Abstract
Liver regeneration after a two-thirds partial hepatectomy (PHx) is a complex process requiring interaction and cooperation of many growth factors and cytokines and cross talk between multiple pathways. Along with hepatocyte growth factor and its receptor MET (HGF-MET), the epidermal growth factor receptor (EGFR) signaling pathway is activated within 60 minutes after PHx. To investigate the role of EGFR in liver regeneration, we used two EGFR-specific short hairpin silencing RNAs to inhibit EGFR expression in regenerating normal rat liver. Suppression of EGFR mRNA and protein was evident in treated rats. There was also a demonstrable decrease but not complete elimination of bromo-deoxyuridine incorporation and mitoses at 24 hours after PHx. In addition, we observed up-regulation of MET and Src as well as activation of the ErbB-3-ErbB-2-PI3K-Akt pathway and down-regulation of STAT 3, cyclin D1, cyclin E1, p21, and C/EBP β. The decrease in the ratio of C/EBP α to C/EBP β known to occur after PHx was offset in shEGFR-treated rats. Despite suppression of hepatocyte proliferation lasting into day 3 after PHx, liver weight restoration occurred. Interestingly, hepatocytes in shEGFR-treated rats were considerably larger when compared with ScrRNA-treated controls. The data indicate that although the MET and EGFR pathways are similar, the contributions made by MET and EGFR are unique and are not compensated by each other or other cytokines.
Partial hepatectomy (PHx), in which two thirds of the rat liver is surgically removed, has been extensively used to study the highly complex phenomenon of liver regeneration. Although hepatocytes in normal adult liver are quiescent and rarely divide, they do retain an astounding ability to reenter the cell cycle and regenerate on surgical insult or injury. PHx in rats/mice results in rapid induction of more than 100 genes that are not expressed in the normal resting liver.1 A rapid up-regulation of genes encoding transcriptional factors like AP1 breakdown of extracellular matrix by uPA and release of pre-existing stores of HGF is observed within 60 minutes of a PHx.2 The hepatocytes leave the quiescent G0 phase and enter the cell cycle. Methods to identify extrahepatic signals leading to liver regeneration have included mitogenic effects on hepatocyte cultures, stimulation of DNA synthesis in the liver of normal (unoperated) animals, and decrease in regeneration-related events in animals genetically or pharmacologically depleted of the agent under study. Of the various agents implicated in liver regeneration, HGF and ligands of the epidermal growth factor receptor (EGFR) are the only ones that stimulate DNA synthesis in hepatocyte cultures maintained in chemically defined media.3 They are also the only ones that stimulate DNA synthesis in the liver of normal mice and rats.4,5,6
HGF and EGF signaling pathways are activated within 60 minutes after a PHx,7,8 as evidenced by tyrosine phosphorylation of MET and EGFR within 30 to 60 minutes after PHx. There is evidence of cross talk and cooperation between MET and EGFR, and it is possible that MET transactivates EGFR.9,10,11,12,13,14 The EGFR family contains four members: ErbB-1, ErbB-2, ErbB-3, and ErbB-4. ErbB-3 is expressed in the adult liver but has no intrinsic kinase activity and relies on ErbB-1 for activity,15 whereas ErbB-4 is not expressed in liver. Recently, both ErbB-2 and ErbB-3 have been shown to play a role in appendix regeneration in zebra fish.16 The four ErbB receptors recognize 11 different but structurally related growth factors that mediate diverse processes like development, cell proliferation, and cell survival.17,18 Some of the ligands of EGFR that also increase after PHx and appear to affect liver regeneration are transforming growth factor α,19 Heparin binding EGF (HB-EGF),20 and amphiregulin.21 There is thus a certain redundancy built in the EGFR pathway with multiple ligands with overlapping functions.
The role of EGFR in embryonic development has been demonstrated by targeted deletion of EGFR. The resulting phenotype was dependent on strain and genetic background with abnormalities in various organs like liver, brain, and kidneys.22,23,24 There have been two recent studies that addressed the role of EGFR in liver regeneration after a PHx. In one study, a monoclonal antibody (mAB) targeting EGFR was used to inhibit EGFR,25 and its impact on liver regeneration was studied. In the second study, effects on liver regeneration after liver-specific perinatal deletion of EGFR were studied.26 In the first study by Van Buren et al,25 inhibiting EGFR had no effect on liver regeneration, whereas in the study performed by Natarajan et al,26 mice lacking EGFR exhibited increased mortality and impaired liver regeneration. However, a number of pitfalls such as histopathological changes of using targeted gene deletions have been recognized that can complicate interpretation of results.27,28,29 To avoid pitfalls resulting from adaptations to gene loss30 and histopathological alterations, we investigated the role of EGFR in liver regeneration in rats after a two-thirds PHx by using short hairpin silencing RNA (shRNA) targeting EGFR, a method we have recently applied for the HGF receptor (c-Met).31 Our data show severe effects on hepatocyte proliferation and compensatory increases in expression of ErbB-3, ErbB-2, MET, and Src. Liver weight was restored in part by increase in hepatocyte size.
Materials and Methods
Animals
Male Fisher 344 rats (150 to 200 gram) were obtained from Charles River Laboratories (Frederick, MD). Animals were allowed access to food and water ad libitum. Metofane was used to anesthetize animals. A two-thirds PHx was performed by resecting median and left lateral lobes.32 For controls, a time-matched sham operation was performed. Sham operations involved a laparotomy and resection of the xyphoid process of the sternum. At defined time points, the animals were anesthetized with Nembutal (Abbott, Chicago, IL), and the remaining lobes were removed. All liver samples were promptly frozen in liquid nitrogen and stored at −80°C. All procedures performed on these rats were approved under the Institutional Animal Care and Use Committee protocol number 0507596A and conducted according to National Institutes of Health guidelines.
Antibodies
The following primary antibodies were obtained from Cell Signaling Technology (Danvers, MA): EGFR rabbit mAB (1:1000, clone C74B9); phosphoEGFR Tyr992 (1:1000); PhosphoEGFR Tyr1068 mouse mAB (1:1000, clone 1H12); ErbB-3 rabbit mAB (1:1000, clone 1B2); phosphoErbB-2 Tyr1248, rabbit antibody (1:1000); ErbB-2 mouse mAB (1:1000, clone 44E7); MET mouse mAB (1:1000, clone 25H2); and PI3Kinase p85 rabbit mAB (1:1000, clone 19H8). The following antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA): ErbB-3 (clone G-4, for co-immunoprecipitation studies); glyceraldehyde-3-phosphate dehydrogenase (GAPDH), mouse mAB (1:1000, clone 0411); Platelet derived growth factor receptor (PDGFR), rabbit polyclonal antibody (1:1000, C-20); MET mouse mAB (clone B-2, for co-immunoprecipitation studies); and Cyclin D1 mouse mAB (1:1000, clone CD1.1). β-Actin antibody was from Millipore (Bedford, MA). Secondary antibodies used for this project were donkey anti-rabbit and donkey anti-mouse (Jackson ImmunoResearch Laboratories, West Grove, PA), used at 1:50,000 dilution.
shRNA Design
Rat EGFR-specific silencing RNAs were designed to down-regulate EGFR expression in regenerating rat livers. Two shRNA sequences, E1 and E2, were based on published rat EGFR, NM_031517 sequence. A mismatch sequence differing from shEGFR 2 sequence was also designed: (1) shEGFR 1: 5′-AGTAACAGGCTCACCCAAC-3′; (2) shEGFR 2: 5′-CACCGTGGAGAGAATCCCT-3′; and (3) mismatch shEGFR: 5′-gACgGTGGAGtGtATCCaT-3′.
As a negative control, a scrambled sequence (Invivogen, San Diego, CA) was also used. All sequences were checked against databases by using the Smith-Waterman algorithm.33
Construction of shRNA Plasmid
Oligonucleotides were designed to contain TCAAGAG loop sequence and were cloned in the Bbs1 site of the shRNA-Hh1-gfp ZEO vector (Invivogen). The plasmid also has a Cytomegalovirus (CMV) enhancer/promoter driven Green Fluorescent Protein (GFP): zeo fusion gene that encodes a red-shifted variant of the jellyfish GFP and resistance to zeocin. The insert was cloned downstream of a RNA polymerase III promoter, the human H1 promoter. It is transcribed into a short double stranded RNA (dsRNA) with a hairpin structure (shRNA) consisting of a 21-bp double stranded region corresponding to the target sequence and a small loop formed by the spacer region. Competent GT116 Escherichia coli (Invivogen) were transformed and selected by growing them in media lysogeny broth (LB) in the presence of 50 μg/ml of zeocin. Positive clones were confirmed by restriction digestion and sequencing.
shRNA Injections
A mixture of two EGFR-specific endotoxin-free shRNA plasmids at 300 μg was diluted in 5% glucose and was complexed with linear Polyethylenimine (In vivo JET PEI; Q-Biogene, Carlsbad, CA) at an N/P ratio of six, following the manufacturer’s protocol. The gene-specific shRNA/PEI complex was injected (in separate experiments) via the superior mesenteric vein 1 day before partial PHx. A second dose of 300 μg/rat was administered at the time of PHx. The mismatch and scrambled plasmids were similarly injected.
Cell Proliferation Index Estimation
Two parameters viz. bromo-deoxyuridine (BrdU) index and mitotic index were estimated following standard protocols. The labeling index corresponds to the ratio between positive nuclei to the total hepatocyte population. Three rats per time point per treatment group were used. Results were expressed as mean (±SEM).
