Abstract
An effective transcriptional response to redox stimuli is of particular importance for Mycobacterium tuberculosis, as it adapts to the environment of host alveoli and macrophages. The M. tuberculosis σ factor σL regulates the expression of genes involved in cell-wall and polyketide syntheses. σL interacts with the cytosolic anti-σ domain of a membrane-associated protein, RslA. Here we demonstrate that RslA binds Zn2+ and can sequester σL in a reducing environment. In response to an oxidative stimulus, proximal cysteines in the CXXC motif of RslA form a disulfide bond, releasing bound Zn2+. This results in a substantial rearrangement of the σL/RslA complex, leading to an 8-fold decrease in the affinity of RslA for σL. The crystal structure of the − 35-element recognition domain of σL, σ4L, bound to RslA reveals that RslA inactivates σL by sterically occluding promoter DNA and RNA polymerase binding sites. The crystal structure further reveals that the cysteine residues that coordinate Zn2+ in RslA are solvent exposed in the complex, thus providing a structural basis for the redox sensitivity of RslA. The biophysical parameters of σL/RslA interactions provide a template for understanding how variations in the rate of Zn2+ release and associated conformational changes could regulate the activity of a Zn2+-associated anti-σ factor.
Abbreviations: RNAP, RNA polymerase; ASD, anti-σ domain; ECF, extracytoplasmic function; Mtb, Mycobacterium tuberculosis; Ec, Escherichia coli; Sco, Streptomyces coelicolor; ZAS, zinc-associated anti-σ; Rsp, Rhodobacter sphaeroides; PDB, Protein Data Bank; PAR, 4-(2-pyridylazo)-resorcinol; SPR, surface plasmon resonance; LC-ESI-MS, liquid chromatography–electrospray ionization–mass spectrometry; MALDI-TOF, matrix-assisted laser desorption/ionization time-of-flight; wt, wild type; DLS, dynamic light scattering; TPEN, N,N,N′,N′-tetrakis(2-pyridylmethyl) ethylenediamine
Keywords: extracytoplasmic function σ factor, zinc binding, redox sensitivity, anti-σ factor
Introduction
The presence of multiple σ factors and the conditional association of these proteins with RNA polymerase (RNAP) are essential components of the prokaryotic transcriptional machinery that is sensitive to a diverse range of developmental and environmental stimuli. The activity of σ factors is often regulated by their association with an anti-σ factor.1,2 Despite poor sequence similarity, the structures of both σ factors, as well as the anti-σ domains (ASDs) of the anti-σ factors, are remarkably well conserved.3–6 σ factors belonging to the extracytoplasmic function (ECF) family often govern changes in transcription in response to environmental stimuli.7,8 Of the 13 σ factors in Mycobacterium tuberculosis (Mtb), 10 belong to the ECF family. A characteristic feature of this bacillus is the long period of dormancy in the host, which is also referred to as the latent phase of mycobacterial growth. It is therefore not surprising that several ECF σ factors are involved in mediating a transcriptional response to oxidative stimuli, in addition to the primary σ factors. The oxidative stress response that partially overlaps with the disulfide response mechanism is remarkably diverse in bacteria, involving chaperones such as the Escherichia coli (Ec) chaperone Hsp33 or receptor proteins such as the anti-σ factor RsrA from Streptomyces coelicolor (Sco).9–11 Since the characterization of Sco RsrA, several anti-σ factors have been noted to have a conserved HXXXCXXC motif, a finding that provided a basis for a class of proteins called the zinc-associated anti-σ (ZAS) factor. The differences between two well-characterized ZAS proteins, Sco RsrA and Rhodobacter sphaeroides (Rsp) ChrR, however suggested that the mechanism by which these ZAS proteins detect oxidative signals and release the bound σ factor is quite diverse. In the case of the Sco σR/RsrA system, Zn2+ binding and σR association are coupled events; Zn2+ release, in tandem with the formation of multiple disulfide bonds, was noted to be a critical feature of the RsrA sensor.12,13 Detection of singlet oxygen by Rsp ChrR, on the other hand, was shown to be localized to a cupin domain and not to the ASD that inactivates σE.3
A search for ZAS homologues in Mtb suggested that three anti-σ factors regulating the ECF σ factors σE, σH, and σL have the HXXXCXXC motif.11 σE and σH respond to oxidative stress in vivo.14,15 With in vitro transcription assays, the anti-σ factor of σL, RslA, was shown to inhibit σL-dependent transcription of the σB gene in a dose-dependent manner.16 It was further noted that many of the genes regulated by σL are involved in processes related to the cell envelope or secreted protein modifications.16,17 Here we describe the structural and biochemical features of Mtb RslA and its interactions with σL. We demonstrate that the ASD of RslA binds Zn2+ and responds to oxidative stress by forming a disulfide bond between the proximal cysteine residues of the CXXC motif. We note that Zn2+ release under oxidizing conditions is a rapid response, while the consequent structural changes in RslA are much slower. Of the two promoter recognition domains in σL, σ4L interacts with RslA with a higher affinity when compared to σ2L. The crystal structure shows that the orientation of domains in the σ4L/RslA complex is different from that in Rsp ChrR, the only other structurally characterized ZAS–σ factor complex. Unlike Rsp ChrR, no structural rearrangements were seen in σ4L upon RslA binding. The Zn2+ binding and biochemical features of RslA mutants suggest that surface accessibility of the conserved CXXC motif plays a more important role in the redox sensitivity of a ZAS protein than the XX dipeptide sequence between the cysteine residues. The structure of the σ4L/RslA complex thus provides a basis for understanding variations in the redox sensitivity of ZAS proteins.
Results
The crystal structure of the σ4L/RslA complex
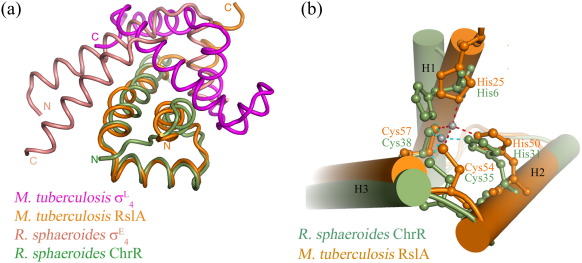
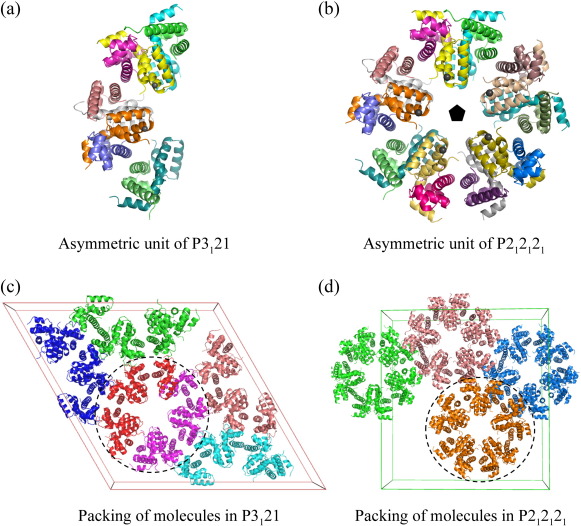
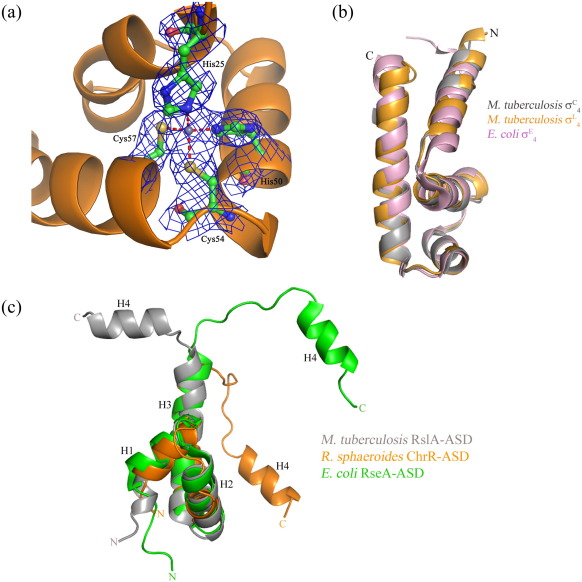
Crystals of the σL/RslA complex diffracted to 3.2 Å, had high mosaicity, and took several months to obtain. These crystals belonged to space group P3121 with unit cell parameters a = 167.46 Å, b = 167.46 Å, and c = 64.65 Å. Subsequently, we noted that these crystals contained only the σ4L domain and RslA; σ2L appeared to have proteolyzed in the course of crystallization. Crystals were readily obtained from an expression construct that had σ4L and RslA. These crystals diffracted to 2.4 Å resolution, and the structure of the σ4L/RslA complex was solved using single-wavelength anomalous dispersion, with data collected at the Zn2+ K-edge [Protein Data Bank (PDB) ID 3HUG]. Data collection and refinement statistics are summarized in Supplementary Table 1. There are 10 molecules of the σ4L/RslA complex in the asymmetric unit. In this arrangement, five heterotetramers of the σ4L/RslA complex pack with a 5-fold symmetry to form a ring-like structure, with the zinc binding region of RslA facing the center of the ring. This arrangement results in a large pore with a diameter of about ∼ 25 Å. A pictorial representation of the packing of the σ4L/RslA complex molecules in the two crystal forms is shown in Supplementary Fig. 1. The 1:1 stoichiometry for the σ4L/RslA complex seen in the crystal structure is consistent with biochemical data (Fig. 1a). About 23 residues in the N-terminus and 22 residues in the C-terminus were disordered in the crystal structure of RslA. σ4L forms a tight complex with RslA involving a buried surface area of ca 1332 Å2. The Zn2+ cofactor in RslA is coordinated by His25, His50, Cys54, and Cys57 (Fig. 1b). Thus, all the conserved residues in the HXXXCXXC motif are involved in Zn2+ binding.
