Abstract
Purpose:
Using Reverse Phase Protein Array (RPPA) we measured protein expression associated with response to primary chemotherapy in patients with advanced-stage high-grade serous ovarian cancer.
Experimental Design:
Tumor samples were obtained from forty-five patients with advanced high-grade serous cancers from the Gynecology Tumor Bank at the British Columbia Cancer Agency. Treatment consisted of platinum-based chemotherapy following debulking surgery. Protein lysates were prepared from fresh frozen tumor samples and 80 validated proteins from signaling pathways implicated in ovarian carcinogenesis were measured by RPPA. Normalization of Ca-125 by the 3rd cycle of chemotherapy was chosen as the primary outcome measure of chemotherapy response. Logistic regression was used for multivariate analysis to identify protein predictors of Ca-125 normalization, and Cox regression to test for the association between protein expression and PFS. A significance level of p ≤ 0.05 was used.
Results:
The mean age at diagnosis was 56.8 years. EGFR, YKL-40 and several TGFβ pathway proteins (c-Jun N-terminal kinase JNK, JNK phosphorylated at residues 183 and 185, PAI-1, Smad3, TAZ) showed significant associations with Ca-125 normalization on univariate testing. On multivariate analysis, EGFR (p < 0.02), JNK (p < 0.01), and Smad3 (p < 0.04) were significantly associated with normalization of Ca-125. Contingency table analysis of pathway-classified proteins revealed that the selection of TGFβ pathway proteins was unlikely due to false discovery (p < 0.007, Bonferroni-adjusted).
Conclusion:
TGFβ pathway signaling likely plays an important role as a marker or mediator of chemoresistance in advanced serous ovarian cancer. On this basis, future studies to develop and validate a useful predictor of treatment failure are warranted.
Keywords: ovarian cancer, functional proteomics, chemotherapy response, reverse phase protein array, Ca-125
Introduction
High-grade serous ovarian cancer is the most common pathologic diagnosis in patients with advanced ovarian cancer. In several randomized trials most patients presenting with advanced disease have high-grade serous cancers.(1, 2) Within this group, the prognosis for patients treated with carboplatin-paclitaxel chemotherapy varies remarkably from a few months to many years, though median times to progression are in the order of 18 months.(3)
It is recognized that performance status, age, grade, residuum, histology, and stage influence outcomes to primary chemotherapy in patients with advanced ovarian cancer. (4, 5) In addition, Ca-125 levels pretreatment and at nadir, normalization of Ca-125 by the 3rd cycle of chemotherapy, and Ca-125 half-life are also significant predictors of progression-free survival and overall survival. (6-9) Despite this knowledge, these factors are only used in a minority of patients to guide primary treatment selection due to limited sensitivity and specificity. Combination platinum/taxane chemotherapy is offered and completed in most patients knowing that up to 20% or more of patients will not have a demonstrable response to therapy and the majority of these patients are destined to progress and succumb to their disease despite therapy.(10)
With the development of modern high-throughput molecular technologies, efforts are being directed to the discovery of biomarkers of treatment response and survival. Markers that function as effective surrogates of these important outcomes have been challenging to identify in ovarian cancer. Ongoing studies in ovarian cancer suggest that there are distinct molecular characteristics that are specific to histological subtypes such as advanced high-grade serous cancers. (11) Molecular classifications may help distinguish various subgroups within high-grade serous cancers that have different outcomes and responses to therapy.
Reverse Phase Protein Array (RPPA) is a new high-throughput technology whereby a multiplexed assessment of protein expression on more than 100 differing proteins can be carried out simultaneously on up to 1000 tumors samples.(12) RPPA can be used as a tool to explore cellular signaling pathway activation that relates to the biology of cancer progression and treatment (13) including the assessment of proteins involved in cellular functions such as growth, proliferation, and apoptosis. We have applied this technology to the study of a well-characterized cohort of patients with advanced high-grade serous cancers from the same institution. Our objective was to determine if we could identify patterns of tumor protein expression that were associated with normalization of Ca-125 by the 3rd cycle of chemotherapy (Ca-125 normalization). This end-point was chosen in order to characterize proteins able to predict response to chemotherapy. Currently there are no biomarkers in clinical use that predict response to first–line chemotherapy. Such a biomarker(s) could facilitate the introduction of investigational drugs much earlier in the disease course particularly in those patients who might be identified as poor responders to conventional treatment, as well as identify potential therapeutic targets for drug development.
Methods
Between November 2000 and May 2006, the Gynecology Tumour Group at Vancouver General Hospital and the British Columbia Cancer Agency collected 221 ovarian cancer tumor samples. The collection and study is of these tissues was approved by the University of British Columbia Ethics Review Board. Tumor samples were collected in the operating room at the time of primary surgery and snap-frozen within 30 minutes after collection in liquid nitrogen. Samples were processed in accordance with tissue bank guidelines and stored at −120°C. All patient samples in the ovarian tumor tissue bank have been subjected to pathology review (BG) to verify the histology and site of origin. Only patients with advanced (Stage 3 or 4) high-grade serous tumors with adequate clinical information were selected for this study. In order to be included in the analysis, patients must have had Ca-125 measurements taken postoperatively within 2 weeks of starting chemotherapy and pretreatment Ca-125 levels must have been at least twice normal (> 70 u/mL). An abnormal Ca-125 was defined as a level greater than 35u/mL. In the majority of cases, Ca-125 measures were available prior to each treatment cycle and normalization prior to the 3rd cycle of chemotherapy (Ca-125 normalization) was used as the primary outcome measure. Though we examined other measures of Ca-125 prognostication such as Ca-125 half-life, Ca-125 normalization (before the third cycle of chemotherapy) was a stronger determinant of outcome than Ca-125 half-life and is simpler to measure.
