Abstract
The equine-associated obligate pathogen Burkholderia mallei was developed by reductive evolution involving a substantial portion of the genome from Burkholderia pseudomallei, a free-living opportunistic pathogen. With its short history of divergence (∼3.5 myr), B. mallei provides an excellent resource to study the early steps in bacterial genome reductive evolution in the host. By examining 20 genomes of B. mallei and B. pseudomallei, we found that stepwise massive expansion of IS (insertion sequence) elements ISBma1, ISBma2, and IS407A occurred during the evolution of B. mallei. Each element proliferated through the sites where its target selection preference was met. Then, ISBma1 and ISBma2 contributed to the further spread of IS407A by providing secondary insertion sites. This spread increased genomic deletions and rearrangements, which were predominantly mediated by IS407A. There were also nucleotide-level disruptions in a large number of genes. However, no significant signs of erosion were yet noted in these genes. Intriguingly, all these genomic modifications did not seriously alter the gene expression patterns inherited from B. pseudomallei. This efficient and elaborate genomic transition was enabled largely through the formation of the highly flexible IS-blended genome and the guidance by selective forces in the host. The detailed IS intervention, unveiled for the first time in this study, may represent the key component of a general mechanism for early bacterial evolution in the host.
Author Summary
It has been known for some time that bacteria undergo genome-reduction when they transition from a free-living state to a constantly host-restricted state. High levels of IS element expansion were also found in these bacteria, and the IS elements were suggested to play a role in genome reductive evolution. Here we provide evidence for stepwise IS actions as the exclusive mechanism that mediates bacterial genomic changes during the early stage of constant host-bacterial association, by unveiling the processes that resulted in the development of B. mallei genome. We show the details of the multi-level interplay of IS elements, which facilitate the wide spread of the IS copies, and the overall mechanics in genome reduction and rearrangement. These processes appeared to operate as chain reactions mediating elaborate genomic transition, without seriously affecting the original gene expression patterns. The absence of differential gene expression in the resulting genome suggests that changes in transcriptional regulation that are often observed in other old bacterial genomes may take place subsequent to the IS-mediated steps, along with gradual nucleotide-level changes.
Introduction
The genomes of host-adapted bacteria, including endosymbionts and obligatory intracellular pathogens, go through reductive evolution [1], [2], [3]. Such changes are partly due to a reduced pressure to maintain genes that are not essential for survival in the host. Similarly, decreased efficiency of purifying selection, resulting from the reduced population size from a restricted life, results in inactivated genes, including beneficial genes, through genetic drift [3]. During the early stage of the genome reduction process, the majority of genes are lost as large chromosomal fragments spanning multiple genes. Such genome reduction has been documented in diverse bacterial groups, including Firmicutes, Chlamydiae, Spirochetes, and γ-Proteobacteria [1], [3], [4], [5], [6], [7]. Most of these bacteria have large expansion of IS elements (insertion sequences), and thus it has been suggested that the IS elements may play an essential role during the genome reduction process [1], [3], [8], [9], [10].
Burkholderia pseudomallei and Burkholderia mallei belong to the ß-Proteobacteria family, and are the causative agents of melioidosis and glanders, respectively [11], [12], [13], [14], [15], [16], [17]. B. mallei has very recently (∼3.5 myr) evolved from a clone of B. pseudomallei through extensive genome reduction [18], [19], accounting for as much as 1.41 Mb or 20% of the genome, as estimated by the size difference between the genomes of B. mallei ATCC 23344 and B. pseudomallei K96243 [18], [20], [21]. Concomitant with this process, B. mallei became constantly associated with mammalian hosts, specifically equines [22], [23], while B. pseudomallei maintains an opportunistic pathogenic lifestyle [17]. Preliminary analyses of the two type strains, B. mallei ATCC 23344 and B. pseudomallei K96243, have suggested that genome reduction and rearrangement in B. mallei were mediated by IS elements that are widely spread throughout the genome [20], [21]. Genes that have been deleted from the B. mallei genome but are maintained in B. pseudomallei include genes that are required for environmental survival. Many of these genes encode metabolic functions for the synthesis of metabolites or the utilization of various sugars and amino acids, without which bacterial propagation in the environment could be significantly hindered [20].
While the genomic reduction during bacterial restriction to their hosts has been well documented [1], [8], [10], most of the stepwise processes have not yet been elucidated. The B. mallei genome has unique significance, as it is much younger than the other genomes in which the genome-reductive evolutionary processes have been most studied to date, including Buchnera (>150 million years) and other much older groups [1], [3], [4], [5], [6], [7]. The studies with these older genomes have been challenging due to the subsequent genomic- and nucleotide-level mutations that accumulated over a long evolutionary history. In this study, we dissected 10 genomes each of B. pseudomallei and B. mallei to understand the early-stage processes that drive genome-reductive evolution in host-associated bacteria.
Results/Discussion
Multiple IS elements with massive proliferation
It is well known that bacteria specialized to a (host) niche, often have a large number of IS elements compared to their free-living relatives [1], [3], [8], [9], [10]. Likewise, by comparing genome sequences, we found that three types of IS elements, ISBma1, ISBma2, and IS407A, were significantly increased in B. mallei compared to B. pseudomallei (Fig. 1A). By contrast, other types of IS, including IS1356, ISBma3, ISBma4, and ISBma5 were found in low copy number in both species of bacteria. These elements appeared to be mostly degenerate evolutionary remnants (i.e., part of the IS disrupted or deleted) of the Burkholderia lineage. ISBma1, ISBma2, and IS407A also had degenerate elements in each species; the ISBma1 elements had the highest levels of degeneration (44%), followed by ISBma2 (20%), and by IS407A (5%) (Fig. 1A).
Figure 1. Expansion of a set of IS elements in the B. mallei genome.
A. Average copy number of IS elements commonly present in B. mallei and B. pseudomallei. Three species of IS elements showed significant proliferation in B. mallei compared with their levels in B. pseudomallei. The number of representative IS elements, both as intact and degenerate (i.e., partly deleted) forms, are shown in graph. B. Distribution of the three types of IS element in the strains of B. mallei. The IS elements ISBma1, ISBma2, and IS407A can be divided into three groups depending on their distribution patterns in the B. mallei strains: 1) “Core” IS elements that are present in all the strains; 2) those IS elements present in more than two strains but not in all; and 3) those elements in only one strain. Groups 2 and 3 are collectively called “accessory IS elements” (for a scaled map with the IS insertion sites in all B. mallei and B. pseudomallei strains, see Fig. S1; for the patterns of genomic rearrangements in the strains of each species, see Fig. S2; for the actual comparative blast data, see Tables S1 and S2).
