Abstract
22q11 deletion syndrome (22q11DS) is characterised by aberrant development of the pharyngeal apparatus and the heart with haploinsufficiency of the transcription factor TBX1 being considered the major underlying cause of the disease. Tbx1 mutations in mouse phenocopy the disorder. In order to identify the transcriptional dysregulation in Tbx1-expressing lineages we optimised fluorescent-activated cell sorting of β-galactosidase expressing cells (FACS-Gal) to compare the expression profile of Df1/Tbx1lacZ (effectively Tbx1 null) and Tbx1 heterozygous cells isolated from mouse embryos. Hes1, a major effector of Notch signalling, was identified as downregulated in Tbx1−/− mutants. Hes1 mutant mice exhibited a partially penetrant range of 22q11DS-like defects including pharyngeal arch artery (PAA), outflow tract, craniofacial and thymic abnormalities. Similar to Tbx1 mice, conditional mutagenesis revealed that Hes1 expression in embryonic pharyngeal ectoderm contributes to thymus and pharyngeal arch artery development. These results suggest that Hes1 acts downstream of Tbx1 in the morphogenesis of pharyngeal-derived structures.
Keywords: Tbx1, Hes1, 22q11, Pharyngeal arch artery, Thymus, Mouse, Microarray, FACS
Introduction
22q11 deletion syndrome (22q11DS) has an incidence of approximately 1 in 4000 live births and classically comprises malformations of the cardiovascular system, thymus gland, parathyroid gland and craniofacial structures (Scambler, 2000). Haploinsufficiency of the transcription factor TBX1 is considered to be the major underlying cause of the disorder. While the penetrance and expression of the deletion phenotype is very varied the cardinal features can be recapitulated by TBX1 mutations in humans (Paylor et al., 2006; Stoller and Epstein, 2005; Yagi et al., 2003; Zweier et al., 2007), mouse (Jerome and Papaioannou, 2001; Lindsay et al., 2001; Merscher et al., 2001), and zebrafish (Piotrowski et al., 2003; Piotrowski and Nusslein-Volhard, 2000). In order to understand the role of Tbx1 in development efforts have been made to identify Tbx1 target genes using “candidate gene” or transcriptomics approaches.
Our previous work has examined the downstream transcriptional events of loss of Tbx1 in dissected pharyngeal tissues (Ivins et al., 2005; Prescott et al., 2005). Several genes were identified and validated as differentially expressed in Tbx1 mutants. In particular we were able to show a dysregulation of genes in the retinoic acid metabolism pathway that are likely to contribute to the phenotype (Ivins et al., 2005; Roberts et al., 2006). However, in these experiments the dissection includes cell types that do not express Tbx1 and thus will either dilute some tissue specific expression changes, or add noise where tissue loss leads to a secondary expression change. We therefore aimed to repeat our analysis using only cells actively expressing from the Tbx1 promoter. One of the Tbx1 null alleles was created with a knock in of ß-galactosidase (Lindsay et al., 2001). We investigated whether a fluorogenic substrate for ß-galactosidase could be used to isolate Tbx1 expressing cells, firstly to further our target identification program, and secondly to demonstrate the general utility of this procedure in light of the thousands of such knock-ins available via the mouse gene trap program. FACS-Gal has been used previously on cultured cells (Laugwitz et al., 2005) but involves preparative procedures that could introduce noise into microarray analyses, and reduce cell viability especially when using early embryos. We therefore examined whether the FACS-Gal procedure could produce good quality microarray data from the material available from mid gestation embryos.
We utilized the Tbx1+/lacZ × Df1/+ cross to obtain Tbx1 null mutants to ensure equivalent fluorescence intensity from the Tbx1lacZ allele in both genotypes. Df1 is a heterozygous deletion of 20 genes including Tbx1 (Lindsay et al., 2001), thus the hemizygously deleted Df1 genes could be used as internal controls to assess the quality of the experiment. It is possible that genes in cis with Tbx1 may have a haploinsufficient effect on gene expression, but Tbx1+/lacZ and Df1/+ embryos have indistinguishable phenotypes and Df1/Tbx1lacZ embryos have exactly the same phenotype as Tbx1lacZ/lacZ mutants (Lindsay et al., 2001) supporting their use as true Tbx1 nulls. Since we were specifically interested in changes due to loss of Tbx1 within cells normally expressing that gene, validation by in situ hybridisation was conducted using Tbx1lacZ/lacZ mutants.
The quality of the microarray data was tested by analysing the down regulation of genes in cis with Tbx1 and reduced to hemizygosity in the Df1 deletion (Lindsay et al., 1999), and by validating selected genes by RT-PCR in an independent sample. We selected the Hes1 transcription factor, an effector of Notch signalling for further analysis. Hes+/− and Hes1−/− mouse embryos displayed a partially penetrant phenotype including pharyngeal arch artery (PAA) hypo/aplasia, outflow tract (OFT) misalignment, thymic agenesis and cleft palate — defects seen in 22q11DS and Tbx1 null embryos. Conditional mutagenesis indicated that lack of Hes1 expression in the neural crest had no significant effect on great vessel development and a mild effect on thymic development. Concurrent ablation of Hes1 expression in pharyngeal ectoderm and neural crest recapitulated much of the thymic phenotype of constitutive nulls, as well as yielding a small proportion of embryos with great vessel defects.
Results
FACS-Gal enrichment of Tbx1-expressing cells
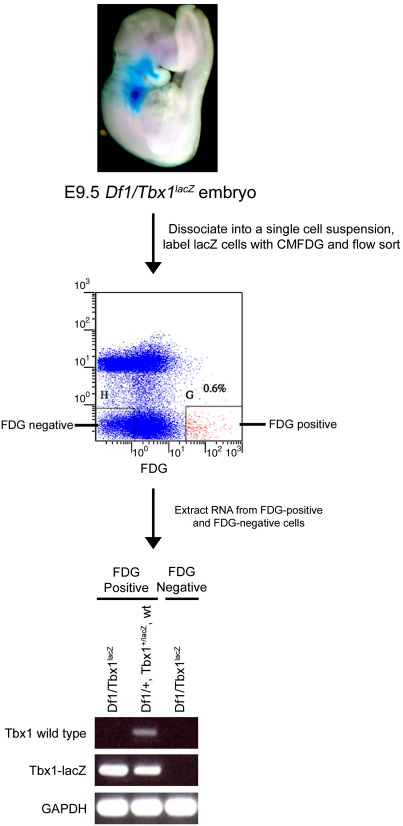
In order to identify and enrich for Tbx1 targets we performed a microarray comparing the gene expression profile of Tbx1-expressing cells from embryonic day 9.5 (E9.5) Tbx1+/lacZ and Df1/Tbx1lacZ (Tbx1-null) embryos. Whole embryos were dissociated at E9.5, a stage at which Tbx1 expression is mainly restricted to the pharyngeal region, as recapitulated by the β-galactosidase reporter (Figs. 1, 2A–C). The Tbx1lacZ allele was generated by inserting a lacZ construct into exon 5 of Tbx1, which encodes part of the DNA-binding domain (Lindsay et al., 2001). Real-time PCR analysis has demonstrated that this allele represents a true null allele (Zhang and Baldini, 2008). The Tbx1lacZ allele was labelled using the fluorogenic substrate, 5-chloromethylfluorescein di-β-d-galactopyranoside (CMFDG) to compare nulls with heterozygous embryos. E9.5 embryos were collected and Df1/Tbx1lacZ embryos were separated by phenotype from the Tbx1+/lacZ;Df1/+;wild type embryo pool for subsequent dissociation into a single cell suspension. After staining with the fluorescent substrate, CMFDG, cells were collected based on fluorescent intensity (above background) and cell viability (Fig. 1). FACS-Gal enrichment yielded an average number of 600 cells per E9.5 embryo. Tbx1 driven lacZ expression was observed in pharyngeal endoderm, ectoderm and core mesoderm at this stage (Figs. 2A–C). Enrichment for Tbx1 expressing cells was examined by RT-PCR using primers that detected only the wild type transcript, and primers that detected the IRES-lacZ. The IRES-lacZ could be detected in FACS-Gal fluorescent cell populations, but the wild type Tbx1 transcript could only be detected in the heterozygote fluorescent population demonstrating we had captured the cells of interest (Fig. 1).
Fig. 1.
CMFDG labelling and analysis of Tbx1LacZ cell enrichment. Example of CMFDG labelling of an E9.5 Df1/Tbx1lacZ embryo separated from the rest of the litter by phenotype. X-gal staining represents expression of the Tbx1lacZ allele. This labelling procedure is also carried out on the Tbx1+/lacZ;Df1/+;wt pool of embryos. RT-PCR analysis indicates that the wild type Tbx1 allele is present only in the FDG-positive cell population containing Df1/+ and Tbx1+/lacZ cells. The Tbx1LacZ allele is only present in FDG-positive cell populations and represents Df1/Tbx1lacZ and Tbx1+/lacZ cells.
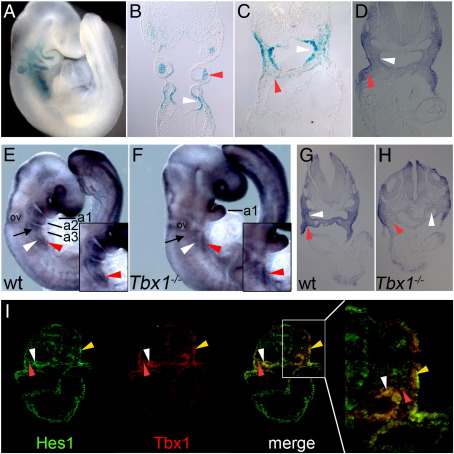
Fig. 2.
Expression of Hes1 in wild type and Tbx1−/− embryos. (A–C) Tbx1LacZ expression at E9.5. (A) Whole embryo, (B) coronal section and (C) transverse section of Tbx1+/LacZ embryo. (D) Transverse section illustrating Hes1 expression at E9.5 by in situ hybridisation. (E–H) Whole mount in situ hybridisation (E–F) and corresponding transverse sections (G–H) showing Hes1 expression is diminished in Tbx1−/−−/− embryos. a indicates arch; ov, otic vesicle. (I) Overlapping expression of Hes1 and Tbx1 as detected by immunofluorescence staining of transverse cryosections of wild type E8.5 embryos. Pharyngeal mesenchyme is indicated by a black arrow, endoderm by a white arrowhead, mesoderm with a red arrowhead and ectoderm with a yellow arrowhead.
Microarray experiments were then carried out using Affymetrix mouse MOE430 v2.0 arrays consisting of over 34 000 well characterised mouse genes. Analysis of the microarray experiments was conducted using Genespring v4.2.1. Microarray experiments were carried out using RNA extracted from 3 pools of Df1/Tbx1lacZ cells and 3 pools of Tbx1+/lacZ cells. The three pools were sorted from independent groups of embryos to provide biological replication. Each pool of cells was obtained from a group of at least 8 E9.5 embryos (ranging in somite number from 17 s to 25 s). Pools of cells were used to minimise variation due to staging differences. (The average number of cells obtained per embryo was 535 cells per Df1/Tbx1lacZ embryo and 646 cells per Tbx1+/lacZ embryo.) Due to the limited number of cells obtained, PCR amplification was used to generate sufficient RNA for microarray analysis.
