Abstract
TMPRSS2, a type II transmembrane serine protease, is highly expressed by the epithelium of the human prostate gland. To explore the regulation and function of TMPRSS2 in the prostate, a panel of monoclonal antibodies with high sensitivity and specificity were generated. Immunodetection showed TMPRSS2 on the apical plasma membrane of the prostate luminal cells and demonstrated its release into semen as a component of prostasomes, organelle-like vesicles that may facilitate sperm function and enhance male reproduction. In prostate cancer cells, TMPRSS2 expression was increased and the protein mislocalized over the entire tumor cell membrane. In both LNCaP prostate cancer cells and human semen, TMPRSS2 protein was detected predominantly as inactive zymogen forms as part of an array of multiple noncovalent and disulfide-linked complexes, suggesting that TMPRSS2 activity may be regulated by unconventional mechanisms. Our data suggested that TMPRSS2, an apical surface serine protease, may have a normal role in male reproduction as a component of prostasomes. The aberrant cellular localization, and increased expression of the protease seen in cancer, may contribute to prostate tumorigenesis by providing access of the enzyme to nonphysiological substrates and binding-proteins.
TMPRSS2 is an androgen responsive gene that encodes a type II transmembrane serine protease (TTSP).1,2,3 The members of the TTSP family share common protein structures including a transmembrane domain at the N terminus, linker regions with a variety of protein–protein interaction domains, and a canonical serine protease domain at the C terminus.4,5,6 TTSPs have been found to play important roles in the development and homeostasis of mammals, and the aberrant expression of TTSP genes are reported to contribute to the etiology of several human disorders, including cancer.7 The importance of TMPRSS2 in vivo remains unclear because homozygous TMPRSS2-null mice are essentially phenotypically normal.8 However, TMPRSS2 was reported to regulate epithelial sodium channel (ENaC) activity in vitro, implying a possible role in epithelial sodium homeostasis.9 TMPRSS2 may play a role in angiogenesis and tubulogenesis in microvesicular endothelial cells, potentially modulating several aspects of prostate tumor biology.10 In addition to its proteolytic activity, TMPRSS2 may also serve as a cell receptor, conducting external signaling or interacting with the extracellular matrix through its extracellular protein binding domains. Overexpression of TMPRSS2 has been demonstrated in poorly differentiated prostate cancer with significant increase in the mRNA level.2,11,12,13 TMPRSS2 was also reported to be involved in the majority of prostate cancer because of the gene fusion of the 5′-untranslational region of TMPRSS2 with ETS family members, which is implicated in the overexpression of ETS genes in the majority of prostate cancer.2,14,15 The coding sequence of TMPRSS2 is not involved in the gene fusion, and as a consequence there is no resultant recombinant protein for the TMPRSS2-ETS gene fusion and the promoter-less copy of TMPRSS2 is silenced, resulting in reduced expression of TMPRSS2 mRNA in those prostate cancer patients with the gene fusion.16
Despite these potentially interesting and important roles for TMPRSS2, characterization of the protein itself remains incomplete and somewhat confusing. Significant discrepancies in the apparent molecular mass of TMPRSS2 protein, with a calculated size around 54 kDa, have been reported in previous studies, with the most commonly sited being around 70 kDa and 32 kDa11,17,18 as the full-length and serine protease domain of TMPRSS2, respectively. In this study, we have generated a panel of mouse monoclonal antibodies (mAbs) directed against TMPRSS2 that are highly sensitive and specific. Using the newly generated TMPRSS2 mAb, we have investigated the pathophysiological role of TMPRSS2 in prostate tissues by comparing its expression and subcellular distribution in prostate tumors and adjacent normal prostate glands. Functional and regulatory aspects of TMPRSS2 biology were also investigated by examining the activation status of the enzyme and the nature of complexes formed between TMPRSS2 and other proteins.
Materials and Methods
Chemicals and Reagents
Peptide-N-glycosidase F (PNGase F) was purchased from New England Biolabs (Beverly, MA), and tunicamycin was from Sigma (St. Louis, MO). Commercial TMPRSS2 antibodies were obtained from Abcam (Cambridge, UK). The matriptase mAb M32 was generated using matriptase–HAI-1 complex as described previously,19 and the epitope recognized by the mAb was determined to be at the third LDL receptor domain of matriptase.20 Human semen samples were purchased from Lee Biosolutions (St. Louis, MI).
Cell Culture Conditions
Human prostate cancer cells were obtained from the Tissue Culture Shared Resource of the Lombardi Comprehensive Cancer Center, Georgetown University Medical Center (Washington, DC). LNCaP human prostate cancer cells were maintained in RPMI 1640 supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin, using a humidified incubator at 37°C with 5% CO2. PC3 and DU145 human prostate cancer cells were routinely maintained in RPMI 1640 supplemented with 5% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin in a humidified incubator at 37°C with 5% CO2.
Generation of TMPRSS2 Monoclonal Antibodies
A recombinant protein antigen consisting of a portion of the TMPRSS2 extracellular domains, tagged at the amino terminus with a Flag tag and at the carboxyl terminus with a His tag, was prepared for monoclonal antibody production. DNA encoding the extracellular region of TMPRSS2 was amplified by PCR using the following primers: forward primer: 5′-TAGAATTCGATTACAAGGATGACGACGATAAGAAGTTCATGGGCAGCAAGTGCTCCAAC-3′; reverse primer: 5′-AACTCGAGCGCCGTCTGCCCTCATTTGTCG-3′. A TMPRSS2 cDNA plasmid (a gift from Dr. Peter Nelson of University of Washington, Seattle, WA) that bears a point mutation R255Q at the canonical activation cleavage site was used as the PCR template. The TMPRSS2 mutant cannot be activated.11 The PCR product was purified, digested with the restriction enzymes EcoRI and XhoI, and cloned into a mammalian secretory expression vector, pSecTag2 (Invitrogen, Carlsbad, CA; Figure 1A). The construct was transfected into HEK293T cells, and stable transfectant pools of cells were selected with 400 μg/ml Zeocin. Transfectants secreting the TMPRSS2 fusion protein were expanded, and conditioned media were then harvested by growing the cells in serum-free DMEM media supplemented with insulin-transferrin-selenium (ITS; Invitrogen, Carlsbad, CA). Conditioned media were collected every other day, and after clarification by centrifugation proteins were precipitated by the addition of ammonium sulfate to 70% saturation. After desalting, the recombinant TMPRSS2 was purified from the concentrated protein solution by nickel column chromatography (through binding to the His-tag), followed by Q column chromatography using an AKTA prime protein purification system (GE Health care Bio-Sciences Corp., Piscataway, NJ). After these procedures, the recombinant protein was purified almost to homogeneity. The purified protein was using to generate monoclonal antibodies using conventional hybridoma technologies.21
Figure 1.
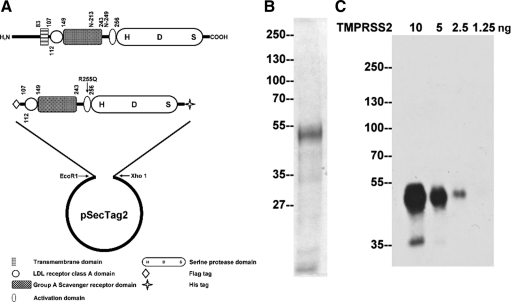
TMPRSS2 recombinant protein and mAbs. A: Expression of TMPRSS2 in mammalian cells. A schematic representation of the structure of TMPRSS2 is presented at the top. The protease consists of 492 amino acids, has a calculated mass of 53,859 Da, and contains a transmembrane domain, a scavenger receptor cysteine-rich (SRCR) domain, a LDLRA domain, and a serine protease domain. The two putative N-glycosylation sites are at N-213 and N-249. The serine protease domain is at the carboxyl terminus, and the active site triad was labeled as H-D-S. A truncated and mutated TMPRSS2 expression construct was generated as described in the Materials and Methods section resulting in the construct shown. B: Purification of TMPRSS2. The recombinant TMPRSS2 protein was produced and purified from HEK 293T cells and resolved by SDS-PAGE and stained by colloidal Coomassie blue to show the protein patterns and to asses the purity of the TMPRSS2 preparation. C: Characterization of a TMPRSS2 monoclonal antibody. To evaluate the sensitivity of the TMPRSS2 monoclonal antibody, AL20, different amounts of purified TMPRSS2 protein as indicated were resolved by SDS-PAGE under reducing and boiled conditions. Immunoblot analysis was conducted with the AL20 monoclonal antibody at 2 μg/ml.
