Abstract
Knowledge of the proteome of the early gymnosperm embryo could provide important information for optimizing plant cloning procedures and for establishing platforms for research into plant development/regulation and in vitro transgenic studies. Compared with angiosperms, it is more difficult to induce somatic embryogenesis in gymnosperms; success in this endeavour could be increased, however, if proteomic information was available on the complex, dynamic, and multistage processes of gymnosperm embryogenesis in vivo. A proteomic analysis of Chinese fir seeds in six developmental stages was carried out during early embryogenesis. Proteins were extracted from seeds dissected from immature cones and separated by two-dimensional difference gel electrophoresis. Analysis with DeCyder 6.5 software revealed 136 spots that differed in kinetics of appearance. Analysis by liquid chromatography coupled to tandem mass spectrometry and MALDI-TOF mass spectrometry identified proteins represented by 71 of the spots. Functional annotation of these seed proteins revealed their involvement in programmed cell death and chromatin modification, indicating that the proteins may play a central role in determining the number of zygotic embryos generated and controlling embryo patterning and shape remodelling. The analysis also revealed other proteins involved in carbon metabolism, methionine metabolism, energy production, protein storage, synthesis and stabilization, disease/defence, the cytoskeleton, and embryo development. The comprehensive protein expression profiles generated by our study provide new insights into the complex developmental processes in the seeds of the Chinese fir.
Keywords: Chinese fir, DIGE, embryogenesis, proteomics, seed development
Introduction
Chinese fir (Cunninghamia lanceolata Lamb. Hook) is one of the most important coniferous evergreen tree species in terms of both industrial and commercial wood supplies in China. It has been cultivated for over 3000 years because of its desirable attributes of rapid growth, wood strength, versatility of its applications, hardiness, and high yield of timber productivity per unit. Chinese fir is the dominant tree species in southern China, with a total area of approximately 4×106 hm2 occupying ∼25% of man-made plantations, and planting is expanding in over 15 southern provinces (Huang et al., 2005). However, the expansion of plantations has been curtailed by the inconsistent availability of genetically improved seeds from year to year. Toward the goal of supplying large numbers of high-quality and genetically superior Chinese fir seedlings, clone propagation through somatic embryogenesis is a potentially large-scale alterative strategy for reforestation.
The immature embryo of Chinese fir is an excellent somatic embryo explant. Before starting an investigation of somatic embryogenesis, however, it is necessary to obtain basic knowledge about zygote development. Embryogenesis is a complex process governed by highly co-ordinated changes in the expression of large protein sets. Protein expression plays a central role in histodifferentiation in the zygote and during seed development. As such, the characterization and functional analysis of proteins involved in seed development offers the possibility of gaining further information on the molecular basis of embryogenesis and histodifferentiation. Proteomics is a powerful tool to examine temporal patterns and simultaneous changes in protein accumulation during development. Although there have been many studies on the proteomics of angiosperm seed or embryo development (Gallardo et al., 2001, 2003, 2007; Pawłowski, 2007), few such reports exist for gymnosperms. However, certain recent studies have reported proteomic analyses of seeds or embryos from woody plants, such as seed development in Araucaria angustifolia (dos Santos et al., 2006), megagametophyte and zygotic embryo development in Cupressus sempervirens L. (Sallandrouze et al., 2002), early somatic embryogenesis in Picea glauca (Lippert et al., 2005), and zygotic embryos and somatic embryos of Pinus strobes (Klimaszewskaa et al., 2004).
The immature embryo has been broadly used to initiate cultures for the study of somatic embryogenesis in conifers (Balbuena et al., 2009). It was found that the developmental stage of the zygotic embryos used as explants could influence the successful rates of somatic embryos convertion. To obtain new insights on Chinese fir seed development and practical ways to improve somatic embryogenesis with proper micro-environmental conditions based on protein expression patterns, a two-dimensional difference gel electrophoresis (2D DIGE)-based proteomic approach was used to analyse the differentially displayed proteins in megagametophytes containing developing embryos during six important developmental phases of early embryogenesis in C. lanceolata. Data are presented here on the dynamics of protein accumulation patterns during seed development. Our results provide a fundamental reference for further studies on the regulation of protein expression during somatic embryogenesis.