Fig. 1.
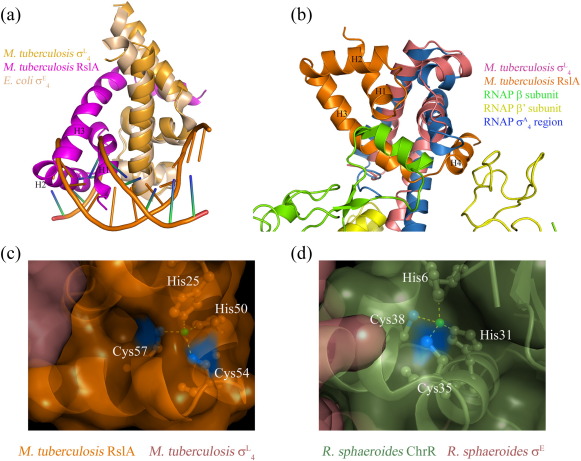
The crystal structure of the − 35-element recognition domain of σL, σ4L, bound to RslA. (a) The interaction of the ASD of Mtb RslA with σ4L when compared with the Rsp σ4E/ChrR complex.3 While the ASDs of Mtb RslA (orange) and Rsp ChrR (green) superpose well, those of Mtb σ4L (magenta) and Rsp σ4E (salmon) do not. (b) Zn2+ coordination in Mtb RslA and Zn2+ coordination in Rsp ChrR are identical.
The structure of the σ4L domain superimposes well with those of Mtb σ4C (PDB ID 2O8X; RMSD of 0.74 Å over 60 Cα)6 and Ec σ4E (PDB ID 1OR7; RMSD of 1.64 Å over 68 Cα)4 (Supplementary Fig. 2). Thus, RslA binding does not lead to a structural rearrangement of σ4L, unlike the case of the Rsp σE/ChrR complex (PDB ID 2Z2S) or the Ec σ4E/AsiA complex (PDB ID 1TLH).3,18 Superposition of the structure of RslA on that of the Rsp σE/ChrR complex shows that only three helices of the four-helical domain of RslA overlap on Rsp ChrR. Helix H4 adopts a different orientation (Supplementary Fig. 2c). This helix (H4) occludes the RNAP β′ coiled-coil binding surface of a σ factor.3,19 The orientation of the σ4 domains in the σ4L/RslA complex is different from that seen in the Rsp σE/ChrR complex (Fig. 1). A possible inhibitory mechanism of RslA can be inferred from the superposition of the σ4L/RslA structure on that of the σ4E/− 35 promoter DNA complex (PDB ID 2H27).20 This superposition revealed several steric clashes with the DNA. Modeling of the σ4L/RslA complex on RNAP (PDB ID 1IW7) also suggested steric clashes of RslA with the β flap–tip–helix of RNAP21,22 (Supplementary Fig. 3a and b). RslA is thus likely to inactivate σL by occluding both the promoter and the RNAP binding regions of σ4L without any structural rearrangements.
Zn2+ release precedes conformational changes in RslA with an oxidative stimulus
To evaluate the role of bound Zn2+ in RslA, we characterized the kinetics of Zn2+ release and conformational changes in RslA with an oxidative stimulus. Several expression constructs were examined to obtain a σL/RslA complex that could be purified and concentrated without aggregation or precipitation. A list of expression constructs examined in this study is compiled in Supplementary Table 2. All the biophysical and biochemical studies reported here were performed on full-length σL (residues 1–177) and on the ASD of RslA (residues 1–108).
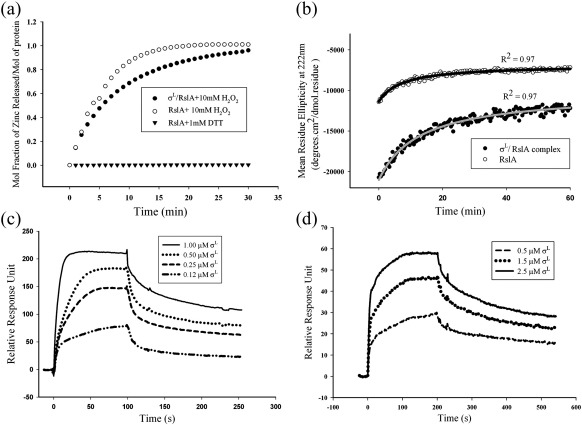
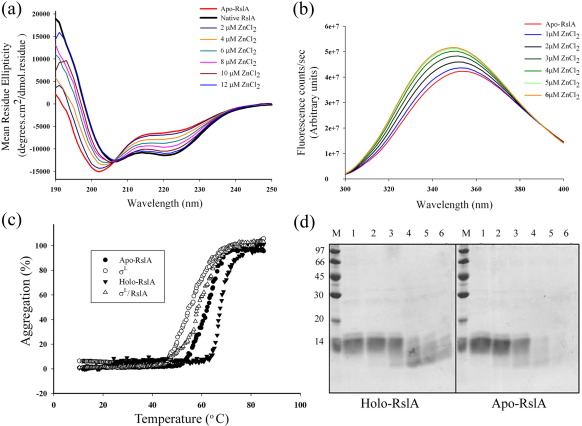
A chromophoric Zn2+-specific chelator, 4-(2-pyridylazo)-resorcinol (PAR), was used to monitor and quantify the release of Zn2+ from RslA and the σL/RslA complex. This assay was performed as described in Materials and Methods (procedure adapted from Bae et al.12). The kinetics of Zn2+ release was monitored after the addition of 10 mM H2O2. Bound Zn2+ is released with a t1/2 of ∼ 3.5 min. This is substantially slower than Sco RsrA, for which the corresponding t1/2 is ∼ 1.8 min.12 The presence of σL also substantially alters the rate of Zn2+ release. The release of bound Zn2+ in the σL/RslA complex (t1/2 ∼ 7 min) is far slower than that of RslA alone (t1/2 ∼ 3.5 min) (Fig. 2a). The rate of Zn2+ release does not significantly differ between the σ4L/RslA complex and the σL/RslA complex. RslA shows substantial changes in its secondary structural composition upon Zn2+ binding. The conformational differences between apo-RslA and holo-RslA were monitored by far-UV circular dichroism (CD) spectroscopy (Fig. 3a). CD spectra of a titration with Zn2+ suggested an increase in the α-helical content of RslA with increasing Zn2+ concentration. When the Zn2+ concentration reached stoichiometric concentrations, no further change in the spectrum of RslA was observed (Fig. 3a). Monitoring the release of Zn2+ by CD measurements (mean residue ellipticity at 222 nm) reveals that the conformational change in the protein occurs at a rate substantially slower than that of Zn2+ release. The t1/2 for the conformational change upon Zn2+ removal is ∼ 7 min for RslA alone, while that for the σL/RslA complex is ∼ 12 min (Fig. 2b). The differences between the rates of Zn2+ release and conformational change thus suggest that while Zn2+ release is a rapid response, structural changes provide a much slower second step of the redox-sensing mechanism in RslA. Reduced and oxidized apo-RslA did not show significant structural changes in CD experiments. It is worth noting in this context that while structural rearrangements are more due to Zn2+ binding in RslA, these changes in Sco RsrA (the other redox-sensitive ZAS protein) are mediated by disulfide bond formation upon oxidation.23
Fig. 2.