Mass Spectrometry Analysis and Identification of Proteins
The protein bands from SDS-gels were excised, washed, destained, and trypsinized overnight at 37°C using standard protocols after dithiothreitol (DTT) reduction and iodoacetamide alkylation. Analysis of tryptic peptides derived from protein samples was performed by liquid chromatography/mass spectrometry by the Proteomics Shared Service at Greenebaum Comprehensive Cancer Center, University of Maryland Baltimore (Baltimore, MD), as described previously.22
Immunohistochemistry
Immunohistochemistry staining was performed using the manufacturer’s standard protocol with minor modification (Dako, CA). Paraffin-embedded human prostate tumor tissue sections were obtained from the Histopathology and Tissue Shared Resource of Georgetown University. The sections were stained using the TMPRSS2 mAb AL20 at a concentration of 2 μg/ml for 1 hour. A negative control slide was stained using the mouse IgG antibody at a concentration of 2 μg/ml for 1 hour simultaneously to ensure staining quality. HRP-labeled anti-mouse IgG and the substrate, diaminobenzidine (DAB), were used for the detection of positive staining. Cell nuclei were counterstained with hematoxylin. Images were captured using an Olympus AH2 Vanox Microscope System (Olympus; Melville, NY).
Immunohistochemistry Staining of Prostate Tissue Microarray for TMPRSS2 and Analysis by AQUA
The construction of the prostate tumor tissue microarray (TMA) was described previously.23 Briefly, formalin-fixed paraffin-embedded prostate specimens from the University of Wisconsin Pathology archives were used with the approval of the Institutional Review Board. The TMA consisted of 41 localized prostate cancers (Pca_local, median Gleason score 7, with a range of 6 to 9), 18 aggressive prostate cancers (Pca_aggr, prostate cancer with lymph node metastasis, median Gleason score 8, with a range of 7 to 9), 18 metastatic prostate cancers (Met, in either lymph nodes or other organs), 24 benign prostate hyperplasia (BPH), 19 high-grade intraepithelial neoplasia (HGPIN), and 48 benign prostate tissues. The median patient age was 62 years (range 37 to 82 years). Cores were 0.6 mm in diameter and spaced 0.8 mm on-center using a Manual Tissue Arrayer (Beecher Instruments, Sun Prairie, WI; Model MTA-1). Each specimen has duplicated cores. Five micron sections of the TMA were used for staining and analysis. Rabbit E-cadherin antibody (Abnova) and Alexa Flour 555 conjugated goat anti-rabbit (Invitrogen) were used to define and visualize epithelial compartment. TMPRSS2 mouse mAb, biotinylated goat anti-mouse (Biocare Medical), streptavidin-HRP (Biocare Medical), and Alexa Fluor 647 tyramide (Invitrogen) were used to detect and visualize TMPRSS2. 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen) was used to define and visualize nuclear compartment.
After staining, the TMA slide was scanned and images were acquired using the AQUA platform (HistoRx, CT). Tissue microarray cores without sufficient epithelium (<5%) or with poor staining quality (i.e., section folding, excess trapping of fluorochrome, etc.) were excluded from analysis. The AQUA_1.5 system (HistoRx, New Haven, CT) was used for TMPRSS2 quantification and analysis. Image acquisition and algorithmic analysis of TMA using automated quantitative analysis have been previously described extensively.23,24
Western Blot Analysis
Proteins for Western blot analysis was prepared from cell cultures by washing the cells three times with PBS followed by lysis with 1% Triton X-100 in PBS. Insoluble debris was removed by centrifugation, and the protein concentration was determined using BCA protein assay reagents (Pierce, Rockford, IL) according to the manufacturer’s instructions. Cell lysates or human semen were diluted with 5× sample buffer in the absence or presence of reducing agent and incubated either at room temperature or 95°C (boiled) for five minutes before SDS-polyacrylamide gel (SDS-PAGE). The proteins were resolved by 7.5% SDS-PAGE, transferred to Protran nitrocellulose membranes (Schleicher and Schuell, Keene, NH), and probed with the desired monoclonal antibodies. The binding of primary antibody was detected using a goat anti-mouse-HRP–conjugated secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA), and Western Lightning Chemiluminescence Reagent Plus (Perkin-Elmer, Boston, MA).
Diagonal SDS-PAGE
LNCaP cell lysates or human seminal plasma samples were diluted with 5× sample buffer and incubated either at room temperature or 95°C for five minutes and then resolved by SDS-PAGE. The gel was sliced into vertical strips, which were then boiled in SDS sample buffer in the absence or presence of DTT. The boiled gel strips were then placed horizontally on the top of fresh SDS-acrylamide gels, and electrophoresed using the same conditions as the first dimension gel, after which the proteins were transferred to nitrocellulose membrane and probed with TMPRSS2 mAb, AL20.
PNGase F Assay
Fifty-microgram samples of protein lysate from LNCaP cells were boiled under denaturing conditions (0.5% SDS, 40 mmol/L DTT) for 5 minutes. The samples were adjusted to 0.05 M sodium phosphate and 1% NP-40. PNGase F (500 U) was added, and the samples were then incubated at 37°C for 1 hour. The reaction was terminated by boiling the samples with 5× SDS sample buffer containing DTT for 5 minutes, followed by SDS-PAGE and immunoblot analysis. Control samples were treated in the same manner except for the addition of the PNGase F.
Analysis of TMPRSS2 and Matriptase Shedding
LNCaP cells were grown in protein-free media for 24 hours, after which the conditioned media was harvested and concentrated up to 100 fold. The cells were lysated with 1% Triton X-100 in PBS and the protein was quantified using BCA protein assay reagents. Both cell lysates and concentrated media were then subjected to immunoblot using TMPRSS2 mAb, AL20.
Preparation of Prostasomes
Human semen samples were centrifuged at 1000g for 15 minutes to remove spermatozoa and cell debris, after which the supernatants were ultracentrifuged at 105,000g for 2 hours. The resulting pellets, which contain prostasomes, were resuspended in PBS containing 1% Triton X-100 and then centrifuged at 21,000g for 15 minutes to remove the insoluble fraction.
Results
Generation and Characterization of TMPRSS2 Monoclonal Antibodies
A schematic representation of the domain structure of wild-type TMPRSS2 and the mutant TMPRSS2 expression construct is shown in Figure 1A. The recombinant TMPRSS2 protein was expressed and purified from the conditioned media as described in the Materials and Methods section. The purified protein is seen as a band of approximately 55-kDa on coomassie blue stained gels (Figure 1B), consistent with the calculated molecular mass of the TMPRSS2 mutant. A minor protein band of approximately 40-kDa was consistently observed and may represent a proteolytic degradation product of the transmembrane protease. Samples of the purified protein were subjected to mass spectrometry–based proteomic analysis. Thirteen tryptic peptides were identified, all of which matched TMPRSS2 (data not shown), confirming the identity of the purified recombinant protein. Monoclonal antibodies directed against TMPRSS2 were generated by conventional hybridoma technologies using the purified TMPRSS2 mutant protein. More than 20 positive clones were selected based on immunodetection of recombinant TMPRSS2 protein (data not shown), of which mAb AL20 was selected having the highest sensitivity and specificity. As shown in Figure 1C, when used at 2 μg/ml, the mAb was able to detect as little as 2.5 ng of recombinant TMPRSS2. These data demonstrate that the TMPRSS2 mAb, AL20, is a sensitive immunological reagent. This antibody was used in the studies described below.