BrdU Incorporation Studies
BrdU was injected intraperitoneally within 1 hour after PHx and then every 24 hours until day of sacrifice. All of the cells that had progressed through the cell cycle in a given animal could be assessed by this cumulative approach. Anti-BrdU was obtained from Amersham Biosciences (Buckinghamshire, UK). BrdU antibody was diluted at 1:100 in a nuclease that was received with the antibody kit. For each animal, at least 10 random fields (original magnification, ×200) were observed and a total of 200 to 400 positively stained nuclei counted. Data were plotted as mean ± SEM.
Mitotic Index
Liver tissues were fixed in formalin, paraffin embedded, sectioned, and stained with H&E. Mitotic index was estimated by counting cells in 10 optical fields on each slide by means of light microscopy at ×200 magnification. Results were plotted as mean ± SEM.
Quantitative RT-PCR for EGFR mRNA
RNA Isolation
Livers were harvested from rats at defined time points with three rats per time point (four separate experiments). Each set included animals subjected to PHx alone, PHx + shRNA, PHx + ScrRNA, PHx + mismatch injected set, or a sham operation. In each set, the time points of animal sacrifice ranged from 0 to 72 hours.
Total RNA was isolated by using TRIzol (Invitrogen, Carlsbad, CA). Briefly, 5 mg of liver tissue frozen in liquid nitrogen was homogenized in 1 ml of TRIzol. RNA was purified by using RNeasy kit from Qiagen (Valencia, CA). The isolated RNA was treated with Turbo DNA-Free (Ambion, Austin, TX) according to the manufacturer’s instructions. RNA was quantified by spectrophotometry at 260 nm, and purity was assessed by optical density 260/280 ratio. The RNA was stored at −80°C.
cDNA Synthesis
Five micrograms of total RNA isolated from liver tissue was converted to cDNA by using random hexamers and reverse transcribed by using Superscript III (Invitrogen) following the manufacturer’s protocol. A no Reverse Transcriptase (RT) control was also included.
Real-Time Reverse Transcriptase (RT-PCR)
PCR Primers used for amplification of EGFR, ErbB-3, and ErbB-2 were obtained from SABiosciences (Frederick, MD). Expression levels of EGFR, ErbB-2, and ErbB-3 were determined by Quantitative Reverse Transcriptase Polymerase Chain Reaction (qRT-PCR) using SYBR green, and levels were normalized relative to expression of GAPDH in each sample. Fold change in gene expression was calculated by using the 2(−ΔΔCt) method.34 Reverse transcribed samples were amplified in parallel on an ABI Prism 7000 SDS instrument (Applied Biosystems, Foster City, CA). Quantitative real-time PCR for each sample was performed in triplicate in a 25-μl reaction with 50 ng of cDNA, two picomoles of each primer, and 1X SYBR Green PCR Master Mix (Applied Biosystems). The standard conditions for real-time PCR were as follows: 2 minutes at 50°C, 10 minutes at 95°C followed by 40 cycles of 15 seconds denaturation at 95°C, and elongation at 60°C for 45 seconds. A dissociation curve analysis was performed at the end of every run. A no RT and a no template controls were also included in every run.
Protein Analyses
Tissue Homogenization
Whole liver cell lysates were prepared by homogenizing 100 mg liver tissue in radioimmunoprecipitation assay buffer (1X PBS, 1% NP-40, 0.1% SDS, pH 7.4) containing protease and phosphatase inhibitor mixtures (Sigma, St. Louis, MO), and then centrifuging at 14,000 × g for 30 minutes. Protein concentration was determined by using bicinchoninic acid assay (Sigma). Supernatants were saved at −80°C.
Western Blot Analyses
Whole liver cell lysates (75 μg in NuPAGE LDS Sample Buffer) were separated by using precast 4% to 12% NuPAGE Bis-Tris gels with 1X MOPS (3-(N-morpholino) propanesulfonic acid; Invitrogen), then transferred to Immobilon-P membranes (Millipore) for 1 hour in transfer buffer containing 10% methanol and 0.005% SDS. Membranes were stained with Ponceau-S to verify loading and transfer efficiency. Membranes were probed with primary and secondary antibodies in Tris-buffered saline Tween 20 containing 5% nonfat mild or 5% bovine serum albumin. All antibodies were used at 1:1000 dilution following the manufacturer’s recommendations. Horseradish-peroxidase conjugated secondary antibodies to mouse and rabbit immunoglobulin was used at 1:50,000 (Chemicon, Temecula, CA). Membranes were processed by using SuperSignal West Pico chemiluminescence substrate (Pierce, Rockford, IL) and exposed to X-ray film (Lab Product Sales, Rochester, NY). Films were scanned, and densitometry readings were measured by using ImageJ (NIH). Band density for each time point (three rats per time point) was averaged, plotted in Excel (Microsoft Corporation, Redmond, WA), and standardized to Actin or GAPDH.
Co-Immunoprecipitation Analyses
Normal-, ScrRNA-, and shEGFR-treated rat liver protein lysates (500 μg) were diluted to final volume of 500 μl and precleared by incubation with 1 μg of hamster IgG (Santa Cruz Biotechnology) and 20 μl of Agarose A/G plus beads (Santa Cruz Biotechnology) for 1 hour at room temperature.
For ErbB-3-phosphoErbB-2-PI3K heterodimerization studies, protein complexes were immunoprecipitated from cleared lysates with anti-Erbb-3 antibody (2 μg/500 μg of total protein lysates, G-4, Santa Cruz Biotechnology) or with control hamster IgG overnight at 4°C followed by 5-hour incubation at 4°C with agarose A/G beads. Immune complexes were washed three times with radioimmunoprecipitation assay buffer followed by resuspension in gel-loading sample buffer and 1X reducing agent (Invitrogen). Seventy-five micrograms of immunoprecipitated samples from shEGFR- and ScrRNA-treated and control livers were then electrophoresed and processed as described above. Blots were probed with phosphoErbB-2 Tyr1248 antibody (Cell Signaling Technology), stripped, and reprobed with p85 antibody (clone 19H8; Cell Signaling Technology). Equal protein loading was ascertained by immunoblotting with an ErbB-3 antibody.
For MET-EGFR heterodimerization studies, whole cell lysates were processed as described above, and protein complexes were immunoprecipitated by using MET mouse mAB (2 μg/500 μg of total protein lysates, clone B-2, Santa Cruz Biotechnology) or with control hamster IgG and processed as described above. Blots were probed with EGFR antibody (1:1000, clone C74B9, Cell Signaling Technology). Equal protein loading was ascertained by immunoblotting with a MET antibody.
Affymetrix Chip Analyses
The Affymetrix oligonucleotide chip specific for the rat (U230.2 A chip) containing 10,000 expressed sequences was used for analyzing changes in global gene expression in EGFR-silenced and ScrRNA-treated rats at 3 hours, 12 hours, and 24 hours after PHx. RNA was pooled from three rats per time point/treatment. Sample preparation, data generation, and analyses were as reported previously.35 Normalization and preprocessing of data were performed by using dChip software. Expression intensities were log transformed, and genes with less than 80% present calls or expression level less than 7 or SD smaller than 0.5 were filtered out. A statistical analyses microarrays package was then used to analyze the data with a false discovery rate of 5%. R software (http://www.r-project.org) was used to generate the hierarchical clustering dendrograms and the heat maps for EGFR and control data set. Clustering was based on similarity of gene expression profiles.36
Lack of Off Target Effects after Treatment with shEGFR
shRNA/small-interfering RNA treatment can often result in unexpected silencing of unrelated genes and induction of interferon response.37 Analysis of expression signatures generated by different shRNAs (in this case, gene specific and scrambled) by DNA microarray is a much more stringent test for investigating off target effects and assessing specificity of shRNA mediated gene silencing.37 Expression intensities of the top 500 expressed genes 3 hours and 24 hours after PHx were plotted, and lines were fitted by statistics through the individual expression values.
We also analyzed the expression pattern of seven genes known to be involved in the interferon response. These were IP10, Oligoadenylate synthetase, cox2, interferon α, interferon β, interleukin-6, and interleukin-12. Expression values derived from the oligo-array for each of these genes for the different treatment groups were plotted.
Pathway-Specific qRT-PCR Arrays
We also performed pathway-specific expression profiling of genes associated with cell cycle, apoptosis, and EGFR/platelet-derived growth factor (PDGF) signaling by using pathway-specific PCR arrays (SuperArray Bioscience, Frederick, MD). These arrays are in a 96-well format and contain 84 pathway-specific genes, 6 house keeping genes, and both positive and negative controls and were used to perform pathway-specific expression profiling by qRT-PCR of genes associated with these pathways.
Assays were performed as recommended by the manufacturer. Total RNA was extracted from shEGFR-treated and untreated control rat liver (n = 3) at day 1 after PHx and was converted to cDNA as described earlier. PCR was performed on ABI Prism 7000 Sequence Detector (Applied Biosystems). Data were analyzed by using the 2(−ΔΔCt) method incorporated in the Excel-based (Microsoft Corporation) PCR Array Data Analysis Template provided by the manufacturer. Fold changes were expressed as log-normalized ratios of values from shEGFR-treated/control liver tissues. A positive value indicated up-regulation and a negative value indicated down-regulation in expression as compared with control animals.
Statistical Analyses
Unpaired Student’s t-test was performed for evaluation of significant differences between the control and shRNA-treated animals by using the Minitab computer program (Minitab, Inc, State College, PA). Data are expressed as mean ± SEM. For real-time PCR and Western blot data, P values were determined by student’s t-test with P < 0.05 being significant.
