GST-Nup98 pulled down endogenous Crm1 only in the presence of RanQ69L-GTP, and these results were confirmed using an in vivo pulldown assay from 293F cells expressing GST-fused Nup98 proteins (Figure 5B). As a control, we used a C-terminal fragment of Nup214 (1864-2090) that directly interacts with Crm1 (Fornerod et al., 1996). In contrast to Nup98, the C-terminal fragment of Nup214 precipitated endogenous Crm1, both in the presence and absence of RanQ69L-GTP (Figure 5A), as shown in a previous report (Kehlenbach et al., 1999). These results are consistent with the observed nuclear colocalization of Crm1 and Nup98 (Figure 4).
Figure 5.
Nup98 associates with Crm1 in a Ran-GTP–dependent manner. (A) Left, bacterially expressed GST, GST-Nup98 (full-length or deletion mutants), and GST-Nup214 FG-repeat region (1864-2090) were purified and analyzed by SDS-PAGE and Coomassie staining. Right, purified GST or GST-fusion proteins bound to glutathione beads were incubated with HeLa cell lysates (300 μg) in the presence or absence of 2 μM RanQ69L-GTP for 3 h at 4°C. The beads were washed, and the associated proteins were eluted and analyzed by immunoblotting using anti-Crm1. Thirty micrograms of whole cell lysate from HeLa cells was used for an input control. Asterisks indicate the full-length GST-fusion proteins. (B) GST-tagged Nup98 or deletion mutants were transiently expressed in HEK293F cells. The cell lysates (500 μg) were prepared and incubated with glutathione beads in the presence or absence of 2 μM RanQ69L-GTP for 3 h. The beads were washed, and associated proteins were eluted and analyzed by immunoblotting using anti-GST (left) or anti-Crm1 (right). Twenty-five micrograms of whole cell lysate from 293F cells was used for an input control. (C) Purified recombinant GST-Nup98 (5 μg) was incubated with the indicated mixtures of Crm1 (200 nM), RanQ69L (1 μM), NES peptide (Rev; 0.6–6 μM) for 2 h at 4°C, and precipitated using glutathione-Sepharose 4B beads. The bound proteins were analyzed by immunoblotting using anti-Crm1 or anti-Ran. Asterisk indicates GST-Nup98, which weakly cross-reacted with the anti-Crm1 antibody.
To determine whether Nup98 directly binds Crm1, we performed an in vitro pulldown assay using recombinant Crm1 and GST-Nup98 proteins. Recombinant Crm1 bound GST-Nup98 in a RanGTP-dependent manner, demonstrating that the interaction between Nup98 and Crm1 is direct (Figure 5C) and that Nup98 interacts with Crm1 in an NES-independent manner in vitro. In addition, when Nup98 deletion mutants were used in this assay, the N-terminal FG-repeat region of Nup98 (1-360 aa), but not the C-terminal non-FG-repeat region (451-880 aa), bound Crm1, albeit considerably less efficiently compared with full-length Nup98 (Figure S5). We also observed a weak interaction between Nup98 and Crm1 in the absence of RanGTP, when a high concentration of NES peptide (6 μM) was present. However, similar interaction was also observed when a mutant NES peptide (M10) was used in the assay (data not shown). Therefore, it is likely that this RanGTP-independent weak interaction is not biologically significant.
RanBP3, a Cofactor for Crm1-mediated Nuclear Export, Regulates the Nup98-Crm1 Interaction
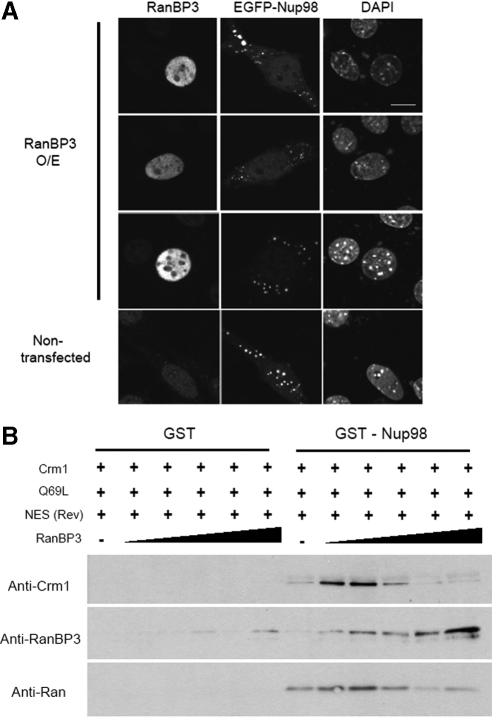
RanBP3 acts as a cofactor for Crm1-mediated nuclear export (Lindsay et al., 2001; Englmeier et al., 2001; Hendriksen et al., 2005), and RanBP3 has a biphasic effect on the Crm1-RanGTP-NES interaction. At low concentrations, RanBP3 stimulates the binding of Crm1 and RanGTP to the NES substrate, but at high concentrations, it prevents their interaction (Englmeier et al., 2001). Therefore, we first examined the effects of RanBP3 on Nup98 dot formation in vivo. When RanBP3 was overexpressed in NIH3T3 cells, the nuclear dots formed by EGFP-Nup98 disappeared (Figure 6A). Thus, overexpression of RanBP3 promotes the disassembly of Crm1-containing nuclear Nup98 dots in vivo, suggesting that at superphysiologic concentrations, RanBP3 may prevent the binding of Nup98 and Crm1.