Materials and methods
Plant materials and seed development stages
Immature cones of C. lanceolata (Lamb.) Hook were collected every 2 weeks from clone No. 4009 at Shao Wu seed orchards in Fujian province in south-eastern China. Cones were kept on ice and delivered to the laboratory within 48 h. Ten developing seeds were chosen randomly from the middle part of each cone, and the developmental stage of each seed was estimated according to the zygotic embryo state under a dissecting microscope. Given that most of the zygotic embryos that were observed in seeds from several cones were of a similar development stage, it was concluded that most of the seeds were at the stage that had been estimated. The developmental stages of the collected seeds were classified as follows: cleavage polyembryony, dominant embryo, columnar embryo, or early cotyledonary (Nagmani et al., 2000; von Arnold et al., 2000). Seeds of a similar developmental stage were placed in a vial and then frozen in liquid nitrogen and stored at –70 °C until use.
Sample preparation
Protein extracts were prepared in triplicate for each developmental stage. About 50 mg of seed material was homogenized with 0.3 ml lysis buffer (7 M urea, 2 M thiourea, 4% CHAPS, 30 mM TRIS, pH 8.5) using a grinding kit (Amersham Biosciences/GE Healthcare, UK). The extracted samples were incubated at room temperature for 20 min and then centrifuged at 15 000 g at 4 °C for 50 min to remove insoluble material. The supernatant was cleared of residual particulates using a protein Clean-up kit (Amersham Biosciences/GE Healthcare). The resulting extract powder was solubilized in lysis buffer. After 1 h of gentle stirring at room temperature, samples were centrifuged at 15 000 g at room temperature for 45 min. Protein concentration was determined with the PlusOne 2-D Quant kit (Amersham Biosciences/GE Healthcare). Aliquots of protein samples were stored in 0.6 ml Eppendorf tubes at –70 °C for further analysis.
Cy dye labelling
Lyophilized protein samples (50 μg) were labelled with 400 pmol of N-hydroxy succinimidyl ester derivatives of cyanine dyes Cy3 or Cy5 in anhydrous dimethylformamide. A pool standard that contained equal quantities of all samples was labelled with Cy2. Labelling reactions were performed on ice for 30 min in the dark, and each reaction was terminated by adding 1 μl of 10 mM lysine (Sigma, St Louis, MO, USA). These samples were mixed, centrifuged briefly in a microcentrifuge, and then incubated on ice in the dark for 10 min. Differentially labelled samples were mixed and combined with an equal volume of 2× sample buffer [7 M urea, 2 M thiourea, 4% (w/v) CHAPS, 2% (w/v) DTT, and 2% (v/v) pH 4–7 Pharmalyte™] for at least 10 min on ice.
Fluorescent two-dimensional electrophoresis analysis
For two-dimensional electrophoresis, proteins were solubilized in a rehydrating solution containing 8 M urea, 2% CHAPS, 20 mM DTT, a trace of bromophenol blue, and 0.5% (v/v) immobilized pH gradient (IPG) buffer (pH 4–7) (Amersham Biosciences/GE Healthcare) in the dark. IPG strips (24 cm, linear pH 4–7, Amersham Biosciences/GE Healthcare) were rehydrated overnight with 450 μl rehydrating solution containing 150 μg proteins. Rehydration and isoelectric focusing were carried out at 20 °C using an IPGphor III (Amersham Biosciences/GE Healthcare), with the following settings: 30 V for 6 h, step; 60 V for 6 h, step; 200 V for 1 h, step; 500 V for 1 h, step; 1000 V for 1 h, step; 4000 V for 1 h, step; 8000 V for 30 min, grad; 8000 V for 6 h, step. After isoelectric focusing, proteins were chemically reduced by incubating the IPG strips with 1% (w/v) DTT and then alkylated with 4% (w/v) iodoacetamide in 10 ml equilibration buffer [6 M urea, 30% (w/v) glycerol, 2% SDS, and 50 mM TRIS-HCl] with gentle shaking for 15 min. After equilibration, the IPG strips were blotted with filter paper for 1 min to remove excess equilibration solution, and gently rinsed for 1 min using SDS-PAGE running buffer (250 mM TRIS pH 8.3, 1.92 M glycine, and 1% SDS). The IPG strips were then transferred to 12.5% SDS-PAGE gels with low fluorescence glass plates for second-dimension electrophoresis with the Ettan Dalt twelve gel system (Amersham Biosciences/GE Healthcare) using SDS-PAGE running buffer. After the sealed agarose cooled down, the second-dimension SDS-PAGE was carried out at 1 W gel−1 for 16 h at 20 °C until the dye front reached the bottom of the gel.