Aberrant Subcellular Distribution and Increased Expression of TMPRSS2 Protein in Prostate Cancer Cells
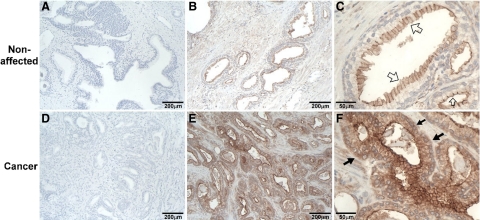
Using the highly sensitive TMPRSS2 mAb, we compared the subcellular localization and expression of TMPRSS2 in prostate cancer cells versus their normal counterparts. Preliminary immunohistochemical staining of prostate tumor tissue sections from seven different patients showed plasma membrane localization of TMPRSS2 in all specimens (Figure 2, A–F), consistent with its integral transmembrane protein nature. In the nonaffected areas of the prostate tumor tissue, which are representative of the normal prostate gland, TMPRSS2 was predominantly detected at the apical plasma membrane of the luminal epithelial cells of the prostate acini (Figure 2, B and C, open arrows). The staining was essentially limited to uppermost portion of plasma membrane either directly exposing to lumen or between the cell–cell contacts in these nonaffected prostate tumor tissues. In the adjacent prostate tumor, however, although the TMPRSS2 was still predominantly localized at the cell surface, the staining was observed to be around the entire cell surface (Figure 2, F, closed arrows). In addition, the expression levels of TMPRSS2 protein in the prostate cancer cells were much higher than in the neighboring prostate luminal epithelial cells (Figure 2, compare E with B).
Figure 2.
Cellular localization of TMPRSS2 in normal and carcinomatous prostate tissues. Paraffin-embedded human prostate tumor tissue sections were stained by immunohistochemistry using TMPRSS2 mAb AL20 (B, C, E, and F) and mouse IgG (A and D). Positive staining for TMPRSS2 was observed as brown precipitates (diaminobenzidine), and the nuclei were counterstained with hematoxylin. The micrographs in A–C were taken from noncancerous acini portions of the prostate tumor (nonaffected) and D–F from the cancer portions. The sizes of bars are indicated in each panel. Open arrows show TMPRSS2 on the apical surface of luminal cells. Closed arrows show the difference of TMPRSS2 protein expression in nonaffected prostate tissue and carcinomatous prostate tissue.
To further examine TMPRSS2 protein levels in more quantitative way and to examine the role of TMPRSS2 in prostate cancer and in tumor progression, a prostate cancer TMA was stained using the TMPRSS2 antibody and protein levels quantitatively determined using the AQUA system. The TMA was constructed with duplicate cores of 41 localized prostate cancers (Pca_local) 18 aggressive prostate cancers (Pca_aggr), 18 metastatic prostate cancers (Met), 24 benign prostate hyperplasia (BPH), 19 high-grade intraepithelial neoplasia (HGPIN), and 48 benign prostate tissues. The reliability of the TMA and AQUA system was tested using a known prostate cancer marker α-methylacyl coenzyme A racemase (AMACR) (data not shown).23 We found that the TMPRSS2 protein levels were higher in proliferative prostate diseases compared with benign prostatic tissue (Table 1). TMPRSS2 levels were significantly higher in BPH and localized and aggressive prostate cancers (mean AQUA scores, 316, 302, and 366, respectively) than benign prostatic tissue (mean AQUA score, 185). However, the elevation of TMPRSS2 protein expression in metastatic cancer and HGPIN (mean AQUA score, 242 and 249, respectively) did not reach statistical significance when compared with that in benign prostatic tissue (Table 1). Taken together, these data suggest that both the expression level and subcellular localization of TMPRSS2 may be deregulated in prostate cancer cells.
Table 1.
AQUA Scores of TMPRSS2
| Dx | Age (y) | GS | N | AQUA score | P value |
|---|---|---|---|---|---|
| Benign | 61.3 | 80 | 185 ± 18 | ||
| BPH | 67.8 | 39 | 316 ± 42 | 0.005* | |
| HGPIN | 62.9 | 36 | 249 ± 25 | 0.097 | |
| Pca_local | 58.2 | 6 to 9 (7) | 72 | 302 ± 25 | 0.003* |
| Pca_aggr | 62.2 | 7 to 9 (8) | 29 | 366 ± 60 | 0.016* |
| Met | 67.6 | 24 | 242 ± 40 | 0.301 |
GS indicates Gleason score; N, core number analyzed; P, significance level using ANCOVA.
P < 0.05.
TMPRSS2 in Prostate Cancer Cell Lines
The expression of TMPRSS2 by prostate cancer cell lines, including LNCaP, PC3, and DU145, has been well characterized at the mRNA level. TMPRSS2 mRNA was consistently detected in LNCaP and not in DU145; PC3 may not express the protease or express at very low level.2,11 TMPRSS2-ETS gene fusion was not found in LNCaP and PC3 cells.15 So as an additional control for the specificity of our antibody, the expression status of TMPRSS2 at the protein level was investigated in these three cancer cell lines. Of these three prostate cancer cell lines, TMPRSS2 protein was only detected in LNCaP cells (Figure 3A) and not in PC3 or DU145 cells (data not shown), consistent with their mRNA expression status shown in previous studies.
Figure 3.
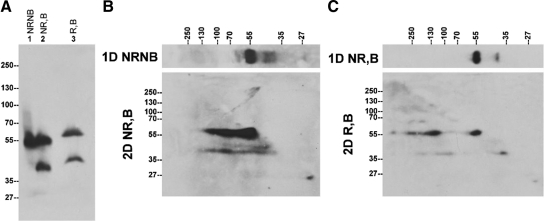
The expression status of endogenous TMPRSS2 protein in human prostate cancer cells. A: TMPRSS2 protein expression in LNCaP prostate cancer cells. LNCaP cells were lysed using PBS containing 1% Triton X-100. Equal amounts of total cellular proteins were mixed with SDS sample buffer in the absence (NR) or presence (R) of reducing agents and incubated either at room temperature (NB) or 95°C (B) for 5 minutes, followed by SDS-PAGE and Western blot analysis using the monoclonal antibody AL20. NRNB indicates nonreducing and nonboiled conditions; NR,B, nonreducing and boiled conditions; R,B, reducing and boiled conditions. B: TMPRSS2 is present in non-covalent-linked complexes in LNCaP cells. LNCaP cell lysates were resolved by diagonal gel electrophoresis under nonreducing and nonboiled conditions in the first dimension and under nonreducing and boiled condition in the second dimension. Immunoblot analysis of the second dimensional gel was performed with the TMPRSS2 mAb AL20. C: TMPRSS2 is also present in disulfide bond-linked complexes in LNCaP cells. LNCaP cell lysates were resolved by diagonal gel electrophoresis under nonreducing and boiled conditions in the first dimension and under reducing and boiled condition in the second dimension. Immunoblot analysis of the second dimensional gel was performed with the TMPRSS2 mAb AL20.
In LNCaP cells, endogenous TMPRSS2 was detected as a major protein band near the 55-kDa marker under nonreducing and nonboiled conditions (Figure 3, lane 1, NRNB). Some additional minor bands were also detected, particularly after longer exposure to X-ray film. Under boiled and non-reducing conditions (Figure 3A, lane 2, NR, B), a second protein band with a size of approximately 40 kDa was seen. The ratio of the 40-kDa species relative to the 56-kDa form varied among different experiments. Under boiled and reducing conditions, TMPRSS2 was detected as two protein bands with apparent sizes of 58 and 42 kDa (Figure 3A, lane 3, R,B). The decreased rate of migration of TMPRSS2 on SDS-PAGE caused by chemical reduction is consistent with the fact that TMPRSS2 contains many cysteine residues, which likely participate in 9 disulfide bonds.