Results
Validation of EGFR Silencing in Regenerating Rat Liver
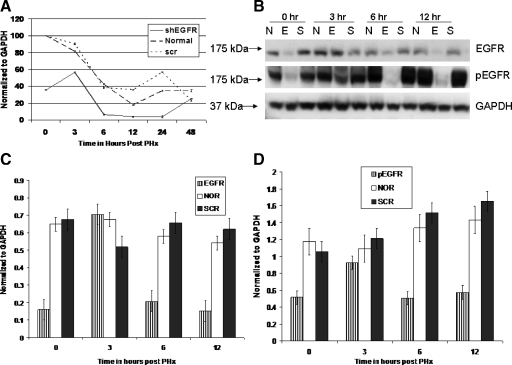
A mixture of the two shEGFR plasmids was complexed with PEI and administered 24 hours before and at the time of PHx.31 A qRT-PCR was performed to estimate levels of EGFR mRNA. Data were normalized to GAPDH. As seen in Figure 1A, significant suppression of EGFR mRNA was evident 24 hours after the first injection (time 0, at the time of PHx). A slight increase was evident at 3 hours after PHx, but the levels were significantly less than ScrRNA-treated and control rats. The levels of EGFR mRNA decreased significantly (P < 0.001) at 6 hours after PHx compared with ScrRNA and untreated controls. The suppression was maintained for 24 hours.
Figure 1.
A: qRT-PCR analysis of EGFR mRNA in shEGFR-treated rats and controls at time points indicated after PHx. Data were normalized to GAPDH. Two injections of the silencing constructs were administered; the first injection was 24 hours before PHx and the second injection was administered at the time of PHx (time 0 on the A, C, and D). We observed significant suppression of EGFR mRNA (P < 0.005) by time 0 that was maintained until 24 hours after PHx. ScrRNA had no effect on EGFR mRNA expression. B: Western blot analysis of total EGFR and PhosphoEGFR in shEGFR-treated rats and controls at indicated time points after PHx. Blot was probed with Tyr 1068 mouse monoclonal phosphoEGFR antibody, stripped, and reprobed with a rabbit polyclonal EGFR antibody. Bands were analyzed by using ImageJ, normalized to GAPDH, and shown in C (EGFR) and D (phospho EGFR). Significant suppression of both EGFR and PhosphoEGFR at 0, 6, and 12 hours after PHx was seen in shEGFR-treated rats. However, for reasons not clear, an up-regulation of EGFR and PhosphoEGFR was seen at three hours after PHx. E, shEGFR-treated rats; N, normal untreated rats; and S, scrambled treated rats.
The reduction in total and phosphoEGFR protein was also quantitated by Western blot analysis followed by densitometry scans by using ImageJ (NIH). Band density for each time point (three rats/point) was averaged, plotted in Excel (Microsoft Corporation), and standardized to GAPDH. Levels of phosphoEGFR were determined by immunoblotting with two different antibodies that detected EGFR phosphorylated at Tyr992 or Tyr1068 as a marker for activated EGFR. A representative immunoblot with phosphoEGFR Tyr1068 is shown in Figure 1B. Highly significant suppression (P < 0.001) in total EGFR (Figure 1C) and phosphoEGFR (Figure 1D) was evident 24 hours after first injection (time 0, at the time of PHx). A significant reduction in total and phosphoEGFR was observed at 6 and 12 hours after PHx. Compared with ScrRNA-treated and control rats, the levels of EGFR/phospho EGFR increased at 3 hours, mirroring the increase in mRNA levels. We cannot explain the up-regulation seen at 3 hours after PHx, but it maybe due to diluting out of the shEGFR plasmid.
Mitotic Index
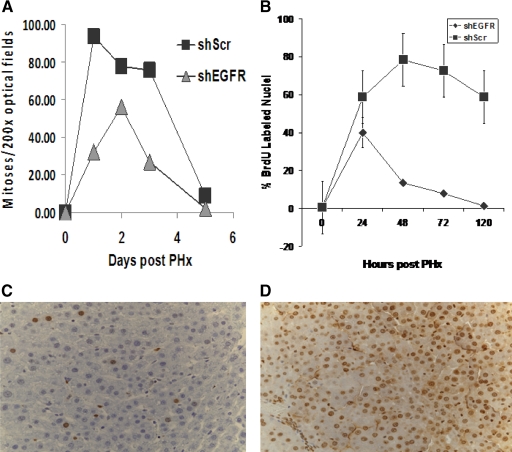
In shEGFR-treated rats there was suppression of mitosis on day 1 (32 mitoses/10 fields) as compared with ∼93 mitoses/10 fields in ScrRNA controls. By day 2, the number of mitoses in shEGFR-treated rats increased to about 56 mitoses/10 fields as compared with 77 mitoses/10 fields in ScrRNA-treated rats (Figure 2A).
Figure 2.
A: Mitotic index was estimated as described in Materials and Methods. At day one, suppression of mitosis was seen in shEGFR-treated rats, compared with controls. B: BrdU index was estimated as described in Materials and Methods. At day one, suppression of BrdU incorporation was seen in shEGFR-treated rats. Nuclei labeled by BRDU immunohistochemistry in rats injected with (C) shRNA and (D) ScrRNA at 48 hours after partial hepatectomy are shown.
BrdU Labeling Index
To further assess the effect of silencing of EGFR on hepatic cellular proliferation, DNA synthesis was monitored by cumulative labeling of hepatocytes by BrdU as described.31 As seen in Figure, 2B, there was significant suppression in BrdU incorporation in shEGFR-treated rats compared with controls. Even though the percentage of BrdU positive hepatocyte nuclei went up in both shRNA- and ScrRNA-treated rats, the percentage of labeled nuclei was less in the shRNA-treated rats and then it dropped considerably below the control levels and did not recover even until the end of the regenerative process. A representative section of nuclei labeled by BrdU of rat livers injected with shRNA or ScrRNA are shown in Figure 2, C and D, respectively.
Expression Patterns for ErbB-3 and ErbB-2
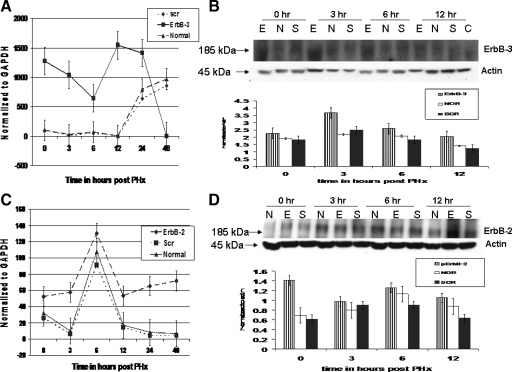
To investigate the possibility of up-regulation of other members of the EGFR family, a qRT-PCR was performed to determine expression levels of ErbB-3 and ErbB-2 in shEGFR-treated rats. As seen in Figure 3A, compared with ScrRNA and control, significant (P < 0.05) up-regulation of ErbB-3 mRNA was observed in rats treated with shEGFR at 3 hours after PHx, and the high levels were maintained until 12 hours after PHx. Increase in ErbB-3 at 6 and 48 hours after PHx has also been previously reported.17
Figure 3.
Analyses of ErbB-3 and ErbB-2 expression in rats treated with shEGFR constructs. Real-time PCR analysis of ErbB-3 mRNA in shEGFR-treated rats. A: Up-regulation of ErbB-3 mRNA at 0, 3, 6, and 12 hours was evident in shEGFR-treated rats compared with controls. The levels decreased dramatically at 24 hours after PHx. Data were normalized to GAPDH by using the 2(−ΔΔCt) method. B: Western blot analysis of total ErbB-3 in shEGFR-treated rats and controls at indicated time points after PHx. Blots were probed with a rabbit ErbB-3 monoclonal antibody. Bands were analyzed by using ImageJ and normalized to Actin. Statistically significant up-regulation of ErbB-3 mRNA and protein was maintained until 12 hours after PHx. C: Real-time RT-PCR analyses of ErbB-2 mRNA in shEGFR-treated rats. A biphasic pattern in ErbB-2 mRNA expression was apparent in shEGFR-treated rats. A sustained increase in ErbB-2 mRNA at 0, 3, and 6 hours after PHx was evident followed by a drop at 12 hours and then an increase at 24 and 48 hours after PHx. Data were normalized to GAPDH. D: Western blot analysis of total ErbB-2 in shEGFR-treated rats and controls at indicated time points after PHx. Blots were probed with a mouse monoclonal ErbB-2 antibody. Bands were analyzed by using ImageJ and normalized to Actin. ErbB-2 protein expression was found to be significantly up-regulated at 0 hours in shEGFR-treated rats compared with ScrRNA-treated and controls. At all other time points, the levels of ErbB-2 in shEGFR-treated rats did not differ significantly from the ScrRNA and untreated controls. E, shEGFR-treated rats; N, normal untreated rats; and S, scrambled treated rats.
We then performed a Western blot analyses of ErbB-3 protein expression in shEGFR-treated, ScrRNA-treated, and control rats. As seen in Figure 3B, a statistically significant (P < 0.05) up-regulation of ErbB-3 at 3 hours after PHx was evident in shEGFR-treated rats compared with controls. The levels were consistently higher than those in ScrRNA and untreated controls at all of the time points examined.