Figure 6.
RanBP3 modulates the Nup98-Crm1 interaction in a biphasic manner. (A) Cells were transfected with EGFP-Nup98 alone or in combination with RanBP3 for 48 h. The cells were then fixed and examined by confocal microscopy. The nuclei were visualized by DAPI staining. Bar, 10 μm. (B) GST-Nup98 (5 μg) was incubated with Crm1 (200 nM), RanQ69L (1 μM), NES (6 μM), and different concentrations of RanBP3 (from left to right: 11, 33, 100, 300, and 900 nM) for 2 h at 4°C and was precipitated using glutathione-Sepharose 4B beads. The bound proteins were analyzed by immunoblotting.
To confirm this hypothesis, we examined the effects of RanBP3 on the RanGTP-dependent binding of Nup98 with Crm1 using the in vitro binding assay. At low concentrations (11–33 nM), RanBP3 enhanced the interaction of Crm1-RanGTP with Nup98 to form the Nup98-RanBP3-Crm1-RanGTP-NES-cargo complex (Figure 6B), but, at a higher concentration, RanBP3 strongly inhibited the binding of Nup98 with Crm1. These results indicate that at physiological concentrations, the RanGTP-dependent interaction between Nup98 and Crm1 is accelerated by RanBP3 and suggest that RanBP3 stimulates the formation of cargo-containing export complexes comprised of Nup98-RanBP3-Crm1-RanGTP-NES-cargo. Of note, at the high concentrations (300–900 nM), when Crm1 was barely detectable in the complex, RanBP3 still bound to Nup98. Collectively, these results suggest that Nup98 and RanBP3 cooperatively function in the assembly of NES cargo-containing export complexes.
Endogenous Nup98 Accumulates in the Nucleolus after Act-D Treatment in a Crm1-dependent Manner
Thus far, we have examined the effects and behavior of exogenously expressed EGFP-Nup98, but endogenous Nup98 does not appear to form nuclear dots to the same extent in NIH 3T3 cells. Therefore, we next wanted to confirm that endogenous Nup98 interacts with Crm1, and we examined the effects of LMB or Act-D treatment on the localization of endogenous Nup98 in NIH 3T3 cells (Figure 7A). Although there were no clear effects of LMB treatment on the localization of endogenous Nup98, Act-D treatment led to Nup98 nucleolar localization and the formation of crescent-like structures in the nucleolus. Importantly, when cells were treated with both LMB and Act-D, the nucleolar Nup98 disappeared.
The nucleolus consists of three major compartments: the granular compartment (GC), the dense fibrillar component (DFC), and the fibrillar center (FC). We attempted to determine the nucleolar region at which Nup98 localized after Act-D treatment, and we examined for the colocalization of Nup98 with nucleophosmin/B23 (NPM) as a marker of the GC, fibrillarin for the DFC, and upstream binding factor (UBF) for the FC. Using this approach, Nup98 colocalized with NPM, but not with fibrillarin or UBF, indicating that Nup98 accumulated at the crescent-like structures of the GC upon Act-D treatment (Figure 7B). Next, we examined the colocalization of Crm1 with Nup98 after drug treatment, and both Nup98 and Crm1 were redistributed and colocalized at the nucleolar crescent-like structures upon Act-D treatment (Figure 7C). In contrast, when cells were treated with both LMB and Act-D, the nucleolar accumulation of both Nup98 and Crm1 was abrogated, indicating that the Nup98-Crm1 complex was retained within the nucleolus in an LMB-sensitive manner. Collectively, these data suggest that an association between Crm1 and NES-cargo was necessary for the association of Nup98 with Crm1 within the nucleolus in living cells.