TMPRSS2 Is Present in Both Noncovalent and Disulfide-Linked Complexes in LNCaP Cells
The appearance of the 40-kDa TMPRSS2 fragment after boiling treatment (Figure 3A, comparing lane 2 with lane 1) suggests that the 40-kDa TMPRSS2 species may be present in some form of noncovalent complexes, which can be dissociated by boiling treatment. Furthermore, the epitope recognized by the TMPRSS2 mAb on the 40-kDa TMPRSS2 species seems to be largely masked in the putative TMPRSS2 complexes, given that the TMPRSS2 mAb only detects these putative complexes very weakly under nonreducing and nonboiled conditions (Figure 3A, lane 1). We therefore set out to test this hypothesis using a two-dimensional, diagonal gel electrophoresis (Figure 3B). In the first dimension under nonboiled and nonreducing conditions, TMPRSS2 was detected as a major 56-kDa protein band with few minor bands with greater or smaller sizes than the 56-kDa TMPRSS2 monomer (Figure 3B, 1D NRNB). In the second dimension under boiled and nonreducing conditions, the major 56-kDa TMPRSS2 protein band was detected at the same size as 56-kDa, suggesting that the 56-kDa species is likely to be a monomer and not in a complex with other proteins. In addition to the major 56-kDa band, the 56-kDa species was also detected at positions corresponding to 80 to 90 kDa and 120 kDa in the first dimensional gel, despite the fact that there was no TMPRSS2 detected at these sizes in first dimensional gel. These data suggest that the 56-kDa TMPRSS2 species can form multiple noncovalent complexes in which the epitope recognized by mAb AL 20 is largely masked. Furthermore, in the second dimensional gel, the 40-kDa TMPRSS2 species was detected as a continuous protein band with corresponding molecular weight from 120 kDa through 40 kDa in the first dimension (Figure 3B). These data suggest that the 40-kDa TMPRSS2 species undergoes similar complex formation to the 56-kDa species. These data indicate that TMPRSS2 is able to form noncovalent complexes.
In addition to the noncovalent complexes, TMPRSS2 also forms disulfide-linked complexes, which can be clearly detected by diagonal gel electrophoresis (Figure 3C). In the first dimension, under boiled and nonreducing conditions, TMPRSS2 was detected as a major 56-kDa band and a minor 40-kDa band, as expected (Figure 3C, 1D NR,B). In the second dimension under boiled and reducing conditions, both the 56- and 40-kDa TMPRSS2 species were detected along the diagonal line (Figure 3C, 2D R,B), suggesting that both are single-chain TMPRSS2 zymogen species. Consistent with the previous result, exposure to the reducing agents resulted in minor changes to their migration rates between the two gel systems. The 56-kDa TMPRSS2 zymogen was also detected in the second dimension in three positions corresponding to the molecular weights of around 130 kDa, 250 kDa, and a very high molecular weight in the first dimensional gel. These data suggest that there are at least three disulfide-linked TMPRSS2 zymogen complexes present in LNCaP cells. Similar disulfide-linked complexes involving the 40-kDa TMPRSS2 species were also detected in this experiment.
TMPRSS2 Is Lightly Glycosylated
Because the calculated mass of TMPRSS2 is 53,859 Da, the 58-kDa TMPRSS2 species detected in LNCaP cells is likely the mature full-length form. Human TMPRSS2 contains two putative N-glycosylation sites, N-213 and N-249,1 and the mouse protein contains three, N-111, N-212, and N-474.3 The difference between the calculated mass of approximately 54 kDa and 58 kDa is most likely attributable to posttranslational modifications, specifically glycosylation. We used two different approaches to investigate the extent to which glycosylation contributes to the apparent mass of endogenous TMPRSS2. Tunicamycin blocks assembly of the GlcNAc-dolichol complex and has been broadly used to study the glycosylation of proteins, and so we cultured LNCaP cells in the presence of tunicamycin for various time periods up to eight hours (Figure 4A). After two hours of tunicamycin treatment a 53-kDa TMPRSS2 species began to appear, and after between four to eight hours of treatment, the 58-kDa TMPRSS2 form had disappeared (Figure 4A). The 53-kDa form is essentially the same as the calculated mass of the protease (Figure 4A). Exposure to tunicamycin also resulted in the disappearance of the 40-kDa TMPRSS2 species and the appearance of a smaller TMPRSS2 species that could be detected using more protein and longer exposure of the blot to X-ray film (Figure 4A, lane 8*). In an alternate approach we made use of the enzyme PNGase F, which cleaves between the innermost N-acetylglucosamine (GlcNAc) and asparagine residues of high mannose glycosylation. Treatment with this enzyme should remove all of the N-glycosylation from proteins such as TMPRSS2. As shown in Figure 4B, digestion with PNGase F resulted in the disappearance of both the 56-kDa and 40-kDa TMPRSS2 and the concomitant appearance of two smaller TMPRSS2 species. Taken together, these data suggest that glycosylation contributes about 4 to 5 kDa to the apparent molecular mass of TMPRSS2, and removal of the N-glycan results in an apparent molecular mass almost identical to the mass calculated based on the amino acid sequence.
Figure 4.
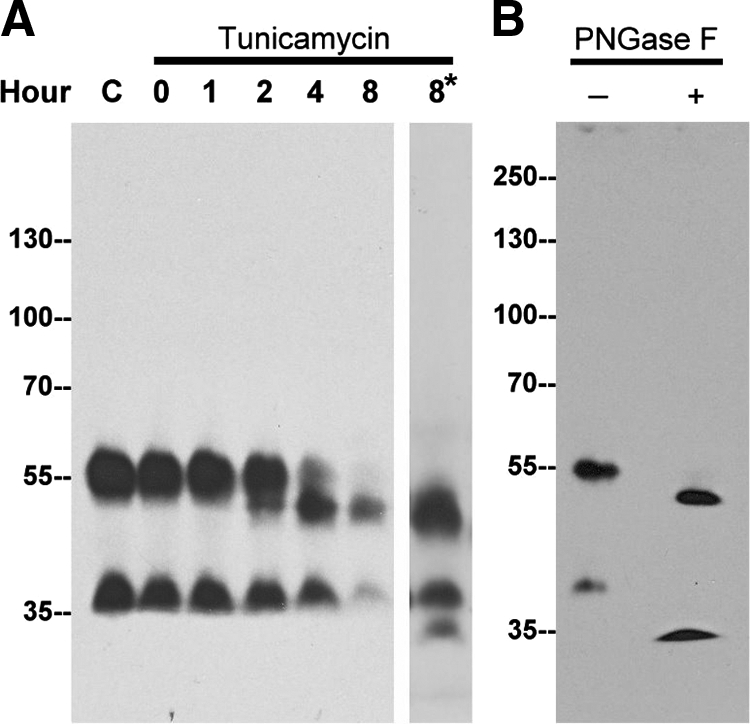
N-glycosylation of TMPRSS2 is responsible for a modest increase in molecular mass. A: LNCaP cells were treated with tunicamycin (5 μg/ml) for the indicated times. Cell lysates were analyzed for TMPRSS2 by immunoblot using the mAb AL20. To facilitate detection of the deglycosylated 40-kDa species, a longer exposure of the immunoblot for the 8-hour time point was taken (8*). B: LNCaP cell lysates were incubated in the absence (−) or presence (+) of PNGase F, and TMPRSS2 was probed with the mAb AL20.
