Abstract
Flower opening is an event accompanied by morphological changes in petals which include elongation, expansion, and outward-curving. Petal cell growth is a fundamental process that underlies such phenomena, but its molecular mechanism remains largely unknown. Suppression subtractive hybridization was performed between petals during the early elongation period (stage 1) and during the opening period (stage 5) in carnation flowers and a pair of subtraction libraries abundant in differentially expressed genes was constructed at each stage. 393 cDNA clones picked up by differential screening out of 1728 clones were sequenced and 235 different cDNA fragments were identified, among which 211 did not match any known nucleotide sequence of carnation genes in the databases. BLASTX search of nucleotide sequences revealed that putative functions of the translational products can be classified into several categories including transcription, signalling, cell wall modification, lipid metabolism, and transport. Open reading frames of 15 selected genes were successfully determined by rapid amplification of cDNA ends (RACE). Time-course analysis of these genes by real-time RT-PCR showed that transcript levels of several genes correlatively fluctuate in petals of opening carnation flowers, suggesting an association with the morphological changes by elongation or curving. Based on the results, it is suggested that the growth of carnation petals is controlled by co-ordinated gene expression during the progress of flower opening. In addition, the possible roles of some key genes in the initiation of cell growth, the construction of the cell wall and cuticle, and transport across membranes were discussed.
Keywords: Carnation, flower opening, gene expression, petal cell growth, suppression subtractive hybridization
Introduction
From a horticultural viewpoint, the manner in which flowers open and senesce is important in order to determine the quality of cut flowers. In cut flowers of carnation, one of the most economically important ornamental plants, climacteric ethylene production induces petal senescence, which therefore determines the vase life of cut flowers in a cultivar-dependent manner (Nukui et al., 2004). The induction of genes involved in ethylene biosynthesis (ten Have and Woltering, 1997), ethylene perception (Shibuya et al., 2002; Iordachescu and Verlinden, 2005), protein degradation (Jones et al., 1995), and lipid degradation (Hong et al., 2000) has been characterized in senescing carnation petals. Transcript accumulation and its inhibition by sucrose are associated with visible senescence symptoms of the petals induced by ethylene (Lawton et al., 1989; Hoeberichts et al., 2007). On the other hand, there are only a few studies on genes associated with petal growth, which is essential for flower opening in carnation.
Flower opening is generally caused by the expansion of petal cells (van Doorn and van Meeteren, 2003). Physiological and molecular aspects of petal cell growth have been described in several species. The significance of the translocation of sugars has been demonstrated in the petal growth of rose (K Yamada et al., 2007) and tobacco (Kwak et al., 2007). In tulip, phosphorylation of plasma membrane intrinsic protein (PIP) accompanies flower opening and four PIP genes have been identified (Azad et al., 2004, 2008). The inhibition of cell expansion by ethylene is accompanied by the suppressed expression of a PIP gene in petals in opening rose flower (Ma et al., 2008). The role of α-expansin, a cell-wall loosening protein, in corolla development has been demonstrated in petunia (Zenoni et al., 2004). In rose, certain members of the multigene families of expansin and xyloglucanendotransglucosylase/hydrolase (XTH) have been proposed as being involved in the increase in cell wall extensibility at the opening stage (Yamada et al., 2009). In Gerbera, GEG, a homologue of the gibberellin-inducible GAST1 gene of tomato, is involved in the determination of cell and organ shape during corolla and carpel development (Kotilainen et al., 1999).
The advantages of collecting ESTs from petal tissues in order to understand the molecular mechanisms of the physiological processes in flower development have been demonstrated in many species (Channelière et al., 2002; Guterman et al., 2002; Ok et al., 2003; van Doorn et al., 2003; Breeze et al., 2004; Hoeberichts et al., 2007; Laitinen et al., 2007; Xu et al., 2007; T Yamada et al., 2007). A proteomic approach has also been applied to a few species (Dafny-Yelin et al., 2005). Suppression subtractive hybridization (SSH) is a useful technique to isolate genes differentially expressed between distinct tissues or developmental stages (Diatchenko et al., 1996). To survey the genes involved in petal growth during the opening of carnation flowers, SSH was performed using carnation petals at two different stages of flower opening. The expression of genes during flower opening of carnation is reported here through the successful identification of more than 200 ESTs that have not been reported previously. The open reading frames (ORFs) of 15 selected genes are confirmed as well as the correlative fluctuation in transcript levels of several genes during flower opening. The roles of putative translational products of some genes are discussed in relation to petal growth.
Materials and methods
Plant materials and incubation
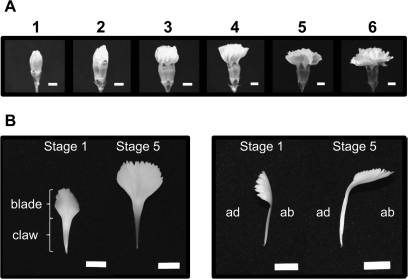
Cut flowers of carnation (Dianthus caryophyllus L. cv. Light Pink Barbara) obtained from a commercial grower in Miyagi prefecture were cut at the end of stems and placed in containers with their cut end in water. They were incubated under constant white fluorescent light (14 μmol m−2 s−1) at 23 °C. Flower opening was categorized into six stages as follows: stage 1, petals just emerged from buds; stage 2, petals elongated vertically; stage 3, petal clusters expanded; stage 4, outer petals start to warp outside; stage 5, outer petals bend outside; stage 6, fully open flower with outer petals at right angles to a stem. Ten outermost petals per flower were collected from ten flowers at each stage to make one sample set for RNA extraction. Three independent sample sets per stage were stored at –80 °C until extraction of RNA.
