Abstract
Conventional approaches for ultrastructural high-resolution imaging of biological specimens induce profound changes in bio-molecular structures. By combining tissue cryo-sectioning with non-destructive atomic force microscopy (AFM) imaging we have developed a methodology that may be applied by the non-specialist to both preserve and visualize bio-molecular structures (in particular extracellular matrix assemblies) in situ. This tissue section AFM technique is capable of: i) resolving nm–µm scale features of intra- and extracellular structures in tissue cryo-sections; ii) imaging the same tissue region before and after experimental interventions; iii) combining ultrastructural imaging with complimentary microscopical and micromechanical methods. Here, we employ this technique to: i) visualize the macro-molecular structures of unstained and unfixed fibrillar collagens (in skin, cartilage and intervertebral disc), elastic fibres (in aorta and lung), desmosomes (in nasal epithelium) and mitochondria (in heart); ii) quantify the ultrastructural effects of sequential collagenase digestion on a single elastic fibre; iii) correlate optical (auto fluorescent) with ultrastructural (AFM) images of aortic elastic lamellae.
Keywords: Atomic force microscopy, Tissue cryo-sections, Collagen fibrils, Elastic fibres, Native biomolecules
1. Introduction
The function of many connective tissues is dependent on the supramolecular structure of their component extracellular matrix (ECM) assemblies (Birk and Bruckner, 2005). Whilst electron microscopy (EM) has made an immense contribution to our knowledge of such tissue ultrastructures, routine EM methods irreversibly denature biomolecules during specimen preparation because of harsh chemical fixation and staining regimes, and during imaging via electron beam interactions (Ruprecht and Nield, 2001). To some extent these limitations are addressed by cryo-EM approaches, which preserve native tissue structure and can provide detailed 3D information (Baumeister et al., 1999). However, the technical challenge of freezing tissues in vitreous ice and the poor signal–noise ratio of the resultant low electron dose images necessitate the use of highly specialised equipment and analysis strategies. Here we present an alternative approach to the ultrastructural imaging of native tissues which uses relatively simple and ubiquitous laboratory equipment; the cryostat and atomic force microscope (AFM), and which provides quantitative morphological data without the need for subsequent, computationally-intensive image analysis.
AFMs are members of a larger family of scanning probe microscopes that record interactions between a probe and a surface. In addition to permitting quantitative, high-resolution, non-destructive imaging with a high signal–noise ratio the instruments can, uniquely, operate in physiologically appropriate solution conditions and manipulate biological structures directly (Bushell et al., 1999; Fotiadis et al., 2002). Consequently, these microscopes are increasingly finding applications within the biological sciences. Although occasionally employed to characterise the morphology of relatively stiff skeletal tissues (Hassenkam et al., 2004; Imer et al., 2006), freeze-etched tissue replicas (Raspanti et al., 1996) and hydrated tissues (Raspanti et al., 2001); the technique is rarely applied directly to thin tissue sections. Where AFM of sections has been reported, the tissues had been previously exposed to fixation and embedding procedures (Li et al., 2008; Matsko and Mueller, 2004). The impact of such chemical fixation protocols on protein structure was, however, recognised in the early 1950s (Fernandezmoran, 1952) and confirmed in recent AFM studies of bulk tissue samples (Jastrzebska et al., 2005). Consequently, tissue freezing and cryo-sectioning, now widely utilized in immuno-cytochemical studies (Akagi et al., 2006), were developed to prevent protein denaturation and larger-scale alterations in structural morphology. Here, we employ these cryo-preservation methods to prepare tissues for non-destructive AFM imaging.