Microinjection of anti-Nup98 Antibodies Inhibits Crm1-mediated Nuclear Protein Export
Our data indicate that Nup98 directly interacts with Crm1 in a RanGTP-dependent manner, and both Nup98 and Crm1 accumulate within the nucleolus after Act-D treatment. The nucleolus is thought to be a route of Crm1-mediated nuclear export (Fornerod et al., 1997; Zolotukhin and Felber, 1999; Daelemans et al., 2005), and, therefore, we attempted to determine whether Nup98 affects the function of Crm1. We examined the effects of a microinjected anti-Nup98 mAb that recognizes the FG-repeat region of Nup98 on the Crm1-mediated nuclear export of proteins. Anti-Nup98 was injected into the nuclei of Hela cells together with GST-GFP-NES, which contained a Rev-derived leucine-rich NES. Fifteen minutes after microinjection, the cells were fixed, and the localization of GST-GFP-NES was examined. As shown in Figure 8A, when GST-GFP-NES was coinjected with control IgG or anti-Nup62, it was exported to the cytoplasm. In contrast, in the anti-Nup98 injected cells, the nuclear export of GST-GFP-NES was strongly suppressed. Because both anti-Nup98 and anti-Nup62 recognize the FG-repeat region of the corresponding nucleoporin (Fukuhara et al., 2005; Fukuhara et al., 2006), the inhibitory effect was specific for anti-Nup98. These results clearly show that Nup98 is directly involved in the Crm1-mediated nuclear export of proteins in vivo.
Figure 8.
Microinjection of anti-Nup98 inhibits NES-dependent nuclear export. (A) HeLa cells were microinjected with control IgG, anti-Nup62, or anti-Nup98 together with GST-GFP-NES and Alexa568-conjugated anti-sheep IgG (injection marker) into the nucleus. After 15 min of incubation at 37°C, the cells were fixed, washed, and mounted for microscopic observation. Bar, 20 μm. (B) HeLa cells were cytoplasmically microinjected with control IgG, anti-Nup62, or anti-Nup98. After 15 min of incubation at 37°C, the cells were fixed and stained with anti-RanBP1. Alexa488-conjugated anti-rat IgG or Alexa568-conjugated anti-rabbit IgG were used to detect rat monoclonal IgG or anti-RanBP1, respectively. Bar, 20 μm. (C) HeLa cells were microinjected with anti-Nup62 or anti-Nup98 in the cytoplasm, together with Alexa568-conjugated anti-sheep IgG (injection marker). After 15 min of incubation at 37°C, the cells were fixed and stained with Alexa488-conjugated anti-rat IgG. Bar, 20 μm.
Nup98 shuttles between the nucleus and the cytoplasm, and it localizes to both the nuclear and cytoplasmic faces of the NPC. Thus, we also examined the effect of cytoplasmic injection of the antibodies on the nuclear import of GST-GFP-NLS. Cytoplasmic injection of anti-Nup98 antibody showed an inhibitory effect on the import of the protein in only ∼30% of the injected cells (Figure S7). In contrast, the export of GST-GFP-NES was strongly inhibited by nuclear injection of this antibody in almost all the injected cells, which suggests that Nup98 primarily acts on the nuclear export of proteins.
Next, we examined the effects of the cytoplasmic injection of the anti-Nup98 antibody on the nuclear export of endogenous RanBP1 (Figure 8B). Interestingly, cytoplasmic injection of anti-Nup98 strongly inhibited the nuclear export of RanBP1, but its nuclear import was not affected. There were no effects of control IgG or anti-Nup62 injection on the localization of endogenous RanBP1. We next examined the behavior of injected antibody. As shown in Figure 8C, cytoplasmically injected anti-Nup62 localized to the nuclear rim or remained in the cytoplasm. In marked contrast, cytoplasmically injected anti-Nup98 efficiently accumulated in the nucleus, and this indicates that the Nup98-antibody complex formed in the cytoplasm, was transported into the nucleus, and inhibited the nuclear export of RanBP1. This data clearly show that Nup98 shuttles between the nucleus and cytoplasm and plays an essential role in Crm1-mediated nuclear export. Collectively, the dynamic behavior of Nup98 strongly suggests that Nup98 functions as a mobile cofactor for Crm1-mediated nuclear export analogous to the role of Npap60/Nup50 in importin α–mediated nuclear import (Lindsay et al., 2002).
DISCUSSION
Nup98 is a mobile nucleoporin that localizes to the nucleoplasm and to both the cytoplasmic and nuclear faces of the NPC (Griffis et al., 2003; Rabut et al., 2004). In this study, we demonstrated that Nup98 interacts with Crm1 through its N-terminal FG-repeat domain in a RanGTP-dependent manner and that Nup98 plays an important role in Crm1-mediated nuclear export. Indeed, our observation that anti-Nup98 efficiently inhibited NES-dependent protein export clearly shows the essential role for Nup98 in Crm1-mediated nuclear export in vivo. Importantly, cytoplasmically injected anti-Nup98 comigrated with Nup98 into the nucleus and efficiently inhibited the nuclear export, but not the nuclear import, of endogenous RanBP1 (Figure 8), although cytoplasmic injection of anti-Nup98 antibody partially affected the importin α/β–mediated import of coinjected substrates (Figure S7). These results indicate that Nup98 plays an essential role in Crm1-mediated protein export rather than protein import and that Nup98 shuttles between the nucleus and cytoplasm. Additionally, these data suggest that the recycled Nup98 is reused for subsequent nuclear protein export events.