Clinical information on study patients was collected both prospectively and retrospectively as part of a formal process to collect and update clinical data on a regular basis. Patients were classified as having macroscopic residuum or no macroscopic residuum as this distinction most clearly separates patients into prognostic risk groups. For the duration of this study combination chemotherapy using carboplatin and paclitaxel was the standard regimen.
Reverse Phase Protein Array
Tissues were processed by RPPA in our laboratory as previously described.(12) Briefly, 5 to 25 mg of tumor tissue was cut from the frozen banked tumor specimen and immediately homogenized in cold lysis buffer (1% Triton X-100, 50mm HEPES, pH 7.4, 150mM NaCl, 1.5mM MgCl2, 1mM EGTA, 100mM NaF, 10mM Na Pyrophosphate, 1mM Na3VO4, 10% glycerol). After centrifugation, the supernatant was removed and total protein concentrations were corrected to a final concentration of 1 ug/uL and boiled in denaturing buffer with SDS (final concentration of 1% SDS),
Samples were serially diluted and spotted on nitrocellulose-coated glass slides (FAST Slides, Schleicher & Schuell BioScience, Inc. USA, Keene, NH) using an Aushon 2470 robotic printer (Aushon Biosystems, Billerica, MA). Printing and staining quality control was performed by reprinting 5 slides. These slides were printed more than eight months after the original 120-slide print run and stained for AKT, cyclin E1 (CCNE1), JNKp183_185, PTEN and GSK3. The Pearson correlation coefficients for these 5 slides were 0.90, 0.95, 0.95, 0.97, and 0.97 respectively. Negative and positive controls were printed on each slide only as a means of assessing printing and staining quality across each slide. Slides are examined microscopically and those with obvious defects in print spot quality or staining consistency are not used. Each arrayed slide was probed with a specific primary antibody followed by a secondary antibody (anti-mouse or anti-rabbit) as a means for signal amplification (Dako, Carpinteria, CA - catalyzed system). Each antibody is validated as previously described (12, 13) and a list of the 80 validated antibodies and others used for staining including the applied antibody dilutions and the source of the antibody is included as Supplement 1. The stained slides were scanned using an HP flatbed scanner then analyzed and quantified using Microvigene software (VigeneTech Inc., Carlisle, MA). Serial dilution-signal intensity curves were generated for each sample. Using the logistic fit model ln(y)=a+(b-a)/(1+exp(c*(d-ln(x)))) logarithmic values were determined for each spot in the serial dilution after background correction. A representative value of the fit of each sample dilution curve relative to a fitted curve generated from all spots on the slide was used to quantify the relative protein expression (Supercurve v.997 Department of Bioinformatics and Computational Biology, M. D. Anderson Cancer Center) of each specific protein or phosphorylated protein counterpart evaluated in each sample.(14) The estimated protein concentrations are normalized by a median polish method and corrected for protein loading using the average expression levels of all measured proteins in each sample.
Statistical Methods
To obtain a global visualization and assessment of tumor protein expression profiles, supervised cluster analysis was performed using Treeview software (University of Glasgow, Scotland) and X- cluster software was used to generate heat maps and cluster groups. Data was analyzed using SPSS software (Version 17, Chicago Illinois). Proteins were classified according to their relevant signaling pathway using the Gene Set Enrichment Analysis Database (GSEA v2.05, www.broad.mit.edu/gsea). The curated Molecular Signatures Database (homo sapiens only) was used to classify each protein by pathway status (See Supplement 2). The Fisher's exact test was used to assess the association between categorical variables, test for significance between cluster analyses of patient groups by Ca-125 normalization status and evaluate false discovery rates by assessing differences in the signaling pathway classification of the identified protein predictors. Means were compared using the Student's t-test, while logistic regression was used for assessing the effect of individual protein expression on Ca-125 normalization. The log-rank test was used to compare survival curves using the Kaplan-Meier method. Significant variables on univariate analysis were subsequently examined as predictors of PFS as continuous variables using Cox regression. Median protein expression levels were used as cut-points for the regression models. A level of significance of 0.05 was used throughout.
Results
Forty-five cases fit the inclusion criteria from the 221 tumor samples collected for RPPA. The majority of patients (87) were excluded based on histological findings that included, non-serous histology, non-ovarian primaries, and low-grade or borderline serous tumor. Another 19 patients had early-stage (I or II) disease. Patients with unclassified tumors and those with mixed tumors were not included. Thirty-three patients were excluded who did not complete or receive chemotherapy, or were treated with neoadjuvant chemotherapy. Patients were also excluded if the pretreatment Ca-125 less than twice normal (10 patients), or they were missing Ca-125 measures at necessary time points (17 patients).