2. Results and discussion
2.1. Visualizing extracellular matrix assemblies in mammalian tissues
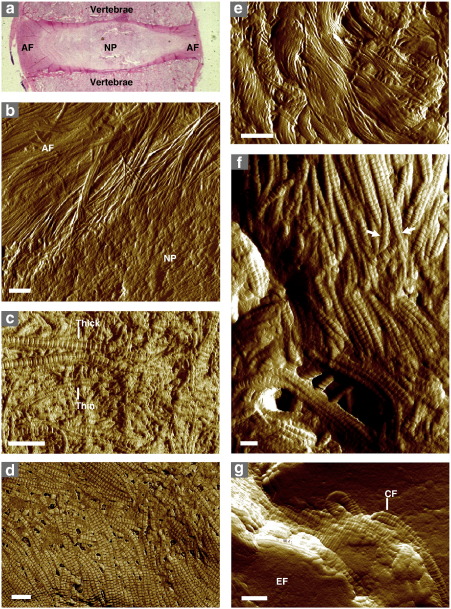
We immobilized tissue samples by freezing in an aqueous embedding medium prior to cryo-sectioning. Freshly-cut 5 μm thick sections were adsorbed directly onto 13 mm diameter glass coverslips, allowed to thaw, air-dried, washed with H2O to remove the support medium, and air-dried again prior to imaging. AFM of these tissue cryo-sections clearly identifies well preserved structural features including the interface between functionally distinct regions in the intervertebral disc (IVD) (Fig. 1a and b), thick and thin type II collagen fibrils embedded within an aggrecan matrix in articular cartilage (AC) (Fig. 1c), collagen fibrils arranged into sheets in the epicardium (Fig. 1d) and “basket-weave” bundles in skin (Fig. 1e). In the elastin-rich murine lung, collagen fibril bundles branch and connect with adjacent bundles (Fig. 1f), whilst in the aorta, collagen fibrils appear to insert into the “granular” elastin-rich core of the elastic lamellae (Fig. 1g), in the manner described previously by Raspanti et al. (2006). Operating the AFM in intermittent contact mode (also known as amplitude modulation or tapping mode) both reduces lateral forces on the sample (Zhong et al., 1993) and facilitates mapping of variations in surface chemistry, and hence composition, via phase imaging (Fig. 1d) (Garcia and Perez, 2002).
Fig. 1.
Morphology of mammalian connective tissues visualized by tissue section AFM. (a and b) The IVD, which acts as a cushion between adjacent vertebrae in the spine, is divided into two distinct regions, an inner, cartilage-like nucleus pulposus (NP), and an outer, type I collagen-rich, tendon-like annulus fibrosus (AF). These regions are clearly visible in low power optical micrographs (haematoxylin and eosin stained) (a) and in tissue section AFM images of the bovine IVD (b). (c) Human AC is composed primarily of thick and thin collagen II fibrils embedded in an aggrecan matrix. Both fibrillar species are identifiable by their 67 nm periodic structure in AFM phase images of cryo-sectioned cartilage. (d) Within ferret cardiac muscle, collagen fibril orientation, which determines the direction of the contractile force, is readily discernible in AFM phase images of the left ventricle epicardium. (e) In amplitude images of human skin cryo-sections, the papillary dermis at the dermal–epidermal junction is characterised by an intercalated “basket-weave” of fibrillar collagen bundles. (f) In the murine lung, individual collagen fibrils (arrows) form laterally associated bundles in which the fibrillar D-periods appear in register. (g) Elastin-rich fibres (EF) within elastic lamellae in the ovine aorta appear granular in amplitude images with an inter-lamellar space that is populated by collagen fibrils (CF). Scale bars = 1 μm (b and e), 500 nm (c and d) and 200 nm (f and g).