RslA binds Zn2+ under reducing conditions. (a) Zn2+ release monitored using the chromophore PAR. (b) Time-course measurements of structural changes in RslA and in the σL/RslA complex monitored by CD. (c) SPR measurements of σL/RslA interactions. RslA forms a complex with σL with a Kd of ca 20 nM under reducing conditions. (d) Interactions of apo-RslA with σL. An 8-fold decrease in binding, corresponding to a Kd of ∼ 150 nM, was observed.
Fig. 3.
Zinc removal with an oxidative stimulus induces conformational changes in RslA and in the σL/RslA complex. (a) CD spectra showing changes in the secondary structure of RslA with increasing concentration of Zn2+ under reducing conditions. (b) Zinc binding monitored by intrinsic tryptophan fluorescence. Zinc binding stabilizes helix H1 of RslA. (c) Zinc binding confers stability to RslA and the σL/RslA complex. Temperature-induced denaturation of σL, RslA, and the σL/RslA complex. As seen by thermal melting profiles, Zn2+ binding confers stability to RslA and the σL/RslA complex. Temperature was scanned between 10 °C and 90 °C, with a dT/dt of 1 °C/min. (d) Zn binding does not confer significant resistance to RslA proteolysis. In this experiment, holo-RslA and apo-RslA were incubated with trypsin (1:2000) for various time points and analyzed on 12% SDS-PAGE. Lane 1, 0 min; lane 2, 5 min; lane 3, 10 min; lane 4, 20 min; lane 5, 30 min; lane 6, 60 min.
The partial unfolding of RslA under oxidizing conditions is also consistent with fluorescent spectroscopy measurements (Fig. 3b). RslA has one Trp residue located in helix H1. The fluorescence spectrum of apo-RslA has an emission maximum at ∼ 354 nm that shifts to ∼ 349 nm upon the addition of Zn2+ under reducing conditions. This change in the spectrum suggests that helix H1 undergoes a significant structural rearrangement upon Zn2+ binding. The preference for Zn2+ as a metal cofactor was noted by changes in the CD spectra when RslA was incubated with other metal ions. Among the other metal ions that were analyzed (viz. Mg2+, Mn2+, Cu2+, Fe2+, Ni2+, and Co2+), none, with the exception of Co2+, could replicate the CD spectrum of Zn2+-bound RslA (data not shown). Even this was only possible when Co2+ was present at a 10-fold molar excess concentration as compared to Zn2+.
Zn2+ removal results in a reduced affinity of RslA for σL
An observation in support of the hypothesis that bound Zn2+ in RslA plays a functional role is that Zn2+ binding substantially influences the ability of RslA to bind σL. The interaction between σL and RslA was examined by surface plasmon resonance (SPR) measurements. An analysis of the SPR data suggests that the σL/RslA complex has a dissociation constant Kd of ca 20 nM (Fig. 2c). This affinity is comparable to that reported for the Sco σR/RsrA complex (ca 10 nM).11 Zn2+-free RslA binds σL with an ∼ 8-fold reduced affinity (Kd ∼ 150 nM), again comparable to the ca 9-fold reduction observed under similar conditions for Sco σR/RsrA (Fig. 2d). The SPR experiments carried out in the presence or in the absence of DTT did not substantially alter the affinity of apo-RslA for σL (data not shown), suggesting that loss of Zn2+ alone (and not disulfide bond formation) plays a role in the interaction of RslA with σL.
Zn2+ binding enhances the structural stability of RslA
Zn2+ binding also confers thermostability to RslA. RslA is a stable protein with a Tagg of 70 °C. Zn2+ binding appears to confer thermostability to this protein as apo-RslA has a Tagg of 60 °C. Binding of RslA also enhances the stability of the σL/RslA complex. Free σL has a Tagg of 56 °C, whereas upon RslA binding, the Tagg of the σL/RslA complex increases to 61 °C (Fig. 3c). Zn2+ binding, however, does not confer significant resistance to RslA proteolysis, as both holo-RslA and apo-RslA show similar profiles in a controlled proteolysis experiment using trypsin (Fig. 3d). This finding is different from that of Sco RsrA, where Zn2+ binding provides significant protection to trypsin digestion.12
Proximal cysteines of the HXXXCXXC motif form a disulfide bond under oxidizing conditions
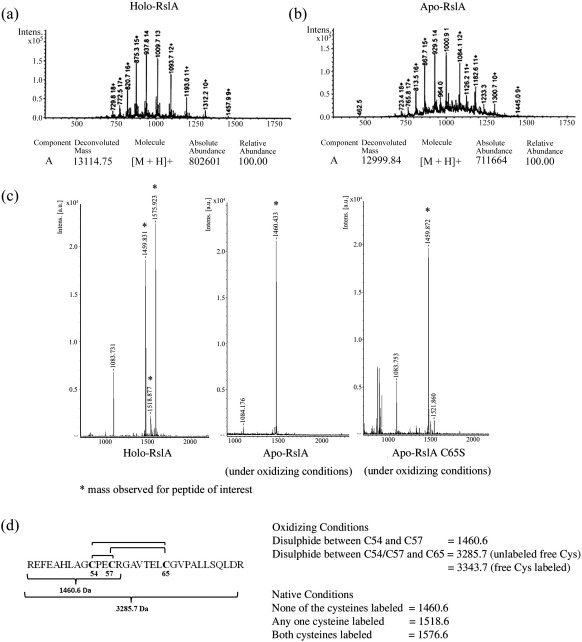
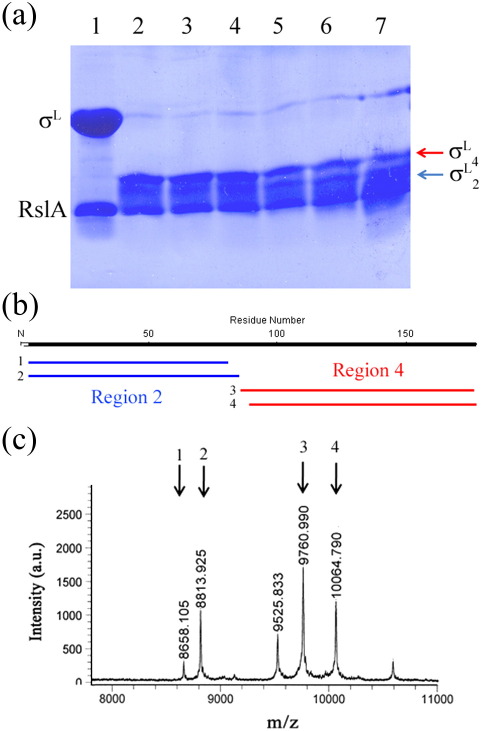
In the crystal structure of the σ4L/RslA complex, Zn2+ is coordinated by His25, His50, Cys54, and Cys57. The last three residues are from the conserved HXXXCXXC motif. This coordination geometry is similar to that reported for Sco RsrA and seen in the crystal structure of Rsp ChrR (Fig. 1b). Based on iodoacetamide labeling of cysteines in holo-RslA and apo-RslA and on subsequent analysis by liquid chromatography–electrospray ionization–mass spectrometry (LC-ESI-MS), we note that oxidized RslA has one disulfide bond, whereas there are three free cysteines in holo-RslA. The expected mass for RslA with all three cysteines modified with iodoacetamide is 13,116.5 Da (Supplementary Fig. 4a). The experimentally determined mass for iodoacetamide-treated holo-RslA is 13,114.75 Da, which is within 2 Da of the calculated mass. The oxidized disulfide-bonded RslA with one modified Cys residue has a theoretical mass of 13,000.39 Da that also compares well with the experimentally determined mass of 12,999.84 Da (Supplementary Fig. 4b). In an effort to determine the identity of the cysteines that form the disulfide bond, cysteines in apo-RslA and holo-RslA were modified using iodoacetamide and subsequently proteolyzed using trypsin. Analysis of matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) spectra corresponding to these samples revealed that Cys54 and Cys57 form a disulfide in oxidized apo-RslA (Supplementary Fig. 4c). That the distal Cys does not participate in Zn2+ binding was verified by characterizing the C65S variant of RslA. The C65S mutant has biochemical properties identical to those of wild-type (wt) RslA. The C54S mutant of RslA, on the other hand, does not bind Zn2+ and has a similar far-UV CD spectrum as Zn2+-free RslA. The spectroscopic features of the C54S mutant thus reaffirm the hypothesis that structural rearrangements are a consequence of Zn2+ removal and are not due to the disulfide bond. RslA thus differs from Sco RsrA, where the disulfide involves the distal cysteines (Cys11 and C44) rather than those in the HXXXCXXC motif, and formation of multiple disulfide bonds results in major structural rearrangements.13 Indeed, a single disulfide bond in Sco RsrA was shown to abolish binding to σR.23