The median age of the population was 56.8 years with a range of 34-84 years. The distribution of stage in the population was as follows: 6 patients with stage 3B disease, 31 with stage 3C, and 8 stage 4 patients. There were 41 patients with macroscopic residuum and 4 patients with no macroscopic residuum. All but 4 patients received paclitaxel/carboplatin chemotherapy following primary debulking surgery. Three patients received carboplatin alone while one patient was treated with carboplatin and gemcitabine. Pretreatment Ca-125 values ranged from 88 – 6300 u/mL. Thirty-three percent of patients had a pretreatment Ca-125 level of less than 200 u/mL, while 51% of patients had values between 200 and 1000 u/mL. There were 25 patients who normalized their Ca-125 before the 3rd cycle of chemotherapy while 20 patients retained abnormal levels.
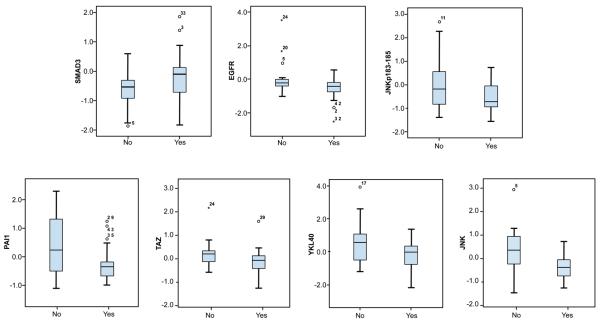
Table 1 shows the proteins associated with Ca-125 normalization on both uni- and multivariate analyses. Expression of these proteins was not associated with stage, residuum, or age with the exception of YKL-40 that correlated with stage (p < 0.05, data not shown). None of the clinical variables were significantly associated with normalization of Ca125, with stage showing a trend (p = 0.06, data not shown). Those proteins that were significant determinants of outcome on univariate analyses and known prognostic clinical factors were then tested by multivariate analyses. Box plots showing the distributions of each protein in accordance with Ca-125 normalization are shown in Figure 1. Proteins showing a significant association with Ca-125 normalization on univariate testing are EGFR, JNK (C-Jun N-terminal kinase), JNKp183_185 (JNK phosphorylated at residues 183 and185), PAI-1 (plasminogen activator inhibitor 1), Smad3, TAZ, and YKL-40, with higher levels associated with failure of Ca-125 normalization, except for Smad3. In contrast, higher Smad3 expression was associated with normalization of Ca-125 levels. On multivariate logistic regression analysis (See Table 1) only EGFR, JNK, and Smad3 were independent determinants of Ca-125 normalization. Table 2 shows the sensitivity and specificity for predicting Ca-125 normalization using EGFR, JNK and Smad3 expression. The expression of these three proteins in the combined regression model had a sensitivity of 80%, with a specificity of 75%, and an overall accuracy of 78% for predicting Ca-125 normalization.
Table 1. Univariate and Multivariate Analyses of Clinical Variables and Proteins on Normalization of Ca-125 and Progression-Free Survival.
| Ca-125 normalization by cycle #31 |
Progression-free survival | ||||
|---|---|---|---|---|---|
| Variables |
|
||||
| Univariate | Multivariate7 | Univariate | |||
|
|
|||||
| p-value | HR2 (95% CI) | p-value | HR6 (95% CI) | p-value | |
| Clinical | |||||
| Age3 | 0.57 | - | - | - | - |
| Stage4 | 0.06 | - | - | - | - |
| Residuum5 | 0.90 | - | - | - | - |
| Pre-treatment Ca-1253 | 0.13 | - | - | - | - |
| Proteins | |||||
| JNK | 0.01 | 6.25 (1.6-25.0) | 0.01 | 1.61 (1.07-2.4) | 0.02 |
| EGFR | 0.05 | 6.67 (1.3-33.3) | 0.02 | 1.50 (0.98-2.3) | 0.06 |
| SMAD3 | 0.04 | 0.14 (0.02-0.9) | 0.04 | - | NS |
| TAZ | 0.04 | - | NS | 1.58 (0.97-2.6) | 0.07 |
| PAI-1 | 0.02 | - | NS | - | NS |
| JNKp183_185 | 0.05 | - | NS | - | NS |
| YKL 40 | 0.04 | - | NS | - | NS |
Abbreviations: HR = hazard ratio; 95% CI = 95% confidence interval; NS = not significant
Notes:
Ca-125 normalization defined as ≤ 35 IU/ml;
Logistic regression hazard ratio > 1 indicates lower chance of normalization of Ca-125 with higher levels of protein expression;
variable analyzed as a continuous variable;
stage 3 vs. 4;
macroscopic vs. no macroscopic;
Hazard ratio > 1 indicates higher risk of progression with higher levels of protein expression;
Only the significant variables in univariate analysis for Ca-125 normalization were taken forward into the multivariate analysis.
Figure 1.
Box plots of protein expression by RPPA and Ca-125 normalization status by 3rd cycle (No vs. Yes). Y-axis values are mean-centered and represent log2 protein expression.