2.2. In situ molecular organisation at nm-length scales
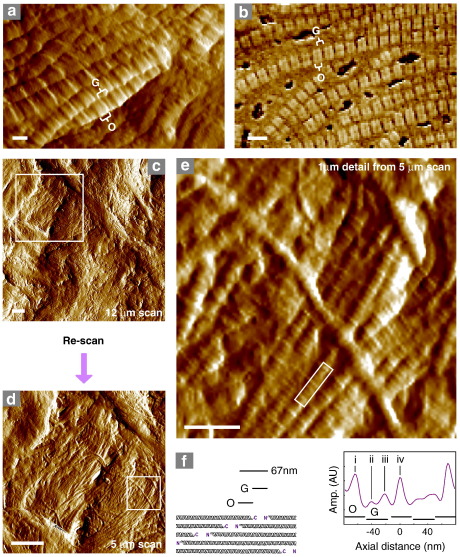
Whilst the organisation of ECM molecules into μm-scale structures is evident at larger scan sizes (Fig. 1), tissue section AFM is also capable of resolving nm-scale structures in situ (Fig. 2). In general tissue cryo-sections are relatively rough. The height range within a 10 μm square AFM image often exceeds 1 μm and this roughness may be due to the tendency of the exposed face to follow the surface of discrete features such as fibrillar collagen bundles and elastic fibres (Figs 1e and f and 2a and b). It seems therefore, that under these sectioning conditions the tissue is fractured rather than cut (Mobius, 2009). Where, for example, the surface of collagen bundles is revealed, the characteristic 67 nm gap/overlap repeat of type I collagen is clearly resolved (Fig. 2a and b). Furthermore, the non-destructive nature of AFM imaging makes it possible to re-scan tissue sections at higher resolution (Fig. 2c and d) revealing finer structural details (Fig. 2e and f). The axial organisation of the collagen fibrils in these regions is reminiscent of the complex banding patterns observed by EM of negatively stained (Chapman et al., 1990) and freeze-etched (Raspanti et al., 1989) tissues suggesting that, at least in some areas, the fracture plane travels through, rather than over, discrete tissue components hence providing information on both internal and external fibril organisation.
Fig. 2.
Extracellular matrix (ECM) ultrastructure in tissue cryo-sections imaged using AFM. (a and b) The gap (G) and overlap (O) regions of type I collagen fibrils are evident in amplitude and phase images of both sheep aorta (a) and ferret epicardium (b). The non-destructive nature of AFM imaging allows areas of interest to be located in initial large scans and re-scanned at higher lateral resolutions. (c–e) Original 12 μm AFM amplitude scan (c), re-scanned 5 μm region (d) and digitally extracted 1 μm detail (e) of a human skin cryo-section. The locations of the re-scanned and extracted regions (c and d) and axial profile (e) are indicated by boxes. (f) The quarter-staggered arrangement of type I collagen monomers induces not only a 67 nm gap/overlap morphology in the fibril but also bands within the repeat which correspond to the position of telopeptide ridges at the gap/overlap interface (peaks i and iv). Minor bands are also evident in the gap region of the AFM amplitude profile (peaks ii and iii). Scale bars = 50 nm (a), 100 nm (b), 1 μm (c and d) and 200 nm (e).
2.3. Cellular organelles and the cell/cell interface
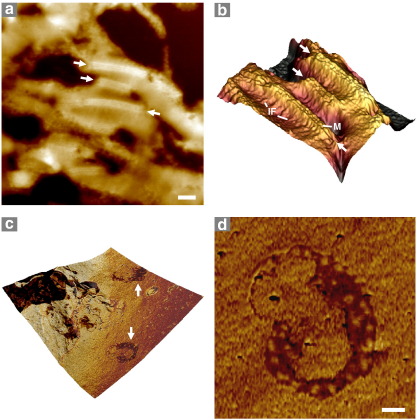
Highly ordered supramolecular assemblies of native biomolecules also play key roles in the function of animal cells. Desmosomes, for example, are intercellular junctions that mediate cell adhesion in epithelial tissues and cardiac muscle (Garrod et al., 2002; Scothern and Garrod, 2008). Al-Amoudi et al. (2007) employed sophisticated cryo-electron tomography to visualize key features of unstained desmosomes within human skin sections. In our high signal–noise ratio AFM images of bovine epidermal desmosomes, similar characteristic structures such as the intermediate filaments and dense midline are readily discernible (Fig. 3a and b). Within the eukaryotic cytoplasm, cellular organelles such as mitochondria possess a unique multi-compartment morphology which is evident in conventional TEM micrographs and AFM images of glutaraldehyde-fixed and osmium tetroxide-stained thin sections of cultured mouse embryonic stem cells (Chen et al., 2005). Tissue section AFM can visualize similar sub-structures of cardiac myocyte mitochondria within unstained ferret ventricular wall (Fig. 3c and d).