Significance of the XX dipeptide in the CXXC motif
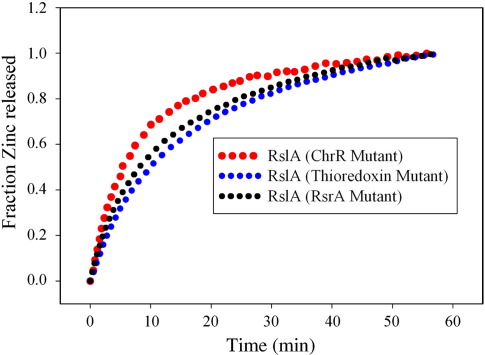
ZAS proteins show substantial variations in their redox sensitivity. The Zn2+ cofactor in the ASD of Rsp ChrR was suggested to play a more structural role rather than to serve as a sensor for singlet oxygen or other oxidative stimuli. To evaluate if these differences could be explained by the sequence of the XX dipeptide in the CXXC motif, we examined mutants of RslA with different dipeptide sequences. In this experiment, the CXXC motif of RslA (CPEC) was mutated to CDEC, CSPC, and CGPC, corresponding to those of Rsp ChrR, Sco RsrA, and Ec thioredoxin, respectively. These mutants of RslA were purified using a procedure similar to that used in wt protein, and the kinetics of Zn2+ release were examined. We note that all dipeptide mutants release Zn2+ in the presence of H2O2. The t1/2 values for Zn2+ release vary, however, ranging from ∼ 6.5 min (Rsp ChrR variant), to ∼ 13 min (Ec thioredoxin), to ∼ 15 min (Sco RsrA), all higher than that observed for wt-RslA (∼ 3.5 min) (Fig. 3; Supplementary Fig. 5).
Interactions of the σL domains with RslA
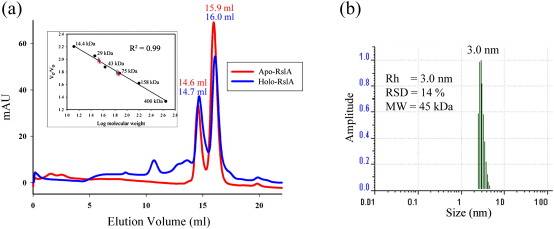
A limited proteolysis experiment with the σL/RslA complex reveals that σL is cleaved into two stable domains (σ2L and σ4L) within a minute of initiating the proteolysis reaction (Supplementary Fig. 6). This is consistent with observations from crystallization trials on the intact σL/RslA complex. However, we note that RslA is protected from proteolysis upon forming a complex with σL. RslA alone, on the other hand, is very susceptible to proteolysis. In an effort to identify the relative contributions of the two domains of σL to complex formation, we monitored the interactions of the two domains with RslA by SPR. To our surprise, σ4L alone did not interact with RslA. Homooligomerization of this domain appears to be the likely cause for this observation (size-exclusion chromatography; data not shown). σ2L, on the other hand, binds RslA, albeit with an ∼ 100-fold reduction in its affinity as compared to intact σL (Kd∼ 2 μM). However, the results of protein–protein interaction experiments using coexpression and copurification were more conclusive. Of the two constructs in the pETDuet-1 vector having σ2L and σ4L in multiple cloning site 1 (Supplementary Table 2, constructs 5 and 6) with a poly-histidine tag at the N-terminus and RslA in multiple cloning site 2, RslA copurified with σ4L, whereas the σ2L/RslA complex could not be obtained. We also note that both apo-RslA and holo-RslA (Supplementary Table 2, construct 7) self-associate into dimeric and tetrameric oligomers in solution. This feature is seen in analytical gel-filtration experiments and is consistent with data from dynamic light scattering (DLS) measurements (Supplementary Fig. 7). A small shift in elution profile was observed in analytical gel-filtration experiments with the apo and holo forms, suggesting that Zn2+-bound RslA adopts a more compact conformation. The self-association of RslA in solution appears similar to the phage-λ-encoded protein AsiA that is also predominantly a dimer in solution. As in the case of σL/RslA and the σ4L/RslA complex, AsiA forms a 1:1 complex with the Ec primary σ factor σ70.24
Discussion
ECF σ factors respond to a variety of environmental signals that include periplasmic stress in Gram-negative bacteria, oxidative or disulfide stress, changes in light intensity, or resistance to cobalt or nickel.2,25,26 The receptor for these diverse signals is often located in the anti-σ factor that responds by releasing the σ factor upon specific environmental cues. Mtb RslA is a ZAS protein with a cytosolic domain (ASD) that interacts with the ECF σ factor σL and a membrane-spanning C-terminal domain. Of the two promoter recognition domains σ2 and σ4, σ4 is often a target for various regulators. Examples of this feature include λcI, Rhas, CRP, MotA, and anti-σ factors.27 An analysis of the structures of some of these σ factor/anti-σ complexes, notably Ec RseA, Rsp ChrR, Salmonella typhimurium FlgM, and Ec Rsd, suggests that these proteins inactivate their cognate σ factors by occluding both the RNAP binding sites and the promoter binding regions.4,28–30 More importantly, Ec AsiA and Rsp ChrR also bring about structural rearrangements in σ4 that affect its promoter binding ability.3,18 In the Ec FecI/FecR σ/anti-σ factor pair, σ4 plays a dominant role in interactions, as in the case of Ec AsiA and Rsd, which interact with σ4A with similar affinities.31,32 The distinct mechanisms of interactions in these two anti-σ factors were reported to have functional implications, with AsiA being a more potent inhibitor of transcription.32 The Mtb σL/RslA complex differs from both the Ec σE/RseA complex and the Rsp σE/ChrR complex, where the σ2 and σ4 domains contribute similarly to the σ/anti-σ complex. Ec RseA, for example, was shown to pull down both domains of Ec σE.4 Instead, the σL/RslA interactions appear closer to that seen in the Sco σR/RsrA or Ec FecI/FecR systems, where one of the promoter binding domains interacts with the anti-σ factor with a significantly higher affinity than the other.31,33
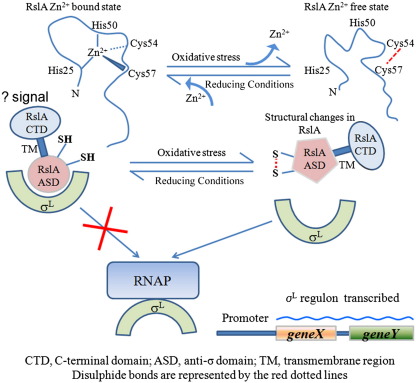