Table 2. Sensitivity and Specificity of Protein Expression Modeling to Predict Ca-125 Normalization (using Smad3, JNK and EGFR).
| Predicted Ca-125 Normalization based on Protein Expression |
Test Characteristics |
|||
|---|---|---|---|---|
| No | Yes | |||
| Observed Ca-125 Normalization by 3rd Cycle |
No | 15 | 5 | Sensitivity - 75% |
| Yes | 5 | 20 | Specificity - 80% | |
A number of methods were used to assess false discovery rates. The expected number of falsely discovered proteins should on average identify 4 by chance (α = 0.05) if testing 80 validated proteins. As a crude measure, the false discovery rate is therefore 57% (4 of 7 identified proteins). Alternatively, the expected number of significant proteins follows a binomial distribution with a “success rate” of 5%. If the t-test is applied 80 times, the probability of identifying 7 or more significant proteins by chance is 0.11. In terms of further assessing the potential for false discovery it is statistically relevant that 5 of the 7 proteins selected were in the TGFβ pathway whereas there were only 6 other proteins that were identified as TGFβ signaling members. Using contingency tables, all validated proteins were assessed according to pathway classification (See Supplement B). Fisher's exact testing revealed a highly significant p-value (p < 0.007, Bonferroni-adjusted) for the enriched identification of signaling proteins in the TGFβ pathway. All pathways were tested and this was the only pathway identified of significance in this analysis.
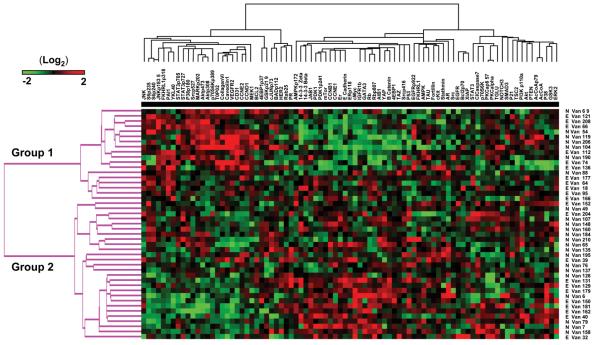
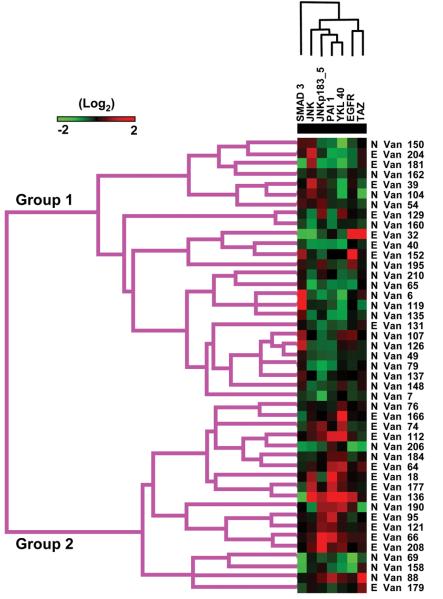
The results of unsupervised clustering of proteins and samples are shown in Figure 2. It is worth noting that overall there are marked differences in protein expression that can be clearly distinguished on cluster analysis in a group of clinically similar high-grade serous cancers. In particular there are two major groups of AHSC identified by changes in protein expression. Group 1 (see Figure 3) identifies a cluster of patients with high cyclin D1 and cyclin E2, pAkt, and stromal markers that include collagen VI, CD31, and VEGFR2. In addition this group contains high expression of many of the proteins associated with a persistently elevated Ca-125 by cycle 3. A second large cluster (Group 2) has high expression of Cyclins B and E1, estrogen receptor alpha (ER) and ER phosphorylated at residue 118, Rb, mammalian target of rapamycin (mTOR), and c-Myc. This latter group may contain markers associated with better outcomes such as ER and those indicative of increased cell cycle progression that may indicate sensitivity to cytotoxic therapy such as cyclins Rb and Myc. Classification of patients into these two groups showed a trend to being associated with their Ca-125 normalization status (Group 1: N=7,E=11; Group 2 N=18, E=9; p= 0.1, Fisher's exact test), with patients that normalized indicated by the prefix “N”, and those that remained elevated with an “E” (Figure 2). Figure 3 shows a supervised clustering analysis using only those proteins identified on univariate testing as predictors of Ca-125 normalization by the 3rd cycle of chemotherapy. There are a number of patients seen with high Smad3, low JNK and pJNK levels, forming a cluster of patients with a significantly higher rate of Ca125 normalization (Group 1: N=18,E=8; Group 2: N=7, E=12; p= 0.04, Fisher's exact test).
Figure 2.
Heat map of unsupervised cluster analysis of proteins and samples by RPPA. Proteins tested by RPPA are listed across the top of the heat map while the patient samples are listed down the right side. An E preceding the sample name indicates an elevated Ca-125 by 3rd cycle of chemotherapy whereas a preceding N indicates a Ca-125 normalization. Cluster trees of patient groups and proteins are shown on the left and top of the heat map respectively. Red indicates increased protein expression while green is low relative to the other samples.
Figure 3.
Heat map of supervised cluster analysis using only proteins associated with Ca-125 normalization by univariate analysis. Proteins tested by RPPA are listed across the top of the heat map while the patient samples are listed down the right side. An E preceding the sample name indicates an elevated Ca- 125 by 3rd cycle of chemotherapy whereas a preceding N indicates Ca-125 normalization. Cluster trees of patient groups and proteins are shown on the left and top of the heat map respectively. Red indicates increased protein expression while green is low relative to the other samples.