To assess the sensitivity of the approach for microarray we examined the expression levels of genes reduced to hemizygosity in this cross. It has been shown previously that the Df1 genes are decreased to approximately 50% of wild type expression levels in Df1/+ heterozygotes and they do not exhibit dosage compensation (Prescott et al., 2005). Seventeen of 20 Df1 genes represented on the array were expressed above threshold levels, and 16 of these 17 were down regulated to an average of 0.611 ± 0.096 (mean ± SD) in Df1/Tbx1lacZ arrays compared to Tbx1+/lacZ arrays (Table 1) indicating the sensitivity of the experiment in detecting downregulated genes. The one of 17 genes upregulated was Es2el, whose upregulation is a previously described artifact secondary to the fact that this gene is at the Df1 deletion breakpoint (Ivins et al., 2005; Prescott et al., 2005).
Table 1.
Df1 genes analysed by microarray.
| Gene | Relative expression in Df1/Tbx1lacZ vs. Tbx1+/lacZ cells |
|---|---|
| Dgcr6 | 0.517 ± 0.068 |
| Zdhhc8 | 0.663 ± 0.140 |
| Ranbp1 | 0.598 ± 0.095 |
| Htf9c | 0.549 ± 0.104 |
| D16h22s680e | 0.563 ± 0.267 |
| Arvcf | 0.490 ± 0.165 |
| Comt | 0.601 ± 0.118 |
| Txnrd2 | 0.531 ± 0.054 |
| Septin 5 | 0.568 ± 0.167 |
| Ufd1l | 0.459 ± 0.091 |
| Slc25a1 | 0.639 ± 0.097 |
| Cdc45l | 0.694 ± 0.206 |
| Prodh | 0.632 ± 0.205 |
| Dgcr8 | 0.807 ± 0.153 |
| Cldn5a | 0.752 ± 0.478 |
| Gnb1la | 0.706 ± 0.325 |
| Vpreb2 (L) | 0.749 ± 0.704 |
| Rtn4r (L) | 0.977 ± 0.631 |
| Tbx1 (L) | 0.324 ± 0.344 |
| Es2el | 1.459 ± 0.238 |
L indicates low signal strength.
These genes did not pass the filtering and statistical analysis applied.
In order to identify non-Df1 deletion genes which were downregulated in Df1/Tbx1lacZ cells, we chose a statistical filter which selected the majority (14/16) of down regulated Df1 genes as having significantly reduced expression. This filter was downregulated to at least 0.83 in 5 out of 6 Df1/Tbx1lacZ arrays compared to Tbx1+/lacZ arrays, with p < 0.05 using the Benjamini and Hochberg false discovery rate correction. This resulted in a total of 1553 unique sequences which were downregulated at least to the same degree as most Df1 genes (Supplementary Table 1). The full dataset is available at ArrayExpress, accession E-MEXP-2096.
Validation of downregulated genes by qRT-PCR
Validation focused on downregulated genes because Tbx1 has been demonstrated to be a transcriptional activator (Ataliotis et al., 2005; Stoller and Epstein, 2005; Xu et al., 2004). Quantitative, real-time PCR (qRT-PCR) was carried out to confirm gene expression changes using RNA amplified from an independent pool of Df1/Tbx1lacZ and Tbx1+/lacZ cells collected by FACS from at least 10 E9.5 Df1/Tbx1lacZ embryos and Tbx1+/lacZ embryos.
Genes were selected for independent validation by qRT-PCR based on their expression pattern, known or potential function in pharyngeal or heart development, and significant downregulation in the microarray experiments. Genes were also chosen based on their previous identification in other Tbx1 microarrays. Altogether, 30 genes downregulated in the absence of Tbx1 were analysed by qRT-PCR; 18 (60%) showed significant (p < 0.05) downregulation in Df1/Tbx1lacZ cells and 12 genes showed no appreciable changes in expression (Table 2).
Table 2.
Genes validated by qRT-PCR.
| Gene | Average relative expression in Df1/Tbx1lacZ cells vs. Tbx1+/lacZ cells (± SD) | Significance (*p < 0.05, **p < 0.01) | Reason for analysis |
|---|---|---|---|
| Nkx2.6 | 0 ± 0 | ** | 1 # |
| Cxcl4 | 0.133 ± 0.047 | ** | 3 |
| Smad7 | 0.145 ± 0.094 | ** | 2, 3 |
| Cdh1 | 0.201 ± 0.125 | ** | 3 |
| Notch3 | 0.249 ± 0.103 | ** | 2 |
| Hes1 | 0.261 ± 0.093 | ** | 2, 3 # |
| Crb3 | 0.386 ± 0.058 | ** | 3 # |
| Sema3c | 0.360 ± 0.170 | ** | 1, 3 |
| Pleckstrin | 0.454 ± 0.019 | ** | 3 ^ |
| Slit2 | 0.529 ± 0.125 | ** | 2 |
| Lztfl1 | 0.562 ± 0.153 | ** | 3 |
| Isl1 | 0.643 ± 0.153 | * | 1 # |
| Dsg2 | 0.656 ± 0.020 | ** | 1, 3 |
| Bmpr1a | 0.712 ± 0.116 | ** | 1 |
| Tbx18 | 0.754 ± 0.140 | * | 1, 3 # |
| Tle4 | 0.783 ± 0.101 | * | 3 |
| Daam1 | 0.797 ± 0.152 | * | 2 |
| Gsk3b | 0.831 ± 0.035 | ** | 1, 2 |
| Dab2 | 0.721 ± 0.185 | # ^ | |
| Pitx1 | 0.814 ± 0.134 | 2 | |
| FoxP1 | 1.025 ± 0.349 | 1 | |
| Socs3 | 1.051 ± 0.052 | 3 | |
| Lrr16 | 1.001 ± 0.130 | 3 | |
| Tmeff1 | 1.032 ± 0.270 | 3 | |
| Ephb2 | 1.264 ± 0.312 | 1, 3 | |
| Tbx20 | 0.981 ± 0.227 | 1 | |
| Pafah1b2 | 0.956 ± 0.176 | 3 | |
| Dkk1 | 1.908 ± 1.208 | 1 3 # | |
| Cxcl12 | 1.048 ± 0.086 | 2 3 | |
| Numb | 1.493 ± 0.879 | 3 |
Values represent average changes in gene expression of n = 3 experiments (except for Dsg2 where n = 2).
-
1.Known role in pharyngeal/heart development.
-
2.Family members with a known role in pharyngeal/heart development in mice or other species.
-
3.Stringently downregulated/multiple probes downregulated.
#Downregulated in Liao et al. (2008).
^Downregulated in Ivins et al. (2005).
The most strikingly altered gene was Nkx2.6, whose expression was completely abolished in Tbx1-null cells by qRT-PCR (Table 2). Other genes which were downregulated greater than 3-fold were Notch3 and Hes1, members of the Notch signalling pathway, Smad7, required for heart development (Chen et al., 2009), and Cxcl4 and Cdh1 whose role in pharyngeal or heart development have yet to be characterised. Downregulation of a number of genes with a known function in the pharyngeal apparatus or the heart (Sema3c, Isl1, Tbx18, Bmpr1a, Dsg2 and Gsk3β) were also validated by qRT-PCR with Sema3c exhibiting the greatest downregulation, to 0.360 ± 0.170, in Tbx1-null cells. The remaining validated genes (Crb3, Plek, Slit2, Lztfl1, Tle4 and Daam1) represent additional, novel factors which have not previously been shown to be involved in development of the pharyngeal system and its derivatives.
To demonstrate the utility of the FACS-Gal approach in identifying genes potentially contributing to Tbx1 loss of function phenotypes, we chose Hes1 for further functional analysis. Targeted mutants were already available, and other Notch pathway genes have previously been implicated in cardiovascular morphogenesis as discussed below.
Hes1 is downregulated in the pharyngeal apparatus of Tbx1 null embryos
In situ hybridization revealed that Hes1 is expressed in the pharyngeal region at E9.5 (Figs. 2D, E, G) and that this expression is diminished, particularly in mesoderm and endoderm of Tbx1−/− embryos of the same stage (Figs. 2F, H), consistent with diminished Hes1 levels in FACS sorted Tbx1−/− cells. Expression of Tbx1 in pharyngeal ectoderm at E8.5 is especially important for the great vessel defects typically seen in 22q11DS (Randall et al., 2009; Zhang et al., 2005). At the protein level, Hes1 is broadly expressed in pharyngeal endoderm, ectoderm and pharyngeal mesenchyme (Fig. 2I) at this stage. Dual staining reveals co-expression of Hes1 and Tbx1 in pharyngeal ectoderm, endoderm and mesoderm (Fig. 2I). Together, these data suggest a possible role for Hes1 in the morphogenesis of the pharyngeal apparatus.
Hes1 mutant embryos have abnormalities of the pharyngeal arch arteries
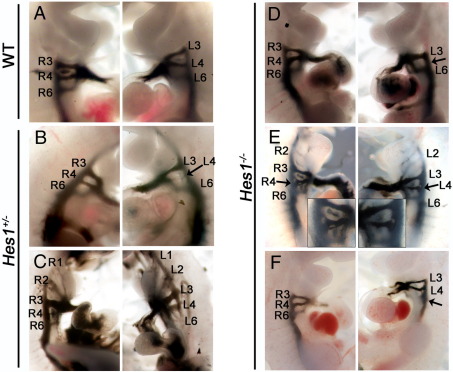
A major feature of Tbx1 heterozygous animals and 22q11DS patients is defective great vessel development secondary to abnormal growth and remodelling of the 4th PAA (Lindsay et al., 2001). We therefore assessed the growth and patency of Hes1 mutant PAAs at E10.5 using ink injection and found a defect in 13/40 (32.5%) Hes1−/− embryos analysed on an MF1 background (Table 3), including hypo/aplasia of the 3rd, 4th (Fig. 3D) and 6th arch arteries (Fig. 3F), persistent 1st and 2nd arch arteries (Fig. 3E and not shown), and aberrant branching of the arch arteries (arrows and inset, Fig. 3E). We also detected the same range of PAA defects in a smaller proportion of heterozygous embryos (13/80, 16%) (Table 3, Figs. 3B, C), although defects involved hypoplasia rather than aplasia.
Table 3.
Frequency of pharyngeal arch artery defects in E10.5 Hes1 mutants.
| WT | Hes1+/− | Hes1−/− | |
|---|---|---|---|
| Total embryos | 77 | 80 | 40 |
| Total normal | 76 | 67 | 27 |
| Total abnormal (%) | 1 (1%) | 13a (16%) | 13a (32.5%) |
| Persistent 1st and/or 2nd PAA | – | 2 | 6 |
| Hypo/aplastic 3rd PAA | – | – | 1 (apl) |
| Hypo/aplastic 4th PAA | 1 (hypo) | 8 (hypo) | 3 (hypo), 3 (apl) |
| Hypo/aplastic 6th PAA | – | – | 1 (hypo) |
| Aberrant branching | – | 4 | 4 |
PAA, pharyngeal arch artery; hypo, hypoplastic; apl, aplastic.