Intriguingly, up to almost 90% of ISBma1, ISBma2, and IS407A (88.5%, 86.1%, and 89.6%, respectively) were found to be present at the corresponding loci in all 10 B. mallei strains, when examined after the rearranged genomic fragments in each strain were aligned against a reference genome of B. pseudomallei K96243 (Fig. 1B; for a scaled map with the IS insertion sites in all B. mallei and B. pseudomallei strains, see Fig. S1; for the patterns of genomic rearrangements in the strains of each species, see Fig. S2; for the actual comparative blast data, see Tables S1 and S2). In contrast to these “core” elements, those elements that were not present in all (singletons and those found in a few strains), collectively referred to as “accessory” elements, were much less common. That the core elements, expected to be associated with the speciation of B. mallei from B. pseudomallei, accounted for most of the elements clearly reflects the common origin of B. mallei strains from a clone of B. pseudomallei [18], [20]. More importantly, it also suggests that further transpositions were significantly slowed after subsequent geographical segregation of the bacteria. There are 13 core elements in B. mallei that have matching IS elements located at the same sites in B. pseudomallei strains (Table 1). These elements were found to be composed of elements of ISBma1 and ISBma2 but not of IS407A. This finding suggests that ISBma1 and ISBma2 have a longer history of association with B. pseudomallei than IS407A does.
Table 1. IS elements matched across strains of B. mallei and B. pseudomallei.
| B. pseudomallei | B. mallei | ||||||||||
| IS nameb | 1a | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | IS namec |
| Bp_chr1_3_ISBma2 | Xd | Chr1_13ab_ISBma2_A | |||||||||
| Bp_chr1_15_ISBma2 | X | Chr1_52_ISBma2_A | |||||||||
| Bp_chr1_27_ISBma2 | X | Chr1_90_ISBma2_A | |||||||||
| Bp_chr1_36_ISBma2 | X | X | X | X | X | X | X | X | X | Chr1_118_ISBma2_A | |
| Bp_chr1_38_ISBma2 | X | Chr1_120_ISBma2_A | |||||||||
| Bp_chr1_45_ISBma2 | X | X | Chr1_147_ISBma2_A | ||||||||
| Bp_chr1_48_ISBma2 | X | Chr1_151ab_ISBma2_A | |||||||||
| Bp_chr1_53_ISBma2 | X | Chr1_159ab_ISBma2_A | |||||||||
| Bp_chr2_12_ISBma2 | X | Chr2_71ab_ISBma2_A | |||||||||
| Bp_chr1_6_ISBma1 | X | Chr1_26_ISBma1_A | |||||||||
| Bp_chr1_49_ISBma1 | X | X | X | X | X | Chr1_153_ISBma1_A | |||||
| Bp_chr1_51_ISBma1 | X | X | Chr1_156_ISBma1_A | ||||||||
| Bp_chr2_11_ISBma1 | X | Chr2_69_ISBma1_A | |||||||||
aStrains of B. pseudomallei are denoted by numbers (1: K96243; 2: 1106a; 3: 1106b; 4: 1710a; 5: 1710b; 6: 406e; 7: 668; 8: Pasteur; 9: S13; 10: 1655).
bNames of the IS elements present in B. pseudomallei can be found in Table S1.
cNames of the IS elements present in B. mallei can be found in Table S2. These elements are present in all strains of B. mallei.
dPresence of the IS element in the strain.
IS-mediated large genomic deletions and rearrangements
Among the three largely expanded elements, we found that IS407A and ISBma2 were associated with almost all of the large genomic deletions and rearrangements in the B. mallei strains (Fig. 2; Figs. S1 and S2; Tables S1 and S2). The only exception to this was a large deletion found in the strain ATCC 23344 and its direct derivatives, FMH, JHU, and GB8 horse 4 [24], between the 43rd and the 44th elements in chromosome 2 (Fig. 2; Table S1). No genomic rearrangement was mediated by features other than the two IS elements. ISBma1, which was significantly increased in B. mallei, was not directly involved in any of the genomic deletions or rearrangements, however as many as 35% of it served as secondary entry points for IS407A. The majority of the core elements of IS407A, 71.8% and 63.3% in chromosomes 1 and 2, respectively, mediated rearrangements, deletions, or both (Fig. 3A). By contrast, accessory elements of IS407A contributed less, but were more active in chromosome 2 than in chromosome 1. By contrast, 50.4% and 53.2% of the core elements of ISBma2 in chromosomes 1 and 2, respectively, contributed to rearrangements and/or deletions, and the accessory elements in both chromosomes were very rarely involved (Figs. 2 and 3). We identified 59 and 28 genomic fragments in chromosomes 1 and 2, respectively, which were encompassed by core elements of IS407A or ISBma2; these core elements mediated genomic rearrangements in at least one strain (Figs. 2; Table S1). We referred to these genomic fragments as BRUs (basic rearrangement units), a set of basic units for genomic reduction and rearrangement in B. mallei. The BRUs formed various rearrangement patterns in the B. mallei strains (Fig. S2A). By contrast, B. pseudomallei strains had little variation in genome arrangement among one another due to low levels of IS elements- a few rearrangements were found but were around non-IS repeat sequences (Fig. S2B).
Figure 2. IS-blended B. mallei genomes.
Locations of ISBma1, ISBma2, and IS407A and large deletions in each B. mallei strain were mapped back to the reference genome of B. pseudomallei K96243 to show their relative positions. For simplicity, these features are not displayed to scale, but are shown in boxes of equal sizes (for a scaled view of the same data, see Fig. S1). The IS elements were numbered in the order in which they appear in the reference chromosomes. The 2nd element in chromosome 1 is denoted by an *, and is the IS407A insertion that disrupted fliP, which encodes a key factor for flagella formation. The numbers in red indicate the IS elements that were disrupted by a neighboring IS407A element. Each BRU is denoted as an open box and by sequential numbers, which are preceded by A or B for chromosome 1 or chromosome 2, respectively. For additional information, see Figure S1 and Tables S1 and S2, in which the IS elements are listed with their names, composed of the sequential numbers, the IS species to which they belong (i.e. ISBma1, ISBma2, and IS407A), and their distribution patterns among the B. mallei strains (i.e., _A: present in all the strains; _B: present in only some strains; and _C: present in a single strain).
Figure 3. Patterns of IS-mediation in genomic deletions and rearrangements.
A. Association of IS407A and ISBma2 with genomic deletions and rearrangements. Core elements played a major role in such genomic modifications compared with the accessory elements. The latter elements had a more active role in chromosome 2 than in chromosome 1. B. A phylogenetic relationship of the B. mallei and B. pseudomallei strains based on the presence of the IS insertions and their association with the genomic deletions and rearrangements. Two strains of B. pseudomallei from Australia, 1655 and 668, are labeled in orange. The B. mallei strains of European origins, NCTC 10247, NCTC 10229, and 2002721280, are labeled in green, while ATCC 23344 and its direct derivatives are shown in red.