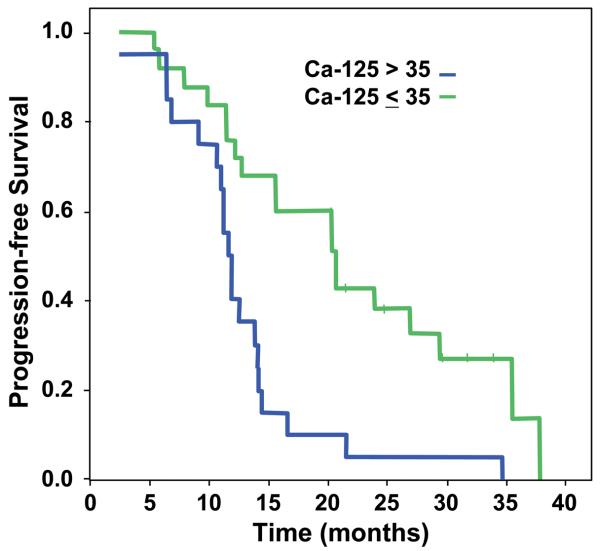
Although our primary interest was to examine the relationship between protein expression and Ca-125 normalization, we also assessed whether these same proteins influenced PFS. As expected, Ca-125 normalization as a clinical predictor significantly influenced PFS times in this cohort (p<0.001, log-rank). Progression-free survival curves by normalization status are shown in Figure 4. There is a marked difference in PFS between the two groups, with those patients who fail to normalize Ca-125 by the third cycle of chemotherapy showing much higher rates of progression. In terms of determining whether the seven proteins of interest associated with Ca-125 normalization also predicted for PFS, we performed univariate Cox regression analyses (Table 1). Higher levels of JNK expression were significantly associated with a poorer progression-free survival (p=0.02), whereas high EGFR and TAZ expression were both of borderline significance (p=0.06,0.07 respectively) for adverse outcomes.
Figure 4.
Progression-free survival and Ca-125 normalization status by the 3rd cycle of chemotherapy.
Discussion
This study identifies a number of candidate proteins that may serve as surrogates of Ca-125 normalization in AHSC. Expression of Smad3, JNK, and EGFR by RPPA are strongly associated with normalization of Ca-125. This analysis was done as a training set and although the study population is small, it was the result of a deliberate strategy to use a homogenous well-characterized group of patients with verification of pathology from a single institution. An obvious advantage to such an approach is to reduce the confounding effects of other important clinical variables such as stage, grade, histology, and treatment on protein expression patterns. The individual proteins that have been identified as predictors of Ca125 normalization represent interesting candidate markers. Subsequent validation in independent sample sets is required to ensure that the current findings do not represent false discoveries from multiple parameter testing (15), and that the selected proteins can be modeled in such a way to effectively predict early treatment failure.
This study also shows marked differences in protein expression and signaling pathway activation profiles within this homogeneous population of advanced serous tumors. This is evident by both the unsupervised and supervised cluster analysis. The potential to exploit these differences for therapeutic purposes is intriguing. Although less studied than mRNA expression profiling, reverse phase protein array is a useful technology for identifying important overexpressed or functionally activated proteins and their pathways as desirable targets for drug development. (13, 16) In this respect RPPA may be more advantageous than expression profiling as protein expression/function may not correlate well with mRNA levels with proteins being the ultimate modulators of cellular function. (17)
We have identified a number of proteins of interest using RPPA that may have relevance from a clinical standpoint. Overexpression of EGFR particularly as a result of EGFR mutation is associated with adverse outcomes in lung cancer. (18) In ovarian cancer, EGFR mutations are detected in less than 4% of patients. (19) There is conflicting information with respect to the prognostic significance of EGFR overexpression in ovarian cancer. For the most part, studies using immunohistochemistry (IHC) have failed to show EGFR expression as a determinant of survival. (20, 21) The difference between these results and our data may reflect a limitation of IHC relative to RPPA. RPPA affords a more objective quantitative measure of protein expression than IHC and is not subject to the potential fixation effects of paraffin embedded tissue on antibody binding characteristics. It will be of future interest to examine EGFR expression by RPPA in relation to outcome and Ca-125 normalization in validation sets.