RanBP3, a cofactor for Crm1-mediated nuclear export, directly binds to Crm1 and facilitates the formation of Crm1-RanGTP-NES-cargo complexes at physiological concentrations, but, at higher concentrations, it inhibits complex formation (Englmeier et al., 2001). Thus, under physiological conditions, RanBP3 functions as a positive regulator of Crm1-mediated nuclear protein export. In contrast, we found that Nup98 efficiently binds Crm1, leading to formation of a Nup98-Crm1-RanGTP-NES-cargo complex, but Nup98 also forms a Nup98-Crm1-RanGTP complex in a RanGTP-dependent manner. These data suggest that Nup98 may facilitate the nuclear export of cargo-free Nup98-Crm1-RanGTP complexes, but, at low concentrations RanBP3 facilitates the formation of the Nup98-RanBP3-Crm1-RanGTP-NES-cargo complex. Therefore, we hypothesize that, under normal conditions, RanBP3 promotes Nup98-RanBP3-Crm1-RanGTP-NES-cargo complex formation to prevent the nuclear export of cargo-free Nup98-Crm1-RanGTP complexes. Furthermore, our in vitro results suggest that RanBP3 can directly associate with Nup98 to prevent its interaction with Crm1 (Figure 6). Further analysis will be required to understand the in vivo relevance of RanGTP-independent binding of RanBP3 to Nup98 to the protein export.
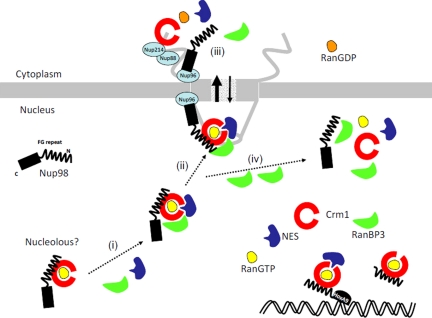
The C-terminal region of Nup98 mediates its association with Nup96 and Nup88 (Hodel et al., 2002; Griffis et al., 2003), suggesting that Nup96 and Nup88 may facilitate the nuclear translocation of the Nup98 complex through the NPC. The dependence of this interaction upon the C-terminus of Nup98 is compatible with the role of the N-terminal FG region in mediating Crm1 interactions and cargo export. Thus, we propose the following model for the function of Nup98 (Figure 9). In the nucleus, Nup98 binds Crm1 in a RanGTP-dependent manner, and RanBP3 promotes formation of the Nup98-RanBP3-Crm1-RanGTP-NES-cargo, leading to the efficient recruitment of NES-cargo to an export complex. The interaction of the C-terminal region of Nup98 with Nup96 and Nup88 promotes engagement of the export complex with the NPC and supports its efficient translocation through the NPC. After translocation, GTP hydrolysis triggers the dissociation of Nup98 from Crm1 and the NES-cargo. Thus, Nup98 functions as a cofactor in the nuclear protein export complex to accelerate complex formation in the nucleus and translocation through the NPC. Because it is known that Nup98 localizes either at the NPC as its component or within the nucleus as a mobile nucleoporin, further analysis will be required to determine whether Nup98 localizing at NPC or soluble nucleoplasmic Nup98 primarily participates in the export of proteins in vivo.
Figure 9.
A model for the role of Nup98 in Crm1-mediated nuclear protein export. Nup98 forms a complex with Crm1 in a RanGTP-dependent manner in the nucleus or nucleolus. RanBP3 facilitates the formation of the NES-cargo–containing Nup98-RanBP3-Crm1-RanGTP complex (i). This complex is then targeted to the NPC (ii) and transported into the cytoplasm. The C-terminal non-FG-repeat domain of Nup98 is involved in the targeting of the export complex to the NPC facilitating the Crm1-dependent translocation of NES-containing proteins through the NPC. After translocation, the export complex dissociates after the conversion of RanGTP to RanGDP (iii). When an excess amount of RanBP3 is present, the NES-cargo containing complex dissociates (iv). Both the FG-repeat fragment of Nup98 and Nup98-HoxA9 fusion interact with Crm1 but are not targeted to the NPC, and both inhibit the function of Crm1.
We identified an association of endogenous Nup98-Crm1 within the GC of the nucleolus after cell treatment with low-dose Act-D (Figure 7), which specifically inhibits RNA polymerase I reducing rRNA synthesis (Perry and Kelley, 1970). Furthermore, the nucleolar Nup98-Crm1 association disappeared upon LMB treatment (Figure 7), indicating that the interaction between NES-cargo and Crm1 is necessary for the association of Nup98 with Crm1 in the nucleolus. The GC is the nucleolar region associated with rRNA processing and ribosomal subunit assembly, suggesting that the Nup98-Crm1 complex may also play a role in the nuclear export of ribosomal subunits and/or the nucleolus may function as a platform for Nup98-Crm1-RanGTP-NES-cargo assembly (Figure 9). These possibilities should be examined in future studies.