Nb. Total WT embryos include additional WT non-littermates examined for the presence of PAA defects.
Significantly different from WT (Fisher's exact test p < 0.01).
Fig. 3.
Intracardiac ink injection of E10.5 Hes1 mutants. (A) In wild type embryos, bilateral pairs of caudal PAAs R3–R6 and L3–L6 are present at E10.5. (B) A proportion of Hes1+/− mutants have hypoplasia of the fourth arch artery (arrow) and persistent first and second arch arteries (C). Hes1−/− mutants also exhibit fourth arch artery aplasia (arrow in D), persistent second arch arteries and aberrant branching of the arch arteries (arrow in E and inset) and sixth arch artery hypoplasia (arrow in F). L indicates left; R, right.
Hes1 mutants exhibit defects in vascular smooth muscle patterning
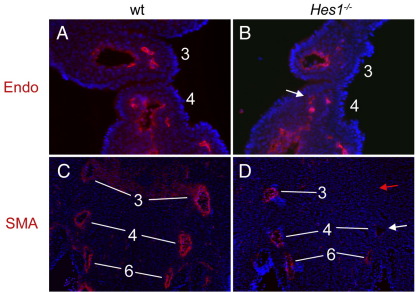
Defective PAA development can occur due to a failure of endothelial tube formation or aberrant growth and remodelling. We first analysed formation of endothelial tubes using Endomucin, an endothelial cell marker. Hes1 mutant embryos with hypoplastic PAAs or PAAs non-patent to ink still retained expression of Endomucin surrounding the arteries even though they were very small at E10.5 (Fig. 4B). Thus, initial formation of the capillary tubes appears to occur normally, although growth of the arteries is affected. The non-endothelial component of the developing PAAs derives from the surrounding neural crest (NC) which differentiates into smooth muscle α-actin (α-SMA)-positive vascular smooth muscle (VSM) surrounding the PAAs (Jiang et al., 2000; Li et al., 2000). Wild type embryos at E11.5 showed strong expression of α-SMA surrounding the 3 rd, 4th and 6th PAAs (Fig. 3C). However, in Hes1−/− mutants which exhibit hypoplastic PAAs, α-SMA staining is absent (PAA3, red arrow in Fig. 4D) or diminished (PAA4, white arrow in Fig. 4D). These results demonstrate that during PAA morphogenesis Hes1 is required for normal VSM development.
Fig. 4.
Hes1 mutants exhibit loss of vascular smooth muscle markers in the developing PAAs. (A, B) Immunofluorescence on coronal E10.5 embryo sections for the endothelial marker, Endomucin (Endo). Endo staining is present in wild type (A) and Hes1−/− embryos with aplastic/non-patent 4th PAAs (arrow in B). (C, D) Immunofluorescence on coronal E11.5 embryo sections for α-SMA. Strong α-SMA expression surrounding the 3 rd, 4th and 6th PAA in wild types (C) is lost in the hypo/aplastic left 3 rd and 4th PAA (red and white arrows in D, respectively) of Hes1−/− mutant embryos. The contralateral vessels, which have formed in this embryo, have normal α-SMA staining.
Hes1−/− mutants exhibit great vessel and intracardiac defects
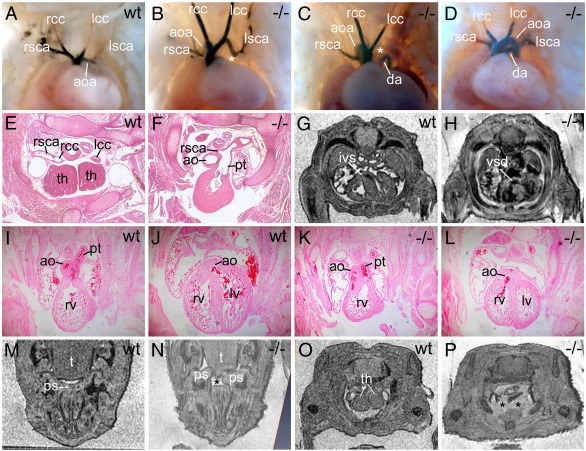
Defective growth and remodelling of the PAAs produces later defects of the great vessels. We examined E14.5 to E15.5 embryos on a pure MF1, and a mixed MF1;C57Bl/6 (MF1;Bl6) background, by MRI, histology and ink injection. A proportion of Hes1−/− embryos exhibited cardiovascular defects (7/40, 17.5%, both genetic backgrounds; 4/23 (17%) MF1;Bl6 and 3/17 (17%) MF1) (Table 4, Fig. 5). We observed defects in OFT alignment in 3/40 (8%) Hes1−/− homozygotes (1/23 (4%) MF1;Bl6 and 2/17 (12%) MF1) (Table 4, Figs. 5K, L) which were accompanied by a ventricular septal defect (VSD) (Fig. 5H). Great vessel defects (4/23 (17%) MF1;Bl6 and 1/17 (6%) MF1) included right-sided aortic arch (RAA) (Figs. 5B, C), interruption of the aortic arch type B (IAA-B) (Fig. 5C) and isolation of the right subclavian artery (I-RSA) (Figs. 5D, F) which branched off the pulmonary trunk instead of the right common carotid. These defects represent aberrant development of the 4th PAA. Some embryos exhibited right-sided ductus arteriosus, a 6th arch artery defect (Fig. 5B). The phenotype of these Hes1−/− mutants is consistent with the earlier PAA defects present at E10.5.
Table 4.
22q11DS-like defects observed at E14.5–E15.5.
| Strain | Genotype | n | Total CV defects | Great vessel defect | OFT defect | Cleft Palate | Thymic hypo/aplasia |
|---|---|---|---|---|---|---|---|
| MF1 | Hes1−/− | 15 | 2 (13%) | 1 | 9 (60%) | 13 (87%) | |
| Hes1+/− | 15 | 1a (7%) | 0 | 0 | 0 | 0 | |
| WT | 2 | 0 | 0 | 0 | 0 | 0 | |
| MF1;C57Bl/6 | Hes1−/− | 23 | 5 (22%) | 4 | 2 | 9 (39%) | 23 (100%) |
| Hes1+/− | 21 | 0 | 0 | 0 | 1 (5%) | 0 | |
| WT | 7 | 0 | 0 | 0 | 0 | 0 |
CV — cardiovascular, OFT — outflow tract, WT — wild type.
Dextroposition of the heart.
Fig. 5.
22q11DS-like defects in E15.5 Hes1−/− mutants. (A–D) Great vessel defects in E15.5 Hes1−/− embryos. (A) Normal wild type (wt) great vessel morphology. (B–D) Hes1−/− (–/–) mutants with RAA and right-sided ductus arteriosus (B), IAA-B and RAA (C), and I-RSA (D). (E) In wt embryos, the right subclavian artery branches off the right common carotid. (F) Hes1−/− mutant in which the right subclavian branches off the pulmonary trunk. (G–L) OFT defects and VSD in Hes1−/− mutants. (G, H) Transverse MRI scans. (I–L) Transverse histological sections. (G) In wild type embryos the interventricular septum separates the right and left ventricles. (H) Hes1−/− mutants exhibit a VSD (dark staining represents blood). (I, J) Consecutive transverse sections of a wild type embryo in which the pulmonary trunk arises from the right ventricle (I) and the aorta joins the left ventricle (J). (K, L) Consecutive sections of a Hes1−/− embryo with double outlet right ventricle in which both the aorta and pulmonary trunk arise from the right ventricle. (M, N) Transverse MRI sections of the palate at E15.5. (M) In wild type embryos the palatal shelves are fused. (N) Hes1−/− mutants exhibit cleft palate where the palatal shelves do not meet in the midline. (O, P) Transverse MRI sections of the thymic lobes which are missing in Hes1−/− mutants (P). ao indicates aorta; aoa, aortic arch; da, ductus arteriosus; ivs, interventricular septum; lcc, left common carotid; lsca, left subclavian artery; lv, left ventricle; ps, palatal shelf; pt, pulmonary trunk; rcc, right common carotid; rsca, right subclavian artery; rv, right ventricle; t, tongue; th, thymic lobe; vsd, ventricular septal defect. Asterisks indicate missing segments.
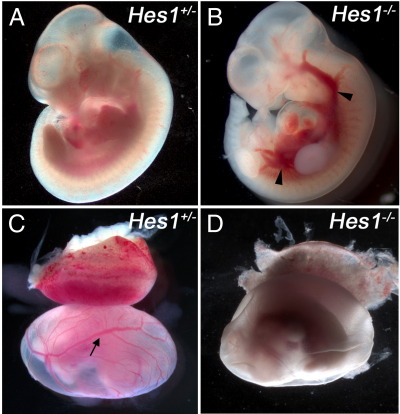
Genetic background appeared to affect the penetrance of the Hes1 mutant phenotype. On an MF1 background, a proportion of Hes1−/− mice exhibit neural tube defects and are also embryonic lethal prior to E15.5 resulting in lower than expected numbers of Hes1−/− mutants according to the predicted Mendelian ratios. However, on a mixed background, no neurulation defects were identified and all genotypes were present at the expected ratios (Supplementary Table 2). This suggests that the proportion of MF1 Hes1−/− homozygotes with a heart defect could be higher if investigated earlier. At E11.5, we noted that a number of Hes1−/− embryos on an MF1 background exhibit body hemorrhage and the yolk sac of these embryos appear to lack large vitelline vessels (Supplementary Fig. 1), suggesting the earlier lethality is due to a more serious defect in cardiovascular development. Thus, the mixed MF1;Bl6 background appears to allow Hes1−/− embryos to survive at least until E15.5.
In contrast to the PAA defects detected in Hes1+/− embryos at E10.5, no great vessel abnormalities were found in Hes1+/− heterozygotes at E15.5 (Table 4) indicating that these defects may largely recover later in development. Tbx1 heterozygotes display a similar background dependent rescue of PAA malformations as embryogenesis proceeds (Taddei et al., 2001). However, one MF1;Bl6 Hes1+/− embryo was slightly growth delayed in comparison to its littermates and had a cleft palate, and another MF1 Hes1+/− embryo had dextroposition of the heart (Table 4) indicating potential haploinsufficiency.