The Shedding of TMPRSS2 into the Extracellular Milieu Is Very Limited in LNCaP Cells
Shedding to the extracellular milieu after the zymogen activation has been observed for other TTSP family members, such as matriptase.25 Taking advantage of the specificity and sensitivity of the TMPRSS2 antibody, we examined whether LNCaP cells release TMPRSS2 into media. Matriptase was used as a positive control for the quality of the conditioned media because this enzyme is known to be activated and shed by LNCaP cells.25 As expected, TMPRSS2 was readily detected in the LNCaP cell lysates but was completely absent from the conditioned media. In contrast to TMPRSS2, matriptase was detected both in the lysates and the conditioned media (Figure 5, A and B). Furthermore, both the latent matriptase at 70 kDa and activated matriptase in HAI-1 complex at 95 kDa were detected in the conditioned media (Figure 5), suggesting that matriptase underwent activation and shedding. These data suggest that shedding of TMPRSS2 appears to be very limited in LNCaP cells.
Figure 5.
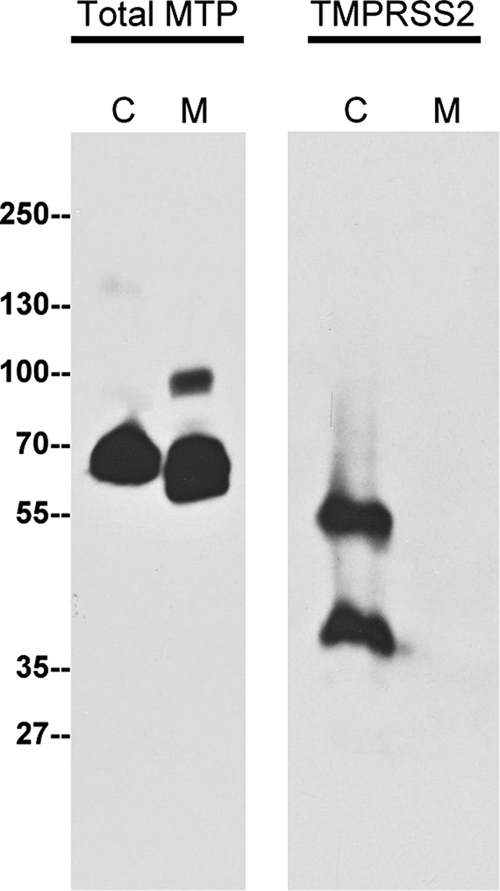
Shedding of TMPRSS2 to the extracellular milieu by LNCaP cells is very limited. LNCaP cells were grown in serum-free media for 24 hours, and then the conditioned medium and cell lysates were harvested. The conditioned medium was concentrated up to 100 fold. The cell lysates (C) and the conditioned medium (M) were analyzed for total matriptase (Total MTP) using the matriptase mAb M32 and TMPRSS2 using the mAb AL20.
TMPRSS2 Is Secreted as Both Noncovalent and Disulfide Bond-Linked Complexes into Human Semen as Well
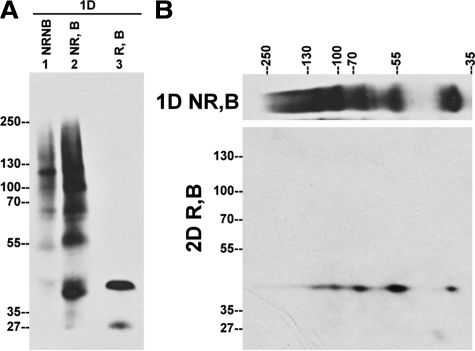
Although TMPRSS2 is apparently not secreted by LNCaP cells (Figure 5), the expression of TMPRSS2 on the apical plasma membrane of the prostatic luminal epithelial cells suggests that TMPRSS2 may be secreted into semen. We therefore examined the expression status of TMPRSS2 in human semen from twenty three healthy men of different ages and races. When the samples were not treated with reducing agents or high temperature (nonreducing and nonboiled conditions), TMPRSS2 from human semen was detected as multiple protein bands of 40 kDa, 56 kDa, 68 kDa, 100 kDa, 120 kDa, and 140 kDa (Figure 6, lane 1, NRNB). These species may be TMPRSS2 complexes because their size greater than that of full-length TMPRSS2. Under boiled and nonreducing conditions, the protease was also detected as multiple protein bands, though the bands were of significantly increased intensity (Figure 6, lane 2, NR,B) suggesting that the heat treatment may either expose the epitope recognized by TMPRSS2 mAb or cause the dissociation of TMPRSS2 complexes, or both. After treating with reducing agents, TMPRSS2 was detected as one major protein band of 42 kDa and a minor band of 27 kDa (Figure 6, lane 3, R,B). The disappearance of most of the higher molecular weight TMPRSS2 species and the appearance of the 42-kDa species after chemical reduction suggests that TMPRSS2 is present in multiple disulfide-linked complexes in semen that are converted to the 42-kDa species after chemical reduction (Figure 6, lane 3, R,B). This hypothesis was further supported by a diagonal gel electrophoresis analysis with the second dimension under reducing conditions (Figure 6). Immunoblot analysis revealed that all of these TMPRSS2 complexes species were converted to the 42-kDa species. These data suggest that TMPRSS2 is secreted into human semen as the 42-kDa species in the form of multiple noncovalent and disulfide-linked complexes. Although a 56-kDa TMPRSS2 species was detected in semen under boiled and nonreducing conditions (Figure 6, lane 2), this 56-kDa species is not the same as the full-length TMPRSS2 detected in LNCaP cells under boiled and nonreducing conditions (Figure 3A, lane 2), because of the fact that the 56-kDa species is a disulfide-linked complex that contains the 42-kDa species. A minor 27-kDa species detected in semen may be a degradation product because its levels vary among different batches of human semen and increases along with the disappearance of the high molecular weight species during the processing of the semen.
Figure 6.
TMPRSS2 is present in multiple complexes in human semen. A: Equal amount of human semen were mixed with SDS sample buffer in the absence (NR) or presence (R) of reducing agents and incubated either at room temperature (NB) or 95°C (B) for 5 minutes, followed by SDS-PAGE and Western blot analysis using TMPRSS2 mAb AL20. B: Human semen was resolved by diagonal gel electrophoresis under nonreducing and boiled conditions in the first dimension and under reducing and boiled conditions in the second dimension. Immunoblot analysis of the second dimensional gel was performed with the TMPRSS2 mAb AL20.
TMPRSS2 Is Secreted as Components of the Prostasome into Human Semen
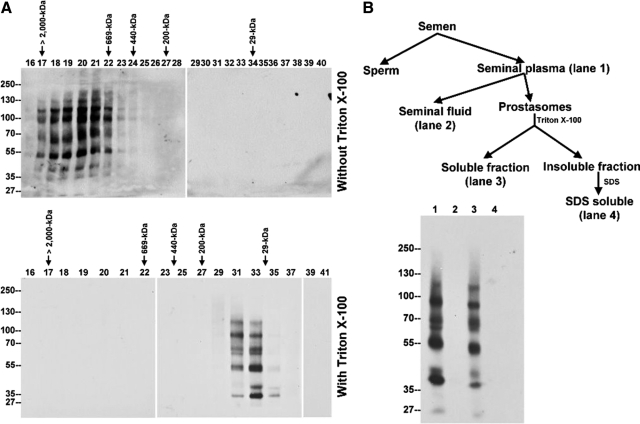
In addition to the formation of multiple protein complexes, we also noticed that semen-derived TMPRSS2 complexes appear to be present in gigantic complex(es) with apparent sizes of up to several million Da, because they eluted together in the void volume of a Superose 6 sizing column (Figure 7A, top). Human semen is known to contain organelle-like membrane vesicles, named prostasomes, which have previously been isolated in the void volume of various sizing columns and by ultracentrifugation,26,27 and so we inferred that TMPRSS2 and its complexes may reside in these membrane vesicles. Two approaches were taken to demonstrate that TMPRSS2 is a component of prostasomes. When seminal plasma was loaded onto Superose 6 sizing column, in the absence of detergent, all of the TMPRSS2 species were found to elute in fractions 17 through 22, which represents the void volume (Figure 7A, top). In the presence of Triton X-100, TMPRSS2 species eluted in fractions 31 through 35, which contain proteins with molecular mass between 200- and 29-kDa (Figure 7A, bottom). The nonionic detergent apparently solubilized the membrane vesicles and released the TMPRSS2 from prostasomes, resulting in the change in the sizing column elution profile. Because prostasomes can be precipitated by ultracentrifugation, we fractionated human semen into 4 different fractions by differential centrifugation as described in Figure 7B. These fractions include seminal plasma (after removal of sperm and cell debris), seminal fluid (after removal of prostasomes), prostasome-Triton X-100 soluble fraction, and prostasome-Triton X-100 insoluble fraction. All of the TMPRSS2 complexes detected in the seminal plasma (Figure 7B, lane 1) were all spun down with the prostasomes and detected in the prostasome Triton-soluble fractions (Figure 7B, lane 3) and not in the seminal fluid fraction (Figure 7B, lane 2). These data suggest that TMPRSS2 is secreted by prostate luminal epithelial cells into semen as components of prostasomes.