Fig. 3.
In situ AFM imaging of mammalian cell organelles. (a and b) AFM height (a 5 μm scan) images of unstained desmosomes (arrows) within the bovine epidermis. (b) 3D surface rendering of an 800 × 600 nm region extracted from panel a. Desmosomes in situ, appear as rigid plate-like structures which project from the surrounding tissue. The dense midline (M) and the intermediate filaments (IF) which connect with the cyto-skeleton are clearly visible. (c) Height and overlaid phase image (2 μm scan) of ferret left ventricle. Mitochondria are easily discernible by phase imaging (arrows and inset [d]). Scale bars = 200 nm (a).
2.4. Visualizing dynamic changes in ECM ultrastructure
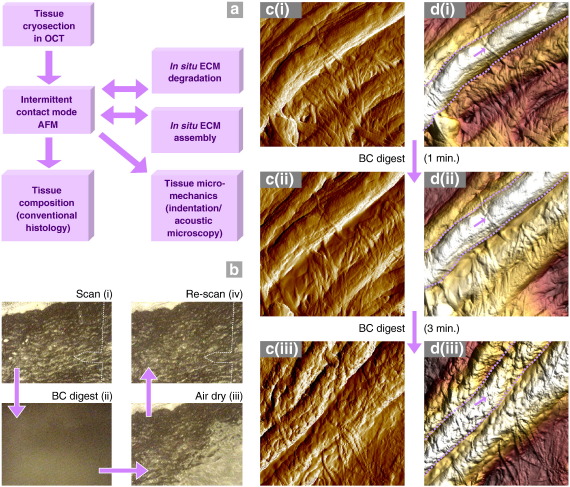
Cryo-sectioning, in combination with intermittent contact mode imaging, preserves native protein structures during both sample preparation and imaging, respectively. Consequently, the same tissue region may be repeatedly imaged by AFM before and after an experimental intervention, or imaged by AFM followed by other microscopical and micromechanical techniques (Fig. 4a). By exploiting these capabilities, tissue section AFM enables the in vitro effects of potential degradative agents, which may act sporadically or at low intensity over many years in vivo, to be compared with the effects of chronic disease-specific pathologies on tissue ultrastructure (Fig. 4).
Fig. 4.
Dynamic applications of tissue section AFM. (a) Following non-destructive AFM imaging, tissue sections may be exposed to catabolic or anabolic environments and subsequently re-scanned to characterise potential degradative or synthetic mechanisms in situ. Alternatively, AFM imaging may be followed by: i) conventional histological and immuno-histochemical techniques, to characterise tissue composition, or ii) functional assays to map tissue mechanical properties. (b–d) Tissue cryo-section of sheep aorta imaged by AFM both before and after, a 1 min and a further 3 min exposure to 1 mg/ml bacterial collagenase (BC). (b) Optical images of the same region of the tissue section: i) after an initial AFM scan, ii) during a 1 min digest and a water wash, iii) during air drying and iv) prior to re-scanning. The position of the AFM tip is indicated by a dotted line. Amplitude (c) and height (d) data (4 μm extracted regions from 5 μm scans) from the same region of a sheep aorta cryo-section before (i) and after exposure to 1 min (ii) and 3 min (iii) bacterial collagenase digests. Large elastic fibres (within dotted lines) and fine interconnecting collagen fibrils (arrows) remained evident following a 1 min digest. Further enzymatic exposure however, induced profound changes in both surface roughness and fibrillar organisation.