In our study we found remarkable associations between Smad/TGFβ signaling proteins and Ca-125 normalization. It is most unlikely that this finding is due to false discovery from multiple parameter testing, as if it were; we would not have expected a statistically significant enrichment of TGFβ pathway proteins by contingency table analysis (p < 0.007, Bonferroni adjusted). Low levels of Smad3 were associated with a failure to normalize Ca-125 measures by the 3rd cycle of chemotherapy suggesting that Smad3 may be acting as a tumor suppressor gene. Smad3 is a transcription factor mediating TGFβ signaling. Smad3 plays a role in gonadal tumorigenesis, (22) and may function as a positive or negative regulator of carcinogenesis depending on the cell type and phosphorylation status. (23, 24) The finding that TAZ is associated with Ca-125 normalization on univariate analysis lends additional support to the role of Smad3. TAZ is involved in Smad3 nucleocytoplasic shuttling. (25) Moreover, PAI-1 is transcriptionally regulated by TGFβ (26), overexpressed in ovarian cancer cells, (27) and modulates cell invasion and metastasis. (27, 28). It is important to note that there is conflicting data with respect to the role of TGFβ/Smad3 signaling and the malignant phenotype. While there is evidence that high Smad3 levels promote epithelial-to-mesenchymal transition in ovarian cancer cells,(29) our data on tumor tissue and other studies indicate that Smad3 may function as a tumor suppressor in more complex interactions that involve important signaling pathways and immune response regulation. (24, 30)
We found that increased JNK expression correlated with both chemoresistance and poorer progression-free survival. JNK phosphorylates the linker region of Smad3 promoting cell growth and invasion in contrast to Smad3 phosphorylation by TGFβ that suppresses the malignant phenotype. (23, 31) This mechanism may in part explain the differential oncogenic effects Smad/TGFβ signaling as shown in a rat hepatocellular carcinoma model. (32) In keeping with this it has been recently reported that SP600125, an inhibitor of JNK, sensitizes mouse ovarian cancer cells to paclitaxel. (33) Furthermore, in a recent study on a distinct cohort of patients from the M. D. Anderson Cancer Center, RPPA analysis of tumor tissues found that increased JNKp183_185 expression was associated with shorter progression-free survival and JNK inhibition either by small interfering RNA (siRNA) or by using a novel JNK inhibitor resulted in increased sensitivity to paclitaxel.(34) Thus JNK appears to play an important role in modulating chemoresistance in ovarian cancer.
This study represents proof-in-principle that RPPA can be used to develop a predictor of chemoresistance in ovarian cancer. It also further demonstrates the usefulness of RPPA as a tool for individual tumor-based proteomics assessment and pathway signaling assessment in patients with serous ovarian cancer. Because of the potential for false discovery, we cannot presently determine which individual proteins should be used in models as predictors. Our future validation studies will focus on those proteins involved primarily in TGFβ pathway signaling, as it is most likely that this pathway plays an important role as a marker or mediator of chemoresistance in high-grade serous ovarian cancer.
Ideally, the earlier one can predict outcome to primary therapy the better. In ovarian cancer, a change in serum Ca-125 levels over the course of primary treatment is a validated marker of chemotherapy response and an important determinant of outcome. (7-9, 35) If we are to develop a test that is applied at the time of initial diagnosis then we require a surrogate for Ca-125 response. It is encouraging that a considerable amount of prognostic information relating to Ca-125 normalization can be captured by proteomic testing at the time of diagnosis by measuring 3 proteins: Smad3, EGFR, and JNK. Future studies should further explore proteomic testing as a predictive tool at the time of diagnosis. On the other hand, if proteomic markers are not robust enough on their own to effectively predict chemoresistance it is possible that they could be used in combination with Ca-125 assessment. Finally, our findings lead to some important considerations relating to clinical trial design. If a proteomic marker for treatment failure is developed for patients with serous ovarian cancer, there will be an opportunity to incorporate investigational drugs earlier into the primary treatment setting for those patients with very adverse outcomes. Basing this type of design on proteomic assessment of individual tumors would represent an important advance in terms of assessing new targeted therapies or those that are designed to disrupt TGFβ pathway signaling in order to enhance the effectiveness of chemotherapy.
Statement of Translational Relevance.
We studied functional proteomic aberrations in tumor lysates from patients with advanced serous ovarian cancers in order to determine predictors of response to primary chemotherapy. Having the ability to determine those patients who benefit from a particular therapy will allow the triage of patients to the most effective approaches. Strikingly, pretreatment assessment of EGFR, YKL-40, and several TGFβ pathway proteins (Smad3, c-Jun N-terminal kinase, PAI-1, TAZ, and phospho-JNK) was associated with normalization of Ca-125 before cycle 3 of chemotherapy. Contingency table analysis showed that the identification of TGFβ pathway proteins is unlikely due to false discovery. Thus, this information provides a sound basis for future studies on outcome prediction and drug development.
Recognizing that over 20% of patients with advanced ovarian cancer do not respond to first-line therapy, the early identification of patients with poor outcomes based on tumor proteomics assessment will also have important implications for future clinical trial design.
Supplementary Material
Acknowledgements
R. Agarwal is funded by a Clinician Scientist Fellowship from Cancer Research UK (CA2757/A5902). G.B. Mills and B.T. Hennessy were supported by NCI P50CA083639 and CCSG P30 CA16672, M.S. Carey was the recipient of a Myer-Levy Fellowship and G.B. Mills, B.T. Hennessy, M.S. Carey, and R. Agarwal were supported by the Kleberg Center for Molecular Markers. The authors would also like to thank Dr. Molli McGahren, Yang Li, and Jane Li for their valuable assistance with this work.