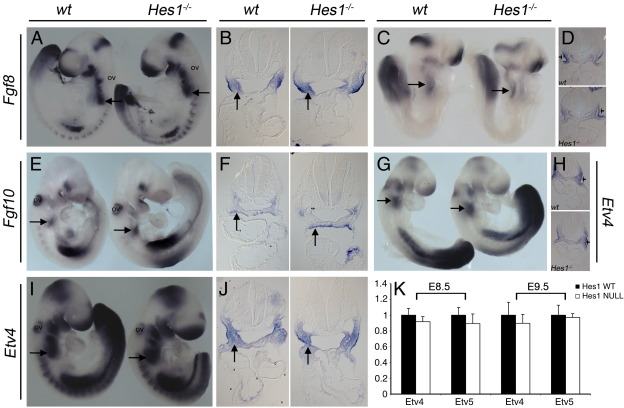
Fgf8 is required for PAA morphogenesis (Frank et al., 2002). Furthermore, dominant negative interference of the Notch pathway in the second heart field gives rise to a defect in epithelial to mesenchymal transition that, in ex vivo culture, can be rescued by Fgf8 (High et al., 2009). Thus, down regulation of Fgf8 may contribute to both the arch artery and outflow tract abnormalities observed in Hes1 nulls. However, no alteration of Fgf8, or Etv4, a readout for Fgf activity (Mao et al., 2009) was detected at E8.5, while Fgf8, Fgf10 and Etv4 appeared unaffected at E9.5 as well (Supplementary Fig. 2). qRT-PCR detected no quantitative change in Etv4 or Etv5 expression at both stages (Supplementary Fig. 2).
MRI of Hes1 mutant embryos reveals additional 22q11DS-like malformations
One of the original descriptions of Hes1 null mutants commented upon lack of thymus gland development (Tomita et al., 1999). To quantify this and survey the anatomy of mutants we used MRI of E15.5 embryos on both a mixed MF1;C57Bl6 and pure MF1 background. We found highly penetrant thymic hypo/aplasia on both backgrounds (Table 4, Fig. 5P). Of particular note is the presence of cleft palate in 9/15 (60%) MF1 Hes1−/− embryos and 9/23, (39%) MF1;C57Bl6 Hes1−/− mutants (Table 4, Fig. 5N), which is observed in Tbx1−/− embryos of the same stage (Jerome and Papaioannou, 2001).
Expression of other notch pathway genes is altered in FACS isolated Tbx1-null cells
Hes1 is the main effector of Notch signalling and Notch pathway genes have previously been implicated in cardiovascular development (High and Epstein, 2008). We therefore analysed expression of Notch pathway genes by qRT-PCR to investigate whether Hes1 may be downregulated in Tbx1 mutants due to aberrant Notch signalling. In Df1/Tbx1lacZ cells, the Notch receptors, Notch3 and Notch4 are downregulated while Notch1 and Notch2 are upregulated. In addition, the Notch ligand, Dll1 and DNA-binding transactivator, Rbpj are also upregulated (Supplementary Fig. 3). These results could suggest that Hes1 may act downstream of Notch3 or Notch4. Upregulation of other Notch pathway members could be due to functional compensation or negative feedback from loss of Hes1. The incomplete penetrance of pharyngeal-derived defects in Hes1 mutants suggest a possible functional redundancy with other Hes-related genes, especially since HeyL;Hey1 double mutants, but not single mutants, have heart defects (Fischer et al., 2007). In addition, Hes1, Hey1 and Hey2 bind to promoters of an overlapping set of genes (M. Gessler, personal communication). In Tbx1-null cells, Hey1 expression is diminished but Hey2 is unchanged and HeyL is upregulated 3.4 fold (Supplementary Fig. 3).
Hes1 is required in embryonic pharyngeal ectoderm for normal thymic development
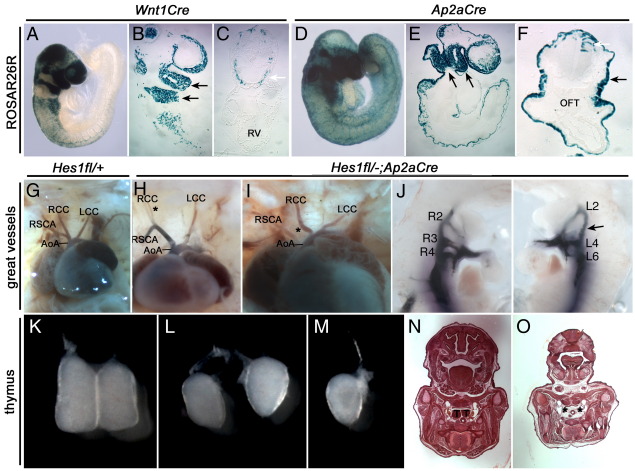
Conditional dominant negative interference of Notch signalling in cardiac neural crest cell progenitors (using Pax3Cre and Wnt1Cre) results in pulmonary stenosis and aortic arch patterning defects (High et al., 2007). Thus, even though Tbx1 is not expressed in neural crest, we investigated Hes1 requirements in these cells as the effect of loss of Tbx1 on Hes1 expression might be indirect. We therefore crossed Hes1+/−;Wnt1Cre mice to Hes1fl/fl mice using a conditional allele for Hes1 (Kita et al., 2007), and assessed defects in pharyngeal structures. Although it can only act as a surrogate for recombination at the Hes1 locus, the ROSAR26R reporter confirmed strong Cre activity in neural crest lineages within this cross (i.e. on the same genetic background (Figs. 6A–C)). Thymic hypo/aplasia was observed in 3 of 21 Hes1fl/− embryos, 2 of 31 embryos hemizygous for Hes1 in neural crest only, and in 10 of 26 (38%) Hes1fl/−;Wnt1Cre embryos. Thus, complete loss of Hes1 in neural crest increased the frequency of thymic hypoplasia; however, there were no instances of thymic aplasia in these crosses. Great vessel defects were not apparent in any embryos (Table 5). In summary, there is not a major requirement for Hes1 in the neural crest lineage although a contributory role in thymus development was detected.
Fig. 6.
22q11DS-like defects in E10.5 and E15.5 Hes1 conditional mutants. (A–F) ROSAR26R reporter crosses. (A–C) Hes1−/fl;Wnt1Cre x ROSAR26R and (D–F) Hes1−/fl;Ap2αCre x ROSAR26R crosses following X-gal staining. (A) E9.5 whole mount image, (B) sagittal section of (A) showing strong recombination in the neural crest of first and second pharyngeal arches (black arrows), and (C) transverse section of (A) showing recombination in the neural crest cells at the second heart field level (white arrow). (D) E9.5 whole mount image, (E) sagittal section of (D) showing strong recombination in the neural crest and ectoderm of first and second pharyngeal arches (black arrows) and (F) transverse section of (D) showing recombination in the ectoderm (black arrow) and neural crest cells (white arrow). (G–I) Great vessel defects in E15.5 Hes1−/fl;Ap2αCre embryos. (G) Normal Hes1+/fl (wild type) great vessel morphology. (H–I) Hes1−/fl;Ap2αCre (Hes1 null in the ectoderm and neural crest) mutants with aberrant branching of the RCC (H) and I-RSA (I). (J) Intracardiac ink injection of E10.5 Hes1−/fl;Ap2αCre mutants displaying persistent second arch arteries (R2, L2) and unilateral third arch artery aplasia (arrow). (K–O) Thymic defects in E15.5 Hes1−/fl;Ap2αCre embryos. (K) Normal thymic lobe morphology in Hes1+/fl (wild type) embryos. (L–M) Hypoplastic lobes (L) and a single, hypoplastic lobe (M) observed in Hes1−/fl;Ap2αCre mutants. (N–O) Transverse section of the thymic lobes in Hes1−/fl;Ap2αCre embryos showing normal development (N) and aplasia (O). aoa indicates the aortic arch; lcc, left common carotid; oft, outflow tract; rcc, right common carotid; rsca, right subclavian artery; rv, right ventricle; t, thymic lobe. Asterisks indicate missing or duplicated segments.
Table 5.
22q11DS-like defects observed at E15.5 in the Hes1 conditional mutants.
| Strain | Genotype | n | Total CV defects | Great vessel defect | OFT defect | Thymic hypo/aplasia |
|---|---|---|---|---|---|---|
| MF1;C57Bl/6 | Hes1fl/−;Wnt1Cre | 26 | 0 | 0 | 0 | 10 (38.5%) |
| Hes1fl/+;Wnt1Cre | 31 | 0 | 0 | 0 | 2 (6.5%) | |
| Hes1fl/− | 21 | 0 | 0 | 0 | 3 (14.3%) | |
| Hes1fl/+ (WT) | 17 | 0 | 0 | 0 | 0 | |
| MF1;C57Bl/6 | Hes1fl/−;Ap2αCre | 32 | 2 (6.25%) | 2 | 0 | 26 (81.3%) |
| Hes1fl/+;Ap2αCre | 20 | 0 | 0 | 0 | 0 | |
| Hes1fl/− | 23 | 0 | 0 | 0 | 2 (8.7%) | |
| Hes1fl/+ (WT) | 27 | 0 | 0 | 0 | 2 (7.4%) |
CV — cardiovascular, OFT — outflow tract, WT — wild type.
Tbx1 is required in pharyngeal epithelia for normal great vessel morphogenesis (Zhang et al., 2005). We have recently refined this to a requirement in pharyngeal ectoderm (Randall et al., 2009). Thus, to investigate the requirement for Hes1 in ectoderm we used the Ap2αCre driver which effects recombination in both ectoderm and neural crest, the results being compared to the Wnt1Cre, neural crest only, conditionals. The ROSAR26R reporter was used to assess recombination within this cross and confirmed strong Cre activity in the embryonic surface ectoderm and neural crest lineages (Figs. 6D–F). 26 of 32 (81%) Hes1fl/−;Ap2αCre as compared to 2 of 23 Hes1fl/− (9%) embryos exhibited thymic hypo/aplasia (Table 5, Figs. 6K–M). This recapitulates much of the thymic malformation seen in the constitutive nulls, (thymic aplasia was noted in seven cases). In two of 36 (6%) Hes1fl/−;Ap2αCre embryos we observed a great vessel defect. One embryo had a branched right common carotid and the other had isolation of the right subclavian artery, abnormalities which occur due to defective development of the 3rd and 4th PAA, respectively (Figs. 6H, I). This frequency of defects does not fully reproduce that seen in constitutive mutants suggesting a modifying effect of genetic background or that great vessel morphogenesis requires Hes1 expression in additional tissues. In summary, ectodermal expression of Hes1 makes a major contribution to thymus development and also plays a role in PAA development. As Ap2αCre effects recombination in both ectoderm and neural crest, a synergistic effect of crest ablation of Hes1 in the absence of ectodermal expression cannot be ruled out.