Subtractive hybridization
Total RNA was extracted from 5 g of fresh petal tissues at stage 1 (the early elongation period) and stage 5 (the opening period) according to Harada et al. (2005). mRNA was isolated from total RNA with a mRNA purification kit (GE Healthcare, Little Chalfont, Buckinghamshire, UK) according to the manufacturer's instructions. cDNA synthesis, digestion with RsaI, hybridization, and PCR amplification were carried out using the PCR-Select cDNA Subtraction Kit (Clontech, Palo Alto, CA, USA) according to the manufacturer's instructions. Forward subtraction was performed using stage-1 cDNA as a tester and stage-5 cDNA as a driver. Reverse subtraction was performed using stage-5 cDNA as a tester and stage-1 cDNA as a driver. PCR products were ligated into pGEM-T Easy Vectors (Promega, Madison, WI, USA) to obtain forward and reverse subtraction libraries. About 3000 colonies each were obtained using a portion of PCR products by suppression subtractive hybridization in both directions and cDNA clones different in abundance between stage 1 and stage 5 were selected using the Differential Screening Kit (Clontech) according to the manufacturer's instructions. An insert cDNA of 864 clones from each library was amplified by colony PCR and blotted on Hybond-N+ membranes (GE Healthcare). 32P-labelled probes were prepared from unsubtracted cDNA from each stage and forward- and reverse-subtracted cDNA and then hybridized with cDNA blotted on membranes. Signals were visualized by BAS-1800II (Fujifilm, Tokyo, Japan) and analysed by Multi Gauge Ver. 2.0 (Fujifilm).
Sequence analysis and annotation
Plasmid DNA was extracted with a QIAprep Spin Miniprep Kit (Qiagen, Hilden, Germany). Nucleotide sequence determination was outsourced to FASMAC, Kanagawa, Japan. Nucleotide sequences were edited and analysed by GENETYX-WIN and a homology search was performed by the BLASTX program on the DNA Data Bank of Japan (DDBJ) website. For annotation of the sequences, the protein function of a gene showing the highest score was adopted when the search hits with an E-value lower than e−10, with a few exceptions in case that enough scores were not obtained because of the position of the sequences. As homologous genes from Arabidopsis was identified in almost all cases, the MIPS Funcat annotation of Arabidopsis was utilized to classify genes. Fourteen categories of biological function that facilitate the classification of genes identified in the present study were set voluntarily.
Rapid amplification of cDNA ends (RACE)
RACE was performed using a GeneRacer Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Gene specific primers were appropriately designed from the nucleotide sequences obtained. cDNA fragments were amplified using the Advantage 2 PCR Enzyme System (Clontech) and subcloned as described above. After sample preparation for sequencing by a BigDye terminator v3.1 Cycle Sequencing Kit (PE Biosystems, Foster City, CA, USA), they were sequenced with an ABI PRISM 310 Genetic Analyser (PE Biosystems).
Real-time RT-PCR
RNA extraction was performed three times per stage using separate sample sets and the three RNA sets per stage were independently used for cDNA synthesis using ReverTra Ace (TOYOBO, Osaka, Japan) according to the manufacturer's instructions. Gene-specific primers were designed mainly from 3′-UTR to give 90–350 bp products (see Supplementary Table S2 at JXB online). PCR was performed with three cDNA sets per stage as templates using the LightCycler FastStart DNA Master SYBR Green I (Roche, Basel, Switzerland) in capillaries. Conditions were 95 °C for 10 min followed by 40 cycles of 1 s at 95 °C, 5 s at 53 °C, and 4–13 s at 72 °C, in which the extension time is dependent on the length of amplificates. The absolute transcript level was calculated using a dilution series of a target sequence on LightCycler Software Ver. 3.5. DcACT1 (accession number, AY007315) was almost constant in transcript level throughout the stages investigated and was used to standardize the transcript level.
Accession numbers
Nucleotide sequences obtained in this work were registered in DDBJ and assigned accession numbers from DK999551 to DK999785 and from AB517644 to AB517658.
Results
Stages of flower opening in carnation flowers and petal growth
There were six stages of carnation flower opening according to flower shape (Fig. 1A). At stage 1, the outermost petals just appeared from the buds and were about 30 mm long (Fig. 1B). They elongated quickly and reached about 36 mm long at stage 2. As the inner petals grew, the buds swelled (stage 3) and the outer petals warped outside (stage 4). The outermost petals then bent at the boundary between the claw and blade (stage 5), when a flower as a whole appeared to be open. At stage 6, the flowers fully opened, and the outermost petals reached 45 mm long and formed an almost 90° angle to a stem (Fig. 1B). From these observations, stages 1 and 2 were regarded as elongation periods and stages 3–6 as opening periods. Because the petal grew differentially depending on its position on the receptacle, ten outermost petals only were sampled for RNA extraction and gene expression analysis.
Fig. 1.
Morphology of an opening carnation flower. (A) Stages of flower opening defined by the flower shape as explained in the Materials and methods. (B) Front (left panel) and side (right panel) views of an outermost petal in stages 1 and 5. ad, Adaxial side; ab, abaxial side. Scale bars=10 mm.
Isolation of cDNA fragments of genes which were expressed differentially between the early elongation period and the opening period
Nearly 3000 colonies each were obtained using a portion of the PCR products by suppression subtractive hybridization (SSH) in both directions and then 864 colonies were chosen from each library. Differential screening of the 864 clones each showed that 69% of clones of the forward subtraction library (FSL) were more abundant in their transcript levels in the FSL than in the reverse subtraction library (RSL), whereas 61% of clones of the RSL were more abundant in the RSL than in the FSL. Clones (226 from the FSL and 167 from the RSL) that showed >2 signal ratios by unsubtracted probes and >4 signal ratios by subtracted probes were finally chosen for sequencing.