Image and data analysis
Gel images were scanned with a Typhoon Trio Variable Mode Imager (Amersham Biosciences/GE Healthcare). Cy2 images were scanned using a 488 nm laser and an emission filter of 520 nm/band pass 40. Cy3 images were scanned using a 532 nm laser and an emission filter of 580 nm/band pass 30. Cy5 images were collected using a 633 nm laser and an emission filter of 670 nm/band pass 30. Prescan was carried out with a pixel size of 500 μm to adjust the photomultiplier tube voltage to give a maximum pixel value of between 60 000 and 80 000 to avoid image saturation. All gels were then scanned using a pixel size of 100 μm to obtain high-resolution images. The images were cropped with ImageQuant TL 2005 software (Amersham Biosciences/GE Healthcare) prior to DeCyder™ 6.5 analysis (Amersham Biosciences/GE Healthcare). The differential in-gel analysis function of DeCyder™ 6.5 was used to co-detect and differentially quantify the protein spots in each gel, which included a Cy2-labelled pooled standard and Cy3/Cy5-labelled samples. Variations between samples, as determined by DeCyder™ 6.5, enabled gel to gel matching of spots according to the internal standard, allowing quantitative comparisons of proteins differentially expressed across multiple gels. The Batch Processor of DeCyder™ 6.5 linked both differential and biological variation analysis to perform all analyses automatically without user intervention, and spot intensities were quantified based on an estimated 3000 spots per gel. To match spots across gels accurately, landmarks were set. One-way analysis of variance was used to reveal proteins that were differentially expressed (P <0.01) among six samples (i.e., stages 1, 2, 3, 4, 5, 6; see the Results and discussion for stage description). A statistical analysis was performed on triplicate measurements for each stage. Hierarchical clustering analysis was performed on the standardized log abundance of spot volumes using Gene Cluster software 3.0 with centred correlation and the average linkage procedure. Phylogenetic trees were constructed/visualized using TreeView software (http://rana.lbl.gov/downloads/TreeView/TreeView_vers_1_60.exe).
Protein identification by mass spectrometry analysis
Pick gels were prepared with sufficient amounts of protein (150 μg) and stained with mass spectrometry (MS)-compatible silver. Spots of interest were located on each gel by comparison with the 2-D DIGE protein pattern. Spots were excised manually from the gels, washed with double-distilled water and then transferred to siliconized Eppendorf tubes. The excised gel pieces were destained and dehydrated with acetonitrile. The proteins were chemically reduced in 10 mM DTT/50 mM NH4HCO3 for 1 h at 56 °C and alkylated in 55 mM iodoacetamide/50 mM NH4HCO3 for 1 h at room temperature. The gel pieces were washed several times in 50 mM NH4HCO3 followed by dehydration with acetonitrile and dried via centrifugation in a Speed-Vac centrifuge. The proteins were digested with trypsin (10 ng μl−1 Promega, Madison, WI, USA) at 37 °C for 16–18 h. The digestion reaction was stopped by the addition of 2 ml 10% trifluoroacetic acid, and the digestion products were desalted with a Millipore ZipTip C18 column. The resulting peptide mixtures were analysed by MS.
The resulting peptide mixtures were dissolved in 0.1% formic acid and separated on a capillary chromatography column connected directly to the source of the linear ion trap LC-MS/MS (LTQ; Thermo Finnigan, San Jose, CA, USA). Each sample was loaded on a C18 trap column (id 150 μM, length 150 mm, Agilent Technologies, Wilmington, DE, USA) for concentration and desalting using an acetonitrile gradient. The mass spectrometer was operated in positive ion mode with a capillary temperature of 170 °C. All data were acquired with the mass spectrometer operating in automatic data-dependent switching mode. Zoom scans were performed on the ten most intense ions to determine the charge state prior to MS/MS analysis.
For analysis via matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) MS, digestion products were desalted with a Millipore ZipTip C18 column. The MALDI-TOF MS was operated in the positive ion delayed extraction reflector mode for highest resolution and mass accuracy. Peptides were ionized with a 337 nm laser, and spectra were acquired at 19 kV acceleration potential with optimized parameters.
Database search
Peptide mass fingerprinting data were interpreted using Mascot (http://www.matrixscience.com) in the non-redundant database of the National Center for Biotechnology Information (NCBI). Taxonomic assignments were based on data available for Pinaceae in that non-redundant database. Our criterion for positive peptide mass fingerprinting was that at least four peptides must be matched. Monoisotopic masses were used, and a mass tolerance of ±100 ppm was allowed. The maximum number of missed cleavages was set at one. Complete carbamidomethylation of cysteines and variable oxidation of methionines (Met) were assumed.