Mtb RslA and Rsp ChrR are the only ZAS proteins for which structures have been determined. Of these, Rsp ChrR has two domains: the ASD and a double-stranded β-helix (cupin) domain. In this case, ASD is involved in binding σE, while the cupin domain serves as the sensor of singlet oxygen levels.3 The other characterized ZAS proteins to date include Mtb RshA and Sco RsrA, both of which are sensitive to redox stimuli.23,34 RslA responds to oxidative stimuli with the release of the Zn2+ cofactor, followed by the formation of a disulfide link between proximal cysteines of the HXXXCXXC motif (Fig. 4). While this disulfide link differs from the other characterized ZAS protein Sco RsrA, it is closer to redox sensor proteins such as Spx, ResA, and Hsp33.35–37 Although there appeared to be some discrepancy in the reports on Zn2+ coordination and disulfide connectivity in Sco RsrA, it appears likely that all the three conserved residues in the HXXXCXXC motif are involved in Zn2+ binding.12,13,23 The Zn2+ coordination geometry is different from Sco RsrA in Rsp ChrR, as the distal Cys11 residue is replaced by a histidine.3 Rsp ChrR does not respond to oxidative stress; instead, it acts as a sensor for singlet oxygen species, with this role performed by a different domain.3 Two probable reasons were cited for the insensitivity of ChrR to oxidative stress: (i) surface inaccessibility of Cys, or (ii) the chemical nature of the Zn2+ ligands. Both Cys residues in the conserved CXXC motif of Mtb RslA are solvent exposed, unlike Rsp ChrR, where only Cys35 is surface accessible while Cys38 is not (Supplementary Fig. 3c and d). This observation is analogous to the Zn2+ binding redox sensor of Hsp33 where three of four Cys residues that coordinate Zn2+ are accessible (PDB ID 1VQ0). The sequence of the XX dipeptide in the CXXC motif has been shown to be crucial in controlling the redox properties of the protein in which it is found.36,38,39 In the present analysis, the XX dipeptide in the CXXC motif of RslA was mutated to the corresponding sequences of the other characterized ZAS proteins Sco RsrA and Rsp ChrR, and of a thioldisulfide oxidoreductase (Ec thioredoxin). In this study, we examined the rate of release of bound Zn2+ with an oxidative stimulus. All the RslA dipeptide variants showed an increase in t1/2 for Zn2+ release when compared to wt-RslA. It thus appears likely that the redox sensitivity in ZAS proteins involves other sequence structure features apart from the dipeptide sequence in the CXXC motif.
Fig. 4.
Schematic of the mechanism by which RslA regulates σL. Please refer to the text of the article for details.
In summary, the crystal structure of the σ4L/RslA complex reveals that RslA inactivates σL by sterically occluding both the promoter and the RNAP binding regions on σ4L. The structure also provides a rationale for the biochemical characteristics of this ZAS protein. These results thus provide a template for understanding the mechanism by which RslA ASD regulates σL, while providing a model for further studies to understand the sequence structure features of ZAS proteins that contribute to redox sensitivity.
Materials and Methods
Cloning, expression, and purification of different σL and anti-σL constructs
The details of the expression constructs used in this study are compiled in Supplementary Table 2. Site-directed mutagenesis was carried out, using a single primer-based PCR method, to obtain point variants of RslA.40 The σ4L/RslA and σL/RslA complexes were purified with a coexpression and copurification strategy using pETDuet-1 (Novagen, Inc.) expression vector. All protein samples were purified using the same protocol. Briefly, after the plasmid had been transformed into Rosetta(DE3)pLysS cells (Novagen, Inc.), the cells were grown in Luria broth with antibiotics (100 μg/ml ampicillin and 30 μg/ml chloramphenicol) to an A600 of 0.5–0.6. The cells were induced with 0.5 mM IPTG. Subsequently, growth temperature was lowered to 290 K, and the cells were grown for 12–18 h before they were spun down. Cells were lyzed by sonication in lysis buffer [20 mM Tris (pH 7.5) and 250 mM NaCl]. Cell-free lysate was incubated with Co-NTA beads and eluted by a gradient of elution buffer [20 mM Tris–HCl (pH 7.5), 250 mM NaCl, and 200 mM imidazole]. Recombinant proteins were further purified by size-exclusion chromatography using a Superdex S-200 column (GE Healthcare) after the affinity chromatography step. For analytical gel-filtration experiments, ∼ 0.2-mg to 0.5-mg protein samples were passed through a Superdex S-200 10/300 GL column equilibrated in 20 mM Tris–HCl (pH 7.5) and 200 mM NaCl at 6 °C. The proteins were then concentrated using a membrane-based centrifugal ultrafiltration system (Amicon-Ultra). The molecular weights of the recombinant proteins were verified by MS on both MALDI-TOF and LC-ESI-MS (Bruker Daltonics, Inc.).
Crystallization and structure determination of σ4L/RslA
Initial crystallization conditions were obtained by hanging-drop vapor diffusion in 24-well plates using a sparse matrix screen (Hampton Research) at room temperature by mixing 2 μl of protein solution with 2 μl of well solution and equilibrating against 400 μl of well solution. Diffraction-quality crystals for the σ4L/RslA complex were obtained by combining 2 μl of protein solution (∼ 10 mg/ml) and 2 μl of well solution [0.1 M 2,2'-(Propane-1,3-diyl iimino)bis[2-(hydroxymethyl)propane-1,3-diol] propane (pH 7.0), 0.8–1.2 M ammonium sulfate, and 0.8–5% polyethylene glycol 8000] at room temperature. Crystals appeared within ∼ 2 days of tray setup and grew to maximum dimensions in 1–2 weeks. The crystals were flash-frozen in a cryoprotectant solution containing 15% glycerol. Diffraction data were collected at wavelengths of 1.282 Å and 0.9762 Å at beamline BM14 of the European Synchrotron Radiation Facility. The data were processed using iMOSFLM41 and scaled using SCALA.42 Zn2+ substructure was located using PHENIX.43 Ten Zn2+ sites were located in the asymmetric unit, with each site corresponding to one molecule of the complex. Subsequent refinement and manual model building were performed using REFMAC44 and Coot.45 TLS refinement was performed using REFMAC.44 The final model contains 1661 residues, 10 Zn2+, 10 SO42−, and 269 water molecules. Analysis of the structure showed 97.8% of the residues in the most favored regions of the Ramachandran plot and 2.2% of the residues in additionally allowed regions (no residues in generously allowed or disallowed regions). Crystals of the σL/RslA complex (protein concentration, ∼ 7 mg/ml) were obtained after a year of setting up the hanging-drop experiment in 0.5 M succinic acid (pH 7.0) and 0.1 M 2,2'-(Propane-1,3-diyl iimino)bis[2-(hydroxymethyl)propane-1,3-diol] (pH 7.0). Upon SDS-PAGE and LC-ESI-MS analysis, we could deduce that σL was proteolyzed with the removal of σ2L. One of these crystals diffracted to 3.2 Å resolution at the home source, and the structure was solved by molecular replacement using the σ4L/RslA complex as search model in PHASER.46 No electron density was observed for σ2L domain in this structure.