2.5. Potential applications of tissue section AFM
AFM imaging may be followed by histological techniques such as fluorescence microscopy that can identify the chemical composition of specific topographical features (Fig. 5), or by non-destructive micro-mechanical mapping techniques such as scanning acoustic microscopy (Akhtar et al., 2009). We also foresee a number of additional applications for tissue section AFM which include; imaging freeze dried or critical point dried specimens to maximize structural preservation and utilizing the ability of the AFM to operate in physiological buffers. This latter approach would enable the quantitation of dynamic changes in tissue structures due to in vitro assembly or degradation. Finally tissue section AFM may be applied to alternative biological materials such as plant tissues.
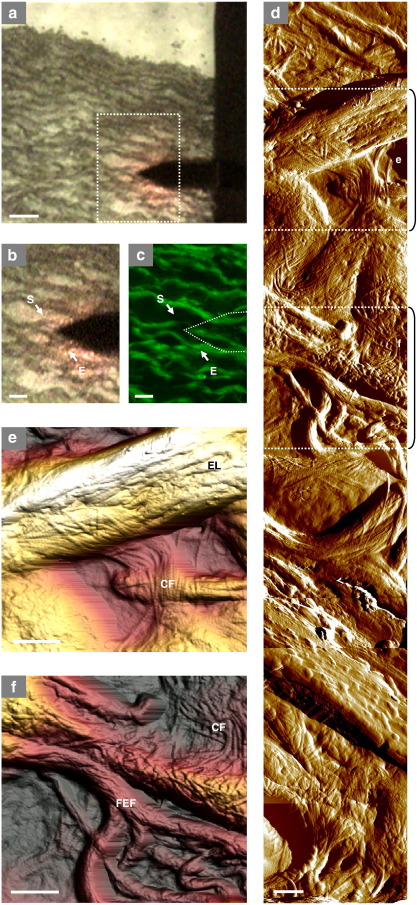
Fig. 5.
Correlation between optical and atomic force microscopy images. (a) Most commercial AFMs possess an optical system, which both assists in microscope alignment and facilitates tip/sample location. The resolution of these systems should be sufficient to localise major features in unstained cryo-sections. In sheep aorta, for example, the tip may be located over features that appear to correspond to elastic lamellae. The composition of these features can be confirmed following AFM imaging by conventional histological methods and imaging the same region by light or fluorescence microscopy. (b) High magnification bright field image of the inset in panel (a). The start (S) and end (E) points of a series of 5 μm AFM scans are indicated by arrows. (c) The same region imaged by fluorescence microscopy, the location of the AFM tip is indicated by a dotted line. (d to f) Variations in elastic fibre diameter (thick elastic lamellae [EL] and fine elastic fibres [FEF]), topology and interaction with collagen fibrils (CF) are evident in a composite image constructed from 8 AFM amplitude scans (5 μm) collected between points S and E (panels b and c) and in AFM height images (e and f) which are denoted by dotted lines and brackets on panel d. Scale bars = 50 μm (a) 20 μm (b and c) and 1 μm (d–f).
