Abstract
The shape of motile cells is determined by many dynamic processes spanning several orders of magnitude in space and time, from local polymerization of actin monomers at subsecond timescales to global, cell-scale geometry that may persist for hours. Understanding the mechanism of shape determination in cells has proved to be extremely challenging due to the numerous components involved and the complexity of their interactions. Here we harness the natural phenotypic variability in a large population of motile epithelial keratocytes from fish (Hypsophrys nicaraguensis) to reveal mechanisms of shape determination. We find that the cells inhabit a low-dimensional, highly correlated spectrum of possible functional states. We further show that a model of actin network treadmilling in an inextensible membrane bag can quantitatively recapitulate this spectrum and predict both cell shape and speed. Our model provides a simple biochemical and biophysical basis for the observed morphology and behaviour of motile cells.
Cell shape emerges from the interaction of many constituent elements—notably, the cytoskeleton, the cell membrane and cell–substrate adhesions—that have been studied in great detail at the molecular level1–3; however, the mechanism by which global morphology is generated and maintained at the cellular scale is not understood. Many studies have characterized the morphological effects of perturbing various cytoskeletal and other cellular components (for example, ref. 4); yet, there have been no comprehensive efforts to try to understand cell shape from first principles. Here we address this issue in the context of motile epithelial keratocytes derived from fish skin. Fish keratocytes are among the fastest moving animal cells, and their motility machinery is characterized by extremely rapid molecular dynamics and turnover5–8. At the same time, keratocytes are able to maintain nearly constant speed and direction during movement over many cell lengths. Their shapes, consisting of a bulbous cell body at the rear attached to a broad, thin lamellipodium at the front and sides, are simple, stereotyped and notoriously temporally persistent9,10. The molecular dynamism of these cells, combined with the persistence of their global shape and behaviour, make them an ideal model system for investigating the mechanisms of cell shape determination.
The relative simplicity of keratocytes has inspired extensive experimental and theoretical investigations into this cell type5–17, considerably advancing the understanding of cell motility. A notable example is the graded radial extension (GRE) model12, which was an early attempt to link the mechanism of motility at the molecular level with overall cell geometry. The GRE model proposed that local cell extension (either protrusion or retraction) occurs perpendicular to the cell edge, and that the magnitude of this extension is graded from a maximum near the cell midline to a minimum towards the sides. Although this phenomenological model has been shown experimentally to describe keratocyte motion, it does not consider what generates the graded extension rates, neither does it explain what determines the cellular geometry in the first place. Thus, even for these simple cells, it has remained unclear how the biochemical and biophysical molecular dynamics underlying motility give rise to large-scale cell geometry. In this work we address this question by exploiting the natural phenotypic variability in keratocytes to measure the relations among cell geometry, actin distribution and motility. On the basis of quantitative observations of a large number of cells, we have developed a model that relates overall cell geometry to the dynamics of actin network treadmilling and the forces imposed on this network by the cell membrane. This model is able to quantitatively explain the main features of keratocyte shapes and to predict the relationship between cell geometry and speed.
Low-dimensional keratocyte shape space
Individual keratocytes assume a variety of cell shapes (Fig. 1a). A quantitative characterization18,19 of a large population of live keratocytes revealed that keratocyte shapes are well described with just four orthogonal modes of shape variability (Fig. 1b), which together account for ~97% of the total variation in shape. Roughly, these modes can be characterized as measures of: the projected cell area (mode 1); whether the cell has a rounded ‘D’ shape or an elongated ‘canoe’ shape (mode 2)11; the angle of the rear of the lamellipodium with respect to the cell body (mode 3); and the left–right asymmetry of the side lobes (mode 4). These shape modes provide a meaningful and concise quantitative description of keratocyte morphology using very few parameters. Specifically, over 93% of the cell-to-cell shape variation can be captured by recording only two parameters per cell: the cell’s position along shape modes 1 and 2, or, essentially equivalently, its projected area and aspect ratio. Two additional parameters are required to describe the detailed shape of the rear of the cell (shape modes 3 and 4). The existence of only a few meaningful modes implies that the phase space in which keratocytes reside is a relatively small subregion of the space of all possible shapes.