Hes1 is required for normal cell proliferation in the pharyngeal ectoderm
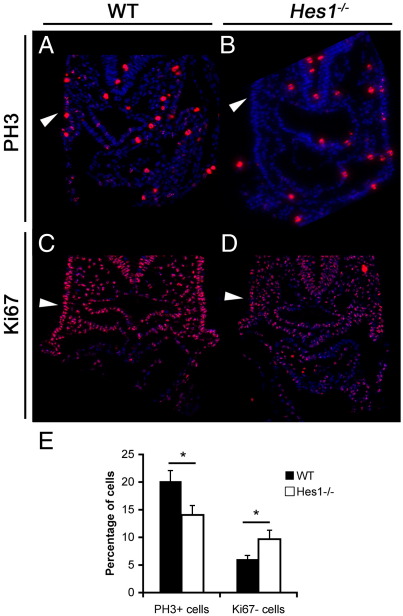
Previous analyses have demonstrated impairment of cell cycle progression in the pharyngeal endoderm and second heart field, but not the myocardium, of Hes1 mutants (Rochais et al., 2009). We therefore asked whether pharyngeal ectoderm was similarly affected. PAA and thymic development requires Tbx1 at stage E8.5 (Xu et al., 2005). At this stage, we observed decreased proliferation in the ectoderm — a reduction in the number of phospho-histone H3 positive cells (from 20 to 14%) in Hes1 nulls compared to wild type controls (Fig. 7; p < 0.02). There was also an increase in the number of Ki67 negative (quiescent) cells (from 6 to 9.6%), (Fig. 7; p < 0.05). There was no alteration in apoptosis in the pharyngeal ectoderm or neural crest at E9.5 (assayed using a caspase3 antibody (not shown)). The sections studied are from the same embryos used by Rochais et al. (2009). Our results indicate that Hes1 is important for proliferation of the pharyngeal ectoderm, which may in turn affect morphogenesis of the thymus and PAAs. Altered proliferation is unlikely to be secondary to decreased Fgf8 signalling at this stage, since expression of Fgf8 itself and downstream readout of Fgf activity are not detectably altered (Supplementary Fig. 2).
Fig. 7.
Proliferation defect in Hes1 mutants. (A–B) Immunohistochemistry with anti-phospho-Histone H3 (PH3) antibody in paraffin sections of E8.5 WT (A) and Hes1−/− (B) embryos showing a decrease in PH3-positive cells within the surface ectoderm of Hes1−/− mutants (arrowhead). (C–D) Immunohistochemistry with anti-Ki67 antibody in sections of E8.5 WT (C) and Hes1−/− (D) embryos showing an increase in the number of Ki67-negative cells in Hes1−/− embryos (arrowhead). (E) Graph comparing the percentage of PH3 positive cells and Ki67 negative cells in the pharyngeal ectoderm of WT embryos and Hes1−/− mutants. A significant decrease in PH3 positive cells (p < 0.05) and increase in Ki67 negative cells (p < 0.05, Student's t-test) is observed in Hes1−/− mutants.
Discussion
Our studies demonstrate that FACS-Gal is a useful technique to enhance the power of transcriptomics studies of murine embryogenesis. FACS-Gal can make use of the large existing repository of β-galactosidase-trapped ES lines now encompassing approximately 33% of all mouse genes (http://www.genetrap.org/index.html). Isolation of the expressing cells of interest will enrich for autonomous effects and reduce noise from non-expressing tissues. Moreover, the existence of Cre-reversible traps opens the way to within group comparisons of flow-sorted cells, one pool having had expression restored using a tamoxifen activated Cre enzyme.
We used FACS-Gal to analyse gene expression changes between Tbx1-null and Tbx1-heterozygous cells and identified a list of potential Tbx1 targets including Hes1. Recently, an independent microarray screen also identified Hes1 as downregulated in dissected Tbx1−/− caudal pharyngeal tissue (Liao et al., 2008). These experiments were aimed at identifying dysregulated genes in the secondary heart field, and were not restricted to Tbx1-expressing cells. Nevertheless there were genes identified in common with those reported here. In particular, Hes1 was downregulated 2.5 fold at E8.75. Four other genes were downregulated by microarray and confirmed by qRT-PCR in both studies: Nkx2.6, Crb3, Isl1, and Dab2. Thus these five genes represent excellent candidates to contribute to the phenotype caused by loss of Tbx1. Nkx2.6 was almost undetectable by qRT-PCR in the Tbx1−/− cells, and missense mutation of the gene has been described in common arterial trunk malformation in humans (Heathcote et al., 2005). Nkx2.6 is expressed transiently in the mouse embryonic pharyngeal endoderm, but Nkx2.6−/− mice are apparently normal (Tanaka et al., 2000). However, Nkx2.5 expression expands into Nkx2.6 expression domains in these mutants suggesting a regulatory compensation by Nkx2.5. Consistent with this hypothesis, Nkx2.6−/−;Nkx2.5−/− embryos had an abnormal, dilated pharynx with reduction in the numbers of endodermal cells, phenotypes not observed in Nkx2.5−/− single mutants (Tanaka et al., 2001). However, the Nkx2.6−/− mouse line is no longer available to pursue the possibility of Tbx1:Nkx2.6 interaction.
We focused on Hes1 as a gene worth investigating for a possible biological role in the morphogenesis of structures requiring Tbx1. Hes1 represents one of the main effectors of Notch signalling and has been shown to play diverse roles in development (Kageyama et al., 2007). Hes1 is known to be involved in thymic morphogenesis (Tomita et al., 1999) and recently, Hes1 has been shown to play a role in the development of the outflow tract (Rochais et al., 2009). However, to date there has been no published role for Hes1 in pharyngeal arch artery or palate development. Numerous members of the Notch pathway play important roles in cardiovascular development, including the Notch ligand, Jag1 and the Notch target genes, Hey1, Hey2 and HeyL (Donovan et al., 2002; Fischer et al., 2004, 2007; Gessler et al., 2002; Kokubo et al., 2005, 2004; Li et al., 1997; Oda et al., 1997; Sakata et al., 2002). These Notch pathway genes are known to be important for processes such as arterial-venous differentiation and targeted Notch pathway mutants exhibit heart defects including VSDs, atrioventricular valve defects, tetralogy of Fallot and cardiomyopathy (Donovan et al., 2002; Fischer et al., 2004, 2007; Gessler et al., 2002; Kokubo et al., 2005, 2004; Sakata et al., 2002). Dominant negative repression of Notch signalling within the neural crest (NC-DNMAML mutants) causes a range of heart defects, predominantly pulmonary stenosis and great vessel defects (High et al., 2007). SHF-specific inhibition of Notch signalling (SHF-DNMAML mutants) also results in great vessel defects as well as OFT alignment and septation defects (High et al., 2009). However, Hes1 is the first Notch effector mutant described as having PAA abnormalities. While it is possible that functional redundancy among related Notch pathway members may explain this inconsistency, our findings reveal that Hes1−/− mutants exhibit a spectrum of PAA-derived malformations similar to those which occur in DNMAML mutants (High et al., 2007) suggesting that Hes1 may play a dose-dependent role in mediating the effects downstream of Notch signalling in PAA development and remodelling. Hes-related genes are expressed in overlapping domains with Hes1 within pharyngeal tissues including the neural crest and pharyngeal endoderm (High et al., 2007). This potential for redundancy may provide an additional explanation for the partially penetrant Hes1 null phenotype we observed. Indeed, we observed a 3.4 fold increase in the level of HeyL expression in Tbx1−/− cells. Hey gene functional redundancy could affect analysis of a genetic interaction between Tbx1 and Hes1 in doubly heterozygous mice. It will therefore be interesting in the future to determine whether DNMAML and Tbx1 mutants show an epistatic relationship during development.
The spectrum of PAA malformations in Hes1 mutants ranges from persistence of the 1st and 2nd arch arteries to hypo/aplasia and aberrant branching of the caudal PAAs. This phenotype is similar to NC ablation phenotypes (Hutson and Kirby, 2007). Thymic and palatal defects are also found in several neural crest gene mutants (Gritli-Linde, 2007; Rodewald, 2008). Hes1+/− heterozygotes exhibit PAA defects at E10.5 however, they exhibit hypoplasia and aberrant PAA branching rather than aplasia. The relatively mild phenotype may represent an intermediate defect in PAA patterning from which embryos recover during development. This may help to explain why the percentage of embryos with PAA defects has decreased at E15.5 in Hes1−/− nulls and why no Hes1+/− heterozygotes appeared with PAA-derived defects later in development. A similar reduction in PAA derivative defects is seen in Tbx1 mice (Lindsay and Baldini, 2001).
Here, we show that Hes1 and Tbx1+/− mutants exhibit an overlapping phenotype, specifically 4th PAA hypo/aplasia, thymic hypo/aplasia and cleft palate. In PAA development, Tbx1 is known to play a non-cell autonomous role in controlling VSM differentiation (Calmont et al., 2009; Lindsay and Baldini, 2001; Zhang et al., 2005). Similarly, Hes1 mutants exhibit a defect in VSM patterning of the PAAs which may be due to decreased NC contribution (Rochais et al., 2009) and/or a failure of the NC to properly differentiate, suggesting that Hes1 may act downstream of Tbx1 to effect its role in PAA morphogenesis. It is possible that Hes1 acts as an indirect target of Tbx1 through downregulation of the Notch signalling pathway. In support of this role, downregulation of Notch signalling within the NC causes a failure of VSM differentiation and SHF-DNMAML mutants exhibit aberrant CNC migration both leading to PAA defects (High et al., 2009, 2007).
Tbx1 is required within the pharyngeal ectoderm for PAA, thymus and palate development (Randall et al., 2009). Tbx1 is not expressed in the NC but we wanted to ascertain what role Hes1 played in this tissue. We specifically ablated Hes1 within the NC using Wnt1Cre. No great vessel defects were observed in these embryos, the only abnormalities involving thymus gland hypoplasia. We therefore used the Ap2αCre driver and a conditional Hes1 allele to test Hes1 requirements within the pharyngeal ectoderm. We observed a low frequency great vessel defect, however thymic hypo/aplasia was prevalent in Ap2αCre conditional Hes1 mutants, suggesting a non-cell autonomous role of Hes1 to organogenesis similar to Tbx1 (Randall et al., 2009). It remains to be determined what role, if any, Notch signalling plays within the pharyngeal ectoderm. However, we surmise that expression of Hes1 in tissues other than ectoderm and neural crest will contribute to cardiovascular morphogenesis because the pharyngeal ectoderm-specific knockout of Hes1 did not recapitulate the spectrum and frequency of defects seen in constitutive Hes1 nulls. In particular, second heart field and pharyngeal endoderm lineages may be important in this regard. Disruption of Notch signalling within the SHF causes OFT alignment and septation defects and great vessel abnormalities (among other cardiovascular malformations). These defects occur due to aberrant signalling between the SHF and the NC leading to a decrease in CNC migration through the pharyngeal arches (High et al., 2009). Hes1 mutants exhibit reduced proliferation within the SHF and show similar CNC migration defects (Rochais et al., 2009). It will be interesting to determine whether SHF-specific Notch deletion mutants also exhibit decreased proliferation of SHF progenitors and to examine the SHF-specific role of Hes1 in OFT morphogenesis. The SHF-DNMAML mutants also had defective endothelial to mesenchymal transition in endocardial cushion explant tissue (High et al., 2009). Addition of exogenous Fgf8 to these cultures was sufficient to rescue the defect. As Fgf8 is also intimately involved in PAA morphogenesis we examined Fgf8 and Fgf10 expression, together with Fgf activity as determined by Etv4/Etv5 expression, in Hes1 null embryos, but no abnormalities were observed.