When the pattern of the IS insertions and their involvement in genome-reductive and rearrangement processes in strains were used to construct a phylogenetic tree, strains sharing a recent common ancestry (e.g., ATCC 23344 and its immediate derivative isolates, FMH, JHU, and GB8 horse 4) or common recent geographical origins (e.g., strains NCTC 10257, NCTC 10229, and 2002721280 from European countries) were grouped together (Fig. 3B). This phylogenic relationship supports the hypothesis that the accessory IS elements, which provided the major determinants for the tree rather than the common core elements, occurred following the speciation and geographical segregation of the B. mallei strains. By contrast, such patterns were not obvious among the B. pseudomallei strains which did not go through IS element expansions; Australian strains 1655 and 668 did not branch separately from the South Asian strains.
The deletions and rearrangements that were mediated by accessory elements were most frequently noted in strains SAVP1 and 2002721280, which lost virulence after successive passages in laboratory cultures [25] (Figs. 2 and S1). Most of the extra deletions in these strains were more prominent in chromosome 2 than in chromosome 1. In SAVP1, an IS407A-mediated deletion removed a major group of virulence genes encoding the animal-type type III secretion system in the BRU B22 (Figs. 2 and S1); this deletion may be a major cause of the avirulence of that strain. By contrast, there is no obvious deletion that may be responsible for the loss of virulence in strain 2002721280. That the strains SAVP1 and 2002721280 obtained deleterious mutations from in vitro culturing suggests that maintenance of the genomic contents in B. mallei requires selective pressure for survival in the host environment. By contrast, the fully virulent strain PRL-20 showed more frequent deletions and rearrangements mediated by accessory elements than other virulent strains. This strain may represent one of the more evolved (more genome-reduced) strains of B. mallei.
Although extra deletions and rearrangements were noted, the actual number of the accessory IS elements was not significantly increased in PRL-20, SAVP1 or 2002721280. Furthermore, none of the direct derivatives of the strain ATCC 23344 (i.e. FMH, JHU, and GB8 horse 4) had new IS insertions (Fig. 2 and Table S1). These ATCC 23344 derivatives also did not have genomic rearrangements; the only change found was a single IS407A-mediated deletion located within the BRU B17 in the strain JHU (Fig. 2 and Table S1). These lines of evidence suggest that B. mallei genomes are structurally flexible with regard to deletions, however perhaps not as much anymore for additional IS transpositions or genomic rearrangements.
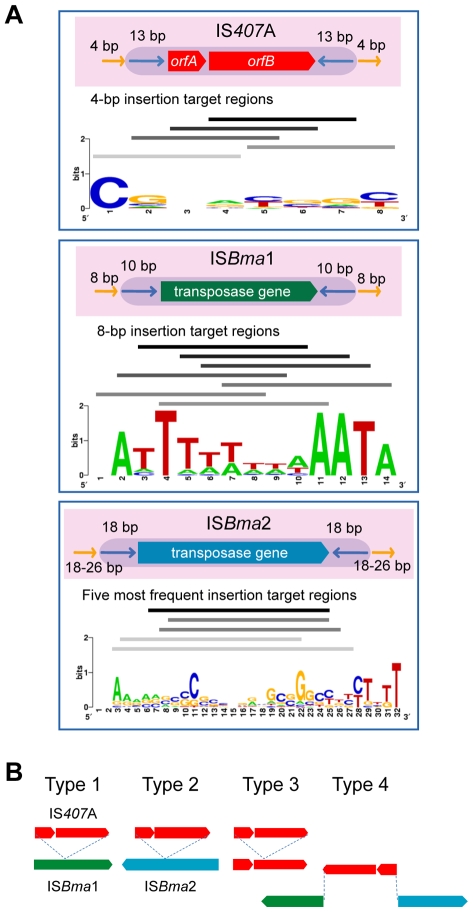
Different insertion target preferences
IS407A elements are known to generate 4-bp target region duplications as direct repeats around them when they transpose [26]. We found that ISBma1 generates 8-bp target region duplications, and that ISBma2 generates longer repeats of various lengths (18–26 bp) (Table 2; for the entire data, see Table S3). In addition to the various lengths of duplications, these target regions of the three types of IS had different nucleotide compositions and patterns. Most notably, the sequences of ISBma1 contained homopolymers of A and/or T in up to 8-bp stretches of nucleotides (Fig. 4A). The target sequences of ISBma2 had a loose pattern in which the GC-rich central region was encompassed by strands of As and Ts on either side. Target sequences of IS407A had the least characteristic composition. It is intriguing to note that each IS element showed different levels of copy number expansion, ISBma1 with the lowest (3.3×), ISBma2 with an increased level (9.5×), and IS407A with the highest (16.7×) (Fig. 1A). Perhaps this difference, at least in part, resulted from the availability of genomic sites suited for insertion targets.
Table 2. Repeat sequence pairs associated with the major IS elements in B. mallei.
| 5′-end | 3′-end | |
| IS407A | ||
| Inverted Repeatsa | TGACCTGCCCCCT b | AGGGGGCAGGTCA |
| Example direct target repeatsc | TGTC | TGTC |
| GTAG | GTAG | |
| TTCG | TTCG | |
| GGGC | GGGC | |
| ATCA | ATCA | |
| ISBma1 | ||
| Inverted Repeats | GGTTCATCGC | GCGATGAACC d |
| Example direct target repeats | AATTTTTT | AATTTTTT |
| AAAAAAAT | AAAAAAAT | |
| AAAAAATA | AAAAAATA | |
| CTTTTTTT | CTTTTTTT | |
| TTTTTCTT | TTTTTCTT | |
| ISBma2 | ||
| Inverted Repeats | CAGATTGCTGACAAACCC | GGGTTCGTCAGCAGTCTG |
| Example direct target repeats | AGCCAGCTTGCGCTGGCT | AGCCAGCTTGCGCTGGCT |
| ACCGCAGCGAAGGCTGCGGT | ACCGCAGCGAAGGCTGCGGT | |
| AATGGGCCGGAAACGGCCCATT | AATGGGCCGGAAACGGCCCATT | |
| AGGCCCGCCGAAGCGGGCTT | GGGCCGTTTCAAACGGCCCG | |
| GGGGCGCTTCGGCGCCCC | GGGGCGCTTCGGCGCCCC | |
aInverted repeats are part of the IS elements, that are present on either side of the ends.
bConsensus sequences of the inverted repeats.
cFive examples of the direct repeat target duplicates, which encompass the IS elements. The entire data can be found in Table S3.
dGCGATGA overlaps with the 3′-terminus of the transposase gene in the upstream.