Figure 1. Keratocyte shapes are described by four primary shape modes.
a, Phase-contrast images of different live keratocytes illustrate the natural shape variation in the population. b, The first four principal modes of keratocyte shape variation, as determined by principal components analysis of 710 aligned outlines of live keratocytes, are shown. These modes—cell area (shape mode 1), ‘D’ versus ‘canoe’ shape (shape mode 2), cell-body position (shape mode 3), and left–right asymmetry (shape mode 4)—are highly reproducible; subsequent modes seem to be noise. For each mode, the mean cell shape is shown alongside reconstructions of shapes one and two standard deviations away from the mean in each direction along the given mode. The variation accounted for by each mode is indicated. (Modes one and two are scaled as in a; modes three and four are 50% smaller.)
To investigate further the role of various molecular processes in determining cell shape, we targeted specific components of the cytoskeleton in live cells with pharmacological agents that affect actin dynamics or myosin activity. The different treatments elicited statistically significant morphological changes (Supplementary Fig. 1), but their extent was rather small. In particular, the natural shape variation in the population (Fig. 1) was substantially larger than the shifts induced by any of the perturbations (Supplementary Fig. 1). Furthermore, whereas the shape of an individual cell can be significantly affected by such perturbations11, the phase space of cell shapes under the perturbations tested was nearly identical to that spanned by the population of unperturbed cells (Supplementary Fig. 1). This led us to focus on the phenotypic variability in unperturbed populations, which, as described, provided significant insight into the underlying mechanisms of shape determination.
Cell shape is dynamically determined
The natural phenotypic variability described presents a spectrum of possible functional states of the system. To better characterize these states, we measured cell speed, area, aspect ratio and other morphological features in a large number of live cells (Fig. 2a) and correlated these traits across the population (Fig. 2b; see also the Supplementary Information). To relate these measures to cellular actin dynamics, we concurrently examined the distribution of actin filaments along the leading edge. To visualize actin filaments in live cells, we used low levels of tetramethylrhodamine (TMR)-derivatized kabiramide C, which at low concentrations binds as a complex with G-actin to free barbed ends of actin filaments20,21, so that along the leading edge the measured fluorescence intensity is proportional to the local density of filaments.
Figure 2. Quantitative and correlative analysis of keratocyte morphology and speed.
a, The distributions of measures across a population of live keratocytes (left panels) are contrasted with values through time for 11 individual cells (right). Within each histogram, the population mean ± one standard deviation is shown by the left vertical bar, whereas the population mean ± the average standard deviation exhibited by individual cells over 5 min is shown by the right bar. b, Significant pair-wise correlations (P <0.05; bootstrap confidence intervals) within a population of keratocytes are diagrammed (left panel). Two additional measures are included: front roughness, which measures the local irregularity of the leading edge, and actin ratio, which represents the peakedness of the actin distribution along the leading edge. The correlations indicate that, apart from size differences, cells lie along a single phenotypic continuum (right panel), from ‘decoherent’ to ‘coherent’. Decoherent cells move slowly and assume rounded shapes with low aspect ratios and high lamellipodial curvatures. The actin network is less ordered, with ragged leading edges and low actin ratios. Coherent cells move faster and have lower lamellipodial curvature. The actin network is highly ordered with smooth leading edges and high actin ratios. c, Phase-contrast images depict a cell transiently treated with DMSO (Supplementary Movie 1), which caused a reversible inhibition of motility and loss of the lamellipodium. Images shown correspond to before (20 s), during (610 s) and two time points after (830 s and 1,230 s) the perturbation. d, Time traces of area, aspect ratio and speed for the cell in c show that shape and speed are regained post perturbation. Dashed lines show time points from c; arrowheads indicate the time of perturbation. e, Area, aspect ratio and speed of nine cells are shown as averages obtained from one-minute windows before, during and after DMSO treatment (shown sequentially from left to right for each cell). The cell shown in c and d is highlighted.
The phenotypic variability in our test population is depicted in the histograms shown in Fig. 2a. We further characterized this variability by following several individual cells over time. Particularly notable was the observation that the projected cell area, although quite variable across the population, was essentially constant for a given cell (Fig. 2a). This suggests that the area, probably determined by the total amount of available plasma membrane or by tight regulation of the membrane surface area, is intrinsic to each cell and constant through time. Individual cells showed larger variability in other measures such as speed and aspect ratio; nevertheless, in every case, individual variability remained smaller than that of the population as a whole (Fig. 2a). The measured properties correlate well across the data set (Fig. 2b and Supplementary Fig. 2), producing a phenotypic continuum that we have described previously11: from rough, slow and rounded ‘decoherent’ cells, to smooth, fast and wide ‘coherent’ cells that exhibit a more pronounced peak in actin filament density at the centre.