Our results suggest that altered regulation of Crm1 may play an important role in the pathogenesis of leukemia. The chromosomal region containing the Nup98 gene frequently translocates during leukemogenesis, generating a variety of chimeric Nup98-fusion genes (Borrow et al., 1996; Nakamura et al., 1996; Cronshaw and Matunis, 2004). We showed that expression of the oncogenic Nup98-HoxA9 fusion protein strongly inhibited the Crm1-dependent nuclear export of RanBP1 (Figure 3), but it is unclear how the perturbed function of Crm1 induced by Nup98-fusion protein expression is involved in leukemogenesis. Recent studies showed that expression of Nup98-HoxA9 leads to the up-regulation of a number of genes that promote the self-renewal of hematopoietic stem cells, and it also down-regulates several genes involved in cellular differentiation (Ghannam et al., 2004; Chung et al., 2006; Takeda et al., 2006). However, multiple cooperative mechanisms are thought to be required for the development of leukemia. Therefore, it is reasonable to speculate that for cellular differentiation to occur, a subset of nuclear factors required for self-renewal must first be excluded from the nucleus. The suppression of Crm1-mediated nuclear protein export by Nup98-HoxA9 fusions may impair the removal of critical nuclear proteins involved in cell proliferation, thereby inhibiting the normal process of differentiation and promoting leukemia. Thus, the Nup98-HoxA9 fusion may have multiple functions, and the FG-repeat region of Nup98 and its fusion partner may cooperatively induce oncogenesis through the dysregulation of two physiological processes, transcriptional control, and nucleocytoplasmic protein transport (Figure 9).
In summary, our results show that Nup98 is a novel cofactor for Crm1-dependent protein export and that Nup98 facilitates the export of cargo-loaded Crm1 in conjunction with RanBP3. Npap60/Nup50 is a cofactor for importin α–mediated nuclear import (Lindsay et al., 2002), and our finding that Nup98 is an important cofactor in nuclear transport suggests diverse roles for mobile Nups in nucleocytoplasmic transport.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Drs. Jun Katahira (Osaka University) and Jan Ellenberg (EMBL) for providing us with pCAG-wtag and pEGFP-Nup214 plasmids, respectively. This work was supported, in part, by the Ministry of Education, Culture, Sports, Science, and Technology of Japan; the Japan Society for the Promotion of Science; and the Takeda Science Foundation.
Abbreviations used:
- Act-D
actinomycin D
- FG
phenylalanine-glycine
- LMB
leptomycin B
- NES
nuclear export signal
- NPC
nuclear pore complex
- Nup
nucleoporin
- RanBP
Ran-binding protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-12-1041) on April 7, 2010.
REFERENCES
- Bastos R., Ribas de Pouplana L., Enarson M., Bodoor K., Burke B. Nup84, a novel nucleoporin that is associated with CAN/Nup214 on the cytoplasmic face of the nuclear pore complex. J. Cell Biol. 1997;137:989–1000. doi: 10.1083/jcb.137.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh N., et al. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J. Cell Biol. 2001;154:1147–1160. doi: 10.1083/jcb.200101081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Efraim I., Gerace L. Gradient of increasing affinity of importin beta for nucleoporins along the pathway of nuclear import. J. Cell Biol. 2001;152:411–417. doi: 10.1083/jcb.152.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff F. R., Klebe C., Kretschmer J., Wittinghofer A., Ponstingl H. RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc. Natl. Acad. Sci. USA. 1994;91:2587–2591. doi: 10.1073/pnas.91.7.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer J., Bonten-Surtel J., Grosveld G. Overexpression of the nucleoporin CAN/NUP214 induces growth arrest, nucleocytoplasmic transport defects, and apoptosis. Mol. Cell. Biol. 1998;18:1236–1247. doi: 10.1128/mcb.18.3.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrow J., et al. The t(7;11)(p15;p15) translocation in acute myeloid leukaemia fuses the genes for nucleoporin NUP98 and class I homeoprotein HOXA9. Nat. Genet. 1996;12:159–167. doi: 10.1038/ng0296-159. [DOI] [PubMed] [Google Scholar]
- Brown C. R., Silver P. A. Transcriptional regulation at the nuclear pore complex. Curr. Opin. Genet. Dev. 2007;17:100–106. doi: 10.1016/j.gde.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Chung K. Y., Morrone G., Schuringa J. J., Plasilova M., Shieh J. H., Zhang Y., Zhou P., Moore M. A. Enforced expression of NUP98-HOXA9 in human CD34(+) cells enhances stem cell proliferation. Cancer Res. 2006;66:11781–11791. doi: 10.1158/0008-5472.CAN-06-0706. [DOI] [PubMed] [Google Scholar]