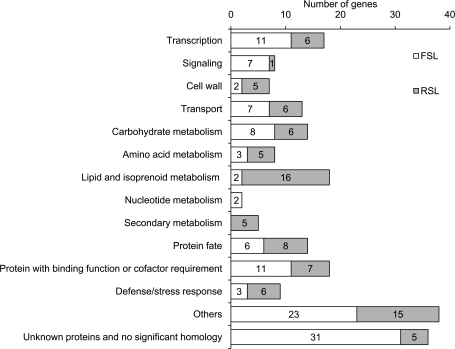
An homology search of the sequences obtained by BLASTX showed that about 40% of the clones were redundant as 158 out of a total of 393 sequences overlapped. Among the residual different 235 clones, 69 clones (29%) appeared more than twice. 133 and 102 different sequences from FSL and RSL were found to represent 116 and 91 putative translational products, respectively. These translational products were classified into 14 functional categories (Fig. 2). The number of translational products from the FSL considerably surpasses that from the RSL in the categories ‘Transcription’, ‘Signalling’, ‘Nucleotide metabolism’, and ‘Unknown proteins and no significant homology’. By contrast, with categories ‘Cell wall’, ‘Lipid and isoprenoid metabolism’, and ‘Secondary metabolism’, proteins from the RSL were superior in number to those from the FSL. Out of total of 235 ESTs identified in this study, 211 did not match any nucleotide sequences of carnation genes in the DDBJ/EMBL-Bank/GenBank. The results of a homology search and details of EST information are available in Supplementary Table S1 at JXB online.
Fig. 2.
Functional classification of ESTs identified by suppression subtractive hybridization. Numerical values represent a number of contigs from the forward subtraction library (FSL) and the reverse subtraction library (RSL) classified into each functional category. When EST clones confirmed or predicted by their nucleotide sequences to be derived from the same gene that gave other EST clones, they were counted as one gene. Classification mainly followed MIPS Funcat annotation.
Identification of open reading frames (ORFs) of selected genes by rapid amplification of cDNA ends (RACE)
5′-upstream or 3′-downstream sequences of selected cDNA fragments were successfully determined by RACE and revealed putative ORFs of 15 genes (Table 1). Nine genes (AB517649, AB517651–AB517658) were selected from the functional categories ‘Cell wall’, ‘Transport’, and ‘Lipid and isoprenoid metabolism’, considering that the related physiological processes could be involved in petal growth or morphology. Six genes (AB517644–AB517648, AB517650) were selected from the categories ‘Transcription’ and ‘Signalling’, expecting that they may serve to overlook regulation of flower opening. Three EST clones, DK999551, DK999552, and DK999553, were revealed to be derived from one gene, AB517644. Similarly, AB517645, AB517648, AB517651, and AB517655 were found to give two or three different EST clones. Amino acid sequences deduced from these genes showed homology to 38–91% to the proteins deposited in public databases.
Table 1.
List of genes analysed by rapid amplification of cDNA ends (RACE) and real-time RT-PCR
| Accession number | Number of deduced amino acid residues | Putative protein function | BLASTX homology |
Average signal intensity ratio |
||
| Accession number [species] | Identity (%) | Unsubtracted | Subtracted | |||
| Forward subtraction library | ||||||
| AB517644 | 573 | GRAS family transcription factor | B9I072 [Populus trichocarpa] | 63 | 19.9 | 466.4 |
| AB517645 | 630 | C3H-type zinc finger protein | A5BK99 [Vitis vinifera] | 41 | 6.2 | 457.6 |
| AB517646 | 249 | JAZ-like protein | B9MT14 [Populus trichocarpa] | 38 | 21.6 | 48.3 |
| AB517647 | 271 | AP2/EREBP family transcription factor | Q9LKK0 [Atriplex hortensis] | 45 | 2.9 | 28.7 |
| AB517648 | 619 | Receptor-like kinase | A5B9Q7 [Vitis vinifera] | 63 | 15.2 | 35.5 |
| AB517649 | 733 | Sugar transporter | B9HPN4 [Populus trichocarpa] | 58 | 4.1 | 10.3 |
| Reverse subtraction library | ||||||
| AB517650 | 238 | Aux/IAA protein | Q8L5G7 [Mirabilis jalapa] | 73 | 3.0 | 60.5 |
| AB517651 | 407 | Pectate lyase | B2BMQ1 [Prunus persica] | 78 | 29.7 | 106.7 |
| AB517652 | 119 | Lipid transfer protein (DcLTP1) | 1803519A [Spinacia oleracea] | 65 | 2.7 | 4.9 |
| AB517653 | 119 | Lipid transfer protein (DcLTP2) | Q9M6B8 [Gossypium hirsutum] | 58 | 4.2 | 31.0 |
| AB517654 | 118 | Lipid transfer protein (DcLTP3) | Q2QCI7 [Vitis vinifera] | 62 | 6.0 | 78.0 |
| AB517655 | 452 | 3-Ketoacyl-CoA synthase | O65677 [Arabidopsis thaliana] | 44 | 20.8 | 116.5 |
| AB517656 | 289 | Plasma membrane intrinsic protein | B2MVY5 [Knorringia sibirica] | 91 | 7.1 | 50.4 |
| AB517657 | 593 | Sodium/calcium exchanger protein | A7P1W8 [Vitis vinifera] | 62 | 11.4 | 118.8 |
| AB517658 | 466 | Auxin influx carrier protein | A9PH79 [Populus trichocarpa] | 84 | 2.2 | 541.6 |
Changes in transcript levels of selected genes in petals during flower opening
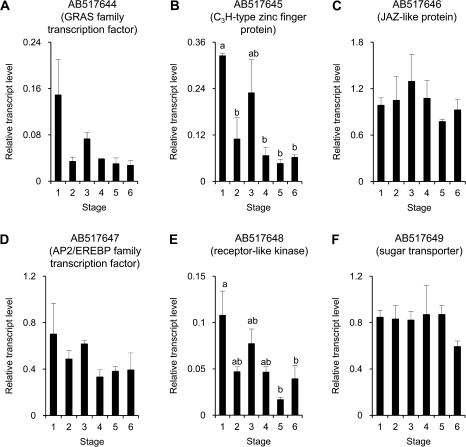
Changes in transcript levels of the selected genes were then investigated in petals during flower opening. Figure 3 shows the results of six genes from the FSL. AB517645 encoding a putative C3H-type zinc finger protein, showed the highest amount of transcript at stage 1. The amount significantly decreased at stage 2, then tended to increase at stage 3, followed by a decrease after stage 4 (Fig. 3B). Similar trends of changes in transcript levels were seen in AB517644, a putative GRAS family protein gene, AB517647, a putative AP2/EREBP transcription factor gene and AB517648, a putative receptor-like kinase gene (Fig. 3A, D, E). Correlation coefficients of changes in transcript levels between these genes (group 1) were more than 0.87. Real-time PCR was predicted to show significant differences in transcript levels between stages 1 and 5 in all cases, but it was not the case for some genes. The transcript level of AB517646, a JAZ-like protein gene, was relatively unchanged although it tended to increase from stage 1 to stage 3 and then to decrease (Fig. 3C). The amount of transcript of AB517649, putatively encoding a sugar transporter, was unchanged until stage 5, followed by a slight decrease at stage 6 (Fig. 3F).