3. Experimental procedures
3.1. Reagents, materials and tissue samples
All reagents were obtained from Sigma-Aldrich Co. Ltd. or BDH Ltd. unless otherwise specified. Metal support stubs and Olympus high aspect ratio etched silicon probes (spring constant ∼ 42 N m− 1) were obtained from Veeco Instruments (Santa Barbara, California, USA). Round borosilicate glass coverslips (13 mm diameter) were obtained from Scientific Laboratory Supplies (Nottingham, Nottinghamshire, UK). Muscovite mica was obtained from Agar Scientific (Stansted, Essex, UK). Optimal cutting temperature (OCT) compound was obtained from Thermo Fisher Scientific Inc. (Loughborough, Leicestershire, UK). Bovine intervertebral disc (IVD) tissue samples (from young adult tails) were sourced from a local abattoir and either processed for conventional histological assessment (formalin fixed and paraffin embedded prior to sectioning and staining with haematoxylin and eosin) or cryo-sectioning and subsequent AFM analysis as detailed below. Human AC tissues were obtained with ethical permission from the South Manchester Research Ethics Committee (REC Ref. 06/Q1403/238) and the University Committee on the Ethics of Research on Human Beings (Ref. 06288). Human skin samples were collected as part of a study into elastic fibres and ageing with ethical approval granted by Salford and Trafford Regional Ethics Committee (Ref. No. 02503). Ferret cardiac muscle, ovine aorta and murine lung were obtained as part of ongoing studies which complied with the United Kingdom Animals (Scientific Procedures) Act of 1986 and had received local ethical approval from the University of Manchester (muscle and aorta) and the Institute of Cancer Research (lung) respectively.
3.2. Cryo-sectioning
IVD, AC, skin, cardiac muscle and descending aorta samples were embedded in OCT aqueous embedding medium within an aluminium foil mould and lowered into isopentane (precooled in liquid N2) until frozen. Murine lung tissue was inflated with 50% OCT by intra-tracheal injection before collection and freezing in 100% OCT on dry ice. All frozen tissue blocks were cryo-sectioned to a nominal thickness of 5 μm, mounted on 13 mm diameter circular glass coverslips and stored at − 80 °C. Prior to AFM imaging, glass coverslip adsorbed tissue cryo-sections were allowed to defrost, air-dried, washed with distilled water to remove excess OCT, and air-dried again. The coverslip, and adsorbed tissue section, may be temporarily or permanently adhered to AFM metal support discs using a drop of water or clear nail varnish respectively.
3.3. AFM
Samples were imaged by intermittent contact mode in air using a Multimode AFM with a Nanoscope IIIa controller and a 12 μm × 12 μm × 3.2 μm (X,Y,Z dimension) E scanner. The instrument was calibrated periodically using a grating with 180 nm deep, 10 μm × 10 μm depressions. Cantilever oscillation frequencies and drive amplitudes were determined by the Nanoscope software. Height, amplitude and phase images at scan sizes of 5 μm or 10 μm were captured at an initial scan rate of 1.97 Hz, integral and proportional gain settings of 0.3 and 0.5 respectively and a setpoint which was lower than that automatically determined on initial tip contact. Surface tracking (and hence image quality) was optimised by dynamically lowering the scan rate, increasing the gains and lowering the setpoint (or conversely increasing the drive amplitude). Given the inherent roughness of most tissue sections it may not be possible to resolve all areas within the same scan, although using alternative instruments and/or piezo scanners with larger X, Y and X ranges would help to alleviate this problem.
3.4. Tissue digestion
Tissue cryo-sections of ovine aorta were imaged by AFM prior to exposure to bacterial collagenase. The position of the AFM probe with relation to the section was recorded in a reflected light optical image. Following this initial analysis, the AFM tip and holder was removed and the coverslip and adsorbed tissue incubated in situ with 0.5 mg/ml bacterial collagenase (type IA), freshly prepared protease inhibitors (2 mM Phenylmethanesulfonyl fluoride, 5 mM N-Ethylmaleimide) and 10 mM CaCl2 in 400 mM NaCl, 50 mM Tris–HCl, pH 7.4 for 1 min at room temperature. Active enzyme was removed by washing with 3 × 300 μl of distilled H2O and the tissue section was allowed to dry prior to AFM imaging of the same region. The tissue section was then subjected to a further 3 min incubation with bacterial collagenase, washed, dried and re-imaged.