To examine the role that the particular history of a given cell has in determining cell morphology, we confronted keratocytes with an acute perturbation—transient treatment with high concentrations of dimethylsulphoxide (DMSO)—which resulted in temporary lamellipodial loss and cell rounding22. We found that cells were able to resume movement (albeit in an arbitrary direction with respect to their orientation before DMSO treatment) and return to their original morphology and speed within minutes (Fig. 2c–e), comparable to the characteristic timescales of the underlying molecular processes such as actin assembly and disassembly and adhesion formation5–8,23. This rapid recovery of pre-perturbation properties suggests that the observed, persistent behaviour of keratocytes is a manifestation of a dynamic system at steady state. Taken together, our results imply that cell shape and speed are determined by a history-independent self-organizing mechanism, characterized by a small number of cellular parameters that stay essentially constant over time (such as available quantities of membrane or cytoskeletal components), independent of the precise initial localization of the components of the motility machinery.
Actin/membrane model explains cell shape
We set out to develop a quantitative physical model of cell shape and movement that could explain this observed spectrum of keratocyte behaviour. Specifically, we sought to describe mechanistically the shape variability captured in the first two principal modes of keratocyte shape (Fig. 1b; comprising over 93% of the total shape variation), setting aside the detailed shape of the cell rear. Two observations—first, that cell area is constant (Fig. 2a), and second, that the density of filamentous actin along the leading edge is graded (Fig. 3a,b)—are central to our proposed mechanism of cell shape regulation. In addition, this mechanism is predicated on the basis of previous observations that the lamellipodial actin network undergoes treadmilling, with net assembly at the leading edge and net disassembly towards the rear8,24,25.
Figure 3. A quantitative model explains the main features of keratocyte shapes.
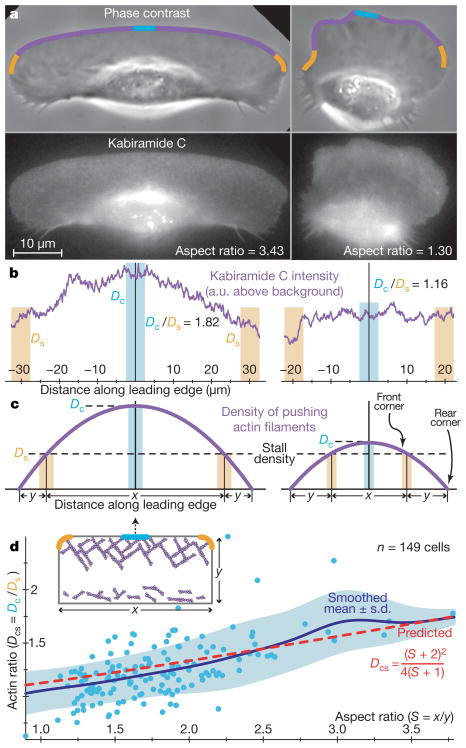
a, Phase-contrast (top) and fluorescence (bottom) images are shown for two live keratocytes stained with TMR-derivatized kabiramide C. The fluorescence intensity reflects the current and past distribution of filament ends, in addition to diffuse background signal from unincorporated probe20. Along the leading edge, the fluorescence intensity is proportional to the local density of actin filaments (see Supplementary Information; 1-μm-wide strips along the leading edge are shown superimposed on the phase-contrast images, with centre and side regions highlighted). b, The average (background-corrected) fluorescence intensity along the strips shown in a is plotted. The cell on the left has a peaked distribution of actin filaments, whereas the actin distribution in the cell on the right is flatter. The ratio of the actin density at the centre (Dc) and sides (Ds; averaged over both sides) of the strip, denoted as Dcs, serves as a robust measure of the peakedness of the distribution. c, The density distribution of pushing actin filaments along the leading edge is approximated as a parabola, with a maximum at the centre. Cells with peaked filamentous actin distributions and, therefore, high Dcs values, have larger regions in which the actin filament density is above the ‘stall’ threshold, and thus have longer protruding front edges (of length x) compared with the length of the stalled/retracting cell sides (y), yielding higher aspect ratios (S =x/y). d, The ratio between actin density at the centre and at the sides, Dcs, is plotted as a function of cell aspect ratio, S. Each data point represents an individual cell. Our model provides a parameter-free prediction of this relationship (red line), which captures the mean trend in the data, plotted as a gaussian-weighted moving average (σ = 0.25; blue line) ± one standard deviation (blue region). Inset: the model of cell shape is illustrated schematically.