Fig. 3.
Changes in transcript levels of six genes, AB517644 (A), AB517645 (B), AB517646 (C), AB517647 (D), AB517648 (E), and AB517649 (F) from the forward subtraction library. Relative transcript levels are calculated by real-time RT-PCR with DcACT1 as a standard. Data are means ±SE of three separate measurements. Significant difference (P <0.05) detected by Tukey's multiple comparison test is shown by different letters above the bars.
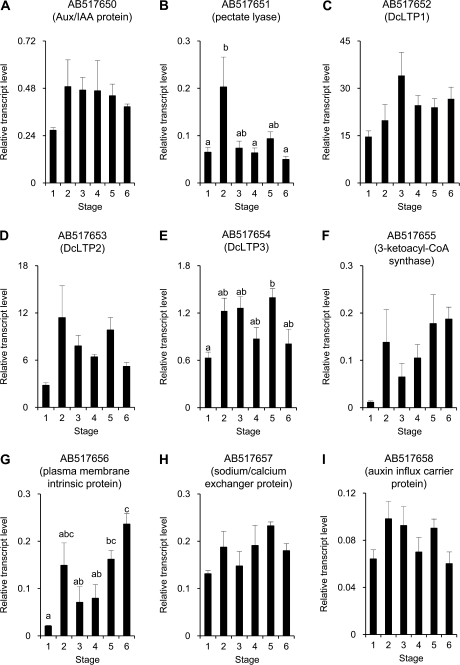
Figure 4 shows changes in transcript levels of genes from the RSL. A significant difference in the transcript level was seen between stages 1 and 5 in AB517654, a putative lipid transfer protein gene (DcLTP3; Fig. 4E). AB517652 (DcLTP2) and AB517658, a putative auxin influx carrier protein gene, resembled AB517654 in changes in transcript levels with a correlation coefficient about 0.9 (Fig. 4D, I; group 2). AB517655 putatively encoding 3-ketoacyl-CoA synthase was quite similar to AB517656, a putative plasma membrane intrinsic protein gene in changes in transcript levels (correlation coefficient=0.94): increased at stage 2, followed by a decrease at stages 3 and 4, and increased again at stages 5 and 6 (Fig. 4F, G; group 3). These two genes in group 3 tended to correlate negatively to the four genes in group 1 in changes in transcript levels. The transcript level of AB517650, a putative Aux/IAA protein gene, increased at stage 2 and thereafter tended to decrease (Fig. 4A). With AB517651 encoding putative pectate lyase, the transcript level significantly increased at stage 2 rather than stage 5 (Fig. 4B). These two genes were rather correlated to the three genes in group 2. The transcript level of AB517657, a putative sodium/calcium exchanger gene, tended to increase at stages 2, 4, and 5, roughly correlated to that of the two genes in group 3 (Fig. 4H). The transcript level of DcLTP1 (AB517652) changed differently from other genes, with the trend to increase from stage 1 to stage 3 and to decrease after stage 4 (Fig. 4C).
Fig. 4.
Changes in transcript levels of nine genes, AB517650 (A), AB517651 (B), AB517652 (C), AB517653 (D), AB517654 (E), AB517655 (F), AB517656 (G), AB517657 (H), and AB517658 (I) from the reverse subtraction library. Relative transcript levels are calculated by real-time RT-PCR with DcACT1 as a standard. Data are means ±SE of three separate measurements. Significant difference (P <0.05) detected by Tukey's multiple comparison test is shown by different letters above the bars.
Discussion
Identification of new carnation ESTs in petals during flower opening
From subtraction libraries representing the early petal elongation period (stage 1) and the opening period (stage 5) in opening carnation flowers, 235 ESTs were successfully identified. As 387 carnation ESTs were found by a search of GenBank or EMBL-Bank, this research resulted in a 1.6-fold increase in the number of published carnation ESTs. Only 7.2% of the sequences identified here appeared in carnation ESTs collected for other purposes in previous studies (Ok et al., 2003; Hoeberichts et al., 2007).
The effectiveness of suppression subtraction hybridization (SSH) is shown by the difference in the number and the kind of translational products of each functional category between the forward subtraction library (FSL) and the reverse subtraction library (RSL). The abundance in ESTs of ‘Transcription’, including transcription factors, ‘Signalling’ including protein kinases, and ‘Unknown proteins and no significant homology’ is distinctive in the FSL. This suggested that these genes are related to uncharacterized mechanisms in the perception of signals and the initiation of flower opening. On the other hand, the RSL gave ESTs of ‘Lipid and isoprenoid metabolism’ and ‘Secondary metabolism’ including some genes known to encode enzymes involved in pigmentation. These findings corresponded to the fact that secondary lipid metabolites including fragrance volatiles (Hudak and Thompson, 1997) and pigment are synthesized as petals mature. The abundance of genes with a low expression level is one of the characteristics of the SSH technique (Diatchenko et al., 1996). More than a 250-fold difference in relative transcript level was observed among two genes analysed by real-time RT-PCR (compare values at stage 5 between Fig. 4C and I).