MS/MS spectra data were searched against the available data for Pinaceae in NCBI using the SEQUEST algorithm as part of the BIOWORKS software suite. Search criteria of SEQUEST queries were as follows: Charge +1, Xcorr ≥1.9; Charge +2, Xcorr ≥2.2; Charge +3, Xcorr ≥3.75; DeICN ≥0.1 (Xcorr: SEQUEST cross-correlation value; DeICN: delta correlation value). The search parameters were based on a precursor-ion mass tolerance of 2 Da and a fragment ion tolerance of 1.0 Da with consideration for Met oxidation and cysteine carbamidomethylation. The estimated experimental Mr/pI was applied to increase the confidence of identification. Assignments annotated as ‘unknown’ were based on sequence similarity as assessed by BLASTP (www.ncbi.nlm.nih.gov/BLAST/).
Results and discussion
DIGE comparison
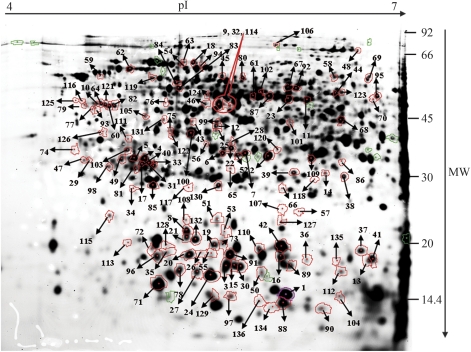
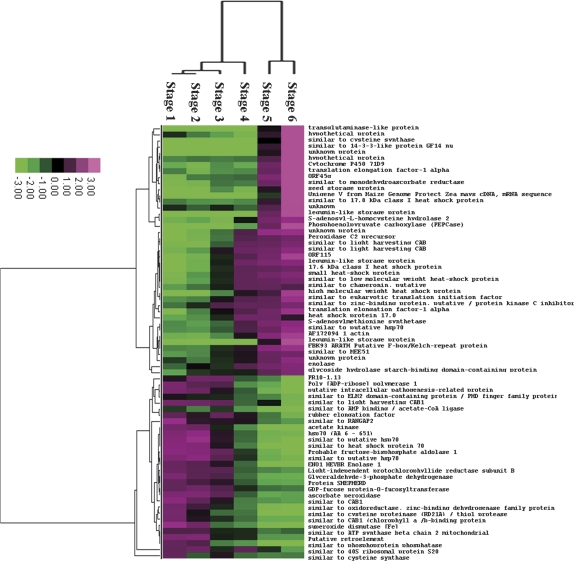
2-D DIGE was used to compare changes in the proteome of Chinese fir seeds during development, which spanned six important developmental phases during early embryogenesis: the cleavage polyembryony-stage seed (stages 1, 2, and 3), dominant embryo-stage seed (stages 4), columnar embryo-stage seed (stage 5), and early cotyledonary-stage seed (stage 6) (Fig. 1). After 2-D DIGE, Typhoon scanning indicated that each gel had approximately 2500 protein spots. The spots between gels were matched by DeCyder™ 6.5 software based on an internal standard. All gel images were matched against a master gel image, allowing comparison of protein abundances across the stages of seed development. The abundance of 136 proteins varied significantly (P <0.01, one-way analysis of variance) during seed development (Fig. 2), and these proteins were subsequently analysed by LC MS/MS and MALDI-TOF. Table 1 presents a list of proteins identified by MS analysis, which were functionally categorized according to the classification reported for Arabidopsis genes (Bevan et al., 1998). Unknown or hypothetical proteins were assigned based on sequence similarity as assessed by BLASTP (www.ncbi.nlm.nih.gov/BLAST/). Twenty-two proteins were assigned as conifer proteins, 27 proteins were identified as the Arabidopsis thaliana homologues, the remaining proteins were classified as homologues of proteins from other plants. It was recently reported that gymnosperms harbour genes, having sequences very similar to genes that regulate angiosperm embryogenesis genes. Moreover, approximately 72% of pine embryo ESTs could be detected in the A. thaliana proteome (Cairney and Pullman, 2007). A comparison of the proteins identified in Chinese fir with those of pine (as deduced from cDNA libraries) that have been implicated in embryogenesis and assigned according to angiosperm homologues, listed in Cairney et al. (2006) revealed only a few identical proteins. Differentially expressed proteins were subjected to hierarchical clustering based on our defined six stages of seed development. These proteins could be grouped into three categories as follows: one class of proteins whose levels increased between stages 5 and 6, a second class of proteins whose levels increased between stages 3 and 6, and a third class of proteins whose levels decreased during seed development (Fig. 3). The functions of some of the differently expressed proteins are discussed below.