We hypothesize that actin polymerization pushes the cell membrane from within, generating membrane tension26. The cell membrane, which has been observed to remain nearly stationary in the cell frame of reference in keratocytes12,14, is fluid and bends easily but is nevertheless inextensible (that is, it can be deformed but not stretched)27. Forces on the membrane at any point equilibrate within milliseconds26 (see Supplementary Information) so that, on the time-scales relevant for motility, membrane tension is spatially homogenous at all points along the cell boundary. At the leading edge, membrane tension imposes an opposing force on growing actin filaments that is constant per unit edge length, so that the force per filament is inversely proportional to the local filament density. At the centre of the leading edge, where filament density is high (Fig. 3a–c), the membrane resistance per filament is small, allowing filaments to grow rapidly and generate protrusion. As filament density gradually decreases towards the cell sides, the forces per filament caused by membrane tension increase until polymerization is stalled at the far sides of the cell, which therefore neither protrude nor retract. At the rear of the cell, where the actin network disassembles, membrane tension, assisted by myosin contraction, crushes the weakened network and moves actin debris forward, thereby retracting the cell rear (Fig. 3d, inset). Membrane tension, which is spatially constant, thus induces a direct coupling between molecular processes occurring at distant regions of the cell and contributes to the global coordination of those processes. The Supplementary Information discusses alternative hypotheses regarding cell shape determination that are inconsistent with our measurements (Supplementary Fig. 3).
This qualitative model can be mathematically specified and quantitatively compared to our data set as follows (see Supplementary Table 1 for a list of model assumptions, and Supplementary Information for further details). As discussed previously (Fig. 1), keratocyte shapes can largely be described by two parameters: shape modes 1 and 2, which essentially correspond to cell area (A) and aspect ratio (S), respectively. Thus, for simplicity, we begin by approximating cells as rectangles with width x and length y (A = xy, S = x/y, and the total leading edge length (front and sides) is ). The observed steady-state centre-peaked distribution of actin filaments along the leading edge (D) can be described as a parabola: , where l is the arc distance along the leading edge (l =0 at the cell midline), β is the total number of nascent actin filaments that branch off from existing growing filaments per cell per second, and γ is the rate of capping of existing filaments (Fig. 3c; see Supplementary Information for derivation). We make the further assumption (described previously) that actin filament protrusion is mechanically stalled by the membrane tension T at the sides of the front of the lamellipodium (l = ± x/2). The force acting on each filament at the sides must therefore be approximately equal to the force required to stall a single actin filament28, fstall, which has been measured29,30, so that: . We find that the peak actin density Dc = D(0) fluctuates more than Ds across the population and in individual cells through time (Supplementary Fig. 4; Supplementary Information), suggesting that most of the shape variation observed correlates with differences in actin dynamics rather than changes in membrane tension.
This simple model provides a direct link between the distribution of filamentous actin and overall cell morphology. From the previous equations, this link can be expressed as a relation between the ratio of actin filament density at the centre (l = 0) versus the sides (l = ± x/2) of the leading edge, denoted Dcs, and the aspect ratio of the cell, S: . Thus, cells with relatively more actin filament density at the centre than the sides (high Dcs) have higher aspect ratios, whereas cells with low Dcs ratios have aspect ratios closer to one. As shown in Fig. 3d, the correlation between Dcs and S in our measurements closely follows this model prediction, which, importantly, involves no free parameters. The model is further supported by perturbation experiments, in which, for example, increasing the capping rate γ (by treatment with cytochalasin D) led to the predicted decrease in cell aspect ratio (Supplementary Fig. 1; Supplementary Information). Remarkably, all the model parameters apart from area can be combined into a single parameter: , which signifies the ratio of the membrane tension to the force needed to stall actin network growth at the centre of the leading edge. This key parameter can be expressed in multiple ways: ; that is, in terms of the membrane tension, filament stall force, and branching and capping rates; in terms of the measurable geometry of the cell alone; or in terms of the actin density ratio and cell geometry (see also Supplementary Fig. 5). Thus, this model describes the basic relation between actin network dynamics at the molecular level and overall actin network structure and shape at the cellular scale using only two biologically relevant parameters: z and A.