However, careful interpretation is needed for the appearance of each EST. Dot blot reverse Northern analysis was performed to screen truly differentially expressed sequences using subtracted and unsubtracted probe sets. Actually, subtracted probes significantly intensified the signal intensity ratio by unsubtracted probes in most cases, suggesting that subtraction worked well. Transcript levels determined by dot blot, however, differ from the results obtained by real-time RT-PCR. For example, real-time RT-PCR showed the same transcript level of AB517649 at stage 1 and stage 5, compared with a 4.1-fold difference in dot blot. The real-time RT-PCR showed only a 1.4-fold difference in the relative transcript level of AB517651 between stage 5 and stage 1, compared with the 29.7-fold difference by dot blot analysis. It is likely that the results of dot blot analysis based on hybridization were affected by related sequences of another member from the same multigene family (Miller et al., 2002). The number of clones that were screened and sequenced may not be enough to evaluate whether the signal intensity ratios of dot blot or the numbers of each clone reflect the degree of difference in the transcript level of each gene. Furthermore, the possibility could not be excluded that important genes other than those identified in this work were obtained from a residual part of the libraries or SSH libraries constructed from carnation petals at other stages.
Fluctuating gene expression along petal growth during flower opening
Three typical patterns of changes in transcript levels investigated by real-time RT-PCR were found: group 1 from the FSL and groups 2 and 3 from the RSL. Group 1 was characterized by higher transcript levels at stages 1 and 3, while a similarity in genes within groups 2 and 3 seems to be caused by trends to peak at stage 2 as well at stages 5 or 6. It should be noted that these genes were not simply up- or down-regulated in an elongation period or an opening period. Considering that stage 2 is a late elongation period and stage 5 is an opening period during which the petals are bending, the common up-regulation in these two stages was probably associated with the rapid change in petal shape. Stages 1 and 3 could be regarded as the preceding stages common for the preparation of morphological changes of petals. Negative correlation in changes in transcript levels between groups 1 and 3 also suggests the existence of some physiological factors co-ordinating gene expression. On the other hand, curving of petals at stage 5 was probably due to the difference in growth regulation between the adaxial and abaxial sides of the petals or between claw, blade, and their border region. Whether the localization of gene expression in horizontal petal growth is different from that in vertical petal growth remains to be solved. Previous studies on gene expression during flower opening in some species focused on the up-regulation of genes in a specific phase (Gookin et al., 2003; van Doorn et al., 2003). From the results of this study, it is proposed that periodic expression of one gene is an important aspect for an understanding of the stepwise reactions in flower opening.
Genes involved in transcriptional regulation and signal transduction
In both the FSL and the RSL, 17 proteins that categorized to ‘Transcription’ including gene silencing and RNA processing were identified. The GRAS family transcription factor (AB517644) belongs to a protein family including DELLA proteins involved in gibberellin (GA) signalling and PAT1 from Arabidopsis involved in phytochrome A signal transduction (Pysh et al., 1999; Bolle et al., 2000). It is likely that the protein encoded by AB517644 is involved in processes other than GA responses because it lacks the DELLA domain common in the GA-related DELLA protein subfamily (Wen and Chang, 2002). Jasmonate ZIM-domain (JAZ) protein has been revealed recently to function as a repressor of jasmonate-induced gene expression (Thines et al., 2007). The primary structure of a JAZ-like protein (AB517646) as a whole shows similarity to JAZ proteins and contains a characteristic TIFY sequence. The crucial role of jasmonate in petal elongation and flower opening has been shown by an Arabidopsis mutant defective in jasmonate biosynthesis (Ishiguro et al., 2001). Aux/IAA protein (AB517650) is a major family of proteins involved in the auxin response. Rapid induction of this protein by auxin, functions as a repressor of gene expression by binding to AUXIN RESPONSIVE FACTOR (ARF) and functional redundancy between multiple isoforms have been demonstrated (Overvoorde et al., 2005). In addition to two contigs appearing in our SSH libraries (DK999685, DK999686), the existence of genes encoding several isoforms of the Aux/IAA protein is suggested by RACE experiments (data not shown), implying an important role of auxin in carnation flower opening through transcriptional regulation. Other kinds of transcriptional regulators including zinc-finger proteins (AB517645, DK999560, DK999687), an AP2/EREBP transcription factor (AB517647), a bHLH transcription factor (DK999562) and a calmodulin-binding transcriptional activator (DK999565) are also candidates for regulators of petal growth in the opening of carnation flowers.
In the category ‘Signalling’ from the FSL, a putative receptor-like kinase (AB517648), protein kinases (DK999572, DK999573, DK999574) and a MAP kinase phosphatase (DK999567) were found. The protein encoded by AB517648 is probably a member of the receptor-like kinase family including a brassinosteroid receptor BRI1 (Morillo and Tax, 2006). The leucine-rich repeat existing in the N-terminal region of AB517648 protein suggests the recognition of some ligand molecules. Moreover, its expression pattern is similar to that of a GRAS family transcription factor (AB517644) and a C3H-type zinc finger protein (AB517645). Protein phosphorylation/dephosphorylation probably plays a role in signal transduction pathways for the initiation of carnation flower opening.
Genes involved in cell wall modification and wax deposition
Nine ESTs related to cell wall proteins were identified. Pectin-related proteins such as pectate lyase (AB517651), pectin methylesterase inhibitor (DK999693), and pectin methylesterase (DK999696) are found in ESTs from the RSL. Pectin is generally believed to reinforce the cell wall construction, where calcium-pectate bridges play an important role and affect cell wall susceptibility to expansin, a cell-wall loosening protein (Zhao et al., 2008). An increase in pectin content has been observed during the development of carnation flowers (de Vetten and Huber, 1990), and may correlate to the predominance of pectin-related ESTs in the RSL. The fluctuation of transcript levels of a putative pectate lyase gene (AB517651) further supports the idea that changes in petal shape by elongation and bending are associated with a modification of the pectin network. Two other kinds of protein, α-expansin (DK999695) and xyloglucan endotransglucosylase/hydrolase (XTH; DK999575, DK999694) are associated with the regulation of extensibility of the cell wall. As is the case of Mirabilis jalapa (Gookin et al, 2003) and rose (Yamada et al., 2009), members from each multigene family are differentially expressed in carnation petals during flower opening, suggesting their possible involvement in petal growth (Y Torii and S Satoh, unpublished data).