3.5. Fluorescence microscopy
The non-destructive nature of AFM imaging facilitates further structural and functional characterization of tissue cryo-sections. Unstained tissue cryo-sections were mounted on glass microscope slides using a droplet of water. The region imaged by AFM was located and fluorescence images were captured using a Fluorescein Isothiocyanate (FITC) filter (488 nmex 520 nmem) on a Leica DM5000B microscope coupled to a fluorescence camera (DFC 350FX) together with fluorescence imaging software (Leica LAS AF). Whilst in this study arterial architecture was visualized by fluorescence microscopy, AFM imaged cryo-sections may be further characterised by alternative histological, immuno-histochemical or micro-mechanical methods.
Acknowledgements
This work was supported by Research into Ageing (Senior Fellowship awarded to MJS), the British Heart Foundation (HKG, MJS and AWT), by a SPARC (Strategic Promotion of Ageing Research Capacity) award (AWT) and by the Medical Research Council (DG, AS, MJS). CEMG is supported in part by the NIHR Manchester Biomedical Research Centre.
References
- Akagi T., Ishida K., Hanasaka T., Hayashi S., Watanabe M., Hashikawa T., Tohyama K. Improved methods for ultracryotomy of CNS tissue for ultrastructural and immunogold analyses. J. Neurosci. Methods. 2006;153:276–282. doi: 10.1016/j.jneumeth.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Akhtar R., Sherratt M.J., Watson R.E.B., Kundu T., Derby B. Mapping the micromechanical properties of cryo-sectioned aortic tissue with scanning acoustic microscopy. Mater. Res. Soc. Symp. Proc. 2009;1132E:Z1103–Z1107. doi: 10.1557/PROC-1132-Z03-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Amoudi A., Diez D.C., Betts M.J., Frangakis A.S. The molecular architecture of cadherins in native epidermal desmosomes. Nature. 2007;450:832–837. doi: 10.1038/nature05994. [DOI] [PubMed] [Google Scholar]
- Baumeister W., Grimm R., Walz J. Electron tomography of molecules and cells. Trends Cell Biol. 1999;9:81–85. doi: 10.1016/s0962-8924(98)01423-8. [DOI] [PubMed] [Google Scholar]
- Birk D.E., Bruckner P. Collagen suprastructures. Top Curr. Chem. 2005;247:185–205. [Google Scholar]
- Bushell G.R., Cahill C., Clarke F.M., Gibson C.T., Myhra S., Watson G.S. Imaging and force-distance analysis of human fibroblasts in vitro by atomic force microscopy. Cytometry. 1999;36:254–264. doi: 10.1002/(sici)1097-0320(19990701)36:3<254::aid-cyto16>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Chapman J.A., Tzaphlidou M., Meek K.M., Kadler K.E. The collagen fibril — a model system for studying the staining and fixation of a protein. Electron Microsc. Rev. 1990;3:143–182. doi: 10.1016/0892-0354(90)90018-n. [DOI] [PubMed] [Google Scholar]
- Chen Y., Cai J.Y., Zhao T., Wang C.X., Dong S., Luo S.Q., Chen Z.W. Atomic force microscopy imaging and 3-D reconstructions of serial thin sections of a single cell and its interior structures. Ultramicroscopy. 2005;103:173–182. doi: 10.1016/j.ultramic.2004.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandezmoran H. The submicroscopic organization of vertebrate nerve fibres — an electron microscope study of myelinated and unmyelinated nerve fibres. Exp. Cell Res. 1952;3:282–359. [Google Scholar]
- Fotiadis D., Scheuring S., Muller S.A., Engel A., Muller D.J. Imaging and manipulation of biological structures with the AFM. Micron. 2002;33:385–397. doi: 10.1016/s0968-4328(01)00026-9. [DOI] [PubMed] [Google Scholar]