References
- 1.du Bois A, Weber B, Rochon J, et al. Addition of epirubicin as a third drug to carboplatin-paclitaxel in first-line treatment of advanced ovarian cancer: a prospectively randomized gynecologic cancer intergroup trial by the Arbeitsgemeinschaft Gynaekologische Onkologie Ovarian Cancer Study Group and the Groupe d'Investigateurs Nationaux pour l'Etude des Cancers Ovariens. J Clin Oncol. 2006;24:1127–35. doi: 10.1200/JCO.2005.03.2938. [DOI] [PubMed] [Google Scholar]
- 2.Piccart MJ, Bertelsen K, James K, et al. Randomized intergroup trial of cisplatin-paclitaxel versus cisplatin-cyclophosphamide in women with advanced epithelial ovarian cancer: three-year results. J Natl Cancer Inst. 2000;92:699–708. doi: 10.1093/jnci/92.9.699. [DOI] [PubMed] [Google Scholar]
- 3.du Bois A, Luck HJ, Meier W, et al. A randomized clinical trial of cisplatin/paclitaxel versus carboplatin/paclitaxel as first-line treatment of ovarian cancer. J Natl Cancer Inst. 2003;95:1320–9. doi: 10.1093/jnci/djg036. [DOI] [PubMed] [Google Scholar]
- 4.Swenerton KD, Hislop TG, Spinelli J, LeRiche JC, Yang N, Boyes DA. Ovarian carcinoma: a multivariate analysis of prognostic factors. Obstet Gynecol. 1985;65:264–70. [PubMed] [Google Scholar]
- 5.du Bois A, Reuss A, Pujade-Lauraine E, Harter P, Ray-Coquard I, Pfisterer J. Role of surgical outcome as prognostic factor in advanced epithelial ovarian cancer: a combined exploratory analysis of 3 prospectively randomized phase 3 multicenter trials: by the Arbeitsgemeinschaft Gynaekologische Onkologie Studiengruppe Ovarialkarzinom (AGO-OVAR) and the Groupe d'Investigateurs Nationaux Pour les Etudes des Cancers de l'Ovaire (GINECO) Cancer. 2009;115:1234–44. doi: 10.1002/cncr.24149. [DOI] [PubMed] [Google Scholar]
- 6.Zorn KK, Tian C, McGuire WP, et al. The prognostic value of pretreatment CA 125 in patients with advanced ovarian carcinoma: a Gynecologic Oncology Group study. Cancer. 2009;115:1028–35. doi: 10.1002/cncr.24084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riedinger JM. Prognostic value of CA125 half-life and early normalization during chemotherapy in advanced ovarian tumors: results of a multicentric French study. Bull Cancer. 2007;94:287–95. [PubMed] [Google Scholar]
- 8.Prat A, Parera M, Peralta S, et al. Nadir CA-125 concentration in the normal range as an independent prognostic factor for optimally treated advanced epithelial ovarian cancer. Ann Oncol. 2008;19:327–31. doi: 10.1093/annonc/mdm495. [DOI] [PubMed] [Google Scholar]
- 9.Fayers PM, Rustin G, Wood R, et al. The prognostic value of serum CA 125 in patients with advanced ovarian carcinoma: an analysis of 573 patients by the Medical Research Council Working Party on Gynaecological Cancer. Int J Gynecol Cancer. 1993;3:285–92. doi: 10.1046/j.1525-1438.1993.03050285.x. [DOI] [PubMed] [Google Scholar]
- 10.Bookman MA. First-line randomized trials: revisiting the Ptolemaic universe. Int J Gynecol Cancer. 2008;18(Suppl 1):47–52. doi: 10.1111/j.1525-1438.2007.01106.x. [DOI] [PubMed] [Google Scholar]
- 11.Kobel M, Huntsman D, Gilks CB. Critical molecular abnormalities in high-grade serous carcinoma of the ovary. Expert Rev Mol Med. 2008;10:e22. doi: 10.1017/S146239940800077X. [DOI] [PubMed] [Google Scholar]
- 12.Tibes R, Qiu Y, Lu Y, et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Molecular cancer therapeutics. 2006;5:2512–21. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 13.Hennessy BT, Lu Y, Poradosu E, et al. Pharmacodynamic markers of perifosine efficacy. Clin Cancer Res. 2007;13:7421–31. doi: 10.1158/1078-0432.CCR-07-0760. [DOI] [PubMed] [Google Scholar]
- 14.Hu J, He X, Baggerly KA, Coombes KR, Hennessy BT, Mills GB. Non-parametric quantification of protein lysate arrays. Bioinformatics. 2007;23:1986–94. doi: 10.1093/bioinformatics/btm283. [DOI] [PubMed] [Google Scholar]
- 15.Coombes KR, Wang J, Baggerly KA. Microarrays: retracing steps. Nature medicine. 2007;13:1276–7. doi: 10.1038/nm1107-1276b. author reply 7-8. [DOI] [PubMed] [Google Scholar]
- 16.Sabatier R, Finetti P, Cervera N, Birnbaum D, Bertucci F. Gene expression profiling and prediction of clinical outcome in ovarian cancer. Critical reviews in oncology/hematology. 2009;72(2):98–109. doi: 10.1016/j.critrevonc.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 17.Stevens EV, Nishizuka S, Antony S, et al. Predicting cisplatin and trabectedin drug sensitivity in ovarian and colon cancers. Molecular cancer therapeutics. 2008;7:10–8. doi: 10.1158/1535-7163.MCT-07-0192. [DOI] [PubMed] [Google Scholar]