Together, these data support a non-cell autonomous role for Hes1 in thymic and, to a lesser extent, PAA morphogenesis and adds to recent data indicating the importance of the embryonic pharyngeal ectoderm in this process. One possibility is that Hes1 regulates an aspect of signalling from the pharyngeal ectoderm to the neural crest or other mesenchymal cell types, as we have postulated for Gbx2 (Calmont et al., 2009). A second possibility is that the moderate decrease in cell proliferation seen in pharyngeal ectoderm of Hes1−/− embryos could result in a decrease in the number of the relevant signalling cell lineages. Despite the comparatively restricted neural crest conditional phenotype, a role for Hes1 in regulating differentiation of the postmigratory NC cannot be excluded since the Ap2αCre driver effects recombination in neural crest as well as ectoderm. Hes1 has previously been shown to play dual roles within the same tissue in maintaining proliferation of stem cells and promoting differentiation depending on the developmental stage (Ohtsuka et al., 2001). We detected reduced proliferation in pharyngeal ectoderm of Hes1 null embryos, but markers for progenitor differentiation are not available for this tissue.
Hes1 has primarily been described as an effector of Notch signalling, but there is evidence that Hes1 also acts independently of Notch through signalling pathways including Shh and Vegf (de la Pompa et al., 1997; Hashimoto et al., 2006; Ingram et al., 2007). Furthermore, Tbx transcription factors have been shown to regulate members of the Hes-related family in heart development (Rutenberg et al., 2006). These results suggest that a conserved regulatory pathway may exist between T-box transcription factors and members of the Hes family. Tbx1 regulation of the Notch pathway has been observed in the developing ear. However, in this situation, loss of Tbx1 results in some members of the Tbx1-dependent cell population switching to a neurogenic fate, an event associated with activation of the Delta-Notch pathway. It is not known whether this is a direct or indirect regulation (Xu et al., 2007).
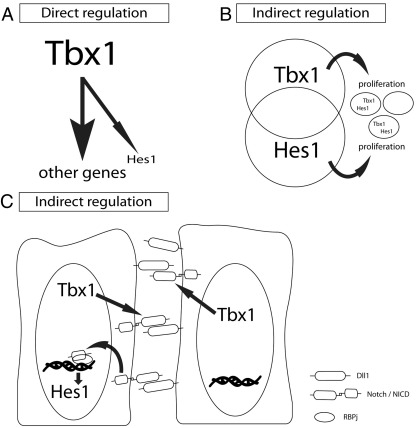
The decreased expression of Hes1 in the Tbx1LacZ expressing, Tbx1 null cells could be explained by several mechanisms (see Supplementary Fig. 4), including direct transcriptional regulation. Transfection assays to date have so far failed to support direct action of Tbx1 on the Hes1 promoter (data not shown), thus the molecular basis of the Hes1 regulation by Tbx1 remains to be elucidated. Since Tbx1 regulates cell proliferation, loss of Tbx1 could reduce the number of Hes1 expressing cells, detected as reduced Hes1 transcript abundance by FACS-GAL. As Hes1 is also involved in controlling cell proliferation, this would result in overlap between the Hes1 and Tbx1 loss of function phenotypes in the absence of a direct regulatory relationship. A third possibility is that Tbx1 directly or indirectly controls expression of other Notch pathway genes (e.g. Notch3, which had reduced expression in the FACS-Gal enriched Tbx1-null cells). In this model, Hes1 would be indirectly downregulated as a consequence.
This work demonstrated the utility of FACS-Gal for identifying genes dysregulated in embryonic models of human disease. Hes1 was validated as downregulated in Tbx1 null embryos and shown to be required for the normal formation of pharyngeal-derived structures. In particular, expression of Hes1 in embryonic pharyngeal ectoderm was shown to be required for normal thymus development and makes a contribution to pharyngeal arch artery morphogenesis. Thus, Hes1 downregulation may contribute to the Tbx1 loss of function phenotype.
Materials and methods
Mice
Tbx1+/lacZ and Df1/+ mice have been described previously and were maintained on a C57BL/6 background (Lindsay et al., 1999, 2001). Hes1+/− mice (Ishibashi et al., 1995) were kindly provided by Francois Guillemot, National Institute of Medical Research, UK and maintained on a MF1 and CD1 background. Wnt1Cre mice (Danielian et al., 1998) were kindly provided by Andrew Copp, Institute of Child Health, University College London, London, UK and maintained on a C57BL/6 background. Ap2αCre mice (Macatee et al., 2003) were obtained from Anne Moon, University of Utah School of Medicine, Salt Lake City, Utah, USA and maintained on a C57BL/6 background. Genotyping of embryos was carried out on yolk sac or tail tip genomic DNA as previously described (Ishibashi et al., 1995; Lindsay et al., 1999, 2001). The Hes1 conditional allele was obtained from R. Kageyama (Kita et al., 2007), and the ROSAR26R reporter originated in the Soriano lab (Soriano, 1999). Mouse work was carried out in accordance with British Home Office regulations.
CMFDG labelling of a single cell suspension
E9.5 embryos were dissected out in DMEM + 25 mM HEPES containing 10% FCS. The embryos were staged by somite counting and Df1/Tbx1lacZ embryos were separated from Df1/+, Tbx1+/lacZ and wild type embryos. CMFDG loading was carried out using the Detectagene green CMFDG lacZ gene expression kit (Molecular Probes). Following three rinses in PBS, embryos were incubated in 0.25% trypsin (Gibco) in PBS for approximately 30 min during which the embryos were dissociated into a single cell suspension by trituration. The single cell suspension was spun down at 300 x g for 3 min and resuspended in pre-warmed (10 min at 37 °C) 50 μM CMDFG, 200 μM verapamil in 5% FCS/DMEM + 25 mM HEPES and incubated for 30 min at 37 °C, 5% CO2. Following incubation, the cells were placed on ice and diluted to approximately 1–3 million cells/mL with 5%FCS/DMEM + 25 mM hepes. 1 mM PETG was added to stop the reaction and 1.5 μM propidium iodide (PI) was added to identify dead cells. Cells were flow-sorted as soon as possible.
FACS analysis
Single cell suspensions were sorted using the Beckman Coulter Epics Altra cell sorter (High Wycombe) and analysed using the Expo2 software (Beckman Coulter) or using the MoFlo cell sorter (Dakocytomation, Fort Collins, Colorado) and analysed using the Summit v4.11 software (Dakocytomation). Cells were gated on the basis of forward scatter and side scatter characteristics to exclude debris. PI was used as a viability stain to exclude all dead cells. Cells which were fluorescent for FDG above background were collected into 10%FCS/DMEM + 25 mM HEPES and placed on ice. At least 50 000 events were acquired for analysis. Sorted cells were immediately spun down at 300 × g for 4 min and resuspended in Trizol (Gibco BRL) for RNA extraction.
RT-PCR
Following RNA extraction, cDNA was synthesized using Superscript Reverse Transcriptase II (Invitrogen). Primers used were as follows: Tbx1-wt, Tbx1ex4-F 5′-TTTGTGCCCGTAGATGACAA-3′ and Tbx1ex6-R 5′-AATCGGGGCTGATATCTGTG-3′; Tbx1LacZ, IreslacZ-F 5′-TCGGTGCACATGCTTTACAT-3′ and IreslacZ-R 5′-GTTTTCCCAGTCACGACGTT-3′.
Microarray
Microarray hybridisation
Six Df1/Tbx1lacZ samples and six Tbx1+/lacZ samples were hybridised onto MOE430 v2 oligonucleotide array chips (Affymetrix) consisting of over 45 000 probe sets representing over 34 000 mouse genes. Purified, biotin-UTP labelled cRNA was fragmented and hybridised onto MOE430 v2 microarray chips according to Affymetrix protocols and this was carried out by the ICH Gene Microarray Centre (London UK). Arrays were scanned using the Affymetrix GeneChip Scanner and array images were captured using the Microarray Analysis Suite v5 software package (MAS, Affymetrix). The report files were examined to establish quality of the hybridisation.
Genespring analysis
Genespring software version 4.2.1 (Silicon Genetics) was used for the comparison of Df1/Tbx1lacZ and Tbx1+/lacZ microarrays. Per chip normalisation used the 50th percentile of all measurements as a positive control for each sample. Measurements for each gene were divided by this synthetic positive control. The bottom tenth percentile was used as a test for correct background subtraction. Per gene normalisation was then carried out, for which all Tbx1+/lacZ samples were normalised to 1 and all Df1/Tbx1lacZ samples were normalised to the Tbx1+/lacZ samples. The data from the mouse MOE430 v.2 microarray chips was then filtered to exclude genes with signals below the raw value of 22. Seventeen out of twenty Df1 genes represented on the chip were expressed above threshold levels. Iterative analyses were then carried out on the normalised data to determine the appropriate filters to apply in order to successfully identify the hemizygously-expressed Df1 genes. Gene lists were statistically analysed using the non-parametric t-test (Wilcoxon–Mann–Whitney/Kruskal–Wallis test) with a p-value cutoff of 0.05 and the Benjamini and Hochberg false discovery rate multiple testing correction was applied. Fourteen out of 17 Df1 genes were downregulated to 0.83 in 5 out of 6 chips hybridised with the Df1/Tbx1lacZ target under these conditions.
Quantitative real-time PCR
LacZ-expressing cells from a pool of at least 10 E9.5 Df1/Tbx1+/− and Tbx1+/− embryos were labelled using the CMFDG labelling kit (Molecular Probes) and collected by FACS. RNA was isolated using Trizol (Gibco BRL) and amplified by PCR-based amplification using the Microarray Target Amplification kit (Roche). Quantitative real-time PCR was carried out on the ABI PRISM 7000 Sequence Detection System (Applied Biosystems). All reactions were performed in triplicate using SYBR green (Qiagen) under the following PCR conditions: 95 °C for 15 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min and expression values were obtained using the DART-PCR analysis excel software which takes into account the amplification efficiency of each gene to obtain relative quantities (Peirson et al., 2003) or by using the ΔΔCt method (Yuan et al., 2008). The qRT-PCR primers used can be found in the supplementary data (Supplementary Table 3).
Mouse embryo analysis
In situ hybridization was performed according to previously published methods (Wilkinson, 1992). Hes1, Fgf8, Fgf10 and Etv4 probes were generated from cDNA plasmid clones. β-galactosidase activity was detected in embryos fixed in formaldehyde using X-gal (Promega) following standard procedures. Indian Ink (Pelikan) was injected into the outflow tract or aorta of embryos using a microinjection needle to visualise the pharyngeal arch arteries or great vessels of the heart. Multi-embryo magnetic resonance imaging (MRI) was carried out as described previously (Schneider et al., 2004) on embryos at E15.5. Following fixation, embryos were dehydrated and embedded in wax. Sections were cut at 8–12 µm and some were counterstained using Eosin. Paraffin or cryosections were used for immunofluorescence, 8–10 µm sections were stained using antibodies against Endomucin (Clone V.7C7 1/50, Santa Cruz), smooth muscle α-actin (Clone 1A4 1/700, Sigma), Hes1 (1/200, R. Kageyama), Tbx1 (1/100, Zymed, Invitrogen), β-galactosidase (1/300, Cappel), Isl1 (1/100, DSHB clones 402D6 and 394D5), phospho-Histone H3 (1/400, Upstate), Ki67 (1/25, Dako) and Caspase 3 (1/100, Cell Signaling). For analysis of cell death using a Caspase 3 antibody, neural crest cells were detected as mesenchymal cells negative for Isl1 or a β-galactosidase-encoding transgene expressed in pharyngeal mesoderm. Proliferation and cell death assays were carried out on Hes1 mutant embryos in a CD1 mutant background. Data were obtained from four or more sections per embryo for three embryos per genotype. Equivalent cell numbers were scored in wild type and mutant embryos. Statistical analyses were carried out using Student's t-test. Dapi or Hoechst was used for counterstaining sections and the preparations were observed using an ApoTome microscope (Zeiss).