Preparation of apo-RslA and zinc-saturated RslA
Apo-RslA was prepared by oxidizing the sample with 1 mM H2O2 in the presence of 5 mM N,N,N′,N′-tetrakis(2-pyridylmethyl) ethylenediamine (TPEN) (a Zn2+ chelator). Excess TPEN and H2O2 were subsequently removed by gel-filtration chromatography. To prepare Zn2+-saturated RslA, we incubated purified RslA with ZnCl2 under reducing conditions overnight at 4 °C. The unbound Zn2+ was removed by several washes of buffer using a centrifugal ultrafiltration system (Amicon-Ultra).
Fluorescence spectroscopy
All fluorescence studies were performed on a JOBIN YVON FlouroMax-3 fluorimeter at room temperature. Fluorescence excitation was set to 280 nm, and emission spectra were recorded from 300 nm to 400 nm at a bandwidth of 1 nm. The excitation and emission slit widths were set to 3 mm and 5 mm, respectively. Each recorded spectrum represents an average of five scans. The spectra were obtained at a protein concentration of 5–10 μM in 5 mM Tris–HCl (pH 7.5) in the presence of 1 mM DTT.
CD spectroscopy
CD experiments were carried out on a JASCO-J715 instrument. All spectra were recorded in 5 mM Tris–HCl buffer adjusted to pH 7.5, with 1 mM DTT, using a 1-mm pathlength cuvette. Protein concentrations were kept in the range of 5–15 μM. Each reported spectrum is an average of five scans.
PAR assay for zinc measurements
The metal chelator PAR, when complexed with free Zn2+, absorbs light intensively at 500 nm.12 The time course of Zn2+ release under oxidizing and reducing conditions was measured. Briefly, Zn2+-saturated samples were prepared as described earlier. For the assay, protein samples were diluted to 5–10 μM in a buffer containing 0.1 mM PAR. All buffers were purged with N2 gas. Nonspecifically bound Zn2+ was trapped by PAR in the absence of oxidation, as monitored by a slight increase in absorbance at 500 nm. When the absorbance had stabilized, 10 mM H2O2 was added, and A500 was monitored every 30 s for 25–50 min at 25 °C.
Limited proteolysis experiments
Limited proteolysis experiments were carried out at room temperature with 1:2000 protease/protein concentration. The samples were incubated for various time points. For MALDI-TOF experiments, reactions were stopped by adding 1% trifluoroacetic acid (final concentration). For SDS-PAGE, analysis reactions were stopped by adding the loading dye solution and by boiling the samples for 5 min.
SPR measurements
The kinetics of interaction between RslA and σL were examined using SPR (BIACORE 2000; GE Healthcare). RslA was immobilized on a CM5 chip (BIACORE; GE Healthcare) at a surface density of 12 ng/mm.2 σL, σ2L, and σ4L at varying concentrations were used as analytes. The first panel of the CM5 chip served as control. Interaction studies were carried out in 20 mM Tris–HCl buffer (pH 7.8), 200 mM NaCl, and 50 mM imidazole. For experiments with apo-RslA, a buffer containing 10 mM H2O2 and 2 mM TPEN was passed on the chip to remove bound Zn2+. A reducing environment was maintained with 1 mM DTT for interaction studies with holo-RslA, although no changes were seen in SPR chromatograms without DTT. For interaction studies with apo-RslA, a buffer without DTT was used. Binding constants were estimated using the BIA-evaluation software (BIACORE; GE Healthcare).
LC-ESI-MS and MALDI analysis of RslA
The cysteines in apo-RslA and holo-RslA were modified using iodoacetamide, which provides a mass difference of 58 Da for each modified Cys residue. Both apo-RslA and holo-RslA samples were incubated with an ∼ 50-fold molar excess of iodoacetamide in 50 mM ammonium bicarbonate (pH 8.0) at 37 °C for 45 min. LC-ESI-MS (Bruker Daltonics, Inc.) was carried out using a C8 analytical column. The differences in mass were within the error range of ± 2Da. For MALDI-TOF MS experiments (Bruker Daltonics, Inc.), the samples were proteolyzed using a 1:2000 trypsin/protein ratio and subsequently desalted using a 1-ml Superdex G-25 column.
Thermal denaturation experiments
Thermal denaturation experiments were carried out on a UV–visible spectrophotometer (JASCO V-530) at a fixed wavelength of 400 nm in a 10-mm pathlength quartz cell. The protein concentrations used varied from 5 μM to 10 μM in 250 mM NaCl and 20 mM sodium phosphate buffer (pH 7.5). The temperature was varied from 10 °C to 90 °C at a rate of 1 °C/min.
Analytical size-exclusion chromatography
For analytical gel-filtration experiments, ∼ 0.2-mg to 3-mg protein samples were passed through a Superdex S-200 10/300 GL column equilibrated in 20 mM Tris–HCl (pH 7.5) and 200 mM NaCl at 6 °C. The flow rate was maintained at 0.3 ml/min.
DLS experiments
DLS experiments were performed using Viscotek 802 DLS in a quartz cuvette (volume, 12 μl) at 22 °C. Protein concentrations were kept in the range of about ∼ 1 mg/ml.
Accession number
Coordinates and structure factors have been deposited in the PDB with PDB ID 3HUG.
Acknowledgements
We thank Mrs. Srilatha for help with SPR experiments, Dr. Hassan Belrhali for help with data collection, and Mrs. Neema Kulkarni for help during the initial phase of the project. We gratefully acknowledge the Department of Science and Technology and the Department of Biotechnology, Government of India, for funding the X-ray facility at the Molecular Biophysics Unit; and the Department of Biotechnology for beam time at station BM14 of the European Synchrotron Radiation Facility. B.G. is an International Senior Research Fellow of the Wellcome Trust, UK.
Edited by R. Huber
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jmb.2010.02.026
Appendix A. Supplementary data
Supplementary Fig. 1.
(a and b) Arrangement of σ4L/RslA complex molecules in an asymmetric unit with P3121 and P212121 crystal forms. All chains are colored differently. Zinc atoms are shown as gray spheres. (c and d) Packing of the σ4L/RslA complex molecules in two different crystal forms. Five molecules of the σ4L/RslA complex in the asymmetric unit of the P3121 crystal form, along with 2-fold crystallographic symmetry, result in a similar packing arrangement as seen in the P212121 crystal form with 10 molecules of the σ4L/RslA complex molecules in an asymmetric unit (dotted circles).
Supplementary Fig. 2.
(a) An (mFo − DFc) electron density map showing the coordination of Zn2+ in the structure of the σ4L/RslA complex. (b) Binding of RslA does not induce structural changes in σ4L. A structural superposition of the crystal structures of region 4 corresponding to ECF σ domains is shown here. (c) Helix H4 of ASDs adopts different conformations, while the other three helices superpose well.
Supplementary Fig. 3.