Fig. 1.
Dissected characteristic zygotic embryo stages of the Chinese fir during seed development. Stages 1, 2, and 3: cleavage polyembryony stage; stage 4: dominant embryo stage; stage 5: columnar embryo stage; stage 6: early cotyledon stage. (This figure is available in colour at JXB online.)
Fig. 2.
Analytical 2-D DIGE gel of Cy2-labelled mixed-sample developing seeds. The marked spots show 136 proteins that were differentially expressed (P <0.01; one-way analysis of variance). MW: apparent molecular mass. (This figure is available in colour at JXB online.)
Table 1.
Proteins identified in developing seeds by LC-MS/MS and MALDI-TOF and functional category according to classification reported for Arabidopsis genes
 |
Fig. 3.
Hierarchical clustering of the differentially expressed proteins identified in this study. Clustering was based on the protein expression levels across stages 1, 2, 3, 4, 5, and 6 of seed development (the grading represents the ratios of protein expression levels).
Role of programmed cell death in gymnosperm seed development
Programmed cell death (PCD) is a part of the mechanism that controls the number and type(s) of cells at a certain location. The archegonium of developing gymnosperm seeds contains a number of embryos, but only one embryo in the polyembryonic seed usually develops and survives. PCD is the major mechanism that eliminates the subordinate embryos in a polyembryonic seed (Filonova et al., 2002). The suspensor, which promotes continuous growth of the embryo and thus is necessary for early embryonic development, begins to degenerate at the early cotyledon stage. This degeneration process is a classic example of developmental PCD (Lombardi et al., 2007). Consistent with a PCD process, two putative PCD-related proteins (spot 57 and spot 74) were down-regulated during seed development, coincident with the elimination of subordinate embryos and of suspensor degeneration. One of these proteins is a cysteine protease (spot 74); notably, in animals, PCD is executed by the caspase family of cysteine proteases. Plants do not have similar caspase homologues but rather express a phylogenetically distant family of cysteine proteinases (Bozhkov et al., 2005). In the early stages of seed development in Brassica napus, a cysteine proteinase gene was found to be expressed only in the inner integument as it undergoes PCD (Wan et al., 2002). That cysteine protease, which is expressed in the early stage in Chinese fir seed development, mediates PCD during embryonic patterning, and thus constitutes a functional link between PCD and embryogenesis in plants (Bozhkov et al., 2005). Another PCD protein is poly [ADP-ribose] polymerase 1 (PARP-1, spot 57), which is also down-regulated during Chinese fir seed development; PARP-1 is a nuclear enzyme that plays a critical role in genomic stability by mediating the DNA damage surveillance network, and it is also a mediator of cell death (Yu et al., 2002). In some animal cell types, PARP may play a role in controlling cell death through NAD depletion. In plants, the function of PARP is less understood. The activation and specific cleavage of PARP are essential for PCD progression induced by heat shock in tobacco suspension cells (Tian et al., 2000). In gymnosperm seed development, another PCD process involves the degeneration of a narrow, unicellular layer that lines the corrosion cavity of the female gametophyte, making room for the actively growing embryo later in seed development (Filonova et al., 2002). In the present study, one PCD-related protein (spot 4, a transglutaminase) accumulated in the columnar embryo stage and early cotyledonary stage, in which the embryo is growing rapidly. Transglutaminases are responsible for the formation of apoptotic bodies in animal PCD. Transglutaminases also function as GTPases and thus can act as effectors in the prevention of PCD (Serafini-Fracassini et al., 2002). Little information is available regarding the involvement of transglutaminase in plant PCD. Recently, however, a transglutaminase was found to participate in PCD and regulate flower life span in the tobacco corolla (Della Mea et al., 2007).