From the comparison of deduced amino acid sequences, three isoforms of putative lipid transfer protein were identified (LTP; the protein structure is compared in Supplementary Fig. S1A at JXB online). LTPs consist of a major family of proteins existing outside of cells and their transcripts have often been found in petals (Kotilainen et al., 1994). Although clear evidence for the in vivo molecular function of LTPs is lacking, LTPs have been proposed to be involved in some physiological processes including defence responses and wax deposition (Yeats and Rose, 2008). On the other hand, Nieuwland et al. (2005) reported an LTP with the capability of enhancing cell wall extension in tobacco, while Tomassen et al. (2007) demonstrated the modification of polygalacturonase activity by an LTP in tomato. These previous studies showed the involvement of LTPs in cell wall modification and hence morphological changes in plant tissues. Transcripts of three LTP genes as a whole were abundant during carnation flower opening, but the temporal patterns of change in transcript level differed among these genes. The present findings showed that multiple LTP genes are present in carnation petals, suggesting their contribution to petal development through the regulation of morphologically important cellular components like the cell wall and cuticle.
Among proteins categorized into ‘Lipid and isoprenoid metabolism’ in the RSL, 3-ketoacyl-CoA synthase (KCS, AB517655) and 3-ketoacyl-CoA reductase (DK999722), components of the elongase system that synthesizes very-long-chain fatty acid (VLCFA), are likely to be involved in wax deposition. Young seedlings of Arabidopsis mutants lacking KCS1 show a retardation of growth (Todd et al., 1999), and VLCFAs synthesized by KCS play an important role in the development of cotton fibre (Qin et al., 2007). The change in the transcript level of AB517655 during carnation flower opening is similar to that of a putative aquaporin gene (AB517656) especially in the highest level at stage 6, suggesting their correlation with water relations in petals. It is possible that up-regulation of the KCS gene meets a demand for cuticular wax during the expansion of the petal epidermis, which leads to the successful opening of double flowers of carnation.
Genes involved in transport
ORFs of four genes were obtained from ESTs representing 11 different proteins categorized into ‘Transport’. The deduced amino acid sequence of a sugar transporter (AB517649) was similar to that of a monosaccharide transporter from Arabidopsis, which is involved in monosaccharide accumulation in vacuoles (Wormit et al., 2006). The transcript level of AB517649 was lower in stage 6 compared with the preceding stages, suggesting its main role until full opening. Plasma membrane intrinsic protein (PIP, AB517656) could be involved in water uptake and thus contribute to petal growth as is the case in tulip (Azad et al., 2004, 2008) and rose (Ma et al., 2008). AB517658 putatively encoded an auxin influx carrier protein which was distinct from the first isoform isolated from carnation cuttings (Oliveros-Valenzuela et al., 2008). The protein encoded by AB517658 may participate in the intercellular movement of auxin in carnation petals, although the action and distribution of endogenous auxin in growing carnation petals has not been demonstrated. AB517657 encoded a putative membrane protein that belongs to the sodium/calcium exchanger (NCX) family (Cai and Lytton, 2004). Although the molecular functions of NCX proteins in plants have not yet been demonstrated, a pair of calcium binding domains (EF-hand) between transmembrane regions, which were conserved among plant species (see Supplementary Fig. S1B at JXB online), suggested its possible involvement in the calcium-related facilitation of cation movement across cellular membranes. The highest expression of AB517657 at stage 5 might be a distinct feature implying its association with petal bending. This is interesting since treatment with calcium is known to promote flower opening in rose (Torre et al., 1999).
Concluding remarks
A search was made for genes differentially expressed in petals of opening carnation flowers and a number of novel carnation ESTs were identified and the expression patterns of some genes potentially involved in petal growth were correlated. The present findings unveiled some of the molecular aspects of flower opening that are distinguishable from the characteristic senescence processes of carnation associated with climacteric ethylene production. Further studies are needed to control petal growth involved in flower opening during postharvest handling of cut flowers of carnation.
Supplementary data
The following supplementary data are available at JXB online.
Supplementary Table S1. List of cDNA clones identified by suppression subtractive hybridization.
Supplementary Table S2. List of primers used for real-time RT-PCR.
Supplementary Fig. S1. ClustalW alignment of deduced amino acid sequences of putative lipid transfer protein genes and putative sodium/calcium exchanger genes.
Supplementary Material
Acknowledgments
This study was supported in part by a Grant-in-Aid (19380024 to SS) for Scientific Research from the Japan Society for the Promotion of Science.