- Clarkson W. D., Kent H. M., Stewart M. Separate binding sites on nuclear transport factor 2 (NTF2) for GDP-Ran and the phenylalanine-rich repeat regions of nucleoporins p62 and Nsp1p. J. Mol. Biol. 1996;263:517–524. doi: 10.1006/jmbi.1996.0594. [DOI] [PubMed] [Google Scholar]
- Cronshaw J. M., Krutchinsky A. N., Zhang W., Chait B. T., Matunis M. J. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronshaw J. M., Matunis M. J. The nuclear pore complex: disease associations and functional correlations. Trends Endocrinol. Metab. 2004;15:34–39. doi: 10.1016/j.tem.2003.11.005. [DOI] [PubMed] [Google Scholar]
- Daelemans D., Costes S. V., Lockett S., Pavlakis G. N. Kinetic and molecular analysis of nuclear export factor CRM1 association with its cargo in vivo. Mol. Cell. Biol. 2005;25:728–739. doi: 10.1128/MCB.25.2.728-739.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasso M. Running on Ran: nuclear transport and the mitotic spindle. Cell. 2001;104:321–324. doi: 10.1016/s0092-8674(01)00218-5. [DOI] [PubMed] [Google Scholar]
- Dasso M. Ran at kinetochores. Biochem. Soc. Trans. 2006;34:711–715. doi: 10.1042/BST0340711. [DOI] [PubMed] [Google Scholar]
- Denning D. P., Patel S. S., Uversky V., Fink A. L., Rexach M. Disorder in the nuclear pore complex: the FG repeat regions of nucleoporins are natively unfolded. Proc. Natl. Acad. Sci. USA. 2003;100:2450–2455. doi: 10.1073/pnas.0437902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englmeier L., Fornerod M., Bischoff F. R., Petosa C., Mattaj I. W., Kutay U. RanBP3 influences interactions between CRM1 and its nuclear protein export substrates. EMBO Rep. 2001;2:926–932. doi: 10.1093/embo-reports/kve200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floer M., Blobel G., Rexach M. Disassembly of RanGTP-karyopherin beta complex, an intermediate in nuclear protein import. J. Biol. Chem. 1997;272:19538–19546. doi: 10.1074/jbc.272.31.19538. [DOI] [PubMed] [Google Scholar]
- Fontoura B. M., Blobel G., Yaseen N. R. The nucleoporin Nup98 is a site for GDP/GTP exchange on ran and termination of karyopherin beta 2-mediated nuclear import. J. Biol. Chem. 2000;275:31289–31296. doi: 10.1074/jbc.M004651200. [DOI] [PubMed] [Google Scholar]
- Fornerod M., Boer J., van Baal S., Morreau H., Grosveld G. Interaction of cellular proteins with the leukemia specific fusion proteins DEK-CAN and SET-CAN and their normal counterpart, the nucleoporin CAN. Oncogene. 1996;13:1801–1808. [PubMed] [Google Scholar]
- Fornerod M., Ohno M., Yoshida M., Mattaj I. W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Frey S., Gorlich D. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell. 2007;130:512–523. doi: 10.1016/j.cell.2007.06.024. [DOI] [PubMed] [Google Scholar]
- Frosst P., Guan T., Subauste C., Hahn K., Gerace L. Tpr is localized within the nuclear basket of the pore complex and has a role in nuclear protein export. J. Cell Biol. 2002;156:617–630. doi: 10.1083/jcb.200106046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M., Asano S., Nakamura T., Adachi M., Yoshida M., Yanagida M., Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- Fukuhara T., Ozaki T., Shikata K., Katahira J., Yoneda Y., Ogino K., Tachibana T. Specific monoclonal antibody against the nuclear pore complex protein, nup98. Hybridoma. 2005;24:244–247. doi: 10.1089/hyb.2005.24.244. [DOI] [PubMed] [Google Scholar]
- Fukuhara T., Sakaguchi N., Katahira J., Yoneda Y., Ogino K., Tachibana T. Functional analysis of nuclear pore complex protein Nup62/p62 using monoclonal antibodies. Hybridoma. 2006;25:51–59. doi: 10.1089/hyb.2006.25.51. [DOI] [PubMed] [Google Scholar]
- Ghannam G., Takeda A., Camarata T., Moore M. A., Viale A., Yaseen N. R. The oncogene Nup98-HOXA9 induces gene transcription in myeloid cells. J. Biol. Chem. 2004;279:866–875. doi: 10.1074/jbc.M307280200. [DOI] [PubMed] [Google Scholar]
- Gorlich D., Mattaj I. W. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- Griffis E. R., Altan N., Lippincott-Schwartz J., Powers M. A. Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol. Biol. Cell. 2002;13:1282–1297. doi: 10.1091/mbc.01-11-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis E. R., Xu S., Powers M. A. Nup98 localizes to both nuclear and cytoplasmic sides of the nuclear pore and binds to two distinct nucleoporin subcomplexes. Mol. Biol. Cell. 2003;14:600–610. doi: 10.1091/mbc.E02-09-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel A., Orjalo A. V., Vincent T., Lachish-Zalait A., Vasu S., Shah S., Zimmerman E., Elbaum M., Forbes D. J. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol. Cell. 2003;11:853–864. doi: 10.1016/s1097-2765(03)00116-3. [DOI] [PubMed] [Google Scholar]