PCD is the main mechanism by which unneeded structures are eliminated and morphology is changed during gymnosperm seed development. The involvement of PCD in the formation and development of somatic embryos in Norway spruce has also been reported (Filonova et al., 2000), and the cysteine protease mcll-Pa executes PCD during Norway spruce embryogesis (Bozhkov et al., 2005). The molecular mechanism by which PCD participates in gymnosperm seed development and somatic embryogenesis remains unclear, however. Based on our data for developing seeds of the Chinese fir, we speculate that there are multiple PCD programmes that maintain the proper balance between cell death and development. Except for the caspase-like protein we identified (cysteine proteinase), the PARP-1 and transglutaminase are new candidates for PCD in gymnosperm seed development. How PCD is regulated by these proteins during seed development in Chinese fir must be investigated further.
Proteins related to embryonic development
Genomic imprinting is a phenomenon in which only a specific maternal or paternal allele is expressed in a developing embryo (Huh et al., 2007). It has been shown that the maternal and paternal contributions to seed development are not equivalent due to genomic imprinting as well as the control over early seed development by the ‘maternal effect’ in plants (Grimanelli et al., 2005). The protein ‘Maternal effect embryo arrest 51’ (spot 92) is involved in the progression of the embryo over time, from zygote formation to the end of seed dormancy. It was found here that this protein—a homologue of the Arabidopsis maternal effect embryo arrest 51 that is responsible for the embryonic development ending in seed dormancy (the reference sequence was derived from AT4G04040)—was up-regulated during seed development. Other reports on various animal and plant species have also described the maternal effect. For example, certain maternal housekeeping proteins are translated during bovine oocyte maturation and early embryo development (Massicotte et al., 2006). The maternal gene SIN1 in the animal egg regulates expression of a diffusible morphogen required for zygotic development and is also required for embryo development in Arabidopsis (Golden et al., 2002). Mutation in the maternal allele of the Arabidopsis gene MEA results in aborted embryos, and this mutation cannot be rescued by pollination with a paternal wild-type gene, implying that MEA promotes embryo development only when expressed maternally (Cairney and Pullman, 2007). There are few reports, however, on the maternal effect in gymnosperms.
Spot 66 was identified as one of the C. lanceolata proteins related to embryo development; this protein is similar to the ELM2 domain–containing protein/PHD finger family. The ELM2 domain is a small domain of unknown function, but it is similar in sequence to domains in Egl-27and MTA1. The ELM2 domain also may be involved in DNA binding, or perhaps may be a protein–protein interaction domain (Solari et al., 1999). Both Egl-27and MTA1 have been cloned in Caenorhabditis elegans, and each of these proteins is in a distinct chromatin regulatory complex; these proteins are required for embryonic patterning, although they are functionally redundant (Solari et al., 1999). The PHD finger, a Cys4-His-Cys3 zinc finger, is found in many regulatory proteins from plants to animals; PHD finger–containing proteins are associated with chromatin-mediated transcriptional regulation (Halbach et al., 2000). Chromatin modification and large-scale remodelling events are necessary for embryo patterning during seed development, and these events control the basic body pattern from fertilization to the heart stage in angiosperm seeds (Baroux et al., 2007). It was found that spot 66 was highly expressed from the cleavage polyembryony stage to the dominant embryo stage and was then down-regulated from the columnar embryo stage to the early cotyledon stage. Accordingly, it is postulated that the spot 66 protein participates in the control of embryo patterning from the cleavage polyembryony stage to the dominant embryo stage.