References
- Azad AK, Katsuhara M, Sawa Y, Ishikawa T, Shibata H. Characterization of four plasma membrane aquaporins in tulip petals: a putative homolog is regulated by phosphorylation. Plant and Cell Physiology. 2008;49:1196–1208. doi: 10.1093/pcp/pcn095. [DOI] [PubMed] [Google Scholar]
- Azad AK, Sawa Y, Ishikawa T, Shibata H. Phosphorylation of plasma membrane aquaporin regulates temperature-dependent opening of tulip petals. Plant and Cell Physiology. 2004;45:608–617. doi: 10.1093/pcp/pch069. [DOI] [PubMed] [Google Scholar]
- Bolle C, Koncz C, Chua N- H. PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes and Development. 2000;14:1269–1278. [PMC free article] [PubMed] [Google Scholar]
- Breeze E, Wagstaff C, Harrison E, Bramke I, Rogers H, Stead A, Thomas B, Buchanan-Wallaston V. Gene expression patterns to define stages of post-harvest senescence in Alstroemeria petals. Plant Biotechnology Journal. 2004;2:155–168. doi: 10.1111/j.1467-7652.2004.00059.x. [DOI] [PubMed] [Google Scholar]
- Cai X, Lytton J. The cation/Ca2+ exchanger superfamily: phylogenetic analysis and structural implications. Molecular Biology and Evolution. 2004;21:1692–1703. doi: 10.1093/molbev/msh177. [DOI] [PubMed] [Google Scholar]
- Channelière S, Rivière S, Scalliet G, et al. Analysis of gene expression in rose petals using expressed sequence tags. FEBS Letters. 2002;515:35–38. doi: 10.1016/s0014-5793(02)02413-4. [DOI] [PubMed] [Google Scholar]
- Dafny-Yelin M, Guterman I, Menda N, et al. Flower proteome: changes in protein spectrum during the advanced stages of rose petal development. Planta. 2005;222:37–46. doi: 10.1007/s00425-005-1512-x. [DOI] [PubMed] [Google Scholar]
- de Vetten NC, Huber DJ. Cell wall changes during the expression and senescence of carnation (Dianthus caryophyllus) petals. Physiologia Plantarum. 1990;78:447–454. [Google Scholar]
- Diatchenko L, Lau Y-FC, Campbell AP, et al. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proceedings of the National Academy of Sciences, USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gookin TE, Hunter DA, Reid MS. Temporal analysis of alpha and beta-expansin expression during floral opening and senescence. Plant Science. 2003;164:769–781. [Google Scholar]
- Guterman I, Shalit M, Menda N, et al. Rose scent: genomic approach to discovering novel floral fragrance-related genes. The Plant Cell. 2002;14:2325–2338. doi: 10.1105/tpc.005207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada T, Satoh S, Yoshioka T, Ishizawa K. Expression of sucrose synthase genes involved in enhanced elongation of pondweed (Potamogeton distinctus) turions under anoxia. Annals of Botany. 2005;96:683–692. doi: 10.1093/aob/mci220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeberichts FA, van Doorn WG, Vorst O, Hall RD, van Wardragen MF. Sucrose prevents up-regulation of senescence-associated genes in carnation petals. Journal of Experimental Botany. 2007;58:2873–2885. doi: 10.1093/jxb/erm076. [DOI] [PubMed] [Google Scholar]
- Hong Y, Wang T-W, Hudak KA, Schade F, Froese CD, Thompson JE. An ethylene-induced cDNA encoding a lipase expressed at the onset of senescence. Proceedings of the National Academy of Sciences, USA. 2000;97:8717–8722. doi: 10.1073/pnas.140213697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudak KA, Thompson JE. Subcellular localization of secondary lipid metabolites including fragrance volatiles in carnation petals. Plant Physiology. 1997;114:705–713. doi: 10.1104/pp.114.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iordachescu M, Verlinden S. Transcriptional regulation of three EIN3-like genes of carnation (Dianthus caryophyllus L. cv. Improved White Sim) during flower development and upon wounding, pollination, and ethylene exposure. Journal of Experimental Botany. 2005;56:2011–2018. doi: 10.1093/jxb/eri199. [DOI] [PubMed] [Google Scholar]
- Ishiguro S, Kawai-Oda A, Ueda J, Nishida I, Okada K. The DEFECTIVE IN ANTHER DEHISCENCE1 gene encodes a novel phospholipase A1 catalysing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. The Plant Cell. 2001;13:2191–2209. doi: 10.1105/tpc.010192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones ML, Larsen PB, Woodson WR. Ethylene-regulated expression of a carnation cysteine proteinase during flower petal senescence. Plant Molecular Biology. 1995;28:505–512. doi: 10.1007/BF00020397. [DOI] [PubMed] [Google Scholar]
- Kotilainen M, Helariutta Y, Elomaa P, Paulin L, Teeri TH. A corolla- and carpel-abundant, non-specific lipid transfer protein gene is expressed in the epidermis and parenchyma of Gerbera hybrida var. Regina (Compositae) Plant Molecular Biology. 1994;26:971–978. doi: 10.1007/BF00028863. [DOI] [PubMed] [Google Scholar]
- Kotilainen M, Helariutta Y, Mehto M, Pöllänen E, Albert VA, Elomaa P, Teeri TH. GEG participates in the regulation of cell and organ shape during corolla and carpel development in Gerbera hybrida. The Plant Cell. 1999;11:1093–1104. doi: 10.1105/tpc.11.6.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak MS, Min SR, Lee S-M, Kim K-N, Liu JR, Paek K-H, Shin JS, Bae JM. A sepal-expressed ADP-glucose pyrophosphorylase gene (NtAGP) is required for petal expansion growth in ‘Xanthi’ tobacco. Plant Physiology. 2007;145:277–289. doi: 10.1104/pp.107.102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laitinen RAE, Pöllänen E, Teeri TH, Elomaa P, Kotilainen M. Transcriptional analysis of petal organogenesis in Gerbera hybrida. Planta. 2007;226:347–360. doi: 10.1007/s00425-007-0486-2. [DOI] [PubMed] [Google Scholar]
- Lawton KA, Huang B, Goldsbrough PB, Woodson WR. Molecular cloning and characterization of senescence-related genes from carnation flower petals. Plant Physiology. 1989;90:690–696. doi: 10.1104/pp.90.2.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N, Xue J, Li Y, Liu X, Dai F, Jia W, Luo Y, Gao J. Rh-PIP2;1, a rose aquaporin gene, is involved in ethylene-regulated petal expansion. Plant Physiology. 2008;148:894–907. doi: 10.1104/pp.108.120154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller NA, Gong Q, Bryan R, Ruvolo M, Turner LA, LaBrie ST. Cross-hybridization of closely related genes on high-density macroarrays. Biotechniques. 2002;32:620–625. doi: 10.2144/02323pf01. [DOI] [PubMed] [Google Scholar]