Shape, speed and lamellipodial radius
To describe cell shape with more accuracy and to relate cell speed to morphology, we must consider the relationship between the growth rate of actin filaments and the magnitude of force resisting their growth. This so-called force–velocity relationship can be used to determine the protrusion rate at the leading edge, and thus cell speed, from the forces exerted by the membrane against the growing lamellipodial actin network. Because membrane tension is the same everywhere along the leading edge, although the filamentous actin density is peaked at the centre of the leading edge, the resistive force per filament increases with distance from the centre. As a result, local protrusion rates decrease smoothly from the centre towards the sides of the leading edge (where, as above, protrusion is stalled). Assuming that protrusion is locally perpendicular to the cell boundary, this implies that the sides of the leading edge lag behind the centre, causing the leading edge to become curved as observed (Fig. 1a; such a relation between geometry and spatially variable protrusion rates was first described in the GRE model12). Thus, keratocytes can be more accurately described as slightly bent rectangles, characterized by the radius of curvature of their leading edge, R, and their overall rate of movement (Fig. 4), in addition to their width and length.
Figure 4. An extended model predicts lamellipodial curvature and the relationship between speed and morphology.
a, The radius of curvature of the leading edge calculated within the model as a function of A and S, , with and , is plotted against the measured radius of curvature (Rm, radius of best-fit circle of the front 40% of the cell). The red dashed line depicts Rc = Rm. b, Cell speed, Vcell, is shown as a function of cell aspect ratio, S. The model prediction (red line; V0 determined empirically) is compared to the trend plotted as a gaussian-weighted moving average (σ =0.25; blue line) ± one standard deviation (blue region), from 695 individual cells (blue points). Purple crosses indicate the mean ± one standard deviation in speed and aspect ratio over 5 min for 11 individual cells (shown in Fig. 2a).
Given a particular force–velocity relation, both cell speed and lamellipodial radius can be expressed, in the context of this model, solely in terms of the parameters A and z. Thus, speed and radius are predicted to vary with cell area and aspect ratio, providing further tests of the model. The exact form of the force–velocity relation for the lamellipodial actin network is unknown. Measurements in branched actin networks, both in motile keratocytes16 and assembled in cytoplasmic extracts31, yielded force–velocity relations that were concave down: that is, the protrusion rate was insensitive to force at weak loads (relative to the stall force), whereas at greater loads the speed decreased markedly. Regardless of its precise functional dependence, as long as the force–velocity relation entails such a monotonic concave-down decrease in protrusion velocity with increasing membrane tension, the predicted trends in cell speed and lamellipodium radius correlate well with our experimental observations (Supplementary Fig. 6). We find good quantitative agreement between the model and our observations using a force–velocity relation given by , where w =8 (Fig. 4).
By combining this force–velocity relation with the geometric formulae of the GRE model, we obtain (see Supplementary Information), which predicts the radius of curvature of a cell’s leading edge from its area and aspect ratio alone. Figure 4a demonstrates the close agreement between the measured and the calculated radii of curvature. At the centre of the leading edge, f = T/Dc; therefore, . Thus, a cell’s speed can be predicted from its aspect ratio, with more canoe-like cells expected to move faster. We find that the trend of the experimental data agrees with our predictions (Fig. 4b), and, in particular, shows the predicted saturation of speed with increasing aspect ratio. We expect cell-to-cell variation in some of the model parameters that determine cell speed such as the concentration of actin monomers and the fraction of pushing actin filaments, as well as in the rate of retrograde actin flow with respect to the substrate13,17. Without detailed per-cell measurements of these, we use constant values that reflect the population mean, allowing correct prediction of population trends, whereas some aspects of cell-to-cell variation remain unexplained.