Figure 7.
TMPRSS2 is expressed as components of the prostasome. A: Seminal plasma was fractionated in the absence (top) or presence (bottom) of 1% Triton X-100 by the Superose 6 sizing column. The Superose 6 fractions were examined by immunoblot analysis using TMPRSS2 mAb AL20. In the absence of Triton X-100, TMPRSS2 was detected in the void volume as indicated by co-elution with blue dextran (top). In the presence of nonionic detergent, TMPRSS2 was eluted in the fractions whose apparent molecular masses were distributed from 200 kDa to 29 kDa as determined by comparison with standard proteins which included blue dextran (>2000 kDa), thyroglobulin (669 kDa), apoferritin (443 kDa), b-amylase (200 kDa), carbonic anhydrase (29 kDa) as indicated (bottom). B: Top: Flow chart of human semen fractionation. Human semen was fractionated by centrifugation at 1,000g for 15 minutes into sperm fractions and seminal plasma. The latter was further fractionated by centrifugation at 105,000g for 2 hours into the insoluble fractions, representing the prostasomes, and the soluble fractions, the seminal fluids. Prostasomes were then lysed with 1% Triton X-100 in PBS, and the insoluble fractions were further extracted by 1% SDS in PBS. Bottom: The four semen fractions were subjected to SDS-PAGE and followed by immunoblot analyses for TMPRSS2. The TTSP, detected in seminal plasma (lane 1), was fractionated into the prostasome Triton X-100–soluble fractions (lane 3), but not in the seminal fluid (lane 2) and the Triton X-100–insoluble fractions (lane 4).
Discussion
TMPRSS2, an androgen responsive gene, is highly expressed in prostate epithelial cells and in human prostate cancers. Here we report new insights into the regulation and function of this TTPS, including (1) its presence in high-molecular-weight complexes, which are SDS-resistant and disulfide-linked, (2) its localization to the apical plasma membrane of prostate luminal epithelial cells, (3) its presence in prostasomes, membranous vesicles in semen, and (4) its increased expression and misregulated subcellular localization in prostate cancer cells.
TMPRSS2 was previously reported to have a mass of either 70 kDa or 65 kDa,11,17,18 which is different from that determined in this study and is greater than the calculated mass. These discrepancies may be attributable to the use of different TMPRSS2 antibodies, which were raised against synthetic peptides or recombinant proteins produced in bacteria. As shown in the supplemental figures (at http://ajp.amjpathol.org), the three commercial TMPRSS2 antibodies we tested, produced against synthetic peptides, appear to be suboptimal for the detection of endogenous TMPRSS2 (see Supplemental Figure S1 at http://ajp.amjpathol.org). In contrast, our mAbs were generated by using TMPRSS2 produced in HEK293T cells and have proven to be highly specific and very sensitive (Figure 1C and Supplemental Figure S1 at http://ajp.amjpathol.org). In our study, the vast majority of TMPRSS2 was detected either at an apparent mass of 58 kDa in LNCaP cells or 42 kDa in semen. The 58-kDa species appears to be the full-length form of the protein and the size is close to the 53,859-Da calculated size of the protease. The small difference between the calculated and observed size is attributable to glycosylation (Figure 4). In LNCaP prostate cancer cells, the 58-kDa species was consistently detected as the major form of TMPRSS2, though the 42-kDa species was also detected and its levels varied from experiment to experiment. In contrast, the 42-kDa TMPRSS2 appears to be the major form detected in human semen and 58-kDa full-length TMPRSS2 was not detected (Figure 6A, lane 3, R,B). The conversion of full-length TMPRSS2 to 42-kDa form may occur before its release. Despite their differential presentation in LNCaP cells and semen, both the 58-kDa and 42-kDa TMPRSS2 forms seem to resemble one another with respect to the formation of complexes. This is particularly evident when comparing the pattern of disulfide-linked complexes containing the 58-kDa versus the 42-kDa form (Figure 3C).
Detection of TMPRSS2 under reducing and boiled conditions as a 58-kDa protein band close to the calculated size of the full-length TMPRSS2 suggests that the TTSP is synthesized as a latent protease, in common with most serine proteases. TMPRSS2 is, however, unusual among TTSPs regarding its formation of multiple complexes. TMPRSS2 can form noncovalent and disulfide-linked complexes both in vitro in LNCaP cells and in vivo in prostatic secretions. The majority of the TMPRSS2 released from complexes is in the latent form, which would not be expected to form stable complexes with endogenous protease inhibitors. The TMPRSS2 complexes are, therefore, apparently not a product of conventional mechanisms governing the zymogen activation and inhibition of active serine proteases. Furthermore, TMPRSS2 has 22 cysteine residues. Although the serine protease domain in TMPRSS2 contains eight Cys residues in conserved positions, the pairing counterpart cysteine for Cys140 is absent. This unpaired Cys in the serine protease domain may be the structural element, which facilitates the formation of disulfide-linked complexes. It will be of great interest to determine whether the extensive formation of TMPRSS2 complexes represents a novel mechanism by which the protease is regulated and to explore the functional significance of complex formation.
Human TMPRSS2 contains 492 amino acid residues. Its activation theoretically involves a proteolytic cleavage between amino acids 255 Arg-Ilu 256, resulting in two fragments with similar sizes of around 27 kDa, held together through a disulfide linkage. Although the exact epitope recognized by our TMPRSS2 mAb has not been determined, the detection of a 27-kDa protein band by the TMPRSS2 mAb under reducing conditions is likely an indication of TMPRSS2 activation. The level of TMPRSS2 activation in LNCaP prostate cancer cells and human semen, however, appears to be very limited since there was no detectable 27-kDa TMPRSS2 protein present under reducing conditions (Figures 3A and 6A).