- Garcia R., Perez R. Dynamic atomic force microscopy methods. Surf. Sci. Rep. 2002;47:197–301. [Google Scholar]
- Garrod D.R., Merritt A.J., Nie Z.X. Desmosomal adhesion: structural basis, molecular mechanism and regulation. Mol. Membr. Biol. 2002;19:81–94. doi: 10.1080/09687680210132476. [DOI] [PubMed] [Google Scholar]
- Hassenkam T., Fantner G.E., Cutroni J.A., Weaver J.C., Morse D.E., Hansma P.K. High-resolution AFM imaging of intact and fractured trabecular bone. Bone. 2004;35:4–10. doi: 10.1016/j.bone.2004.02.024. [DOI] [PubMed] [Google Scholar]
- Imer R., Akiyama T., De Rooji N.F., Stolz M., Aebi U., Friederich N.F., Koenig U., Wirz D., Daniels A.U., Staufer U. Development of atomic force microscope for arthroscopic knee cartilage inspection. Jpn. J. Appl. Phys. 2006;45:2319–2323. [Google Scholar]
- Jastrzebska M., Barwinski B., Mroz I., Turek A., Zalewska-Rejdak J., Cwalina B. Atomic force microscopy investigation of chemically stabilized pericardium tissue. Eur. Phys. J. E. 2005;16:381–388. doi: 10.1140/epje/i2004-10093-1. [DOI] [PubMed] [Google Scholar]
- Li X.H., Ji T., Hu J., Sun J.L. Optimization of specimen preparation of thin cell section for AFM observation. Ultramicroscopy. 2008;108:826–831. doi: 10.1016/j.ultramic.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Matsko N.B., Mueller M. AFM of biological material embedded in epoxy resin. J. Struct. Biol. 2004;146:334–343. doi: 10.1016/j.jsb.2004.01.010. [DOI] [PubMed] [Google Scholar]
- Mobius W. Cryopreparation of biological specimens for immunoelectron microscopy. Ann. Anat-Anat. Anz. 2009;191:231–247. doi: 10.1016/j.aanat.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Raspanti M., Alessandrini A., Gobbi P., Ruggeri A. Collagen fibril surface: TMAFM, FEG-SEM and freeze etching observations. Microsc. Res. Tech. 1996;35:87–93. doi: 10.1002/(SICI)1097-0029(19960901)35:1<87::AID-JEMT8>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Raspanti M., Congiu T., Guizzardi S. Tapping-mode atomic force microscopy in fluid of hydrated extracellular matrix. Matrix Biol. 2001;20:601–604. doi: 10.1016/s0945-053x(01)00174-3. [DOI] [PubMed] [Google Scholar]
- Raspanti M., Ortolani F., Ruggeri A., Marchini M. Collagen fibril surface structures: freeze-etching data and computer modelling. Int. J. Biol. Macromol. 1989;11:77–80. doi: 10.1016/0141-8130(89)90046-9. [DOI] [PubMed] [Google Scholar]
- Raspanti M., Protasoni M., Manelli A., Guizzardi S., Mantovani V., Sala A. The extracellular matrix of the human aortic wall: ultrastructural observations by FEG-SEM and by tapping-mode AFM. Micron. 2006;37:81–86. doi: 10.1016/j.micron.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Ruprecht J., Nield J. Determining the structure of biological macromolecules by transmission electron microscopy, single particle analysis and 3D reconstruction. Prog. Biophys. Mol. Biol. 2001;75:121–164. doi: 10.1016/s0079-6107(01)00004-9. [DOI] [PubMed] [Google Scholar]
- Scothern A., Garrod D. Introduction to Electron Microscopy for Biologists vol. 88. Elsevier Academic Press Inc; San Diego: 2008. Visualization of desmosomes in the electron microscope; pp. 347–366. [DOI] [PubMed] [Google Scholar]
- Zhong Q., Inniss D., Kjoller K., Elings V.B. Fractured polymer silica surface studied by tapping mode atomic-force microscopy. Surf. Sci. 1993;290:L688–L692. [Google Scholar]