Discussion
We have used correlative approaches to map quantitatively the functional states of keratocyte motility from a large number of observations of morphology, speed and actin network structure in a population of cells. This data set provided the basis for and constraints on a quantitative model of cell shape that requires only two cell-dependent parameters; these parameters are measurable from cell geometry alone and are closely related to the two dimensions of a phase space that accounts for over 93% of all keratocyte shape variation. Although conceptually quite straightforward, our model describes connections between dynamic events spanning several orders of magnitude in space and time and is, to our knowledge, the first quantitative approach relating molecular mechanisms to cell geometry and movement. The model is able to explain specific properties of keratocyte shape and locomotion on the basis of a coupling of tension in the cell membrane to the dynamics of the treadmilling network of actin filaments. Overall, the picture is very simple: actin network treadmilling (characterized by the z parameter) drives from within the forward protrusion of an inextensible membrane bag (characterized in two dimensions by its total area). Such a scenario was suggested over a decade ago32, but prior to this work had never been tested. Furthermore, this basic mechanism seems to be sufficient to explain the persistent and coordinated movement of keratocytes without incorporating regulatory elements such as microtubules, morphogens or signalling molecules33, suggesting that, at least in keratocytes, these elements are dispensable or redundant.
The model highlights the important regulatory role of membrane tension in cell shape determination: actin assembly at the leading edge and disassembly at the cell rear are both modulated by forces imposed on the actin network by the membrane. Moreover, because membrane tension is constant along the cell boundary, it effectively couples processes (such as protrusion and retraction) that take place in spatially distinct regions of the cell. On the basis of our results, we estimate the membrane tension in motile keratocytes to be on the order of 100 pN μm−1 (see Supplementary Information), similar to the results of experiments that estimated membrane tension from the force on a tether pulled from the surface of motile fibroblasts34.
Our model does not specifically address adhesion or the detailed shape of the cell rear (captured in shape modes 3 and 4; Fig. 1b). Nevertheless, adhesive contacts to the substrate are obviously essential for the cell to be able to generate traction and to move forward. We assume implicitly that the lamellipodial actin network is attached to the substrate, which allows polymerization to translate into cellular protrusion. This assumption is consistent with experimental evidence indicating that the actin network in the keratocyte lamelli-podium is nearly stationary with respect to the substrate8,13,17. The rear boundary of the cell is also implicit in our model, and is set by the position of the ‘rear corners’ of the lamellipodium: the locations at which the density of actin filaments actively pushing against the cell membrane falls to zero. Thus, we do not address the possible contribution of myosin contraction in retracting the cell rear and disassembling the actin network7,26 (see Supplementary Information).
Our results emphasize that careful quantitative analysis of natural cell-to-cell variation can provide powerful insight into the molecular mechanisms underlying complex cell behaviour. A rapidly moving keratocyte completely rebuilds its cytoskeleton and adhesive structures every few minutes, generating a cell shape that is both dynamically determined and highly robust. This dynamic stability suggests that shape emerges from the numerous molecular interactions as a steady-state solution, without any simple central organizing or bookkeeping mechanism. In this work, we relied on several decades of detailed mechanistic studies on the molecular mechanisms involved to derive a physically realistic model for large-scale shape determination. This model is directly and quantitatively coupled to the molecular-scale dynamics and has surprising predictive power. As individual functional modules within cells are unveiled at the molecular level, understanding their large-scale integration is becoming an important challenge in cell biology. To this end, we propose that the biologically rich cell-to-cell variability present within all normal populations represents a fruitful but currently underused resource of mechanistic information regarding complex processes such as cell motility.
METHODS SUMMARY
Cell culture
Keratocytes were isolated from the scales of the Central American cichlid H. nicaraguensis and were cultured as described previously11. TMR-derivatized kabiramide C was added to cells in culture medium for 5 min and subsequently washed20. DMSO treatment consisted of either application of 2–5 μl DMSO directly onto cells or addition of 10% DMSO to the culture medium.
Microscopy
Cells were imaged in a live-cell chamber at room temperature (~23 °C) on a Nikon Diaphot300 microscope using a ×60 lens (numerical aperture, 1.4). To obtain velocity information, for each coverslip, 15–30 randomly chosen cells were imaged twice, 30 s apart. Time-lapse movies of individual cells were acquired at 10-s intervals.
Shape analysis
Cell morphology was measured from manually defined cell shapes, as described previously11,19. ‘Shape modes’ were produced by performing principal components analysis on the population of cell shapes after mutual alignment.