Acknowledgments
We thank Francois Guillemot for providing the Hes1 mice and the plasmid for in situ hybridisation; Ryoichiro Kageyama kindly provided the Hes1 floxed allele and antibody. Albert Basson provided the Fgf10 plasmid probe; we would like to thank Nipurna Jina and Mike Hubank (UCL-ICH microarray centre) for their guidance.
Sources of funding. This work was supported by the British Heart Foundation (P.J.S., S.B.) and was supported by the European Commission under the FP7 CardioGeNet project (Grant No. HEALTH-2007-B-223463) (P.J.S, R.K. and S.B.), a University College London Graduate Research Scholarship (K.L.V.B) and a Child Health Research Appeal Trust studentship (I.P.). FR acknowledges the support of Inserm and the ISHR-ES/Servier Research fellowship.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ydbio.2010.01.020.
Appendix A. Supplementary data
Supplementary Fig. 1
Supplementary Fig. 2
Supplementary Fig. 3
Supplementary Fig. 4
1567 Genes downregulated (including Df1 genes) to 0.83 in 5 out of 6 Df1/Tbx1lacZ chips compared to Tbx1+/lacZ chips
Frequency of genotypes obtained at E14.5-E15.5 from Hes1+/- intercrosses.
qRT-PCR primers.
References
- Ataliotis P., Ivins S., Mohun T.J., Scambler P.J. XTbx1 is a transcriptional activator involved in head and pharyngeal arch development in Xenopus laevis. Dev. Dyn. 2005;232:979–991. doi: 10.1002/dvdy.20276. [DOI] [PubMed] [Google Scholar]
- Calmont A., Ivins S., Van Bueren K.L., Papangeli I., Kyriakopoulou V., Andrews W.D., Martin J.F., Moon A.M., Illingworth E.A., Basson M.A., Scambler P.J. Tbx1 controls cardiac neural crest cell migration during arch artery development by regulating Gbx2 expression in the pharyngeal ectoderm. Development. 2009;136:3173–3183. doi: 10.1242/dev.028902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Chen H., Zheng D., Kuang C., Fang H., Zou B., Zhu W., Bu G., Jin T., Wang Z., Zhang X., Chen J., Field L.J., Rubart M., Shou W., Chen Y. Smad7 is required for the development and function of the heart. J. Biol. Chem. 2009;284:292–300. doi: 10.1074/jbc.M807233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian P.S., Muccino D., Rowitch D.H., Michael S.K., McMahon A.P. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr. Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- de la Pompa J.L., Wakeham A., Correia K.M., Samper E., Brown S., Aguilera R.J., Nakano T., Honjo T., Mak T.W., Rossant J., Conlon R.A. Conservation of the Notch signalling pathway in mammalian neurogenesis. Development. 1997;124:1139–1148. doi: 10.1242/dev.124.6.1139. [DOI] [PubMed] [Google Scholar]
- Donovan J., Kordylewska A., Jan Y.N., Utset M.F. Tetralogy of fallot and other congenital heart defects in Hey2 mutant mice. Curr. Biol. 2002;12:1605–1610. doi: 10.1016/s0960-9822(02)01149-1. [DOI] [PubMed] [Google Scholar]
- Fischer A., Schumacher N., Maier M., Sendtner M., Gessler M. The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev. 2004;18:901–911. doi: 10.1101/gad.291004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A., Steidl C., Wagner T.U., Lang E., Jakob P.M., Friedl P., Knobeloch K.P., Gessler M. Combined loss of Hey1 and HeyL causes congenital heart defects because of impaired epithelial to mesenchymal transition. Circ. Res. 2007;100:856–863. doi: 10.1161/01.RES.0000260913.95642.3b. [DOI] [PubMed] [Google Scholar]
- Frank D.U., Fotheringham L.K., Brewer J.A., Muglia L.J., Tristani-Firouzi M., Capecchi M.R., Moon A.M. An Fgf8 mouse mutant phenocopies human 22q11 deletion syndrome. Development. 2002;129:4591–4603. doi: 10.1242/dev.129.19.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessler M., Knobeloch K.P., Helisch A., Amann K., Schumacher N., Rohde E., Fischer A., Leimeister C. Mouse gridlock: no aortic coarctation or deficiency, but fatal cardiac defects in Hey2 −/− mice. Curr. Biol. 2002;12:1601–1604. doi: 10.1016/s0960-9822(02)01150-8. [DOI] [PubMed] [Google Scholar]
- Gritli-Linde A. Molecular control of secondary palate development. Dev. Biol. 2007;301:309–326. doi: 10.1016/j.ydbio.2006.07.042. [DOI] [PubMed] [Google Scholar]
- Hashimoto T., Zhang X.M., Chen B.Y., Yang X.J. VEGF activates divergent intracellular signaling components to regulate retinal progenitor cell proliferation and neuronal differentiation. Development. 2006;133:2201–2210. doi: 10.1242/dev.02385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heathcote K., Braybrook C., Abushaban L., Guy M., Khetyar M.E., Patton M.A., Carter N.D., Scambler P.J., Syrris P. Common arterial trunk associated with a homeodomain mutation of NKX2.6. Hum. Mol. Genet. 2005;14:585–593. doi: 10.1093/hmg/ddi055. [DOI] [PubMed] [Google Scholar]
- High F.A., Epstein J.A. The multifaceted role of Notch in cardiac development and disease. Nat. Rev. Genet. 2008;9:49–61. doi: 10.1038/nrg2279. [DOI] [PubMed] [Google Scholar]
- High F.A., Zhang M., Proweller A., Tu L., Parmacek M.S., Pear W.S., Epstein J.A. An essential role for Notch in neural crest during cardiovascular development and smooth muscle differentiation. J. Clin. Invest. 2007;117:353–363. doi: 10.1172/JCI30070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- High F.A., Jain R., Stoller J.Z., Antonucci N.B., Lu M.M., Loomes K.M., Kaestner K.H., Pear W.S., Epstein J.A. Murine Jagged1/Notch signaling in the second heart field orchestrates Fgf8 expression and tissue-tissue interactions during outflow tract development. J. Clin. Invest. 2009;119:1986–1996. doi: 10.1172/JCI38922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutson M.R., Kirby M.L. Model systems for the study of heart development and disease. Cardiac neural crest and conotruncal malformations. Semin. Cell Dev. Biol. 2007;18:101–110. doi: 10.1016/j.semcdb.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram W.J., McCue K.I., Tran T.H., Hallahan A.R., Wainwright B.J. Sonic Hedgehog regulates Hes1 through a novel mechanism that is independent of canonical Notch pathway signalling. Oncogene. 2008;27:1489–1500. doi: 10.1038/sj.onc.1210767. [DOI] [PubMed] [Google Scholar]
- Ishibashi M., Ang S.L., Shiota K., Nakanishi S., Kageyama R., Guillemot F. Targeted disruption of mammalian hairy and Enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix–loop–helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 1995;9:3136–3148. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- Ivins S., Lammerts van Beuren K., Roberts C., James C., Lindsay E., Baldini A., Ataliotis P., Scambler P.J. Microarray analysis detects differentially expressed genes in the pharyngeal region of mice lacking Tbx1. Dev. Biol. 2005;285:554–569. doi: 10.1016/j.ydbio.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Jerome L.A., Papaioannou V.E. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat. Genet. 2001;27:286–291. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- Jiang X., Rowitch D.H., Soriano P., McMahon A.P., Sucov H.M. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- Kageyama R., Ohtsuka T., Kobayashi T. The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development. 2007;134:1243–1251. doi: 10.1242/dev.000786. [DOI] [PubMed] [Google Scholar]
- Kita A., Imayoshi I., Hojo M., Kitagawa M., Kokubu H., Ohsawa R., Ohtsuka T., Kageyama R., Hashimoto N. Hes1 and Hes5 control the progenitor pool, intermediate lobe specification, and posterior lobe formation in the pituitary development. Mol. Endocrinol. 2007;21:1458–1466. doi: 10.1210/me.2007-0039. [DOI] [PubMed] [Google Scholar]
- Kokubo H., Miyagawa-Tomita S., Tomimatsu H., Nakashima Y., Nakazawa M., Saga Y., Johnson R.L. Targeted disruption of hesr2 results in atrioventricular valve anomalies that lead to heart dysfunction. Circ. Res. 2004;95:540–547. doi: 10.1161/01.RES.0000141136.85194.f0. [DOI] [PubMed] [Google Scholar]
- Kokubo H., Miyagawa-Tomita S., Nakazawa M., Saga Y., Johnson R.L. Mouse hesr1 and hesr2 genes are redundantly required to mediate Notch signaling in the developing cardiovascular system. Dev. Biol. 2005;278:301–309. doi: 10.1016/j.ydbio.2004.10.025. [DOI] [PubMed] [Google Scholar]
- Laugwitz K.L., Moretti A., Lam J., Gruber P., Chen Y., Woodard S., Lin L.Z., Cai C.L., Lu M.M., Reth M., Platoshyn O., Yuan J.X., Evans S., Chien K.R. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005;433:647–653. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Krantz I.D., Deng Y., Genin A., Banta A.B., Collins C.C., Qi M., Trask B.J., Kuo W.L., Cochran J., Costa T., Pierpont M.E., Rand E.B., Piccoli D.A., Hood L., Spinner N.B. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat. Genet. 1997;16:243–251. doi: 10.1038/ng0797-243. [DOI] [PubMed] [Google Scholar]
- Li J., Chen F., Epstein J.A. Neural crest expression of Cre recombinase directed by the proximal Pax3 promoter in transgenic mice. Genesis. 2000;26:162–164. doi: 10.1002/(sici)1526-968x(200002)26:2<162::aid-gene21>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Liao J., Aggarwal V.S., Nowotschin S., Bondarev A., Lipner S., Morrow B.E. Identification of downstream genetic pathways of Tbx1 in the second heart field. Dev. Biol. 2008;316:524–537. doi: 10.1016/j.ydbio.2008.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay E.A., Baldini A. Recovery from arterial growth delay reduces penetrance of cardiovascular defects in mice deleted for the DiGeorge syndrome region. Hum. Mol. Genet. 2001;10:997–1002. doi: 10.1093/hmg/10.9.997. [DOI] [PubMed] [Google Scholar]
- Lindsay E.A., Botta A., Jurecic V., Carattini-Rivera S., Cheah Y.C., Rosenblatt H.M., Bradley A., Baldini A. Congenital heart disease in mice deficient for the DiGeorge syndrome region. Nature. 1999;401:379–383. doi: 10.1038/43900. [DOI] [PubMed] [Google Scholar]