- Hendriksen J., Fagotto F., van der Velde H., van Schie M., Noordermeer J., vFornerod M. RanBP3 enhances nuclear export of active β-catenin independently of CRM1. J. Cell Biol. 2005;171:785–797. doi: 10.1083/jcb.200502141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks G. R., Raikhel N. V. Protein import into the nucleus: an integrated view. Annu. Rev. Cell Dev. Biol. 1995;11:155–188. doi: 10.1146/annurev.cb.11.110195.001103. [DOI] [PubMed] [Google Scholar]
- Hodel A. E., Hodel M. R., Griffis E. R., Hennig K. A., Ratner G. A., Xu S., Powers M. A. The three-dimensional structure of the autoproteolytic, nuclear pore-targeting domain of the human nucleoporin Nup98. Mol. Cell. 2002;10:347–358. doi: 10.1016/s1097-2765(02)00589-0. [DOI] [PubMed] [Google Scholar]
- Hu T., Guan T., Gerace L. Molecular and functional characterization of the p62 complex, an assembly of nuclear pore complex glycoproteins. J. Cell Biol. 1996;134:589–601. doi: 10.1083/jcb.134.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto M., Mori C., Kojidani T., Bunai F., Hori T., Fukagawa T., Hiraoka Y., Haraguchi T. Two distinct repeat sequences of Nup98 nucleoporins characterize dual nuclei in the binucleated ciliate tetrahymena. Curr. Biol. 2009;19:843–847. doi: 10.1016/j.cub.2009.03.055. [DOI] [PubMed] [Google Scholar]
- Kalab P., Weis K., Heald R. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 2002;295:2452–2456. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- Kasper L. H., Brindle P. K., Schnabel C. A., Pritchard C. E., Cleary M. L., van Deursen J. M. CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol. Cell. Biol. 1999;19:764–776. doi: 10.1128/mcb.19.1.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehlenbach R. H., Dickmanns A., Kehlenbach A., Guan T., Gerace L. A role for RanBP1 in the release of CRM1 from the nuclear pore complex in a terminal step of nuclear export. J. Cell Biol. 1999;145:645–657. doi: 10.1083/jcb.145.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer D., Wozniak R. W., Blobel G., Radu A. The human CAN protein, a putative oncogene product associated with myeloid leukemogenesis, is a nuclear pore complex protein that faces the cytoplasm. Proc. Natl. Acad. Sci. USA. 1994;91:1519–1523. doi: 10.1073/pnas.91.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo N., Matsumori N., Taoka H., Fujiwara D., Schreiner E. P., Wolff B., Yoshida M., Horinouchi S. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. USA. 1999;96:9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay M. E., Holaska J. M., Welch K., Paschal B. M., Macara I. G. Ran-binding protein 3 is a cofactor for Crm1-mediated nuclear protein export. J. Cell Biol. 2001;153:1391–1402. doi: 10.1083/jcb.153.7.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay M. E., Plafker K., Smith A. E., Clurman B. E., Macara I. G. Npap60/Nup50 is a tri-stable switch that stimulates importin-alpha:beta-mediated nuclear protein import. Cell. 2002;110:349–360. doi: 10.1016/s0092-8674(02)00836-x. [DOI] [PubMed] [Google Scholar]
- Loiodice I., Alves A., Rabut G., Van Overbeek M., Ellenberg J., Sibarita J. B., Doye V. The entire Nup107-160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol. Biol. Cell. 2004;15:3333–3344. doi: 10.1091/mbc.E03-12-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., et al. Fusion of the nucleoporin gene NUP98 to HOXA9 by the chromosome translocation t(7;11)(p15;p15) in human myeloid leukaemia. Nat. Genet. 1996;12:154–158. doi: 10.1038/ng0296-154. [DOI] [PubMed] [Google Scholar]
- Nigg E. A. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- Ossareh-Nazari B., Bachelerie F., Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- Perry R. P., Kelley D. E. Inhibition of RNA synthesis by actinomycin D: characteristic dose-response of different RNA species. J. Cell. Physiol. 1970;76:127–139. doi: 10.1002/jcp.1040760202. [DOI] [PubMed] [Google Scholar]