Met metabolism is critical for the amino acid metabolic pathway of seed filling
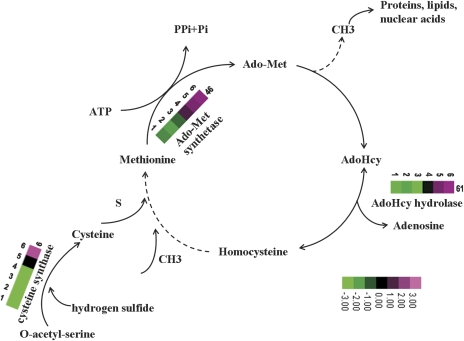
The Met metabolic pathway is essential for cell viability in that it provides building blocks for proteins and generates the precursor to S-adenosylmethionine (Ado-Met), which is the main methyl group-donating compound in cells. There have been reports that Met metabolism has an important role in seed development and germination, as illustrated by proteomic analyses of developing Medicago truncatula seeds (Gallardo et al., 2003, 2007) and of Arabidopsis seed germination and priming (Gallardo et al., 2001). In the present study, consistent with the high demand for protein synthesis in seed filling, two spots involved in Met metabolism, namely, Ado-Met synthase (spot 46), and S-adenosyl-l-homocysteine (AdoHcy) hydrolase (spot 61), were highly expressed between stage 4 and stage 6. Two spots, 6 and 86, were found to be similar to cysteine synthase, which is responsible for the formation of cysteine from O-acetyl-serine and hydrogen sulphide. Cysteine is first reduced by a sulphur-containing compound and serves as the sulphur donor for Met (Chronis and Krishnan, 2003). Ado-Met synthase is a key enzyme in plant metabolism, catalysing the biosynthesis of Ado-Met from Met and ATP. Ado-Met is not only the primary methyl-group donor but also a source of amino-alkyl and amino groups used for the biosynthesis of amino acids, polyamines, and the hormone ethylene, and it also provides the methylene group used in the biosynthesis of fatty acids that accumulate in many types of seeds (Bouvier et al., 2006). AdoHcy hydrolase catalyses the production of AdoHcy, which is formed after donation of the activated methyl group of AdoMet to an acceptor. AdoHcy is the product of all Ado-Met-dependent biological transmethylation reactions and is a potent competitive inhibitor of methyltransferases that are crucial to cell growth and development (Turner et al., 2000). The continuous resynthesis of Ado-Met from ATP and Met is made possible by the recycling of adenosine and homocysteine derived from AdoHcy. Thus, AdoHcy is an important intermediate in maintaining proper Ado-Met levels (Weretilnyk et al., 2001). The high accumulation of Ado-Met synthase, similar to AdoHcy hydrolase, parallels the accumulation of the major seed storage proteins, and therefore these enzymes play a key role in providing the building blocks for proteins, polyamines, and ethylene during the early cotyledonary stage of seeds. In Fig. 4, a model is proposed for the involvement of Met metabolism in the various stages of seed development based on our results for the protein expression patterns observed during the development of Chinese fir seeds as well as specific features of Met biosynthesis/metabolism (Ravanel et al., 1998), which indicated that the metabolism of Met and Cys is critical for storage protein synthesis during seed filling. A similar observation was made in a proteomic analysis of developing M. truncatula (Gallardo et al., 2007).
Fig. 4.
Methionine metabolic pathway during Chinese fir seed filling. The proteins identified in our study as being significantly up- or down-regulated are shown with the corresponding grading.
Developmental expression of enzymes related to carbon metabolism in seeds
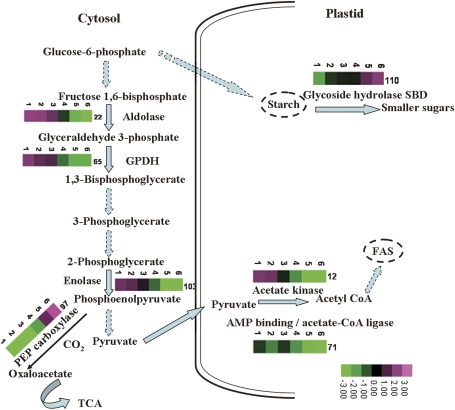
The enzymes and substrates involved in carbon metabolism in seeds contribute to the synthesis of starch. Several enzymes related to carbon metabolism (spot 22, probably fructose-bisphosphate aldolase; spot 65, glyceraldehyde-3-phosphate dehydrogenase; spot 67, GDP-fucose protein-O-fucosyltransferase; spot 103, ENO1_HEVBR enolase) were down-regulated as seed development proceeded (Fig. 5). These enzymes were highly expressed in stage 1 of developing Chinese fir seeds. These proteins are probably involved in supplying carbon substrates for the synthesis of energy storage compounds under conditions that limit CO2 fixation. The proteins ‘similar to ATP synthase beta chain 2 mitochondrial’ (spot 83) and ‘similar to CAB1’ (chlorophyll a/b-binding protein 1; spots 69, 75, 80) also mainly accumulated in stage 1. Embryonic photosynthesis has been proposed to play a role in providing ATP and NADPH for the synthesis of storage compounds during seed development (Gallardo et al., 2003). Another enzyme, glycoside hydrolase starch-binding domain-containing protein (spot 110), was up-regulated during seed development. Glycoside hydrolases catalyse the hydrolysis of glycosidic linkages to generate smaller polysaccharides/monosaccharides. The starch-binding domain belongs to the carbohydrate binding module family 20, members of which accelerate the rate of hydrolysis of insoluble starch. AMY1-SBD (α-amylase 1-starch-binding domain-containing protein) is a type of glycoside hydrolase starch-binding domain-containing protein that hydrolyses barley starch granules 15-fold faster than recombinant AMY1 at low concentration (Juge et al., 2006). A similar observation was reported in developing seeds of soybean (Adams et al., 1980). In stage 1, starch accumulates in transient reserves, which are later utilized in seed development. The up-regulation of AMY1-SBD is possibly responsible for starch mobilization.