- Lindsay E.A., Vitelli F., Su H., Morishima M., Huynh T., Pramparo T., Jurecic V., Ogunrinu G., Sutherland H.F., Scambler P.J., Bradley A., Baldini A. Tbx1 haploinsufficiency in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410:97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- Macatee T.L., Hammond B.P., Arenkiel B.R., Francis L., Frank D.U., Moon A.M. Ablation of specific expression domains reveals discrete functions of ectoderm- and endoderm-derived FGF8 during cardiovascular and pharyngeal development. Development. 2003;130:6361–6374. doi: 10.1242/dev.00850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J., McGlinn E., Huang P., Tabin C.J., McMahon A.P. Fgf-dependent Etv4/5 activity is required for posterior restriction of Sonic Hedgehog and promoting outgrowth of the vertebrate limb. Dev. Cell. 2009;16:600–606. doi: 10.1016/j.devcel.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merscher S., Funke B., Epstein J.A., Heyer J., Puech A., Lu M.M., Xavier R.J., Demay M.B., Russell R.G., Factor S., Tokooya K., Jore B.S., Lopez M., Pandita R.K., Lia M., Carrion D., Xu H., Schorle H., Kobler J.B., Scambler P., Wynshaw-Boris A., Skoultchi A.I., Morrow B.E., Kucherlapati R. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell. 2001;104:619–629. doi: 10.1016/s0092-8674(01)00247-1. [DOI] [PubMed] [Google Scholar]
- Oda T., Elkahloun A.G., Pike B.L., Okajima K., Krantz I.D., Genin A., Piccoli D.A., Meltzer P.S., Spinner N.B., Collins F.S., Chandrasekharappa S.C. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat. Genet. 1997;16:235–242. doi: 10.1038/ng0797-235. [DOI] [PubMed] [Google Scholar]
- Ohtsuka T., Sakamoto M., Guillemot F., Kageyama R. Roles of the basic helix–loop–helix genes Hes1 and Hes5 in expansion of neural stem cells of the developing brain. J. Biol. Chem. 2001;276:30467–30474. doi: 10.1074/jbc.M102420200. [DOI] [PubMed] [Google Scholar]
- Paylor R., Glaser B., Mupo A., Ataliotis P., Spencer C., Sobotka A., Sparks C., Choi C.H., Oghalai J., Curran S., Murphy K.C., Monks S., Williams N., O'Donovan M.C., Owen M.J., Scambler P.J., Lindsay E. Tbx1 haploinsufficiency is linked to behavioral disorders in mice and humans: implications for 22q11 deletion syndrome. Proc. Natl. Acad. Sci. U. S. A. 2006;103:7729–7734. doi: 10.1073/pnas.0600206103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirson S.N., Butler J.N., Foster R.G. Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res. 2003;31:e73. doi: 10.1093/nar/gng073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski T., Nusslein-Volhard C. The endoderm plays an important role in patterning the segmented pharyngeal region in zebrafish (Danio rerio) Dev. Biol. 2000;225:339–356. doi: 10.1006/dbio.2000.9842. [DOI] [PubMed] [Google Scholar]
- Piotrowski T., Ahn D.G., Schilling T.F., Nair S., Ruvinsky I., Geisler R., Rauch G.J., Haffter P., Zon L.I., Zhou Y., Foott H., Dawid I.B., Ho R.K. The zebrafish van gogh mutation disrupts tbx1, which is involved in the DiGeorge deletion syndrome in humans. Development. 2003;130:5043–5052. doi: 10.1242/dev.00704. [DOI] [PubMed] [Google Scholar]
- Prescott K., Ivins S., Hubank M., Lindsay E., Baldini A., Scambler P. Microarray analysis of the Df1 mouse model of the 22q11 deletion syndrome. Hum. Genet. 2005;116:486–496. doi: 10.1007/s00439-005-1274-3. [DOI] [PubMed] [Google Scholar]
- Randall V., McCue K., Roberts C., Kyriakopoulou V., Beddow S., Barrett A.N., Vitelli F., Prescott K., Shaw-Smith C., Devriendt K., Bosman E., Steffes G., Steel K.P., Simrick S., Basson M.A., Illingworth E., Scambler P.J. Great vessel development requires biallelic expression of Chd7 and Tbx1 in pharyngeal ectoderm in mice. J. Clin. Invest. 2009;119:3301–3310. doi: 10.1172/JCI37561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts C., Ivins S., Cook A.C., Baldini A., Scambler P.J. Cyp26 genes a1, b1 and c1 are down-regulated in Tbx1 null mice and inhibition of Cyp26 enzyme function produces a phenocopy of DiGeorge Syndrome in the chick. Hum. Mol. Genet. 2006;15:3394–3410. doi: 10.1093/hmg/ddl416. [DOI] [PubMed] [Google Scholar]
- Rochais F., Dandonneau M., Mesbah K., Jarry T., Mattei M.G., Kelly R.G. Hes1 is expressed in the second heart field and is required for outflow tract development. PLoS ONE. 2009;4:e6267. doi: 10.1371/journal.pone.0006267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodewald H.R. Thymus organogenesis. Annu. Rev. Immunol. 2008;26:355–388. doi: 10.1146/annurev.immunol.26.021607.090408. [DOI] [PubMed] [Google Scholar]
- Rutenberg J.B., Fischer A., Jia H., Gessler M., Zhong T.P., Mercola M. Developmental patterning of the cardiac atrioventricular canal by Notch and Hairy-related transcription factors. Development. 2006;133:4381–4390. doi: 10.1242/dev.02607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata Y., Kamei C.N., Nakagami H., Bronson R., Liao J.K., Chin M.T. Ventricular septal defect and cardiomyopathy in mice lacking the transcription factor CHF1/Hey2. Proc. Natl. Acad. Sci. U. S. A. 2002;99:16197–16202. doi: 10.1073/pnas.252648999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scambler P.J. The 22q11 deletion syndromes. Hum. Mol. Genet. 2000;9:2421–2426. doi: 10.1093/hmg/9.16.2421. [DOI] [PubMed] [Google Scholar]
- Schneider J.E., Bose J., Bamforth S.D., Gruber A.D., Broadbent C., Clarke K., Neubauer S., Lengeling A., Bhattacharya S. Identification of cardiac malformations in mice lacking Ptdsr using a novel high-throughput magnetic resonance imaging technique. BMC Dev. Biol. 2004;4:16. doi: 10.1186/1471-213X-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Stoller J.Z., Epstein J.A. Identification of a novel nuclear localization signal in Tbx1 that is deleted in DiGeorge syndrome patients harboring the 1223delC mutation. Hum. Mol. Genet. 2005;14:885–892. doi: 10.1093/hmg/ddi081. [DOI] [PubMed] [Google Scholar]
- Taddei I., Morishima M., Huynh T., Lindsay E.A. Genetic factors are major determinants of phenotypic variability in a mouse model of the DiGeorge/del22q11 syndromes. Proc. Natl. Acad. Sci. U. S. A. 2001;98:11428–11431. doi: 10.1073/pnas.201127298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Yamasaki N., Izumo S. Phenotypic characterization of the murine Nkx2.6 homeobox gene by gene targeting. Mol. Cell. Biol. 2000;20:2874–2879. doi: 10.1128/mcb.20.8.2874-2879.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Schinke M., Liao H.S., Yamasaki N., Izumo S. Nkx2.5 and Nkx2.6, homologs of Drosophila tinman, are required for development of the pharynx. Mol. Cell. Biol. 2001;21:4391–4398. doi: 10.1128/MCB.21.13.4391-4398.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K., Hattori M., Nakamura E., Nakanishi S., Minato N., Kageyama R. The bHLH gene Hes1 is essential for expansion of early T cell precursors. Genes Dev. 1999;13:1203–1210. doi: 10.1101/gad.13.9.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson D. Whole mount in situ hybridisation of vertebrate embryos. In: Wilkinson D., editor. In situ Hybridisation: a Practical Approach. IRL Press; Oxford: 1992. pp. 75–84. [Google Scholar]
- Xu H., Morishima M., Wylie J.N., Schwartz R.J., Bruneau B.G., Lindsay E.A., Baldini A. Tbx1 has a dual role in the morphogenesis of the cardiac outflow tract. Development. 2004;131:3217–3227. doi: 10.1242/dev.01174. [DOI] [PubMed] [Google Scholar]
- Xu H., Cerrato F., Baldini A. Timed mutation and cell-fate mapping reveal reiterated roles of Tbx1 during embryogenesis, and a crucial function during segmentation of the pharyngeal system via regulation of endoderm expansion. Development. 2005;132:4387–4395. doi: 10.1242/dev.02018. [DOI] [PubMed] [Google Scholar]
- Xu H., Viola A., Zhang Z., Gerken C.P., Lindsay-Illingworth E.A., Baldini A. Tbx1 regulates population, proliferation and cell fate determination of otic epithelial cells. Dev. Biol. 2007;302:670–682. doi: 10.1016/j.ydbio.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi H., Furutani Y., Hamada H., Sasaki T., Asakawa S., Minoshima S., Ichida F., Joo K., Kimura M., Imamura S., Kamatani N., Momma K., Takao A., Nakazawa M., Shimizu N., Matsuoka R. Role of TBX1 in human del22q11.2 syndrome. Lancet. 2003;362:1366–1373. doi: 10.1016/s0140-6736(03)14632-6. [DOI] [PubMed] [Google Scholar]
- Yuan J.S., Wang D., Stewart C.N., Jr. Statistical methods for efficiency adjusted real-time PCR quantification. Biotechnol. J. 2008;3:112–123. doi: 10.1002/biot.200700169. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Baldini A. In vivo response to high-resolution variation of Tbx1 mRNA dosage. Hum. Mol. Genet. 2008;17:150–157. doi: 10.1093/hmg/ddm291. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Cerrato F., Xu H., Vitelli F., Morishima M., Vincentz J., Furuta Y., Ma L., Martin J.F., Baldini A., Lindsay E. Tbx1 expression in pharyngeal epithelia is necessary for pharyngeal arch artery development. Development. 2005;132:5307–5315. doi: 10.1242/dev.02086. [DOI] [PubMed] [Google Scholar]
- Zweier C., Sticht H., Aydin-Yaylagul I., Campbell C.E., Rauch A. Human TBX1 missense mutations cause gain of function resulting in the same phenotype as 22q11.2 deletions. Am. J. Hum. Genet. 2007;80:510–517. doi: 10.1086/511993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 3
1567 Genes downregulated (including Df1 genes) to 0.83 in 5 out of 6 Df1/Tbx1lacZ chips compared to Tbx1+/lacZ chips
Frequency of genotypes obtained at E14.5-E15.5 from Hes1+/- intercrosses.
qRT-PCR primers.