- Powers M. A., Forbes D. J., Dahlberg J. E., Lund E. The vertebrate GLFG nucleoporin, Nup98, is an essential component of multiple RNA export pathways. J. Cell Biol. 1997;136:241–250. doi: 10.1083/jcb.136.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers M. A., Macaulay C., Masiarz F. R., Forbes D. J. Reconstituted nuclei depleted of a vertebrate GLFG nuclear pore protein, p97, import but are defective in nuclear growth and replication. J. Cell Biol. 1995;128:721–736. doi: 10.1083/jcb.128.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard C. E., Fornerod M., Kasper L. H., van Deursen J. M. RAE1 is a shuttling mRNA export factor that binds to a GLEBS-like NUP98 motif at the nuclear pore complex through multiple domains. J. Cell Biol. 1999;145:237–254. doi: 10.1083/jcb.145.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyhtila B., Rexach M. A gradient of affinity for the karyopherin Kap95p along the yeast nuclear pore complex. J. Biol. Chem. 2003;278:42699–42709. doi: 10.1074/jbc.M307135200. [DOI] [PubMed] [Google Scholar]
- Rabut G., Doye V., Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat. Cell Biol. 2004;6:1114–1121. doi: 10.1038/ncb1184. [DOI] [PubMed] [Google Scholar]
- Radu A., Moore M. S., Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- Rasala B. A., Orjalo A. V., Shen Z., Briggs S., Forbes D. J. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc. Natl. Acad. Sci. USA. 2006;103:17801–17806. doi: 10.1073/pnas.0608484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichelt R., Holzenburg A., Buhle E. L., Jr, Jarnik M., Engel A., Aebi U. Correlation between structure and mass distribution of the nuclear pore complex and of distinct pore complex components. J. Cell Biol. 1990;110:883–894. doi: 10.1083/jcb.110.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexach M., Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- Richards S. A., Lounsbury K. M., Carey K. L., Macara I. G. A nuclear export signal is essential for the cytosolic localization of the Ran binding protein, RanBP1. J. Cell Biol. 1996;134:1157–1168. doi: 10.1083/jcb.134.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout M. P., Aitchison J. D., Suprapto A., Hjertaas K., Zhao Y., Chait B. T. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukegawa J., Blobel G. A nuclear pore complex protein that contains zinc finger motifs, binds DNA, and faces the nucleoplasm. Cell. 1993;72:29–38. doi: 10.1016/0092-8674(93)90047-t. [DOI] [PubMed] [Google Scholar]
- Suntharalingam M., Wente S. R. Peering through the pore: nuclear pore complex structure, assembly, and function. Dev. Cell. 2003;4:775–789. doi: 10.1016/s1534-5807(03)00162-x. [DOI] [PubMed] [Google Scholar]
- Taddei A. Active genes at the nuclear pore complex. Curr. Opin. Cell Biol. 2007;19:305–310. doi: 10.1016/j.ceb.2007.04.012. [DOI] [PubMed] [Google Scholar]
- Takeda A., Goolsby C., Yaseen N. R. NUP98-HOXA9 induces long-term proliferation and blocks differentiation of primary human CD34+ hematopoietic cells. Cancer Res. 2006;66:6628–6637. doi: 10.1158/0008-5472.CAN-06-0458. [DOI] [PubMed] [Google Scholar]
- Takeda E., Hieda M., Katahira J., Yoneda Y. Phosphorylation of RanGAP1 stabilizes its interaction with Ran and RanBP1. Cell Struct. Funct. 2005;30:69–80. doi: 10.1247/csf.30.69. [DOI] [PubMed] [Google Scholar]
- Tran E. J., Wente S. R. Dynamic nuclear pore complexes: life on the edge. Cell. 2006;125:1041–1053. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]
- Vasu S., Shah S., Orjalo A., Park M., Fischer W. H., Forbes D. J. Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J. Cell Biol. 2001;155:339–354. doi: 10.1083/jcb.200108007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente S. R., Rout M. P., Blobel G. A new family of yeast nuclear pore complex proteins. J. Cell Biol. 1992;119:705–723. doi: 10.1083/jcb.119.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Matunis M. J., Kraemer D., Blobel G., Coutavas E. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J. Biol. Chem. 1995;270:14209–14213. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]
- Wu X., Kasper L. H., Mantcheva R. T., Mantchev G. T., Springett M. J., van Deursen J. M. Disruption of the FG nucleoporin NUP98 causes selective changes in nuclear pore complex stoichiometry and function. Proc. Natl. Acad. Sci. USA. 2001;98:3191–3196. doi: 10.1073/pnas.051631598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama N., et al. A giant nucleopore protein that binds Ran/TC4. Nature. 1995;376:184–188. doi: 10.1038/376184a0. [DOI] [PubMed] [Google Scholar]
- Zolotukhin A. S., Felber B. K. Mutations in the nuclear export signal of human ran-binding protein RanBP1 block the Rev-mediated posttranscriptional regulation of human immunodeficiency virus type 1. J. Biol. Chem. 1997;272:11356–11360. doi: 10.1074/jbc.272.17.11356. [DOI] [PubMed] [Google Scholar]
- Zolotukhin A. S., Felber B. K. Nucleoporins nup98 and nup214 participate in nuclear export of human immunodeficiency virus type 1 Rev. J. Virol. 1999;73:120–127. doi: 10.1128/jvi.73.1.120-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.