In addition to the unusual complex formation, the targeting of the protease to the apical plasma membrane and its shedding as a component of prostasomes into semen provides insights into the physiological roles of TMPRSS2. Like some other prostasome proteins, which were also detected on the apical plasma membrane of the prostate luminal cells,28 the presence of TMPRSS2 both on the apical plasma membrane of luminal cells in the prostate gland and in the prostasomes suggests that TMPRSS2 may be a marker for prostasomes and its physiological functions may be associated with this organelle-like structure. ENaC, a putative TMPRSS2 substrate, is also localized at the apical plasma membrane.9 If TMPRSS2 can be converted into an active enzyme at the apical membrane, the protease may activate ENaC to regulate sodium homeostasis within the prostate gland. The presence of TMPRSS2 in prostasomes suggests that it may also regulate prostasome functions. Prostasomes have been proposed to be involved in protection of the spermatozoa in the acidic milieu of the vagina,29,30 stabilization of sperm plasma membrane and delay of acrosome reaction,31,32,33,34 enhancement of sperm motility,30,35,36,37,38,39 protection of spermatozoa from phagocytosis and complement attack, and inhibition of viral activity. Many of the proposed biological functions of the prostasomes facilitate sperm function and enhance male reproduction. However, the loss of TMPRSS2 in TMPRSS2-deficient mice did not adversely influence fertility, reduce survival, result in prostate hyperplasia or carcinoma, or alter prostatic luminal epithelial cell regrowth after castration and androgen replacement.8 It remains to be determined whether the lack of phenotype in the TMPRSS2-deficient mouse is attributable to the fact that TMPRSS2 is not involved in these processes, or redundancy with other proteases fulfilling the functions of TMPRSS2, or reflects difference between humans and mice.
TMPRSS2 recently received a great deal of attention as a participant in gene fusions of the promoter regions of the protease with the ETS family members found in the majority of human prostate cancers.2,14 This gene fusion is a frequent and early event in prostate cancer pathogenesis and may promote cancer progression into a more aggressive phenotype through the engagement of the downstream targets of ETS, including plasminogen activation pathway that mediates cancer invasion and metastasis.40 Although the gene fusion should not affect the function of the TMPRSS2 gene product as a transmembrane serine protease, the gene fusion appears to reduce the expression of TMPRSS2 at mRNA levels in those prostate tumors harboring TMPRSS2/ERG.16 Although the expression of TMPRSS2 transcript is decreased in prostate cancer cells with TMPRSS2/ETS fusions, several studies have shown that TMPRSS2 expression is higher in prostate cancer tissue than in normal prostate and increases with Gleason score, tumor grade, and stage.2,11,12,13,16,41 Furthermore, our current study further confirms the increased expression of TMPRSS2 in prostate cancer at the protein level using the AQUA system to quantitate immunohistochemistry staining in a prostate TMA. Therefore, TMPRSS2 may contribute to prostate cancer not only by the increased expression but also through aberrant subcellular localization attributalbe to the loss of epithelial polarity in the transformed cells. This latter abnormality may allow the protease to inappropriately gain access to and/or activate some cancer-promoting substrates, which its normal subcellular localization would preclude under normal physiological conditions.
Footnotes
Address reprint requests to Chen-Yong Lin, Ph.D., Greenebaum Cancer Center, Department of Biochemistry and Molecular Biology, University of Maryland Baltimore, BRB 10-027, 655 W. Baltimore Street, Baltimore, MD 21201; or Wei Huang, M.D., Departments of Pathology and Laboratory Medicine, University of Wisconsin, 600 Highland Avenue, Madison, WI 53792. E-mail: cylin@som.umaryland.edu or whuang23@wisc.edu.
Supported by National Cancer Institute (NCI) grants RO1 CA 096851 and RO1 CA 104944 (to C.-Y.L.) and Taiwan National Science Council grant NSC 97-2320-B-00-052-MY3 (to M.-S.L.).
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Paoloni-Giacobino A, Chen H, Peitsch MC, Rossier C, Antonarakis SE. Cloning of the TMPRSS2 gene, which encodes a novel serine protease with transmembrane. LDLRA, and SRCR domains and maps to 21q223. Genomics. 1997;44:309–320. doi: 10.1006/geno.1997.4845. [DOI] [PubMed] [Google Scholar]
- Lin B, Ferguson C, White JT, Wang S, Vessella R, True LD, Hood L, Nelson PS. Prostate-localized and androgen-regulated expression of the membrane-bound serine protease TMPRSS2. Cancer Res. 1999;59:4180–4184. [PubMed] [Google Scholar]
- Jacquinet E, Rao NV, Rao GV, Zhengming W, Albertine KH, Hoidal JR. Cloning and characterization of the cDNA and gene for human epitheliasin. Eur J Biochem. 2001;268:2687–2699. doi: 10.1046/j.1432-1327.2001.02165.x. [DOI] [PubMed] [Google Scholar]
- Hooper JD, Clements JA, Quigley JP, Antalis TM. Type II transmembrane serine proteases. Insights into an emerging class of cell surface proteolytic enzymes. J Biol Chem. 2001;276:857–860. doi: 10.1074/jbc.R000020200. [DOI] [PubMed] [Google Scholar]
- Netzel-Arnett S, Hooper JD, Szabo R, Madison EL, Quigley JP, Bugge TH, Antalis TM. Membrane anchored serine proteases: a rapidly expanding group of cell surface proteolytic enzymes with potential roles in cancer. Cancer Metastasis Rev. 2003;22:237–258. doi: 10.1023/a:1023003616848. [DOI] [PubMed] [Google Scholar]
- Szabo R, Wu Q, Dickson RB, Netzel-Arnett S, Antalis TM, Bugge TH. Type II transmembrane serine proteases. Thromb Haemost. 2003;90:185–193. doi: 10.1160/TH03-02-0071. [DOI] [PubMed] [Google Scholar]
- Bugge TH, Antalis TM, Wu Q. Type II transmembrane serine proteases. J Biol Chem. 2009;284:23177–23181. doi: 10.1074/jbc.R109.021006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TS, Heinlein C, Hackman RC, Nelson PS. Phenotypic analysis of mice lacking the Tmprss2-encoded protease. Mol Cell Biol. 2006;26:965–975. doi: 10.1128/MCB.26.3.965-975.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson SH, Hirsh A, Li DC, Holloway G, Chao J, Boucher RC, Gabriel SE. Regulation of the epithelial sodium channel by serine proteases in human airways. J Biol Chem. 2002;277:8338–8345. doi: 10.1074/jbc.M105044200. [DOI] [PubMed] [Google Scholar]
- Aimes RT, Zijlstra A, Hooper JD, Ogbourne SM, Sit ML, Fuchs S, Gotley DC, Quigley JP, Antalis TM. Endothelial cell serine proteases expressed during vascular morphogenesis and angiogenesis. Thromb Haemost. 2003;89:561–572. [PubMed] [Google Scholar]
- Afar DE, Vivanco I, Hubert RS, Kuo J, Chen E, Saffran DC, Raitano AB, Jakobovits A. Catalytic cleavage of the androgen-regulated TMPRSS2 protease results in its secretion by prostate and prostate cancer epithelia. Cancer Res. 2001;61:1686–1692. [PubMed] [Google Scholar]
- Vaarala MH, Porvari K, Kyllonen A, Lukkarinen O, Vihko P. The TMPRSS2 gene encoding transmembrane serine protease is overexpressed in a majority of prostate cancer patients: detection of mutated TMPRSS2 form in a case of aggressive disease. Int J Cancer. 2001;94:705–710. doi: 10.1002/ijc.1526. [DOI] [PubMed] [Google Scholar]