Supplementary Material
Acknowledgments
We thank C. Lacayo, C. Wilson and M. Kozlov for discussion, and P. Yam, C. Lacayo, E. Braun and T. Pollard for comments on the manuscript. K.K. is a Damon Runyon Postdoctoral Fellow supported by the Damon Runyon Cancer Research Foundation, and a Horev Fellow supported by the Taub Foundations. A.M. is supported by the National Science Foundation grant number DMS-0315782 and the National Institutes of Health Cell Migration Consortium grant number NIGMS U54 GM64346. J.A.T. is supported by grants from the National Institutes of Health and the American Heart Association.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions Z.P., K.K., E.L.B., G.M.A. and J.A.T. designed the experiments. K.K., G.M.A., E.L.B. and Z.P. performed the experiments. Z.P. together with K.K., A.M., G.M.A. and E.L.B. analysed the data. A.M. together with K.K., Z.P., E.L.B., G.M.A. and J.A.T. developed the model. G.M. provided the kabiramide C probe. Z.P., K.K., A.M. and J.A.T. wrote the paper. All authors discussed the results and commented on the manuscript.
Author Information Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Carlier MF, Pantaloni D. Control of actin assembly dynamics in cell motility. J Biol Chem. 2007;282:23005–23009. doi: 10.1074/jbc.R700020200. [DOI] [PubMed] [Google Scholar]
- 2.Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- 3.Zaidel-Bar R, Cohen M, Addadi L, Geiger B. Hierarchical assembly of cell-matrix adhesion complexes. Biochem Soc Trans. 2004;32:416–420. doi: 10.1042/BST0320416. [DOI] [PubMed] [Google Scholar]
- 4.Bakal C, Aach J, Church G, Perrimon N. Quantitative morphological signatures define local signaling networks regulating cell morphology. Science. 2007;316:1753–1756. doi: 10.1126/science.1140324. [DOI] [PubMed] [Google Scholar]
- 5.Anderson KI, Cross R. Contact dynamics during keratocyte motility. Curr Biol. 2000;10:253–260. doi: 10.1016/s0960-9822(00)00357-2. [DOI] [PubMed] [Google Scholar]
- 6.Lee J, Jacobson K. The composition and dynamics of cell–substratum adhesions in locomoting fish keratocytes. J Cell Sci. 1997;110:2833–2844. doi: 10.1242/jcs.110.22.2833. [DOI] [PubMed] [Google Scholar]
- 7.Svitkina TM, Verkhovsky AB, McQuade KM, Borisy GG. Analysis of the actin–myosin II system in fish epidermal keratocytes: mechanism of cell body translocation. J Cell Biol. 1997;139:397–415. doi: 10.1083/jcb.139.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Theriot JA, Mitchison TJ. Actin microfilament dynamics in locomoting cells. Nature. 1991;352:126–131. doi: 10.1038/352126a0. [DOI] [PubMed] [Google Scholar]
- 9.Euteneuer U, Schliwa M. Persistent, directional motility of cells and cytoplasmic fragments in the absence of microtubules. Nature. 1984;310:58–61. doi: 10.1038/310058a0. [DOI] [PubMed] [Google Scholar]
- 10.Goodrich HB. Cell behavior in tissue cultures. Biol Bull. 1924;46:252–262. [Google Scholar]
- 11.Lacayo CI, et al. Emergence of large-scale cell morphology and movement from local actin filament growth dynamics. PLoS Biol. 2007;5:e233. doi: 10.1371/journal.pbio.0050233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J, Ishihara A, Theriot JA, Jacobson K. Principles of locomotion for simple-shaped cells. Nature. 1993;362:167–171. doi: 10.1038/362167a0. [DOI] [PubMed] [Google Scholar]
- 13.Jurado C, Haserick JR, Lee J. Slipping or gripping? Fluorescent speckle microscopy in fish keratocytes reveals two different mechanisms for generating a retrograde flow of actin. Mol Biol Cell. 2005;16:507–518. doi: 10.1091/mbc.E04-10-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kucik DF, Elson EL, Sheetz MP. Cell migration does not produce membrane flow. J Cell Biol. 1990;111:1617–1622. doi: 10.1083/jcb.111.4.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grimm HP, Verkhovsky AB, Mogilner A, Meister JJ. Analysis of actin dynamics at the leading edge of crawling cells: implications for the shape of keratocyte lamellipodia. Eur Biophys J. 2003;32:563–577. doi: 10.1007/s00249-003-0300-4. [DOI] [PubMed] [Google Scholar]