(a) RslA sterically occludes the promoter recognition region of σ4L. The σ4L/RslA complex was superposed on the crystal structure of the σ4E/− 35 promoter DNA complex (PDB ID 2H27).20 (b) Docking of the σ4L/RslA complex onto Thermus thermophilus holo-RNAP (PDB ID 1IW7).22 Docking of RslA on polymerase reveals several steric clashes of RslA with a helical stretch of the β-subunit. (c) Surface representation of the region containing bound Zn2+ in the ASDs of RslA and ChrR. The sulfur atoms of the cysteine residues (Zn2+ binding) are shown in blue. Both cysteines in the CXXC motif in Mtb RslA are accessible to the solvent. The solvent-accessible cysteines in Mtb RslA provide a structural rationale for the redox sensitivity of this ASD. (d) A surface representation of Rsp ChrR in a similar orientation as Mtb RslA in (c). The Cys38 of the CXXC motif is occluded by Arg41 (Rsp ChrR numbering).
Supplementary Fig. 4.
Oxidation involves the formation of a disulfide bond in RslA. (a) LC-ESI-MS spectrum of holo-RslA. (b) LC-ESI-MS spectrum of apo-RslA. (c) Mass spectra of peptides containing the zinc-coordinating residues of RslA. RslA samples were subjected to iodoacetamide labeling, followed by trypsin digestion, to identify the cysteines involved in the disulfide link under oxidizing conditions. (d) Theoretical trypsin digestion profile and probable disulfide connectivity in the region of RslA with three cysteine residues. A list of masses expected for various peptides under oxidizing and native conditions after iodoacetamide treatment is given at the right-hand side of (d).
Supplementary Fig. 5.
Zn2+ release kinetics for the XX dipeptide mutants of RslA.
Supplementary Fig. 6.
(a) Limited proteolysis of the σL/RslA complex. σL binding to RslA confers protection from proteolysis. The complex was incubated with trypsin (1:2000) for various time points and analyzed on 12% SDS-PAGE. Lane 1, 0 min; lane 2, 5 min; lane 3, 10 min; lane 4, 20 min; lane 5, 30 min; lane 6, 60 min. (b) σ2L and σ4L were mapped onto the σL sequence. These data suggest that σL binding makes the ASD of RslA resistant to proteolysis. It therefore appears likely that proteolytic removal of the ASD may not be a mechanism to release free σL from the σL/RslA complex. (c) Representative MALDI-TOF spectrum of the proteolyzed sample. MALDI-TOF peaks marked in the spectrum correspond to the σ2L or σ4L domains of σL (top).
Supplementary Fig. 7.
(a) Analytical gel-filtration profile of holo-RslA and apo-RslA suggests that RslA exists as dimer and tetramer in solution. Inset: The calibration curve obtained using standard molecular weight markers. Red and blue stars in the inset correspond to the observed molecular weights of apo-RslA and holo-RslA. (b) DLS experiments suggest a hydrodynamic radius (Rh) of 3.0 nm with a high relative standard deviation of ~ 14%. The dimeric and tetrameric oligomers of RslA could not be resolved by DLS experiments.
Supplementary Fig. 8.
Sequence comparison of representative ZAS proteins. Sco RsrA, Streptomyces coelicolor RsrA; Mtb RshA, Mycobacterium tuberculosis RshA; bstylac,Bacillus subtilisYlac σ factor; MSM, anti-σ factor from Mycobacterium smegmatis str. MC;2Rsp ChrR, Rhodobacter sphaeroides ChrR; Mtb RV1222, Mycobacterium tuberculosis anti-σE, Mycobacterium abscessus hypothetical protein MAB_2164; Mtb RslA, Mycobacterium tuberculosis RslA. The asterisk on the top of sequence alignment represents probable residues involved in zinc binding. The residues shown against a red background are involved in Zn2+ coordination (either cysteine or histidine). The conserved zinc binding motif residues are shaded yellow. The sequences of XX dipeptide in conserved CXXC motif in anti-σ factors are shown in pink. The secondary structural elements shown are based on the crystal structure of RslA (PDB ID 3HUG).
Data collection, phasing, and refinement statistics.
Expression constructs of σL and RslA used in this study.
References
- 1.Butcher G.B., Mascher T., Helmann J.D. Bacterial Physiology: A Molecular Approach. Springer-Verlag; Berlin, Germany: 2008. pp. 233–261. [Google Scholar]
- 2.Helmann J.D. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 2002;46:47–110. doi: 10.1016/s0065-2911(02)46002-x. [DOI] [PubMed] [Google Scholar]
- 3.Campbell E.A., Greenwell R., Anthony J.R., Wang S., Lim L., Das K. A conserved structural module regulates transcriptional responses to diverse stress signals in bacteria. Mol. Cell. 2007;27:793–805. doi: 10.1016/j.molcel.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell E.A., Tupy J.L., Gruber T.M., Wang S., Sharp M.M., Gross C.A., Darst S.A. Crystal structure of Escherichia coli sigmaE with the cytoplasmic domain of its anti-sigma RseA. Mol. Cell. 2003;11:1067–1078. doi: 10.1016/s1097-2765(03)00148-5. [DOI] [PubMed] [Google Scholar]
- 5.Campbell E.A., Muzzin O., Chlenov M., Sun J.L., Olson C.A., Weinman O. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol. Cell. 2002;9:527–539. doi: 10.1016/s1097-2765(02)00470-7. [DOI] [PubMed] [Google Scholar]
- 6.Thakur K.G., Joshi A.M., Gopal B. Structural and biophysical studies on two promoter recognition domains of the extra-cytoplasmic function sigma factor sigma(C) from Mycobacterium tuberculosis. J. Biol. Chem. 2007;282:4711–4718. doi: 10.1074/jbc.M606283200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manganelli R., Provvedi R., Rodrigue S., Beaucher J., Gaudreau L., Smith I. Sigma factors and global gene regulation in Mycobacterium tuberculosis. J. Bacteriol. 2004;186:895–902. doi: 10.1128/JB.186.4.895-902.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith I. Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin. Microbiol. Rev. 2003;16:463–496. doi: 10.1128/CMR.16.3.463-496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ilbert M., Graf P.C., Jakob U. Zinc center as redox switch—new function for an old motif. Antioxid. Redox Signal. 2006;8:835–846. doi: 10.1089/ars.2006.8.835. [DOI] [PubMed] [Google Scholar]
- 10.Jakob U., Eser M., Bardwell J.C. Redox switch of hsp33 has a novel zinc-binding motif. J. Biol. Chem. 2000;275:38302–38310. doi: 10.1074/jbc.M005957200. [DOI] [PubMed] [Google Scholar]
- 11.Kang J.G., Paget M.S., Seok Y.J., Hahn M.Y., Bae J.B., Hahn J.S. RsrA, an anti-sigma factor regulated by redox change. EMBO J. 1999;18:4292–4298. doi: 10.1093/emboj/18.15.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bae J.B., Park J.H., Hahn M.Y., Kim M.S., Roe J.H. Redox-dependent changes in RsrA, an anti-sigma factor in Streptomyces coelicolor: zinc release and disulfide bond formation. J. Mol. Biol. 2004;335:425–435. doi: 10.1016/j.jmb.2003.10.065. [DOI] [PubMed] [Google Scholar]
- 13.Zdanowski K., Doughty P., Jakimowicz P., O'Hara L., Buttner M.J., Paget M.S., Kleanthous C. Assignment of the zinc ligands in RsrA, a redox-sensing ZAS protein from Streptomyces coelicolor. Biochemistry. 2006;45:8294–8300. doi: 10.1021/bi060711v. [DOI] [PubMed] [Google Scholar]
- 14.Manganelli R., Voskuil M.I., Schoolnik G.K., Dubnau E., Gomez M., Smith I. Role of the extracytoplasmic-function sigma factor sigma(H) in Mycobacterium tuberculosis global gene expression. Mol. Microbiol. 2002;45:365–374. doi: 10.1046/j.1365-2958.2002.03005.x. [DOI] [PubMed] [Google Scholar]