We also determined the expression of ErbB-2 mRNA by qRT-PCR. Expression of ErbB-2 mRNA showed a similar pattern in shRNA-treated, ScrRNA-treated, and untreated controls until 12 hours, with a sharp increase at 6 hours after PHx, followed by a drop at 12 hours after PHx. However, the ErbB-2 mRNA expression was consistently higher in shRNA-treated rats. The mRNA levels in ScrRNA-treated and untreated control rats decreased further at 24 hours, whereas in treated rats the expression levels increased (Figure 3C). A statistically significant (P < 0.05) up-regulation of ErbB-2 mRNA at 3 hours after PHx was evident in shEGFR-treated rats.
ErbB-2 protein levels in treated and control rats were also examined by Western blot analyses as described in Materials and Methods. The ErbB-2 protein levels also displayed a similar pattern of expression in treated and control rats (Figure 3D). A statistically significant up-regulation (P < 0.05) at 0 hours after PHx (24 hours after first injection) was seen in shEGFR-treated rats. The levels were higher than ScrRNA-treated and controls at the time points examined. Compensatory up-regulation in ErbB-2 expression on inactivation of EGFR has also been observed in colon cancer cells.38
Co-Immunoprecipitation Analyses
We performed co-immunoprecipitation studies to investigate heterodimer formation between MET and EGFR and also between ErbB-3-phosphoErbB-2-PI3K. Briefly, whole liver lysates prepared from shEGFR- and ScrRNA-treated and untreated controls were immunoprecipitated with a MET antibody followed by immunoblotting with an EGFR antibody. In a second set of experiments, whole liver lysates were immunoprecipitated with an ErbB-3 antibody and immunoblotted with phosphoErbB-2 and an antibody against the p85 regulatory subunit of PI3K. Control hamster IgG was used in both experiments.
EGFR-MET Interactions
Cross talk between MET and EGFR has been documented in a number of studies.9,10,11,12,13,14 We decided to investigate the interaction between MET and EGFR after PHx in shEGFR-, ScrRNA-treated, and control rats by carrying out an immunoprecipitation analyses. Briefly, total rat liver lysates from treatment and control groups were immunoprecipitated by using a MET mouse mAB (clone B2; Santa Cruz Biotechnology) or control hamster IgG and probed with an EGFR antibody (clone C74B9, Cell Signaling Technology).
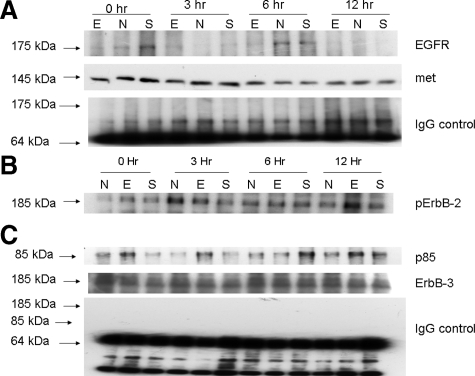
As seen in Figure 4A, more EGFR could be co-immunoprecipitated with MET in ScrRNA and normal untreated controls at 0 and 6 hours after PHx as compared with shEGFR-treated rats that had lower levels of EGFR-MET complex. Interestingly, an increase in MET-EGFR complex was evident at 3 hours after PHx in shEGFR-treated rats, paralleling the observed increase in EGFR mRNA and protein seen at this time point (Figure 1). We were, however, unable to detect ErbB-3 after immunoblotting with ErbB-3 antibody at the time points examined (data not shown).
Figure 4.
Co-immunoprecipitation analyses in treated and control rats. A: MET-EGFR co-immunoprecipitation. Cell lysates were immunoprecipitated with a mouse monoclonal MET antibody and probed with an EGFR antibody as described in Materials and Methods. An increase in MET-co-immunoprecipitated EGFR was evident in ScrRNA and untreated controls compared with shEGFR-treated rats at 0 and 6 hours after PHx. An increase MET-EGFR complex was, however, seen at three hours after PHx samples in shEGFR-treated rats. MET loading control. B: ErbB-3-phosphoErbB-2-PI3K co-immunoprecipitation. ErbB-3-phosphoErbB-2 heterodimer formation was investigated by carrying out co-immunoprecipitation studies. Total liver lysates were immunoprecipitated with an ErbB-3 antibody and probed with a phosphoErbB-2 antibody that detects phosphorylated Tyr 1248 residue. An increase in phosphoErbB-2-ErbB-3 complex was evident in shEGFR-treated rats at 0, 6, and 12 hours after PHx. C: The activation of PI3K-Akt pathway by ErbB-3-phosphoErbB-2 heterodimers was investigated by probing the ErbB-3 immunoblot by a PI3K p85 antibody as described in Materials and Methods. An increase in immunoprecipitable p85 was observed in shEGFR-treated rats at 0, 3, and 12 hours after PHx, compared with controls. ErbB-3 loading control. E, shEGFR-treated rats; N, normal untreated rats; and S, scrambled treated rats.
ErbB-2-ErbB-3-PI3K Interactions
The up-regulation of ErbB-3 and ErbB-2 in shEGFR-treated rats raised the possibility of formation of ErbB-3-ErrbB-2 heterodimers and a compensatory recruitment of PI3K-Akt pathway. To test this hypothesis, ErbB-3 was immunoprecipitated from liver lysates from shEGFR, ScrRNA, and untreated control rat livers with an ErbB-3 antibody (Santa Cruz Biotechnology) or control IgG and probed with phosphoErbB-2 Tyr1248 antibody (Cell Signaling Technology) followed by an antibody that recognizes the p85 regulatory subunit of PI3K (Cell Signaling Technology).
As seen in Figure 4B, an increase in co-immunoprecipitated phosphoErbB-2 was observed at 0, 6, and 12 hours after PHx in shEGFR-treated rats. The blot was stripped and reprobed with PI3K p85 antibody. As seen in Figure 4C, a band at 85 kDa was observed at 0 hours, 3 hours, and 12 hours after PHx in shEGFR-treated rats. This indicated that inhibiting EGFR resulted in an up-regulation of ErbB-3 and ErbB-2 expression, an increase in formation of heterodimerization and recruitment of PI3K-Akt pathway. We were not able to detect MET after immunoblotting with an MET antibody at the time points examined.
ErbB proteins in response to binding of their ligands form homo- or heterodimers followed by trans-autophosphorylation of the ErbB tyrosine kinase domains and initiation of intracellular signaling pathways. For instance, ligand binding to ErbB-3 results in (inactive) homodimerization of ErbB-339 and active ErbB-3-ErbB-2 heterodimerization.15,40 Similar changes in receptor dimerization patterns have also been observed in non-small-cell lung cancer cell line where inactivation of EGFR resulted in stimulation of ErbB-2 and ErbB-3 heterodimer formation41 and recruitment of PI3K-Akt pathway.42,43,44
Microarray Analysis
ShEGFR Cluster Analysis
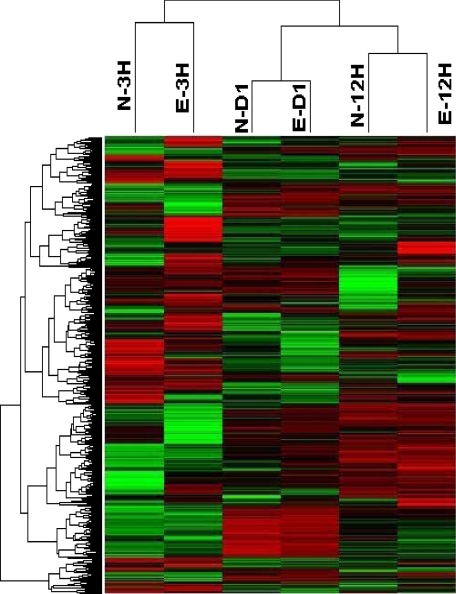
We investigated differentially regulated genes in shEGFR-treated rats by carrying out a microarray analysis at 3, 12, and 24 hours after PHx by using Affymetrix U230.2 chip containing 10,000 expressed sequences. Normalization and preprocessing of data from eight chips were performed by using dChip software. Expression intensities were log transformed, and genes with an expression level less than 7 or SD lower than 0.5 were filtered. Unsupervised hierarchical clustering of the 1231 genes according to similarity in pattern of gene expression produced six clusters. As seen in Figure 5, gene expression profiles cluster according to time after PHx. Even though there were differences in gene expression between shRNA- and ScrRNA-treated rats (Table 1), the differences affected specific gene categories and not the overall pattern, resulting in the clustering shown in Figure 5. This probably indicates that shEGFR treatment did not dramatically alter the gene expression pattern in contrast to that observed in c-met silenced rats (unpublished observation).
Figure 5.
Hierarchical clustering of microarray data obtained from shEGFR and control rats at the time points indicated after PHx. Similarity in pattern of gene expression indicated clustering of genes with similar function. N-3H, normal three hours after PHx; E-3H, shEGFR, three hours after PHx; N-D1, normal day one after PHx; E-D1, shEGFR, day one; N-12 H, normal 12 hours after PHx; and E-12 H, shEGFR, 12 hours after PHx.
Table 1.