Figure 4. The main IS elements and their insertion target sequences.
A. The structures of ISBma1, ISBma2, and IS407A and their duplicated target sequences in the B. mallei genomes. IS407A has a pair of two transposase genes, orfA and orfB, while ISBma1 and ISBma2 each contain a single transposase gene. These genes in each type of IS element are flanked by inverted repeats, which are denoted by blue arrows, and then by duplicated insertion target sequences, denoted by yellow arrows. Sequence logo displays [42] of the duplicated insertion target sequences are shown below the corresponding IS element. Solid lines above the Sequence logo display represent the specific regions to which actual target sequences matched, with the most abundant groups at the top. B. Patterns of a disruption of one type of core IS elements by another in B. mallei. Colored solid arrows represent transposases in each IS element. Four types of pattern were found (see Table S1 for details).
There were concordant patterns of disruption of the core elements of one type by another, in that ISBma1 and ISBma2 were intersected by transposed IS407A (Fig. 4B), while the reverse (IS407A disrupted by ISBma1 or ISBma2) was not found. A possible explanation for these insertion patterns may be that ISBma1 and ISBma2 could not transpose into IS407A due to the lack of sites suited for their rather uncommon target preferences, while IS407A did not have this problem. Consistent with this hypothesis, ISBma1 and ISBma2 also did not have self-disrupted elements, while there were several self-disrupted IS407A elements. The involvement of the three IS elements with different target sites increased the total number of IS insertions in the genome. Furthermore, this increase led to further spread of IS407A, because ISBma1 and ISBma2 provided neutral insertion points for the element. This in turn directly improved the efficiency of IS407A-mediated recombination in the genome, resulting in more sophisticated deletions and rearrangements.
We estimated that 83.7% of IS407A and 65.6% of ISBma2 elements in the B. mallei genomes lost their matching target duplicates, while all of the elements from intact ISBma1 elements were maintained (Table S3). Almost all of the IS407A (see Table S3 for details) and all of the ISBma2 elements that contained matching repeats were not involved in genomic rearrangements in B. mallei. This indicates that recombination among the elements were the major cause of the loss of the matching target duplicates.
Nucleotide-level mutations in B. mallei
B. mallei still has a high nucleotide-level identity (99%) to B. pseudomallei. Consistent with this, there was no AT-biased genome deviation in B. mallei, unlike that seen in many old symbionts or obligatory host-associated pathogens [1], [3]. Although the overall identity is still very high, significant nucleotide-level divergence exists, especially at the SSRs (simple sequence repeats), where there are intrinsically high mutation rates [27]. These SSRs were abundant in both B. mallei and B. pseudomallei at corresponding sites in the genomes. However, there were more genes that were disrupted by frameshift mutations in B. mallei compared to B. pseudomallei (Table S4). Most of these disrupted genes were commonly present in all B. mallei strains, reflecting the clonal origin of the strains. Some of these gene disruptions may have contributed to better adaptation of the bacteria (increased persistence) in the host environment or simply became obsolete [28]. One of the most characteristic loss of function or of surface structure in B. mallei is the loss of flagella [20]. A gene essential for flagellum biogenesis, fliP, [29] in the strain ATCC23344 was disrupted by a 65-kb fragment flanked by IS407A elements, and this mutation completely turned B. mallei flagella-less. This disruption in fliP is present in all B. mallei strains (Table S1; Fig. 2, between BRUs A2 and A3), implying the significance of losing flagella in the evolution of host-restricted B. mallei. The loss of flagella has been noted in other bacteria, including Bordetella pertussis and Bordetella parapertussis during their host specialization, derived from the strains of Bordetella bronchiseptica, [9] and Yersinia pestis during its conversion from a gut to a systemic pathogen [30]. Additional disrupted genes not present in all strains were found at approximately the same levels as in B. pseudomallei, suggesting that there were no significant increases in mutation rates in B. mallei after geographical segregation. There also was no significant level of erosion of these, so called, pseudogenes by purifying selection at levels high enough to contribute to the actual genome size reduction (data not shown).
Genomic potential for gene expression divergence
The extensiveness of the genome-wide reduction and rearrangements as well as additional nucleotide-level mutations may suggest that there is a potential for altered gene expression patterns in B. mallei. A total of 341 potential regulatory genes survived the general IS-mediated genomic reduction in B. mallei (not taking into account the diverse strain-specific deletions that occurred after speciation). Among these genes, only a small fraction (about 10) in each strain had deleterious (e.g. frameshift, null, or IS-insertion) mutations (for the list of the genes, see Table S5). In addition, none of the predicted operons in B. mallei, which correspond to the putative operons previously found in B. pseudomallei K96243 [31], were disrupted by IS elements (data not shown). We also estimated the potential for changes in promoters. There were 2,473 upstream sequences of genes, many of which may overlap or contain promoters, in the reference genome of B. pseudomallei K96243 that have homologous sequences (with at least 95% identity over at least 95% of their lengths) in all other strains of B. pseudomallei. We found that up to 99% of these sequences also matched the corresponding regions in B. mallei ATTC23344 at the same homology levels (see Table S6 for the list of the 2,473 upstream sequences, associated gene information, and the blast data). Together, all these data from the analyses of the conserved genomic regions suggest that there is only a low potential for the genes in B. mallei to have significantly divergent gene expression patterns from B. pseudomallei.
By contrast, there were 56 genes with putative regulatory functions that were lost along with the commonly deleted genomic fragments of the B. mallei genome. These genes include potential global regulatory genes, such as those encoding a quorum-sensing system (genes BPSS1176 and BPSS1180 in the reference genome of B. pseudomallei K96243), a two-component regulatory system (the pair BPSS1994 and BpSS1995 in B. pseudomallei K96243), and a number of regulators of various families (Table S7). Whether the loss of any of these 56 regulatory genes affects the expression of the remaining genes in the B. mallei genome was yet to be examined.