- 15.Manganelli R., Voskuil M.I., Schoolnik G.K., Smith I. The Mycobacterium tuberculosis ECF sigma factor sigmaE: role in global gene expression and survival in macrophages. Mol. Microbiol. 2001;41:423–437. doi: 10.1046/j.1365-2958.2001.02525.x. [DOI] [PubMed] [Google Scholar]
- 16.Dainese E., Rodrigue S., Delogu G., Provvedi R., Laflamme L., Brzezinski R. Posttranslational regulation of Mycobacterium tuberculosis extracytoplasmic-function sigma factor sigma L and roles in virulence and in global regulation of gene expression. Infect. Immun. 2006;74:2457–2461. doi: 10.1128/IAI.74.4.2457-2461.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hahn M.Y., Raman S., Anaya M., Husson R.N. The Mycobacterium tuberculosis extracytoplasmic-function sigma factor SigL regulates polyketide synthases and secreted or membrane proteins and is required for virulence. J. Bacteriol. 2005;187:7062–7071. doi: 10.1128/JB.187.20.7062-7071.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lambert L.J., Wei Y., Schirf V., Demeler B., Werner M.H. T4 AsiA blocks DNA recognition by remodeling sigma70 region 4. EMBO J. 2004;23:2952–2962. doi: 10.1038/sj.emboj.7600312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anthony J.R., Newman J.D., Donohue T.J. Interactions between the Rhodobacter sphaeroides ECF sigma factor, sigma(E), and its anti-sigma factor, ChrR. J. Mol. Biol. 2004;341:345–360. doi: 10.1016/j.jmb.2004.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lane W.J., Darst S.A. The structural basis for promoter - 35 element recognition by the group IV sigma factors. PLoS Biol. 2006;4:e269. doi: 10.1371/journal.pbio.0040269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murakami K.S., Masuda S., Darst S.A. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science. 2002;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- 22.Vassylyev D.G., Sekine S., Laptenko O., Lee J., Vassylyeva M.N., Borukhov S., Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 23.Li W., Bottrill A.R., Bibb M.J., Buttner M.J., Paget M.S., Kleanthous C. The role of zinc in the disulphide stress-regulated anti-sigma factor RsrA from Streptomyces coelicolor. J. Mol. Biol. 2003;333:461–472. doi: 10.1016/j.jmb.2003.08.038. [DOI] [PubMed] [Google Scholar]
- 24.Urbauer J.L., Adelman K., Urbauer R.J., Simeonov M.F., Gilmore J.M., Zolkiewski M., Brody E.N. Conserved regions 4.1 and 4.2 of sigma(70) constitute the recognition sites for the anti-sigma factor AsiA, and AsiA is a dimer free in solution. J. Biol. Chem. 2001;276:41128–41132. doi: 10.1074/jbc.M106400200. [DOI] [PubMed] [Google Scholar]
- 25.Gruber T.M., Gross C.A. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- 26.Tibazarwa C., Wuertz S., Mergeay M., Wyns L., van Der Lelie D. Regulation of the cnr cobalt and nickel resistance determinant of Ralstonia eutropha (Alcaligenes eutrophus) CH34. J. Bacteriol. 2000;182:1399–1409. doi: 10.1128/jb.182.5.1399-1409.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dove S.L., Darst S.A., Hochschild A. Region 4 of sigma as a target for transcription regulation. Mol. Microbiol. 2003;48:863–874. doi: 10.1046/j.1365-2958.2003.03467.x. [DOI] [PubMed] [Google Scholar]
- 28.Campbell E.A., Westblade L.F., Darst S.A. Regulation of bacterial RNA polymerase sigma factor activity: a structural perspective. Curr. Opin. Microbiol. 2008;11:121–127. doi: 10.1016/j.mib.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patikoglou G.A., Westblade L.F., Campbell E.A., Lamour V., Lane W.J., Darst S.A. Crystal structure of the Escherichia coli regulator of sigma70, Rsd, in complex with sigma70 domain 4. J. Mol. Biol. 2007;372:649–659. doi: 10.1016/j.jmb.2007.06.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sorenson M.K., Ray S.S., Darst S.A. Crystal structure of the flagellar sigma/anti-sigma complex sigma(28)/FlgM reveals an intact sigma factor in an inactive conformation. Mol. Cell. 2004;14:127–138. doi: 10.1016/s1097-2765(04)00150-9. [DOI] [PubMed] [Google Scholar]
- 31.Mahren S., Enz S., Braun V. Functional interaction of region 4 of the extracytoplasmic function sigma factor FecI with the cytoplasmic portion of the FecR transmembrane protein of the Escherichia coli ferric citrate transport system. J. Bacteriol. 2002;184:3704–3711. doi: 10.1128/JB.184.13.3704-3711.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma U.K., Chatterji D. Differential mechanisms of binding of anti-sigma factors Escherichia coli Rsd and bacteriophage T4 AsiA to E. coli RNA polymerase lead to diverse physiological consequences. J. Bacteriol. 2008;190:3434–3443. doi: 10.1128/JB.01792-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li W., Stevenson C.E., Burton N., Jakimowicz P., Paget M.S., Buttner M.J. Identification and structure of the anti-sigma factor-binding domain of the disulphide-stress regulated sigma factor sigma(R) from Streptomyces coelicolor. J. Mol. Biol. 2002;323:225–236. doi: 10.1016/s0022-2836(02)00948-8. [DOI] [PubMed] [Google Scholar]
- 34.Song T., Dove S.L., Lee K.H., Husson R.N. RshA, an anti-sigma factor that regulates the activity of the mycobacterial stress response sigma factor SigH. Mol. Microbiol. 2003;50:949–959. doi: 10.1046/j.1365-2958.2003.03739.x. [DOI] [PubMed] [Google Scholar]
- 35.Ilbert M., Horst J., Ahrens S., Winter J., Graf P.C., Lilie H., Jakob U. The redox-switch domain of Hsp33 functions as dual stress sensor. Nat. Struct. Mol. Biol. 2007;14:556–563. doi: 10.1038/nsmb1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewin A., Crow A., Hodson C.T., Hederstedt L., Le Brun N.E. Effects of substitutions in the CXXC active-site motif of the extracytoplasmic thioredoxin ResA. Biochem. J. 2008;414:81–91. doi: 10.1042/BJ20080356. [DOI] [PubMed] [Google Scholar]
- 37.Nakano S., Erwin K.N., Ralle M., Zuber P. Redox-sensitive transcriptional control by a thiol/disulphide switch in the global regulator, Spx. Mol. Microbiol. 2005;55:498–510. doi: 10.1111/j.1365-2958.2004.04395.x. [DOI] [PubMed] [Google Scholar]
- 38.Chivers P.T., Laboissiere M.C., Raines R.T. The CXXC motif: imperatives for the formation of native disulfide bonds in the cell. EMBO J. 1996;15:2659–2667. [PMC free article] [PubMed] [Google Scholar]
- 39.Quan S., Schneider I., Pan J., Von Hacht A., Bardwell J.C. The CXXC motif is more than a redox rheostat. J. Biol. Chem. 2007;282:28823–28833. doi: 10.1074/jbc.M705291200. [DOI] [PubMed] [Google Scholar]
- 40.Shenoy A.R., Visweswariah S.S. Site-directed mutagenesis using a single mutagenic oligonucleotide and DpnI digestion of template DNA. Anal. Biochem. 2003;319:335–336. doi: 10.1016/s0003-2697(03)00286-0. [DOI] [PubMed] [Google Scholar]
- 41.Leslie A.G.W. Recent changes to the mosflm package for processing film and image plate data. Jt. CCP4+ ESF-EAMCB Newsl. Protein Crystallogr. 1992;26 [Google Scholar]
- 42.CCP4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 43.Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. Sect. D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 44.Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 45.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data collection, phasing, and refinement statistics.
Expression constructs of σL and RslA used in this study.