Genes Significantly Regulated after shEGFR Treatment at 24 Hours after PHx
| Genes | Up-down regulation (comparing to normal PHx control group) |
|---|---|
| Growth regulated | |
| AKT 1 | 2.98 |
| Cyclin E1 | −6.68 |
| GSK3b | 2.6 |
| Cyclin A1 | −4.47 |
| Cyclin D1 | −5.05 |
| Cyclin A2 | −10.55 |
| Cyclin B1 | −13.93 |
| Cyclin B21 | −11.23 |
| Rad 17 | −7.34 |
| P21 | −3.57 |
| NFkb1 | 2.98 |
| Cyclin D2 | −2.03 |
| c-met | 6.89 |
| CDK2 | −2.06 |
| Cdkn2a | −3.82 |
| Cdkn1b | 2.36 |
| ATM | −2.37 |
| Chek 1 | −3.96 |
| Brca 1 | −3.34 |
| P53 | 4.91 |
| E2F1 | −4.06 |
| Mcm 4 | −9.85 |
| Mdm2 | −3.59 |
| PCNA | −3.25 |
| Wee1 | −7.77 |
| Src | 5.51 |
| PDGFA | 6.06 |
| PDGFRa | 3.12 |
| PDGFB | 2.86 |
| Braca1 | −3.31 |
| Stat3 | −13.77 |
| Cyclin C | −2.03 |
| Hras | −4.48 |
| Kras | −2.24 |
| LTA | 3.25 |
| LTBR | 9.70 |
| MAPK 10 | 2.48 |
| MAPK 1 | 2.13 |
| Apoptosis related | |
| Caspase 3 | −5.31 |
| MADD | 2.325 |
| BID | 6.90 |
| Bak 1 | 12.66 |
| FADD | 7.76 |
| DFFA | 11.89 |
| DAXX | 7.752 |
| BCL2 | 2.11 |
| Bid3 | 2.44 |
| Caspase 7 | 3.23 |
| Caspase 2 | 7.28 |
| BAD | 4.66 |
| BAX | 2.59 |
| Bcl2a1 | −4.52 |
| Bcl2l11 | 4.36 |
| Bcl2l1 | 12.68 |
| Bok | 3.34 |
| Bik | 2.36 |
| Caspase 9 | 7.28 |
| Birc5 | −9.28 |
| Fas | −2.29 |
The rat RT2 Profiler PCR Array (SABiosciences, Frederick, MD) that profiles the expression of 84 key genes involved apoptosis, cell cycle, and EGFR/PDGF signaling was used. RT-PCR was performed by using a 96-well format by using cDNA from day 1 after PHx shEGFR-treated and control rat liver (n = 3). Genes that showed at least twofold up- or down-regulation are shown. Data were normalized to the average of five house keeping genes provided on the plate. Negative numbers indicate a fold decrease, whereas positive numbers reflect a fold increase of the ratio of the gene expression of shEGFR to control untreated rats.
Analyses of Off Target Effects after shRNA Treatment
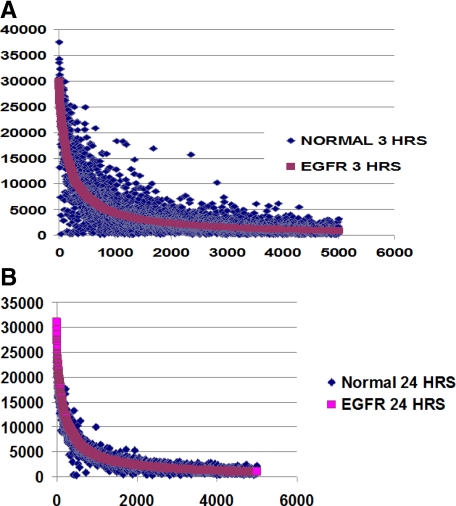
Microarray data generated at 3 and 24 hours after PHx were analyzed for off target effects as described in Materials and Methods. As shown in Figure 6, A and B, the individual line fitted for the shRNA treatment group lay entirely on top of the expression points derived from the ScrRNA-treated rats indicating that the overall patterns of gene expression after PHx were not significantly affected by shRNA treatment at the time points analyzed. We also plotted expression values derived from the oligo-array for IP10, Oligoadenylate synthetase, cox2, interferon α, interferon β, interleukin-6, and interleukin-12 known to be associated with interferon response. There was not much of a difference in the expression of these genes among the different treatment groups, and shRNA treatment did not up-regulate expression of interferon response genes (data not shown).
Figure 6.
Analyses of off target effects after shEGFR treatment. Expression intensity values obtained from microarray data generated at three and 24 hours after PHx (A and B) in treatment and control groups were plotted as described in Material and Methods. The gene expression pattern between these groups does not significantly differ, indicating absence of global off target effects.
Pathway-Specific qRT-PCR
After PHx, expression of a large number of genes is altered. Changes in expression patterns of genes involved in cell cycle, stress response, and growth regulation have been reported in a number of studies.1,27 Analysis of global gene expression in rats treated with shEGFR by microarray indicated dysregulation of many genes involved in cell cycle and growth arrest.
To analyze expression of specific genes associated with apoptosis, cell cycle, and EGFR/PDGF pathways, we performed real-time PCR-based gene specific assays by using commercially available pathway-specific gene arrays. Genes that exhibited at least twofold up or down-regulation compared with controls are listed in Table 1. Particularly relevant was the down-regulation seen in the cyclins, p21, CDK2, Rad 17, PCNA, ras, IGFBP1, and C/EBP-β. STAT3, a key protein downstream of EGFR signaling pathway,45,46 was also down-regulated in shEGFR-treated rats. Inhibition of EGFR by Gefitinib in NSCLC cells also resulted in complete inhibition of STAT3.47 STAT3 has been shown to up-regulate expression of genes involved in apoptosis and proliferation, like Birc548 and cyclin D1,49 and down-regulate p53.50 In shEGFR-treated rats, Birc5 and cyclin D1 were significantly down-regulated, whereas p53 was up-regulated. Some of the genes that were found to be up-regulated in treated rats are c-Src, PDGFa, PDGFRa, PDGFb, nuclear factor κB, C/EBP-α, and p27.
Interestingly, proapoptotic genes like Bad, Bak1, Bax, Bid, etc., were up-regulated in shEGFR-treated rats when compared with controls, whereas prosurvival genes like Birc5 and Bcl2a1 were down-regulated. A similar profile was obtained after inhibition of EGFR by PD153035 (a reversible chemical inhibitor of EGFR) in a human cervical carcinoma cell line.51
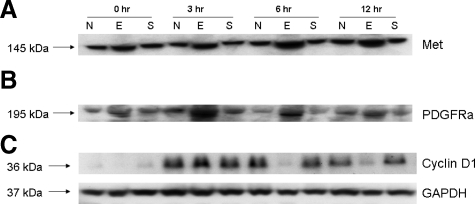
We also performed Western blot analyses to validate some of the differentially regulated genes like MET, PDGFRa, and cyclin D1 identified by using pathway-specific qRT-PCR. As seen in Figure 7, A–C, an increase in MET and PDGFRa was observed at 0, 3, 6, and 12 hours after PHx in shEGFR-treated rats, whereas a reduction in cyclin D1 was observed at 0, 6, and 12 hours after PHx in shEGFR-treated rats. However, higher levels of cyclin D1 were observed at 3 hours after PHx in shEGFR-treated rats, similar to that observed for EGFR and phosphoEGFR. We are presently conducting a complete phosphoproteome analyses to better understand and identify pathways that are differentially regulated after treatment with shEGFR.
Figure 7.
Western blot analyses of some of the relevant targets like MET, PDGFRa, and cyclin D1 that were found to be differentially regulated by qRT-PCR analyses were validated by Western blot analyses as described in Materials and Methods. As seen in A, an up-regulation of c-met at 0, 3, 6, and 12 hours after PHx in shEGFR-treated rats compared with controls was evident. B: An increase in PDGFRa was also evident at 0, 3, 6, and 12 hours after PHx in shEGFR-treated rats compared with controls. C: A reduction in cyclin D1, a downstream target of EGFR, was also observed at 0, 6, and 12 hours with an increase at three hours after PHx in shEGFR-treated rats. The increase in Cyclin D1 at 3 hours after PHx paralleled the increase observed with EGFR/PhosphoEGFR in shEGFR-treated rats. E, shEGFR-treated rats; N, normal untreated rats; and S, scrambled treated rats. GAPDH loading control.
The suppression of expression of cyclin E1 (associated with progression from G1 to S phase)52 and fivefold elevation of the transforming growth factor-β receptor II in the livers of shEGFR-treated rats was significant. The decrease in the ratio of C/EBP-α to C/EBP-β known to occur after PHx53 was offset in the animals treated with shEGFR. The up-regulation of p27 and down-regulation of CDK2 after treatment with shEGFR have also been observed in a prostate cancer cell line treated with an anti-EGFR monoclonal antibody.54
Hepatocyte Cell Size
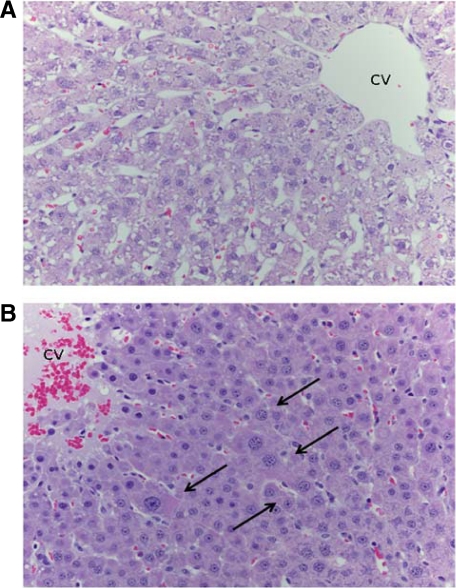
Given the restoration of liver weight in the light of much diminished hepatocyte proliferation, we investigated the possibility that restoration of liver weight may have in part been due to hepatocyte hypertrophy. Average diameter of hepatocytes was estimated from at least 200 hepatocytes of liver sections prepared from shEGFR-treated and ScrRNA control regenerating rat liver at day 5 after PHx. As seen in Figure 8, A and B, hepatocytes in shEGFR-treated rats were at least 30% larger than ScrRNA-treated controls, and many of them had larger nuclei.