Similar gene expression profiles in B. mallei and B. pseudomallei
To experimentally estimate the possible transcriptomic divergence between B. pseudomallei and B. mallei, we infected female BALB/c mice with B. mallei ATCC 23344 or B. pseudomallei K96243, employing the previously established aerosol models of acute glanders and melioidosis [32]. Gene expression was compared in the bacteria that colonized the lungs and the spleens of the mice. Both B. mallei- and B. pseudomallei-challenged animals showed increases in the bacterial loads within these organs over time, with B. pseudomallei having slightly faster growth rates (Fig. 5). In our experience, B. pseudomallei also grew faster than B. mallei in vitro (data not shown). Unlike the mice infected by B. mallei, sampling the B. pseudomallei-challenged animals after 72 hr was not possible due to animal mortality from the more rapid disease progression. When gene expression profiles in the spleens and lungs were compared between B. mallei and B. pseudomallei at middle- (i.e., 24 hr for both bacteria) and late stages (i.e., 48 hr for B. pseudomallei and 72 hr for B. mallei) of infection (a total of four comparison pairs), conserved B. mallei and B. pseudomallei orthologs showed nearly identical patterns with high Pearson correlation coefficient (R) values ranging from 0.94 to 0.97, regardless of the host tissue type (Fig. 5). Therefore, there was no indication of significant modifications of the expression schemes in the genes required by B. mallei to thrive in BALB/c mice compared to those in B. pseudomallei. This is consistent with the findings of our previous gene expression studies in culture and in vivo, which also showed similar gene expression patterns in B. mallei and B. pseudomallei [20], [33], [34], [35]. These data suggest that, during the early stage, genomic reduction proceeds conservatively, not seriously affecting the indigenous gene expression patterns. In contrast to B. mallei, most of the transcription units in the insect symbiont Buchnera were altered, most likely due to complex genomic alterations accumulated over a long period of time [2].
Figure 5. Comparison of in vivo gene expression patterns between B. mallei and B. pseudomallei.
A. Bacterial loads within the lung and the spleen of B. mallei and B. pseudomallei aerosol-challenged mice. Bacterial loads are reported as the average cfu/g of tissue from two animals challenged with either B. mallei or B. pseudomallei. Due to animal mortality in the B. pseudomallei experimental group, only organ loads from 24 hr and 48 hr are reported. B. Histograms depicting the relative gene expression of B. mallei and B. pseudomallei in infected mouse organs. Comparison data from middle- and late stages of infection in two tissue types, spleen and lung, are displayed. Average ratios in the log2 scale, standard deviations of the ratios (SD), and the Pearson correlation coefficients (R's) between the B. mallei and B. pseudomallei samples are shown.
Conclusions
In this study, we unveiled the mechanics of genomic deletions and rearrangements that occur in the early stage of bacterial specialization in the host, by conducting comparative analyses of B. mallei and its parental species, B. pseudomallei. It became clear that stepwise IS intervention was the main driving force mediating a large genomic reduction in B. mallei. Expansion of ISBma1 and ISBma2 in a clone of B. pseudomallei set the stage for the wide spread of IS407A, allowing its proliferation to sites, to which the element itself may rarely target. Actual genomic deletions and rearrangements occurred through recombination reactions mainly among IS407A and also among ISBma2 (Fig. 2). These processes achieved highly efficient deletions of dispensable genomic regions, causing only small disruptions to the portions of the genome that were maintained. This was possible due to the guidance by selective forces in the host and via the intrinsic flexibility of the compactly IS-blended genome. The B. mallei genome currently appears to still be structurally flexible with regard to deletions but is now less flexible with regard to genomic rearrangements and additional transpositions. This may indicate that the genomic evolution in B. mallei has been moving into a second stage, in which large-scale genomic alterations are reduced and nucleotide-level erosion has become more important. On the other hand, a large number of genes disrupted by frameshift mutations in SSRs were found in the B. mallei genome. The loss of function encoded by these genes and of flagella via disruption in fliP by IS407A (Table S1), could be part of the adaptive evolution for survival in the host environment, which will eventually lead to genome size reduction by erosion over time. Widespread relics of IS elements found in diverse symbionts and obligate pathogens [1], [3], [8] clearly suggest that a similar sequential IS intervention, modeled in Figure 6, may illustrate a general mechanism, by which elaborate genome transition occurs during early bacterial evolution after establishing constant association with the host.
Figure 6. A proposed general model for the bacterial genome-reductive evolution in a specialized niche.
Massive expansion of (multiple types of) IS elements may set the stage for extensive genome-reductive evolution in bacteria. When multiple elements are involved, expansion of some elements (e.g. ISMinor1 and ISMinor2) may lead to further spread of a major element (e.g. ISMajor), by providing additional insertion sites in the regions, where the major element itself may rarely target. Gene deactivations by intersecting IS insertions can take place and extensive genomic deletions and rearrangements can occur through recombination reactions among the homologous IS copies. These processes can result in highly efficient deletions of dispensable genomic regions via the intrinsic high flexibility of the compactly IS-blended genome, guided by selective forces in the host. Slow and steady nucleotide-level mutations can accumulate after the IS-mediated genomic changes, eventually also contributing to the genome reduction and divergence in transcriptional regulatory patterns over time.
Materials and Methods
Ethics statement
All research involving live animals was conducted in compliance with the Animal Welfare Act and other federal statutes and regulations relating to animals and experiments involving animals and adhered to the principles stated in the Guide for the Care and Use of Laboratory Animals, National Research Council, 1996. All mouse experiments conducted in the USAMRIID (US Army Medical Research Institute of Infectious Diseases) were approved by the Association for Assessment and Accreditation of Laboratory Animal Care International.
Sequencing and annotation
The type strains for B. mallei (ATCC23344) [20] and B. pseudomallei (K96243) [21] were previously sequenced. Strains FMH, JHU, and GB8 horse 4 were direct derivatives of strain ATCC 23344 after passages in the human or horse, and these strains were also sequenced previously [24]. B. mallei strains NCTC10229, NCTC10247, and SAVP1 were sequenced with full closure and manually annotated as previously described [20]. The remaining three strains (2002721280, ATCC10399, and PRL-20) were sequenced to 8× Sanger sequence coverage by the whole genome shotgun method [36] without closure, and assembled using the Celera Assembler [37], and contigs were oriented by alignment to the reference strain ATCC23344 using PROMER [38]. ORFs were predicted and annotated automatically using GLIMMER [39], [40]. Pseudo-chromosomes were constructed from the ordered scaffolds, using manual examination where necessary. Similarly, B. pseudomallei strains 1106a, 1710b, and 668 were sequenced with full closure and manual annotation, while 1655, 406e, S13, and Pasteur 6068 were sequenced without closure and annotated automatically.