- Morillo SA, Tax FE. Functional analysis of receptor-like kinases in monocots and dicots. Current Opinion in Plant Biology. 2006;9:460–469. doi: 10.1016/j.pbi.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Nieuwland J, Feron R, Huisman BAH, Fasolino A, Hilbers CW, Derksen J, Mariani C. Lipid transfer proteins enhance cell wall extension in tobacco. The Plant Cell. 2005;17:2009–2019. doi: 10.1105/tpc.105.032094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nukui H, Kudo S, Yamashita A, Satoh S. Repressed ethylene production in gynoecium of long-lasting flowers of the carnation ‘White Candle’: role of the gynoecium in carnation flower senescence. Journal of Experimental Botany. 2004;55:641–650. doi: 10.1093/jxb/erh081. [DOI] [PubMed] [Google Scholar]
- Ok SH, Park HM, Kim JY, Bahn SC, Bae JM, Suh MC, Jeung J-U, Kim K-N, Shin JS. Identification of differentially expressed genes during flower development in carnation (Dianthus caryophyllus) Plant Science. 2003;165:291–297. [Google Scholar]
- Oliveros-Valenzuela M, Reyes D, Sánchez-Bravo J, Acosta M, Nicolás C. Isolation and characterization of a cDNA clone encoding an auxin influx carrier in carnation cuttings. Expression in different organs and cultivars and its relationship with cold storage. Plant Physiology and Biochemistry. 2008;46:1071–1076. doi: 10.1016/j.plaphy.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Overvoorde PJ, Okushima Y, Alonso JM, et al. Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. The Plant Cell. 2005;17:3282–3300. doi: 10.1105/tpc.105.036723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN. The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. The Plant Journal. 1999;18:111–119. doi: 10.1046/j.1365-313x.1999.00431.x. [DOI] [PubMed] [Google Scholar]
- Qin Y-M, Hu C-Y, Pang Y, Kastaniotis AJ, Hiltunen JK, Zhu Y-X. Saturated very-long-chain fatty acids promote cotton fiber and Arabidopsis cell elongation by activating ethylene biosynthesis. The Plant Cell. 2007;19:3692–3704. doi: 10.1105/tpc.107.054437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya K, Nagata M, Tanikawa N, Yoshioka T, Hashiba T, Satoh S. Comparison of mRNA levels of three ethylene receptors in senescing flowers of carnation (Dianthus caryophyllus L.) Journal of Experimental Botany. 2002;53:399–406. doi: 10.1093/jexbot/53.368.399. [DOI] [PubMed] [Google Scholar]
- ten Have A, Woltering EJ. Ethylene biosynthetic genes are differentially expressed during carnation (Dianthus caryophyllus L.) flower senescence. Plant Molecular Biology. 1997;34:89–97. doi: 10.1023/a:1005894703444. [DOI] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He AY, Howe GA, Browse J. JAZ repressor proteins are targets of the SCFCOI1complex during jasmonate signalling. Nature. 2007;448:661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- Todd J, Post-Beittenmiller D, Jaworski JG. KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. The Plant Journal. 1999;17:119–130. doi: 10.1046/j.1365-313x.1999.00352.x. [DOI] [PubMed] [Google Scholar]
- Tomassen MMM, Barrett DM, van der Valk HCPM, Woltering EJ. Isolation and characterization of a tomato non-specific lipid transfer protein involved in polygalacturonase-mediated pectin degradation. Journal of Experimental Botany. 2007;58:1151–1160. doi: 10.1093/jxb/erl288. [DOI] [PubMed] [Google Scholar]
- Torre S, Brorchov A, Halevy AH. Calcium regulation of senescence in rose petals. Physiologia Plantarum. 1999;107:214–219. [Google Scholar]
- van Doorn WG, Balk PA, van Houwelingen AM, Hoeberichts FA, Hall RD, Vorst O, van der Schoot C, van Wardragen MF. Gene expression during anthesis and senescence in Iris flowers. Plant Molecular Biology. 2003;53:845–863. doi: 10.1023/B:PLAN.0000023670.61059.1d. [DOI] [PubMed] [Google Scholar]
- van Doorn WG, van Meeteren U. Flower opening and closure: a review. Journal of Experimental Botany. 2003;54:1801–1812. doi: 10.1093/jxb/erg213. [DOI] [PubMed] [Google Scholar]
- Wen C-K, Chang C. Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. The Plant Cell. 2002;14:87–100. doi: 10.1105/tpc.010325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wormit A, Trentmann O, Feifer I, Lohr C, Tjaden J, Meyer S, Schmidt U, Martinoia E, Neuhaus HE. Molecular identification and physiological characterization of a novel monosaccharide transporter from Arabidopsis involved in vacuolar sugar transport. The Plant Cell. 2006;18:3476–3490. doi: 10.1105/tpc.106.047290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Gookin T, Jiang C-Z, Reid M. Genes associated with opening and senescence of Mirabillis jalapa flowers. Journal of Experimental Botany. 2007;58:2193–2201. doi: 10.1093/jxb/erm058. [DOI] [PubMed] [Google Scholar]
- Yamada K, Ito M, Oyama T, Nakada M, Maesaka M, Yamaki S. Analysis of sucrose metabolism during petal growth of cut roses. Postharvest Biology and Technology. 2007;43:174–177. [Google Scholar]
- Yamada K, Takahashi R, Fujitani C, Mishima K, Yoshida M, Joyce DC, Yamaki S. Cell wall extensibility and effect of cell-wall-loosening proteins during rose flower opening. Journal of the Japanese Society for Horticultural Science. 2009;78:242–251. [Google Scholar]
- Yamada T, Ichimura K, Kanekatsu M, van Doorn WG. Gene expression in opening and senescing petals of morning glory (Ipomoea nil) flowers. Plant Cell Report. 2007;26:823–835. doi: 10.1007/s00299-006-0285-4. [DOI] [PubMed] [Google Scholar]
- Yeats TH, Rose JKC. The biochemistry and biology of extracellular plant lipid-transfer proteins (LTPs) Protein Science. 2008;17:191–198. doi: 10.1110/ps.073300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenoni S, Reale L, Tornielli GB, et al. Downregulation of the Petunia hybrida α-expansin gene PhEXP1 reduces the amount of crystalline cellulose in cell walls and leads to phenotypic changes in petal limbs. The Plant Cell. 2004;16:295–308. doi: 10.1105/tpc.018705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Yuan S, Wang X, Zhang Y, Zhu H, Lu C. Restoration of mature etiolated cucumber hypocotyl cell wall susceptibility to expansin by pretreatment with fungal pectinases and EGTA in vitro. Plant Physiology. 2008;147:1874–1885. doi: 10.1104/pp.108.116962. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.