Figure 8.
Hepatocyte enlargement observed in rats at day five after PHx. A: Liver from rats treated with scrambled RNA. B: Liver of rats treated with shEGFR. Enlarged hepatocytes are shown by arrows. CV, central vein.
It is likely that the increase in hepatocyte cell size was a result of recruitment of PI3K-AKT pathway by ErbB-3-ErbB-2 heterodimer. Interestingly, pathway-specific qRT-PCR analyses also indicated a more than twofold up-regulation in Akt1 and GSK3b, also involved in determination of cell size.55
Discussion
Liver regeneration is a complex process involving extensive gene reprogramming controlled by signaling involving several redundant pathways. Genetic ablation models have been used to investigate and identify genes that are essential and critical for liver regeneration; however, several studies have indicated that there is no one single gene that is essential for liver regeneration. Ablating of any specific gene will delay regeneration and not block it, and the restitution of the liver mass always occurs.1,56
The MET and EGFR receptors are activated within 30 to 60 minutes of a PHx. MET and EGFR are the two major mitogenic receptor tyrosine kinases for hepatocytes. We had earlier reported a complete block in hepatocyte proliferation at 24 hours after PHx, after silencing of MET by using a transient nonviral vector in regenerating rat liver.31 Silencing of MET at the time of PHx resulted in complete suppression of mitoses and dramatic suppression of BrdU incorporation at 24 hours after PHx. This was followed by a rebound in mitosis that coincided with disappearance/inactivation of the vector used to express c-met targeting short hairpin RNA.31
In this study, we investigated the effects on regeneration after silencing of EGFR. Silencing of EGFR by shRNA resulted in suppression of both mRNA and protein. Cellular proliferation parameters like mitosis and BrdU incorporation were also affected. The effect was not as profound at 24 hours as was seen with c-met silencing. However, the effect was more prolonged, and there was no “rebound” effect. Yet liver regeneration as assessed by return of liver weight to normal was not affected.
This could indicate that despite cross talk and overlap between the MET and EGFR pathways, the downstream signals provided by MET and EGFR are unique and cannot be compensated by each other.
Recently a study using an inducible Mx-cre line for targeted deletion of EGFR in adult mice liver indicated that EGFR was required for efficient liver regeneration.26 Mice lacking EGFR in the liver showed histopathological alterations, increased mortality, and delayed regeneration. However, it has been pointed out that mortality observed in mice after gene deletion needs to be interpreted carefully.27 The histopathological alterations seen in the liver made it difficult to interpret whether the decrease in hepatocyte proliferation was a consequence of the histopathological changes or the elimination of EGFR. The results obtained in the current study differ in many respects: our study was performed in normal rats without any pre-existing pathology often seen in transgenic animals, and the approach used to silence EGFR was acute in that it did not provide an opportunity for the liver to adapt to the gene loss by introduction of secondary changes. We also did not see any mortality associated with suppression of EGFR. We also found an increase in MET, ErbB-2, Src, and ErbB-3 in EGFR silenced rats and a reduction in cyclin D1. Functional compensation by MET and members of the EGFR family after inactivation of EGFR has been demonstrated in cell culture.38,41,57 Also, during embryonic development, no observable hepatic abnormalities are seen in EGFR knockouts, possibly due to recruitment of ErbB-2 and ErbB-3.1
Compensatory up-regulation of ErbB-3 in presence of inhibitors that target the other ErbB family members has been observed both in vitro and in vivo.58,59,60,61 It is possible that the up-regulation of MET, PDGFRA, Src, ErbB-3, and ErbB-2 observed in the current study in EGFR-silenced rats probably compensated for absence of EGFR. We believe that up-regulation of MET in shEGFR-treated rats may be particularly significant because MET has been shown to activate the ErbB-3 and PI3K pathway57 and the c-Src Kinase pathway.62 Also both ErbB-363 and MET64 have been shown in vitro to interact with Src to compensate for loss of EGFR. Src is also capable of modulating ErbB-3 and ErbB-2 heterodimerization.65
Analysis of gene expression data obtained from MET and EGFR silenced rats pointed out to some important differences. A dramatic up-regulation in Caspase 3 and fas was observed in MET-silenced rats.31 In contrast, Caspase 3 and fas were down-regulated in shEGFR-treated rats.
It should be mentioned that although levels of EGFR mRNA and protein are reduced significantly, they do not disappear completely. This was also true in our previous work with MET.31 This can be due to issues with transfection efficiency that differs for specific hepatic cell types. As shown in our previous study using a comparable plasmid targeting HGF and MET, hepatocytes receive most of the plasmid, whereas endothelial cells are minimally affected, and both cell types express the EGFR, albeit more intensely in hepatocytes.31 In addition, EGFR is always phosphorylated in the resting liver,8 so there will be already a background EGFR activity at the time PHx is done and before the second dose of shEGFR is administered.
The observed differences of effects at 24 hours after PHx between knockdown of c-met (complete elimination of cell cycle events)31 and knockdown of EGFR (partial suppression of cell cycle activities) are not likely to be due to differences in amount and intensity of knockdown because EGFR in this study seems to be more intensely suppressed than MET in our previous study.31
The associated increase in ErbB3 and ErbB-2 may result in compensatory effects mediated between MET and ErbB3 dimerization, as has been reported in lung cancer cell lines.57 We, however, were not able to detect MET association with ErbB-3 in co-immunoprecipitation studies at the time points examined. This raises the interesting question in view of suppression of BrdU incorporation: why was no delay in liver regeneration evident? Regeneration of liver (increase in cell mass in absence of cell proliferation) after a PHx has been observed in Skp-2-deficient mice66 after introduction into hepatocytes of SOC1 and SOC3, both negative regulators of EGFR and MET-mediated STAT3 phosphorylation,14 in STAT3-deficient mice,55 in cyclin E2-deleted mice,67 and in rats and mice subjected to irradiation and to administration of pyrrolizidine alkaloids (such as retrorcine).68 In all these cases, liver mass was restored by an increase in hepatocyte size. As shown in Figure 8, the size of hepatocytes at day 5 after PHx in shEGFR-treated rats seems to be considerably larger than scrambled treated controls. In this context, the down-regulation of Cyclin E167 and recruitment of PI3K-Akt pathway by ErbB-3-ErbB-2 heterodimer in shEGFR-treated rats may be significant.
Comparison with our previous study with suppressed expression of MET indicates similar but not identical profiles for shMET- and shEGFR-treated rats (unpublished observation). The effects at 24 hours after partial hepatectomy are more severe with MET knockdown, suggesting that the MET contributions cannot be compensated by EGFR at that time point. On the other hand, the effects of EGFR knockdown are much more prolonged, also suggesting that there are unique signaling contributions generated by EGFR at the later stages during regeneration not compensated by MET. In this regard, it is of interest that most of the expression of EGFR ligands in liver during regeneration occurs at later times (after 12 hours),1 and thus the unique signaling contributions of EGFR may be more relevant at the later times of liver regeneration. It should be also stated that the overall processing and trafficking of EGFR is well investigated and very complex,69,70 and we are not addressing issues of this type but rather the overall effect of RNA silencing of EGFR on liver regeneration.
It thus seems that after inhibition of EGFR by RNA interference, proapoptotic genes were up-regulated, and genes associated with cellular proliferation were down-regulated. However a compensatory up-regulation in ErbB-3, ErbB-2, MET, and Src resulted in an increase in formation of ErbB-3-ErbB-2 heterodimers, which in turn activated the PI3K-Akt survival pathway and restoration of liver mass by hepatic hypertrophy.71
To conclude, EGFR mRNA expression and protein was significantly reduced in the livers of rats treated with shEGFR compared with scrambled treated and untreated controls. ShEGFR treatment also had an effect on cell proliferation markers like mitotic index and BrdU nuclear labeling index. However, we did not observe any mortality associated with silencing of EGFR, and no delay in regeneration was observed in EGFR-silenced rats.
It is clear that EGFR signaling pathway networks and liver regeneration have a lot of inbuilt redundancies. No one single gene seems to block regeneration completely (except maybe MET), and compensatory alternate pathways are activated. It would be interesting to perform simultaneous acute silencing of MET and EGFR/ErbB-3/ErbB-2 to study its effect on liver regeneration.
Acknowledgments
We thank Dr. Udayan Apte for help with running Western blots during the early phase of the study.
Footnotes
Address reprint requests to George K. Michalopoulos, M.D., Ph.D., Department of Pathology, School of Medicine, University of Pittsburgh, S-410 Biomedical Science Tower, Pittsburgh, PA 5261. E-mail: michalopoulosgk@upmc.edu.
Supported by NIH grants CA35373 and CA103958, and the Rangos Fund for Enhancement of Research in Pathology and Pathology Post-Doctoral Research Training Program (PPRTP) award to S.P.