- 18.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:13306–11. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schilder RJ, Sill MW, Chen X, et al. Phase II study of gefitinib in patients with relapsed or persistent ovarian or primary peritoneal carcinoma and evaluation of epidermal growth factor receptor mutations and immunohistochemical expression: a Gynecologic Oncology Group Study. Clin Cancer Res. 2005;11:5539–48. doi: 10.1158/1078-0432.CCR-05-0462. [DOI] [PubMed] [Google Scholar]
- 20.Lee CH, Huntsman DG, Cheang MC, et al. Assessment of Her-1, Her-2, And Her-3 expression and Her-2 amplification in advanced stage ovarian carcinoma. Int J Gynecol Pathol. 2005;24:147–52. doi: 10.1097/01.pgp.0000152026.39268.57. [DOI] [PubMed] [Google Scholar]
- 21.de Graeff P, Crijns AP, Ten Hoor KA, et al. The ErbB signalling pathway: protein expression and prognostic value in epithelial ovarian cancer. British journal of cancer. 2008;99:341–9. doi: 10.1038/sj.bjc.6604471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Q, Graff JM, O'Connor AE, Loveland KL, Matzuk MM. SMAD3 regulates gonadal tumorigenesis. Molecular endocrinology (Baltimore, Md. 2007;21:2472–86. doi: 10.1210/me.2007-0147. [DOI] [PubMed] [Google Scholar]
- 23.Matsuzaki K. Smad3 phosphoisoform-mediated signaling during sporadic human colorectal carcinogenesis. Histology and histopathology. 2006;21:645–62. doi: 10.14670/HH-21.645. [DOI] [PubMed] [Google Scholar]
- 24.Millet C, Zhang YE. Roles of Smad3 in TGF-beta signaling during carcinogenesis. Critical reviews in eukaryotic gene expression. 2007;17:281–93. doi: 10.1615/critreveukargeneexpr.v17.i4.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Varelas X, Sakuma R, Samavarchi-Tehrani P, et al. TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nature cell biology. 2008;10:837–48. doi: 10.1038/ncb1748. [DOI] [PubMed] [Google Scholar]
- 26.Samarakoon R, Higgins PJ. Integration of non-SMAD and SMAD signaling in TGF-beta1-induced plasminogen activator inhibitor type-1 gene expression in vascular smooth muscle cells. Thrombosis and haemostasis. 2008;100:976–83. [PMC free article] [PubMed] [Google Scholar]
- 27.Koensgen D, Mustea A, Denkert C, Sun PM, Lichtenegger W, Sehouli J. Overexpression of the plasminogen activator inhibitor type-1 in epithelial ovarian cancer. Anticancer research. 2006;26:1683–9. [PubMed] [Google Scholar]
- 28.Lin SW, Ke FC, Hsiao PW, Lee PP, Lee MT, Hwang JJ. Critical involvement of ILK in TGFbeta1-stimulated invasion/migration of human ovarian cancer cells is associated with urokinase plasminogen activator system. Experimental cell research. 2007;313:602–13. doi: 10.1016/j.yexcr.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 29.Do TV, Kubba LA, Du H, Sturgis CD, Woodruff TK. Transforming growth factor-beta1, transforming growth factor-beta2, and transforming growth factor-beta3 enhance ovarian cancer metastatic potential by inducing a Smad3-dependent epithelial-to-mesenchymal transition. Mol Cancer Res. 2008;6:695–705. doi: 10.1158/1541-7786.MCR-07-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Looyenga BD, Hammer GD. Genetic removal of Smad3 from inhibin-null mice attenuates tumor progression by uncoupling extracellular mitogenic signals from the cell cycle machinery. Molecular endocrinology (Baltimore, Md. 2007;21:2440–57. doi: 10.1210/me.2006-0402. [DOI] [PubMed] [Google Scholar]
- 31.Sekimoto G, Matsuzaki K, Yoshida K, et al. Reversible Smad-dependent signaling between tumor suppression and oncogenesis. Cancer research. 2007;67:5090–6. doi: 10.1158/0008-5472.CAN-06-4629. [DOI] [PubMed] [Google Scholar]
- 32.Nagata H, Hatano E, Tada M, et al. Inhibition of c-Jun NH2-terminal kinase switches Smad3 signaling from oncogenesis to tumor- suppression in rat hepatocellular carcinoma. Hepatology (Baltimore, Md. 2009;49:1944–53. doi: 10.1002/hep.22860. [DOI] [PubMed] [Google Scholar]
- 33.Renlund N, Pieretti-Vanmarcke R, O'Neill FH, Zhang L, Donahoe PK, Teixeira J. c-Jun N-terminal kinase inhibitor II (SP600125) activates Mullerian inhibiting substance type II receptor-mediated signal transduction. Endocrinology. 2008;149:108–15. doi: 10.1210/en.2007-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vivas-Mejia P, Benito JM, Fernandez A, et al. c-Jun-NH2-kinase-1 inhibition leads to antitumor activity in ovarian cancer. Clin Cancer Res. 16:184–94. doi: 10.1158/1078-0432.CCR-09-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riedinger JM, Eche N, Basuyau JP, Dalifard I, Hacene K, Pichon MF. Prognostic value of serum CA 125 bi-exponential decrease during first line paclitaxel/platinum chemotherapy: a French multicentric study. Gynecol Oncol. 2008;109:194–8. doi: 10.1016/j.ygyno.2008.01.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.