- Vaarala MH, Porvari KS, Kellokumpu S, Kyllonen AP, Vihko PT. Expression of transmembrane serine protease TMPRSS2 in mouse and human tissues. J Pathol. 2001;193:134–140. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH743>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Hermans KG, van MR, van DH, Jenster G, van Weerden WM, Trapman J. TMPRSS2:ERG fusion by translocation or interstitial deletion is highly relevant in androgen-dependent prostate cancer, but is bypassed in late-stage androgen receptor-negative prostate cancer. Cancer Res. 2006;66:10658–10663. doi: 10.1158/0008-5472.CAN-06-1871. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- Mwamukonda K, Chen Y, Ravindranath L, Furusato B, Hu Y, Sterbis J, Osborn D, Rosner I, Sesterhenn IA, McLeod DG, Srivastava S, Petrovics G. Quantitative expression of TMPRSS2 transcript in prostate tumor cells reflects TMPRSS2-ERG fusion status. Prostate Cancer Prostatic Dis. 2010;13:47–51. doi: 10.1038/pcan.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas JM, True L, Hawley S, Matsumura M, Morrissey C, Vessella R, Nelson PS. The androgen-regulated type II serine protease TMPRSS2 is differentially expressed and mislocalized in prostate adenocarcinoma. J Pathol. 2008;215:118–125. doi: 10.1002/path.2330. [DOI] [PubMed] [Google Scholar]
- Wilson S, Greer B, Hooper J, Zijlstra A, Walker B, Quigley J, Hawthorne S. The membrane-anchored serine protease. TMPRSS2, activates PAR-2 in prostate cancer cells. Biochem J. 2005;388:967–972. doi: 10.1042/BJ20041066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Anders J, Johnson M, Dickson RB. Purification and characterization of a complex containing matriptase and a Kunitz-type serine protease inhibitor from human milk. J Biol Chem. 1999;274:18237–18242. doi: 10.1074/jbc.274.26.18237. [DOI] [PubMed] [Google Scholar]
- Oberst MD, Williams CA, Dickson RB, Johnson MD, Lin CY. The activation of matriptase requires its noncatalytic domains, serine protease domain, and its cognate inhibitor. J Biol Chem. 2003;278:26773–26779. doi: 10.1074/jbc.M304282200. [DOI] [PubMed] [Google Scholar]
- Lin CY, Wang JK, Torri J, Dou L, Sang QA, Dickson RB. Characterization of a novel, membrane-bound, 80-kDa matrix-degrading protease from human breast cancer cells. Monoclonal antibody production, isolation, and localization. J Biol Chem. 1997;272:9147–9152. [PubMed] [Google Scholar]
- Tseng IC, Chou FP, Su SF, Oberst M, Madayiputhiya N, Lee MS, Wang JK, Sloane DE, Johnson M, Lin CY. Purification from human milk of matriptase complexes with secreted serpins: mechanism for inhibition of matriptase other than HAI-1. Am J Physiol Cell Physiol. 2008;295:C423–C431. doi: 10.1152/ajpcell.00164.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren M, Twohig M, Pier T, Eickhoff J, Lin CY, Jarrard D, Huang W. Protein expression of matriptase and its cognate inhibitor HAI-1 in human prostate cancer: a tissue microarray and automated quantitative analysis. Appl Immunohistochem Mol Morphol. 2008;17:23–30. doi: 10.1097/PAI.0b013e31817c3334. [DOI] [PubMed] [Google Scholar]
- Rahrmann EP, Collier LS, Knutson TP, Doyal ME, Kuslak SL, Green LE, Malinowski RL, Roethe L, Akagi K, Waknitz M, Huang W, Largaespada DA, Marker PC. Identification of PDE4D as a proliferation promoting factor in prostate cancer using a Sleeping Beauty transposon-based somatic mutagenesis screen. Cancer Res. 2009;69:4388–4397. doi: 10.1158/0008-5472.CAN-08-3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomiya KI, Lee MS, Tseng IC, Zuo H, Barndt RJ, Johnson MD, Dickson RB, Lin CY. Matriptase activation and subsequent shedding with HAI-1 is induced by steroid sex hormones in human prostate cancer cells, but not in breast cancer cells. Am J Physiol Cell Physiol. 2006;291:C40–C49. doi: 10.1152/ajpcell.00351.2005. [DOI] [PubMed] [Google Scholar]
- Arienti G, Carlini E, De Cosmo AM, Di PP, Palmerini CA. Prostasome-like particles in stallion semen. Biol Reprod. 1998;59:309–313. doi: 10.1095/biolreprod59.2.309. [DOI] [PubMed] [Google Scholar]
- Carlsson L, Nilsson O, Larsson A, Stridsberg M, Sahlen G, Ronquist G. Characteristics of human prostasomes isolated from three different sources. Prostate. 2003;54:322–330. doi: 10.1002/pros.10189. [DOI] [PubMed] [Google Scholar]
- Nilsson BO, Jin M, Einarsson B, Persson BE, Ronquist G. Monoclonal antibodies against human prostasomes. Prostate. 1998;35:178–184. doi: 10.1002/(sici)1097-0045(19980515)35:3<178::aid-pros3>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- Arienti G, Carlini E, Palmerini CA. Fusion of human sperm to prostasomes at acidic pH. J Membr Biol. 1997;155:89–94. doi: 10.1007/s002329900160. [DOI] [PubMed] [Google Scholar]
- Arienti G, Carlini E, Nicolucci A, Cosmi EV, Santi F, Palmerini CA. The motility of human spermatozoa as influenced by prostasomes at various pH levels. Biol Cell. 1999;91:51–54. [PubMed] [Google Scholar]
- Arienti G, Carlini E, Polci A, Cosmi EV, Palmerini CA. Fatty acid pattern of human prostasome lipid. Arch Biochem Biophys. 1998;358:391–395. doi: 10.1006/abbi.1998.0876. [DOI] [PubMed] [Google Scholar]
- Carlini E, Palmerini CA, Cosmi EV, Arienti G. Fusion of sperm with prostasomes: effects on membrane fluidity. Arch Biochem Biophys. 1997;343:6–12. doi: 10.1006/abbi.1997.9999. [DOI] [PubMed] [Google Scholar]
- Cross NL. Human seminal plasma prevents sperm from becoming acrosomally responsive to the agonist, progesterone: cholesterol is the major inhibitor. Biol Reprod. 1996;54:138–145. doi: 10.1095/biolreprod54.1.138. [DOI] [PubMed] [Google Scholar]
- Cross NL, Mahasreshti P. Prostasome fraction of human seminal plasma prevents sperm from becoming acrosomally responsive to the agonist progesterone. Arch Androl. 1997;39:39–44. doi: 10.3109/01485019708987900. [DOI] [PubMed] [Google Scholar]
- Carlsson L, Ronquist G, Stridsberg M, Johansson L. Motility stimulant effects of prostasome inclusion in swim-up medium on cryopreserved human spermatozoa. Arch Androl. 1997;38:215–221. doi: 10.3109/01485019708994880. [DOI] [PubMed] [Google Scholar]
- Fabiani R, Johansson L, Lundkvist O, Ulmsten U, Ronquist G. Promotive effect by prostasomes on normal human spermatozoa exhibiting no forward motility due to buffer washings. Eur J Obstet Gynecol Reprod Biol. 1994;57:181–188. doi: 10.1016/0028-2243(94)90298-4. [DOI] [PubMed] [Google Scholar]
- Fabiani R, Johansson L, Lundkvist O, Ronquist G. Enhanced recruitment of motile spermatozoa by prostasome inclusion in swim-up medium. Hum Reprod. 1994;9:1485–1489. doi: 10.1093/oxfordjournals.humrep.a138735. [DOI] [PubMed] [Google Scholar]
- Fabiani R, Johansson L, Lundkvist O, Ronquist G. Prolongation and improvement of prostasome promotive effect on sperm forward motility. Eur J Obstet Gynecol Reprod Biol. 1995;58:191–198. doi: 10.1016/0028-2243(94)01997-5. [DOI] [PubMed] [Google Scholar]
- Stegmayr B, Ronquist G. Promotive effect on human sperm progressive motility by prostasomes. Urol Res. 1982;10:253–257. doi: 10.1007/BF00255932. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Laxman B, Varambally S, Cao X, Yu J, Helgeson BE, Cao Q, Prensner JR, Rubin MA, Shah RB, Mehra R, Chinnaiyan AM. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia. 2008;10:177–188. doi: 10.1593/neo.07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X, He H, Ye Y, Dai Q, Han Z, Liang Y, Zhong W. Association of TMPRSS2 and KLK11 gene expression levels with clinical progression of human prostate cancer. Med Oncol. 2010;27:145–151. doi: 10.1007/s12032-009-9185-0. [DOI] [PubMed] [Google Scholar]