References
- Michalopoulos GK. Liver regeneration. J Cell Physiol. 2007;213:286–300. doi: 10.1002/jcp.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- Patijn GA, Lieber A, Schowalter DB, Schwall R, Kay MA. Hepatocyte growth factor induces hepatocyte proliferation in vivo and allows for efficient retroviral-mediated gene transfer in mice. Hepatology. 1998;28:707–716. doi: 10.1002/hep.510280317. [DOI] [PubMed] [Google Scholar]
- Bucher NL. Liver regeneration: an overview. J Gastroenterol Hepatol. 1991;6:615–624. doi: 10.1111/j.1440-1746.1991.tb00921.x. [DOI] [PubMed] [Google Scholar]
- Bucher NL, Patel U, Cohen S. Hormonal factors concerned with liver regeneration. Ciba Found Symp. 1977;55:95–107. doi: 10.1002/9780470720363.ch5. [DOI] [PubMed] [Google Scholar]
- Yang J, Chen S, Huang L, Michalopoulos GK, Liu Y. Sustained expression of naked plasmid DNA encoding hepatocyte growth factor in mice promotes liver and overall body growth. Hepatology. 2001;33:848–859. doi: 10.1053/jhep.2001.23438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mars WM, Liu ML, Kitson RP, Goldfarb RH, Gabauer MK, Michalopoulos GK. Immediate early detection of urokinase receptor after partial hepatectomy and its implications for initiation of liver regeneration. Hepatology. 1995;21:1695–1701. [PubMed] [Google Scholar]
- Stolz DB, Mars WM, Petersen BE, Kim TH, Michalopoulos GK. Growth factor signal transduction immediately after two-thirds partial hepatectomy in the rat. Cancer Res. 1999;59:3954–3960. [PubMed] [Google Scholar]
- Agarwal S, Zerillo C, Kolmakova J, Christensen JG, Harris LN, Rimm DL, Digiovanna MP, Stern DF. Association of constitutively activated hepatocyte growth factor receptor (Met) with resistance to a dual EGFR/Her2 inhibitor in non-small-cell lung cancer cells. Br J Cancer. 2009;100:941–949. doi: 10.1038/sj.bjc.6604937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo M, Stolz DB, Esplen JE, Dorko K, Michalopoulos GK, Strom SC. Cross-talk between epidermal growth factor receptor and c-Met signal pathways in transformed cells. J Biol Chem. 2000;275:8806–8811. doi: 10.1074/jbc.275.12.8806. [DOI] [PubMed] [Google Scholar]
- Presnell SC, Stolz DB, Mars WM, Jo M, Michalopoulos GK, Strom SC. Modifications of the hepatocyte growth factor/c-met pathway by constitutive expression of transforming growth factor-alpha in rat liver epithelial cells. Mol Carcinog. 1997;18:244–255. [PubMed] [Google Scholar]
- Scheving LA, Stevenson MC, Taylormoore JM, Traxler P, Russell WE. Integral role of the EGF receptor in HGF-mediated hepatocyte proliferation. Biochem Biophys Res Commun. 2002;290:197–203. doi: 10.1006/bbrc.2001.6157. [DOI] [PubMed] [Google Scholar]
- Xu KP, Yu FS. Cross talk between c-Met and epidermal growth factor receptor during retinal pigment epithelial wound healing. Invest Ophthalmol Vis Sci. 2007;48:2242–2248. doi: 10.1167/iovs.06-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki E, Kondo Y, Iimuro Y, Naka T, Son G, Kishimoto T, Fujimoto J, Tsutsui H, Nakanishi K. Demonstration of cooperative contribution of MET- and EGFR-mediated STAT3 phosphorylation to liver regeneration by exogenous suppressor of cytokine signalings. J Hepatol. 2008;48:237–245. doi: 10.1016/j.jhep.2007.08.020. [DOI] [PubMed] [Google Scholar]
- Michalopoulos GK, Khan Z. Liver regeneration, growth factors, and amphiregulin. Gastroenterology. 2005;128:503–506. doi: 10.1053/j.gastro.2004.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Munoz A, Rajadhyksha S, Gilmour D, van Bebber F, Antos C, Rodriguez Esteban C, Nusslein-Volhard C, Izpisua Belmonte JC. ErbB2 and ErbB3 regulate amputation-induced proliferation and migration during vertebrate regeneration. Dev Biol. 2009;327:177–190. doi: 10.1016/j.ydbio.2008.12.012. [DOI] [PubMed] [Google Scholar]
- Carver RS, Stevenson MC, Scheving LA, Russell WE. Diverse expression of ErbB receptor proteins during rat liver development and regeneration. Gastroenterology. 2002;123:2017–2027. doi: 10.1053/gast.2002.37060. [DOI] [PubMed] [Google Scholar]
- Linggi B, Carpenter G. ErbB receptors: new insights on mechanisms and biology. Trends Cell Biol. 2006;16:649–656. doi: 10.1016/j.tcb.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Fausto N, Mead JE. Regulation of liver growth: protooncogenes and transforming growth factors. Lab Invest. 1989;60:4–13. [PubMed] [Google Scholar]
- Mitchell C, Nivison M, Jackson LF, Fox R, Lee DC, Campbell JS, Fausto N. Heparin-binding epidermal growth factor-like growth factor links hepatocyte priming with cell cycle progression during liver regeneration. J Biol Chem. 2005;280:2562–2568. doi: 10.1074/jbc.M412372200. [DOI] [PubMed] [Google Scholar]
- Berasain C, Garcia-Trevijano ER, Castillo J, Erroba E, Lee DC, Prieto J, Avila MA. Amphiregulin: an early trigger of liver regeneration in mice. Gastroenterology. 2005;128:424–432. doi: 10.1053/j.gastro.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Sibilia M, Kroismayr R, Lichtenberger BM, Natarajan A, Hecking M, Holcmann M. The epidermal growth factor receptor: from development to tumorigenesis. Differentiation. 2007;75:770–787. doi: 10.1111/j.1432-0436.2007.00238.x. [DOI] [PubMed] [Google Scholar]
- Miettinen PJ, Berger JE, Meneses J, Phung Y, Pedersen RA, Werb Z, Derynck R. Epithelial immaturity and multiorgan failure in mice lacking epidermal growth factor receptor. Nature. 1995;376:337–341. doi: 10.1038/376337a0. [DOI] [PubMed] [Google Scholar]
- Dackor J, Strunk KE, Wehmeyer MM, Threadgill DW. Altered trophoblast proliferation is insufficient to account for placental dysfunction in Egfr null embryos. Placenta. 2007;28:1211–1218. doi: 10.1016/j.placenta.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Buren G, 2nd, Yang AD, Dallas NA, Gray MJ, Lim SJ, Xia L, Fan F, Somcio R, Wu Y, Hicklin DJ, Ellis LM. Effect of molecular therapeutics on liver regeneration in a murine model. J Clin Oncol. 2008;26:1836–1842. doi: 10.1200/JCO.2007.11.6566. [DOI] [PubMed] [Google Scholar]
- Natarajan A, Wagner B, Sibilia M. The EGF receptor is required for efficient liver regeneration. Proc Natl Acad Sci USA. 2007;104:17081–17086. doi: 10.1073/pnas.0704126104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fausto N, Campbell JS, Riehle KJ. Liver regeneration. Hepatology. 2006;43:S45–S53. doi: 10.1002/hep.20969. [DOI] [PubMed] [Google Scholar]
- Muller U. Ten years of gene targeting: targeted mouse mutants, from vector design to phenotype analysis. Mech Dev. 1999;82:3–21. doi: 10.1016/s0925-4773(99)00021-0. [DOI] [PubMed] [Google Scholar]
- Wu H, Wade M, Krall L, Grisham J, Xiong Y, Van Dyke T. Targeted in vivo expression of the cyclin-dependent kinase inhibitor p21 halts hepatocyte cell-cycle progression, postnatal liver development and regeneration. Genes Dev. 1996;10:245–260. doi: 10.1101/gad.10.3.245. [DOI] [PubMed] [Google Scholar]
- Gkretsi V, Apte U, Mars WM, Bowen WC, Luo JH, Yang Y, Yu YP, Orr A, St-Arnaud R, Dedhar S, Kaestner KH, Wu C, Michalopoulos GK. Liver-specific ablation of integrin-linked kinase in mice results in abnormal histology, enhanced cell proliferation, and hepatomegaly. Hepatology. 2008;48:1932–1941. doi: 10.1002/hep.22537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paranjpe S, Bowen WC, Bell AW, Nejak-Bowen K, Luo JH, Michalopoulos GK. Cell cycle effects resulting from inhibition of hepatocyte growth factor and its receptor c-Met in regenerating rat livers by RNA interference. Hepatology. 2007;45:1471–1477. doi: 10.1002/hep.21570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins GM. A. RM: Experimental pathology of the liver, 1: restoration of the liver of the white rat following partial surgical removal. Arch Pathol. 1931;12:186–202. [Google Scholar]
- Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Michalopoulos GK, Bowen WC, Mule K, Luo J. HGF-, EGF-, and dexamethasone-induced gene expression patterns during formation of tissue in hepatic organoid cultures. Gene Expr. 2003;11:55–75. doi: 10.3727/000000003108748964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semizarov D, Frost L, Sarthy A, Kroeger P, Halbert DN, Fesik SW. Specificity of short interfering RNA determined through gene expression signatures. Proc Natl Acad Sci USA. 2003;100:6347–6352. doi: 10.1073/pnas.1131959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Learn PA, Krishnegowda N, Talamantez J, Kahlenberg MS. Compensatory increases in Her-2/neu activation in response to EGFR tyrosine kinase inhibition in colon cancer cell lines. J Surg Res. 2006;136:227–231. doi: 10.1016/j.jss.2006.07.046. [DOI] [PubMed] [Google Scholar]