Comparative genomic analyses with B. mallei and B. pseudomallei
For the analyses of genomic deletions and rearrangements in B. mallei and B. pseudomallei, 5,799 predicted protein sequences from the B. pseudomallei type strain K96243 were compared with the nucleotide sequences of the genomes of B. mallei (ATCC 23344, 2002721280, ATCC 10399, FMH, JHU, GB8 horse 4, PRL-20, NCTC 10229, NCTC 10247, SAVP1) and the other strains of B. pseudomallei (1106a, 1106b, 1655, 1710a, 1710b, 406e, 668, Pasteur, S13) using tblastn (http://blast.wustl.edu). For the mapping of the insertions of ISBma1, ISBma2, and IS407A in the genomes of B. mallei and B. pseudomallei, the entire sequences of the IS elements were searched against the 20 genomes using blastn (http://blast.wustl.edu). For the analysis of association of the IS elements with genomic deletions and rearrangements in B. mallei and of the target sequences in the genomes, strain ATCC 23344 represented all of its immediate derivatives, FMH, JHU, and GB8 horse 4, to avoid redundancy in the data, because the three strains showed identical patterns. To compare the patterns of genome rearrangements in the B. mallei strains, the positions of the BRUs in each strain of B. mallei relative to B. pseudomallei K96243 were visualized using a genome-comparative software tool ACT ([41]; http://www.sanger.ac.uk/Software/ACT), and the displays were compared in parallel among the strains.
We also examined B. mallei and B. pseudomallei for intergenic regions that potentially containing promoters, putative regulatory genes, and disruptions of putative operons to estimate the possibility of causing gene expression divergence. For intergenic region comparisons, up to 100 bp upstream of the start codon, or up to as much as available if the neighboring gene was closer, of the genes that contain at least 50 bp of an untranslated upstream region were retrieved from the genome of B. pseudomallei K96243. Then, these sequences (2,268 and 1,566 from chromosomes 1 and 2, respectively) were searched against the genomes of B. mallei and B. pseudomallei using blastn (http://blast.wustl.edu), and the length-match as well as the identity values of the orthologous regions were calculated. Putative operons reported by Rodrigues et al. from the genome of B. pseudomallei K6243 [31] were used to match the orthologous gene clusters in the genome of B. mallei ATCC 23344, and these gene clusters were examined for any disruptions caused by IS elements. All the genome sequences of B. mallei and B. pseudomallei used in this study are available through the Pathema web site (http://pathema.jcvi.org/cgi-bin/Burkholderia/PathemaHomePage.cgi) at the J. Craig Venter Institute (http://www.jcvi.org/).
Construction of the phylogenetic tree
A phylogenetic tree was constructed with the strains of B. mallei and B. pseudomallei based on the insertion patterns of and the role played in the genomic deletions and rearrangements by the three major IS elements, ISBma1, ISBma2, and IS407A. All the data used are shown in Tables S1 and S2 and Figure 2. Bootstrapped maximum parsimony trees were calculated using the PAUP package with default parameters, and a consensus tree was produced from the bootstrap replicates. Branches with bootstrap scores of less than 50 were collapsed in the tree.
Determination of the target sequence patterns
Among the duplicated target regions encompassing the IS elements ISBma1, ISBma2, and IS407A, those regions that had perfectly matching sequences were first collected. Then, among the sequences from unmatched pairs, those that occurred in more than two strains were assumed to be un-mutated valid sequences and, therefore, were added to the data pool for the analysis. Strain ATCC 23344 represented all its direct derivatives (FMH, JHU, and GB8 horse 4) in this analysis to avoid redundancy in the data. The collected sequences were aligned with Clustal X, and the alignments were graphically visualized using Sequence logos [42].
Mouse infection, bacterial load estimation, and RNA preparation
Exposure of mice to bacterial aerosol was performed as described by Roy et al. [43]. Fresh overnight cultures of B. pseudomallei DD503 [44] and B. mallei ATCC 23344 were prepared in LB or in LBG (LB supplemented with 4% glycerol), respectively, at 37°C with aeration (250 rpm). Thirty female BALB/c mice six to eight weeks old (National Cancer Institute, Frederick, MD, USA) were infected with these bacteria: nine mice each with B. pseudomallei and B. mallei for the gene expression studies, and six mice each for the bacterial load assays. The mice infected with B. mallei received an inhaled dose of 7.2×103 cfu (7.2×LD50), and those mice infected with B. pseudomallei received 1.8×104 cfu (18×LD50), as estimated by colony counting on agar plates. The infected mice were provided with rodent feed and water ad libitum and maintained on a 12-hr light cycle. After 24 and 48 hr (for both B. mallei and B. pseudomallei) or 72 hr (for B. mallei) of infection, five mice from each point in time were euthanized in a CO2 chamber, and their spleens and lungs were removed. Due to animal mortality, a 72 hr point in time was not possible for B. pseudomallei. The organs from two randomly picked mice were saved for bacterial load estimations, and the rest were homogenized in 1 ml of Trizol (Invitrogen Corp., Carlsbad, CA, USA) using a Tissue-Tearor (BioSpec Products, Bartlesville, OK, USA). Total RNA was purified according to the manufacturer's recommendations (Invitrogen Corp., Carlsbad, CA, USA). The bacterial load in the mouse organs was estimated as described by Ulrich and DeShazer [32].
RNA labeling and microarray analysis
Total RNA, both bacterial and mouse, from the same organ types from three mice was pooled to compensate for potential individual variation. These pooled RNA samples were used for the experiments without further purification of the bacterial RNA because RNA from mice does not cross-hybridize to the B. mallei microarray at a level affecting the legitimate interactions between the B. mallei array and the Burkholderia transcriptome [35]. The B. mallei whole genome array used in this study for both B. mallei and the closely related B. pseudomallei (average gene identity at the nucleotide level of 99%) was described in detail previously [33]. The B. mallei- and B. pseudomallei-infected organ samples were paired for the hybridization reactions based on early and late pathological states. A total of eight hybridization reactions or four different comparisons were performed, each of which was replicated in flip-dye pairs and the final ratios were calculated as log2 (B. pseudomallei gene expression intensity/B. mallei gene expression intensity). Labeling of the probes, slide hybridization, and slide scanning were carried out as previously described [35]. The independent TIFF slide images from each channel were analyzed using TIGR Spotfinder to assess the relative expression levels, and the data were normalized using a local regression technique LOWESS (LOcally WEighted Scatterplot Smoothing) with the MIDAS software (<http://www.jcvi.org/cms/research/software>, The J. Craig Venter Institute, Rockville, MD, USA). The resulting data were averaged from triplicate genes on each microarray and from duplicate flip-dye arrays for each experiment.
Supporting Information
Graphical alignments of the genomes of B. mallei and B. pseudomallei. Genomes from strains of B. mallei and B. pseudomallei are aligned for close comparisons of the relative positions of IS elements and large deletions. Based on the genome of B. pseudomallei K96243 (displayed in the middle of the alignments), corresponding regions in B. mallei genomes (denoted by the upper green blocks) and those of the other B. pseudomallei genomes (denoted by the lower blue blocks) are displayed. Locations of ISBma1, ISBma2, and IS407A are denoted by red, yellow, and pink lines, respectively, in each strain.