- 16.Prass M, Jacobson K, Mogilner A, Radmacher M. Direct measurement of the lamellipodial protrusive force in a migrating cell. J Cell Biol. 2006;174:767–772. doi: 10.1083/jcb.200601159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vallotton P, et al. Tracking retrograde flow in keratocytes: news from the front. Mol Biol Cell. 2005;16:1223–1231. doi: 10.1091/mbc.E04-07-0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cootes TF, Taylor CJ, Cooper DH, Graham J. Active shape models — their training and application. Comput Vis Image Underst. 1995;61:38–59. [Google Scholar]
- 19.Pincus Z, Theriot JA. Comparison of quantitative methods for cell-shape analysis. J Microsc. 2007;227:140–156. doi: 10.1111/j.1365-2818.2007.01799.x. [DOI] [PubMed] [Google Scholar]
- 20.Petchprayoon C, et al. Fluorescent kabiramides: new probes to quantify actin in vitro and in vivo. Bioconjug Chem. 2005;16:1382–1389. doi: 10.1021/bc050006j. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka J, et al. Biomolecular mimicry in the actin cytoskeleton: mechanisms underlying the cytotoxicity of kabiramide C and related macrolides. Proc Natl Acad Sci USA. 2003;100:13851–13856. doi: 10.1073/pnas.2233339100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanger JW, Gwinn J, Sanger JM. Dissolution of cytoplasmic actin bundles and the induction of nuclear actin bundles by dimethyl sulfoxide. J Exp Zool. 1980;213:227–230. doi: 10.1002/jez.1402130210. [DOI] [PubMed] [Google Scholar]
- 23.Watanabe N, Mitchison TJ. Single-molecule speckle analysis of actin filament turnover in lamellipodia. Science. 2002;295:1083–1086. doi: 10.1126/science.1067470. [DOI] [PubMed] [Google Scholar]
- 24.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 25.Wang YL. Exchange of actin subunits at the leading edge of living fibroblasts: possible role of treadmilling. J Cell Biol. 1985;101:597–602. doi: 10.1083/jcb.101.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozlov MM, Mogilner A. Model of polarization and bistability of cell fragments. Biophys J. 2007;93:3811–3819. doi: 10.1529/biophysj.107.110411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheetz MP, Sable JE, Dobereiner HG. Continuous membrane–cytoskeleton adhesion requires continuous accommodation to lipid and cytoskeleton dynamics. Annu Rev Biophys Biomol Struct. 2006;35:417–434. doi: 10.1146/annurev.biophys.35.040405.102017. [DOI] [PubMed] [Google Scholar]
- 28.Schaus TE, Borisy G. Performance of a population of independent filaments in lamellipodial protrusion. Biophys J. doi: 10.1529/biophysj.107.125005. (in the press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Footer MJ, Kerssemakers JW, Theriot JA, Dogterom M. Direct measurement of force generation by actin filament polymerization using an optical trap. Proc Natl Acad Sci USA. 2007;104:2181–2186. doi: 10.1073/pnas.0607052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kovar DR, Pollard TD. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci USA. 2004;101:14725–14730. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parekh SH, Chaudhuri O, Theriot JA, Fletcher DA. Loading history determines the velocity of actin-network growth. Nature Cell Biol. 2005;7:1219–1223. doi: 10.1038/ncb1336. [DOI] [PubMed] [Google Scholar]
- 32.Mitchison TJ, Cramer LP. Actin-based cell motility and cell locomotion. Cell. 1996;84:371–379. doi: 10.1016/s0092-8674(00)81281-7. [DOI] [PubMed] [Google Scholar]
- 33.Ridley AJ, et al. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 34.Raucher D, Sheetz MP. Cell spreading and lamellipodial extension rate is regulated by membrane tension. J Cell Biol. 2000;148:127–136. doi: 10.1083/jcb.148.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gander W, Golub GH, Strebel R. Least-squares fitting of circles and ellipses. BIT. 1994;34:558–578. [Google Scholar]
- 36.Wilson CA, Theriot JA. A correlation-based approach to calculate rotation and translation of moving cells. IEEE Trans Image Process. 2006;15:1939–1951. doi: 10.1109/tip.2006.873434. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.