- Sliwkowski MX, Schaefer G, Akita RW, Lofgren JA, Fitzpatrick VD, Nuijens A, Fendly BM, Cerione RA, Vandlen RL, Carraway KL., 3rd Coexpression of erbB2 and erbB3 proteins reconstitutes a high affinity receptor for heregulin. J Biol Chem. 1994;269:14661–14665. [PubMed] [Google Scholar]
- Stortelers C, van der Woning SP, Jacobs-Oomen S, Wingens M, van Zoelen EJ. Selective formation of ErbB-2/ErbB-3 heterodimers depends on the ErbB-3 affinity of epidermal growth factor-like ligands. J Biol Chem. 2003;278:12055–12063. doi: 10.1074/jbc.M211948200. [DOI] [PubMed] [Google Scholar]
- Maegawa M, Takeuchi K, Funakoshi E, Kawasaki K, Nishio K, Shimizu N, Ito F. Growth stimulation of non-small cell lung cancer cell lines by antibody against epidermal growth factor receptor promoting formation of ErbB2/ErbB3 heterodimers. Mol Cancer Res. 2007;5:393–401. doi: 10.1158/1541-7786.MCR-06-0303. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Janne PA, Mermel C, Pearlberg J, Mukohara T, Fleet C, Cichowski K, Johnson BE, Cantley LC. ErbB-3 mediates phosphoinositide 3-kinase activity in gefitinib-sensitive non-small cell lung cancer cell lines. Proc Natl Acad Sci USA. 2005;102:3788–3793. doi: 10.1073/pnas.0409773102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sithanandam G, Smith GT, Fields JR, Fornwald LW, Anderson LM. Alternate paths from epidermal growth factor receptor to Akt in malignant versus nontransformed lung epithelial cells: erbB3 versus Gab1. Am J Respir Cell Mol Biol. 2005;33:490–499. doi: 10.1165/rcmb.2005-0049OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltoff SP, Carraway KL, 3rd, Prigent SA, Gullick WG, Cantley LC. ErbB3 is involved in activation of phosphatidylinositol 3-kinase by epidermal growth factor. Mol Cell Biol. 1994;14:3550–3558. doi: 10.1128/mcb.14.6.3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesnelle KM, Boehm AL, Grandis JR. STAT-mediated EGFR signaling in cancer. J Cell Biochem. 2007;102:311–319. doi: 10.1002/jcb.21475. [DOI] [PubMed] [Google Scholar]
- Yu H, Jove R. The STATs of cancer: new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- Haura EB, Zheng Z, Song L, Cantor A, Bepler G. Activated epidermal growth factor receptor-Stat-3 signaling promotes tumor survival in vivo in non-small cell lung cancer. Clin Cancer Res. 2005;11:8288–8294. doi: 10.1158/1078-0432.CCR-05-0827. [DOI] [PubMed] [Google Scholar]
- Kim DJ, Kataoka K, Rao D, Kiguchi K, Cotsarelis G, Digiovanni J. Targeted disruption of Stat3 reveals a major role for follicular stem cells in skin tumor initiation. Cancer Res. 2009;69:7587–7594. doi: 10.1158/0008-5472.CAN-09-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie K, Lang C, Devgan G, Azare J, Berishaj M, Gerald W, Kim YB, Paz K, Darnell JE, Albanese C, Sakamaki T, Pestell R, Bromberg J. Cyclin D1 is transcriptionally regulated by and required for transformation by activated signal transducer and activator of transcription 3. Cancer Res. 2006;66:2544–2552. doi: 10.1158/0008-5472.CAN-05-2203. [DOI] [PubMed] [Google Scholar]
- Niu G, Wright KL, Ma Y, Wright GM, Huang M, Irby R, Briggs J, Karras J, Cress WD, Pardoll D, Jove R, Chen J, Yu H. Role of Stat3 in regulating p53 expression and function. Mol Cell Biol. 2005;25:7432–7440. doi: 10.1128/MCB.25.17.7432-7440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodworth CD, Michael E, Marker D, Allen S, Smith L, Nees M. Inhibition of the epidermal growth factor receptor increases expression of genes that stimulate inflammation, apoptosis, and cell attachment. Mol Cancer Ther. 2005;4:650–658. doi: 10.1158/1535-7163.MCT-04-0238. [DOI] [PubMed] [Google Scholar]
- Zschemisch NH, Liedtke C, Dierssen U, Nevzorova YA, Wustefeld T, Borlak J, Manns MP, Trautwein C. Expression of a cyclin E1 isoform in mice is correlated with the quiescent cell cycle status of hepatocytes in vivo. Hepatology. 2006;44:164–173. doi: 10.1002/hep.21224. [DOI] [PubMed] [Google Scholar]
- Greenbaum LE, Cressman DE, Haber BA, Taub R. Coexistence of C/EBP alpha, beta, growth-induced proteins and DNA synthesis in hepatocytes during liver regeneration: implications for maintenance of the differentiated state during liver growth. J Clin Invest. 1995;96:1351–1365. doi: 10.1172/JCI118170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng D, Fan Z, Lu Y, DeBlasio T, Scher H, Mendelsohn J. Anti-epidermal growth factor receptor monoclonal antibody 225 up-regulates p27KIP1 and induces G1 arrest in prostatic cancer cell line DU145. Cancer Res. 1996;56:3666–3669. [PubMed] [Google Scholar]
- Haga S, Ogawa W, Inoue H, Terui K, Ogino T, Igarashi R, Takeda K, Akira S, Enosawa S, Furukawa H, Todo S, Ozaki M. Compensatory recovery of liver mass by Akt-mediated hepatocellular hypertrophy in liver-specific STAT3-deficient mice. J Hepatol. 2005;43:799–807. doi: 10.1016/j.jhep.2005.03.027. [DOI] [PubMed] [Google Scholar]
- Fausto N. Lessons from genetically engineered animal models. V. Knocking out genes to study liver regeneration: present and future. Am J Physiol. 1999;277:G917–G921. doi: 10.1152/ajpgi.1999.277.5.G917. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, Kosaka T, Holmes AJ, Rogers AM, Cappuzzo F, Mok T, Lee C, Johnson BE, Cantley LC, Janne PA. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- Sergina NV, Moasser MM. The HER family and cancer: emerging molecular mechanisms and therapeutic targets. Trends Mol Med. 2007;13:527–534. doi: 10.1016/j.molmed.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergina NV, Rausch M, Wang D, Blair J, Hann B, Shokat KM, Moasser MM. Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature. 2007;445:437–441. doi: 10.1038/nature05474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baselga J, Swain SM. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer. 2009;9:463–475. doi: 10.1038/nrc2656. [DOI] [PubMed] [Google Scholar]
- Wheeler DL, Huang S, Kruser TJ, Nechrebecki MM, Armstrong EA, Benavente S, Gondi V, Hsu KT, Harari PM. Mechanisms of acquired resistance to cetuximab: role of HER (ErbB) family members. Oncogene. 2008;27:3944–3956. doi: 10.1038/onc.2008.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzone M, Comoglio PM. The Met pathway: master switch and drug target in cancer progression. FASEB J. 2006;20:1611–1621. doi: 10.1096/fj.06-5947rev. [DOI] [PubMed] [Google Scholar]
- Contessa JN, Abell A, Mikkelsen RB, Valerie K, Schmidt-Ullrich RK. Compensatory ErbB3/c-Src signaling enhances carcinoma cell survival to ionizing radiation. Breast Cancer Res Treat. 2006;95:17–27. doi: 10.1007/s10549-005-9023-9. [DOI] [PubMed] [Google Scholar]
- Mueller KL, Hunter LA, Ethier SP, Boerner JL. Met and c-Src cooperate to compensate for loss of epidermal growth factor receptor kinase activity in breast cancer cells. Cancer Res. 2008;68:3314–3322. doi: 10.1158/0008-5472.CAN-08-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizawar RC, Miyake T, Parsons SJ. c-Src modulates ErbB2 and ErbB3 heterocomplex formation and function. Oncogene. 2007;26:3503–3510. doi: 10.1038/sj.onc.1210138. [DOI] [PubMed] [Google Scholar]
- Minamishima YA, Nakayama K, Nakayama K. Recovery of liver mass without proliferation of hepatocytes after partial hepatectomy in Skp2-deficient mice. Cancer Res. 2002;62:995–999. [PubMed] [Google Scholar]
- Nevzorova YA, Tschaharganeh D, Gassler N, Geng Y, Weiskirchen R, Sicinski P, Trautwein C, Liedtke C. Aberrant cell cycle progression and endoreplication in regenerating livers of mice that lack a single E-type cyclin. Gastroenterology. 2009;137:691–703. doi: 10.1053/j.gastro.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laconi S, Pillai S, Porcu PP, Shafritz DA, Pani P, Laconi E. Massive liver replacement by transplanted hepatocytes in the absence of exogenous growth stimuli in rats treated with retrorsine. Am J Pathol. 2001;158:771–777. doi: 10.1016/s0002-9440(10)64019-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley HS. Trafficking of the ErbB receptors and its influence on signaling. Exp Cell Res. 2003;284:78–88. doi: 10.1016/s0014-4827(03)00002-8. [DOI] [PubMed] [Google Scholar]
- Orth JD, Krueger EW, Weller SG, McNiven MA. A novel endocytic mechanism of epidermal growth factor receptor sequestration and internalization. Cancer Res. 2006;66:3603–3610. doi: 10.1158/0008-5472.CAN-05-2916. [DOI] [PubMed] [Google Scholar]
- Haga S, Ozaki M, Inoue H, Okamoto Y, Ogawa W, Takeda K, Akira S, Todo S. The survival pathways phosphatidylinositol-3 kinase (PI3-K)/phosphoinositide-dependent protein kinase 1 (PDK1)/Akt modulate liver regeneration through hepatocyte size rather than proliferation. Hepatology. 2009;49:204–214. doi: 10.1002/hep.22583. [DOI] [PubMed] [Google Scholar]