(1.87 MB TIF)
The patterns of genomic rearrangements in B. mallei. A. Syntenic relationship among the B. mallei strains. Re-localization of BRUs in B. mallei strains (relative to B. pseudomallei strain K96243 as the reference) is shown with the comparative genomics display tool ACT (Wellcome Trust Sanger Institute) using blastn data. Blue and red connecting lines between the genomes indicate the same and opposite directions, respectively, of the corresponding BRUs relative to each other. B. Comparisons among the B. pseudomallei strains. Each strain was compared to the reference genome of strain K96243.
(2.42 MB TIF)
Comparison of B. mallei genomes with that of B. pseudomallei strain K96243.
(8.84 MB PDF)
Comparison of B. pseudomallei genomes with that of strain K96243.
(9.59 MB PDF)
Specifics of the IS elements IS407A, ISBma1, and ISBma2 in the B. mallei strains.
(1.56 MB PDF)
Disrupted genes that resulted from SSR-mediated frameshift mutations in B. mallei and B. pseudomallei.
(0.34 MB PDF)
Potential regulatory genes that survived the genomic reduction in B. mallei.
(0.18 MB PDF)
Comparison of the upstream regions of genes containing potential promoters.
(2.92 MB PDF)
Putative regulatory genes deleted from B. mallei as IS-bounded genomic fragments.
(0.04 MB PDF)
Footnotes
The authors have declared that no competing interests exist.
This work was supported by grants 2009K000516 from the 21C Frontier Microbial Genomics & Applications Center Program, 20090058514 from the Basic Research Program, and the K20601000002-09E0100-00210 from the Korea Foundation for International Cooperation of Science & Technology (KICOS), all of which are from the Ministry of Education, Science & Technology in the Republic of Korea to HSK. Sequencing and annotation of most of the Bp and Bm strains was supported in whole or part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services under contract number NIAID N01-AI30071. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Moran NA. Tracing the evolution of gene loss in obligate bacterial symbionts. Current Opinion in Microbiology. 2003;6:512–518. doi: 10.1016/j.mib.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 2.Moran NA, Mira A. The process of genome shrinkage in the obligate symbiont Buchnera aphidicola. Genome Biology. 2001;2:research0054.0051–0054.0012. doi: 10.1186/gb-2001-2-12-research0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moran NA, Plague GR. Genomic changes following host restriction in bacteria. Current Opinion in Genetics & Development. 2004;14:627–633. doi: 10.1016/j.gde.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Nilsson AI, Koskiniemi S, Eriksson S, Kugelberg E, Hinton JCD, et al. Bacterial genome size reduction by experimental evolution. Proc Natl Acad Sci U S A. 2005;102:12112–12116. doi: 10.1073/pnas.0503654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sallstrom B, Andersson SGE. Genome reduction in the [alpha]-Proteobacteria. Current Opinion in Microbiology. 2005;8:579–585. doi: 10.1016/j.mib.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Batut J, Andersson SGE, O'Callaghan D. The evolution of chronic infection strategies in the [alpha]-proteobacteria. Nat Rev Micro. 2004;2:933–945. doi: 10.1038/nrmicro1044. [DOI] [PubMed] [Google Scholar]
- 7.Moran NA, McLaughlin HJ, Sorek R. The Dynamics and Time Scale of Ongoing Genomic Erosion in Symbiotic Bacteria. Science. 2009;323:379–382. doi: 10.1126/science.1167140. [DOI] [PubMed] [Google Scholar]
- 8.Mira A, Pushker R, Rodríguez-Valera F. The Neolithic revolution of bacterial genomes. Trends in Microbiology. 2006;14:200–206. doi: 10.1016/j.tim.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 9.Parkhill J, Sebaihia M, Preston A, Murphy LD, Thomson N, et al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat Genet. 2003;35:32–40. doi: 10.1038/ng1227. [DOI] [PubMed] [Google Scholar]
- 10.Treangen TJ, Abraham A-L, Touchon M, Rocha EPC. Genesis, effects and fates of repeats in prokaryotic genomes. FEMS Microbiology Reviews. 2009;33:539–571. doi: 10.1111/j.1574-6976.2009.00169.x. [DOI] [PubMed] [Google Scholar]
- 11.Dance D. Ecology of Burkholderia pseudomallei and the interactions between environmental Burkholderia spp. and human-animal hosts. Acta Trop. 2000;74:159–168. doi: 10.1016/s0001-706x(99)00066-2. [DOI] [PubMed] [Google Scholar]
- 12.Dharakul T, Songsivilai S. The many facets of melioidosis. Trends Microbiol. 1999;7:138–140. doi: 10.1016/s0966-842x(99)01477-8. [DOI] [PubMed] [Google Scholar]
- 13.McGilvray C. The transmission of glanders from horse to man. Can J Public Health. 1944;35:268–275. [Google Scholar]
- 14.Benenson A. Control of Communicable Diseases Manual. Washington, DC: American Public Health Association; 1995. [Google Scholar]
- 15.DeShazer D, Waag D. Lindler L, Lebeda F, Korch GW, editors. Glanders: New Insights into an Old Disease. Biological Weapons Defense: Infectious Diseases and Counterbioterrorism The Humana Press Inc. 2004. pp. 209–237.
- 16.Cheng AC, Currie BJ. Melioidosis: epidemiology, pathophysiology, and management. Clin Microbiol Rev. 2005;18:383–416. doi: 10.1128/CMR.18.2.383-416.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inglis TJJ, Sagripanti JL. Environmental factors that affect the survival and persistence of Burkholderia pseudomallei. Appl Environ Microbiol. 2006;72:6865–6875. doi: 10.1128/AEM.01036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Godoy D, Randle G, Simpson A, Aanensen D, Pitt T, et al. Multilocus sequence typing and evolutionary relationships among the causative agents of Melioidosis and Glanders, Burkholderia pseudomallei and Burkholderia mallei. J Clin Microbiol. 2003;41:2068–2079. doi: 10.1128/JCM.41.5.2068-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin CH, Bourque G, Tan P. A Comparative Synteny Map of Burkholderia Species Links Large-Scale Genome Rearrangements to Fine-Scale Nucleotide Variation in Prokaryotes. Mol Biol Evol. 2008;25:549–558. doi: 10.1093/molbev/msm282. [DOI] [PubMed] [Google Scholar]
- 20.Nierman WC, DeShazer D, Kim HS, Tettelin H, Nelson KE, et al. Structural flexibility in the Burkholderia genome. Proc Natl Acad Sci USA. 2004;101:14246–14251. doi: 10.1073/pnas.0403306101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holden MTG, Titball RW, Peacock SJ, Cerdeño-Tárraga AM, Atkins T, et al. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc Natl Acad Sci USA. 2004;101:14240–14245. doi: 10.1073/pnas.0403302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilkinson L. Glanders: medicine and veterinary medicine in common pursuit of a contagious disease. Med Hist. 1981;25:363–384. doi: 10.1017/s0025727300034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whitlock GC, Estes DM, Torres AG. Glanders: off to the races with Burkholderia mallei. FEMS Microbiol Lett. 2007;277:115–122. doi: 10.1111/j.1574-6968.2007.00949.x. [DOI] [PubMed] [Google Scholar]
- 24.Romero C, DeShazer D, Feldblyum T, Ravel J, Woods D, et al. Genome sequence alterations detected upon passage of Burkholderia mallei ATCC 23344 in culture and in mammalian hosts. BMC Genomics. 2006;7:228. doi: 10.1186/1471-2164-7-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schutzer SE, Schlater LR, Ronning CM, DeShazer D, Luft BJ, et al. Characterization of clinically-attenuated Burkholderia mallei by whole genome sequencing: candidate strain for exclusion from Select Agent lists. PLoS ONE. 2008;3:e2058. doi: 10.1371/journal.pone.0002058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeShazer D, Waag DM, Fritz DL, Woods DE. Identification of a Burkholderia mallei polysaccharide gene cluster by subtractive hybridization and demonstration that the encoded capsule is an essential virulence determinant. Microbial Pathogenesis. 2001;30:253–269. doi: 10.1006/mpat.2000.0430. [DOI] [PubMed] [Google Scholar]
- 27.Levinson G, Gutman GA. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- 28.Deitsch KW, Lukehart SA, Stringer JR. Common strategies for antigenic variation by bacterial, fungal and protozoan pathogens. Nat Rev Micro. 2009;7:493–503. doi: 10.1038/nrmicro2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malakooti J, Ely B, Matsumura P. Molecular characterization, nucleotide sequence, and expression of the fliO, fliP, fliQ, and fliR genes of Esherichia coli. J Bacteriol. 1994;176:189–197. doi: 10.1128/jb.176.1.189-197.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parkhill J, Wren BW, Thomson NR, Titball RW, Holden MTG, et al. Genome sequence of Yersinia pestis, the causative agent of plague. Nature. 2001;413:523–527. doi: 10.1038/35097083. [DOI] [PubMed] [Google Scholar]
- 31.Rodrigues F, Sarkar-Tyson M, Harding SV, Sim S-H, Chua H-H, et al. Global map of growth-regulated gene expression in Burkholderia pseudomallei, the causative agent of melioidosis. J Bacteriol. 2006;188:8178–8188. doi: 10.1128/JB.01006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ulrich RL, DeShazer D. Type III secretion: a virulence factor delivery system essential for the pathogenicity of Burkholderia mallei. Infect Immun. 2004;72:1150–1154. doi: 10.1128/IAI.72.2.1150-1154.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tuanyok A, Kim HS, Nierman WC, Yu Y, Dunbar J, et al. Genome-wide expression analysis of iron regulation in Burkholderia pseudomallei and Burkholderia mallei using DNA microarrays. FEMS Microbiol Lett. 2005;252:327–335. doi: 10.1016/j.femsle.2005.09.043. [DOI] [PubMed] [Google Scholar]
- 34.Moore RA, Reckseidler-Zenteno S, Kim H, Nierman B, Yu Y, et al. Contribution of gene loss to the pathogenic evolution of Burkholderia pseudomallei and Burkholderia mallei. Infect Immun. 2004;72:4172–4187. doi: 10.1128/IAI.72.7.4172-4187.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim H, Schell MA, Yu Y, Ulrich RL, Sarria SH, et al. Bacterial genome adaptation to niches: Divergence of the potential virulence genes in three Burkholderia species of different survival strategies. BMC Genomics. 2005;6:174. doi: 10.1186/1471-2164-6-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 37.Myers EW, Sutton GG, Delcher AL, Dew IM, Fasulo DP, et al. A whole-genome assembly of Drosophila. Science. 2000;287:2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- 38.Delcher AL, Phillippy A, Carlton J, Salzberg SL. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002;30:2478–2483. doi: 10.1093/nar/30.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salzberg SL, Delcher AL, Kasif S, White O. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 1998;26:544–548. doi: 10.1093/nar/26.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carver TJ, Rutherford KM, Berriman M, Rajandream M-A, Barrell BG, et al. ACT: the Artemis comparison tool. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 42.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucl Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roy CJ, Hale M, Hartings JM, Pitt L, Duniho S. Impact of inhalation exposure modality and particle size on the respiratory deposition of ricin in BALB/c mice. Inhal Toxicol. 2003;15:619–638. doi: 10.1080/08958370390205092. [DOI] [PubMed] [Google Scholar]
- 44.Moore RA, DeShazer D, Reckseidler S, Weissman A, Woods DE. Efflux-mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob Agents Chemother. 1999;43:465–470. doi: 10.1128/aac.43.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Graphical alignments of the genomes of B. mallei and B. pseudomallei. Genomes from strains of B. mallei and B. pseudomallei are aligned for close comparisons of the relative positions of IS elements and large deletions. Based on the genome of B. pseudomallei K96243 (displayed in the middle of the alignments), corresponding regions in B. mallei genomes (denoted by the upper green blocks) and those of the other B. pseudomallei genomes (denoted by the lower blue blocks) are displayed. Locations of ISBma1, ISBma2, and IS407A are denoted by red, yellow, and pink lines, respectively, in each strain.
(1.87 MB TIF)
The patterns of genomic rearrangements in B. mallei. A. Syntenic relationship among the B. mallei strains. Re-localization of BRUs in B. mallei strains (relative to B. pseudomallei strain K96243 as the reference) is shown with the comparative genomics display tool ACT (Wellcome Trust Sanger Institute) using blastn data. Blue and red connecting lines between the genomes indicate the same and opposite directions, respectively, of the corresponding BRUs relative to each other. B. Comparisons among the B. pseudomallei strains. Each strain was compared to the reference genome of strain K96243.
(2.42 MB TIF)
Comparison of B. mallei genomes with that of B. pseudomallei strain K96243.
(8.84 MB PDF)
Comparison of B. pseudomallei genomes with that of strain K96243.
(9.59 MB PDF)
Specifics of the IS elements IS407A, ISBma1, and ISBma2 in the B. mallei strains.
(1.56 MB PDF)
Disrupted genes that resulted from SSR-mediated frameshift mutations in B. mallei and B. pseudomallei.
(0.34 MB PDF)
Potential regulatory genes that survived the genomic reduction in B. mallei.
(0.18 MB PDF)
Comparison of the upstream regions of genes containing potential promoters.
(2.92 MB PDF)
Putative regulatory genes deleted from B. mallei as IS-bounded genomic fragments.
(0.04 MB PDF)