Fig. 5.
Kinetics of carbon metabolism in developing seeds. Proteins identified as being significantly up- or down-regulated are shown with the corresponding grading.
Information on seed development contributes importantly to our understanding of the molecular and physiological bases of embryogenesis (Balbuena et al., 2009; Silveira et al., 2008). For example, the identification of the PCD proteins will be helpful in establishing a mechanistic link between PCD and embryo patterning; the proteins of highest abundance, namely legumin-like storage protein, are further used for plantlet growth. The up-regulated expression of these proteins might constitute a marker for maturation during seed development. Thus, growth-stimulating agents that induce protein expression when added to the culture medium, are necessary for successful somatic embryo maturation. Our results provide new insights into the complex processes involved in Chinese fir seed development as well as information on both the practical means of further improving somatic embryogenesis through medium manipulation and the protein markers useful for screening based on protein expression. A more spatio-temporally detailed investigation will provide a better understanding of the biological processes involved in seed and embryo development.
Acknowledgments
This work was supported by the Chinese National Forest Bureau's ‘948’ grant No. 2006-4-C02 to Professor Jisen Shi and by the Forest Genetics and Biotechnology of the Open Research Project (No. FGB 200801). We are very grateful to Shanghai Applied Protein Technology Co. Ltd. for kind technical support for LC-MS/MS and MALDI-TOF MS.
Glossary
Abbreviations
- Cy
cyanine dye
- DIGE
difference gel electrophoresis
- LC-MS/MS
liquid chromatography-mass spectrometry/mass spectrometry
- MALDI-TOF-MS
matrix-assisted laser desorption ionization time-of-flight mass spectrometry
- PARP-1
poly [ADP-ribose] polymerase 1
- PCD
programmed cell death
References
- Adams CA, Rinne RW, Fjerstad MC. Starch deposition and carbohydrate activities in developing and germinating soybean seed. Annals of Botany. 1980;45:577–582. [Google Scholar]
- Balbuena TS, Silveira V, Junqueira M, Dias LL, Santa-Catarina C, Shevchenko A, Floh EI. Changes in the 2-DE protein profile during zygotic embryogenesis in the Brazilian Pine (Araucaria angustifolia) Journal of Proteomics. 2009;72:337–352. doi: 10.1016/j.jprot.2009.01.011. [DOI] [PubMed] [Google Scholar]
- Baroux C, Pien S, Grossniklaus U. Chromatin modification and remodeling during early seed development. Current Opinion in Genetics and Development. 2007;17:473–479. doi: 10.1016/j.gde.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Bevan M, Bancroft I, Bent E, et al. Analysis of 19 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature. 1998;391:485–488. doi: 10.1038/35140. [DOI] [PubMed] [Google Scholar]
- Bouvier F, Linka N, Isner JC, Mutterer J, Weber AP, Camara B. Arabidopsis SAMT1 defines a plastid transporter regulating plastid biogenesis and plant development. The Plant Cell. 2006;18:3088–3105. doi: 10.1105/tpc.105.040741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozhkov PV, Suarez MF, Filonova LH, Daniel G, Zamyatnin AA, Jr., Rodriguez-Nieto S, Zhivotovsky B, Smertenko A. Cysteine protease mcII-Pa executes programmed cell death during plant embryogenesis. Proceedings of the National Academy of Sciences, USA. 2005;102:14463–14468. doi: 10.1073/pnas.0506948102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairney J, Pullman GS. The cellular and molecular biology of conifer embryogenesis. New Phytologist. 2007;176:511–536. doi: 10.1111/j.1469-8137.2007.02239.x. [DOI] [PubMed] [Google Scholar]
- Cairney J, Zheng L, Cowels A, et al. Expressed sequence tags from loblolly pine embryos reveal similarities with angiosperm embryogenesis. Plant Molecular Biology. 2006;62:485–501. doi: 10.1007/s11103-006-9035-9. [DOI] [PubMed] [Google Scholar]
- Chronis D, Krishnan HB. Sulphur assimilation in soybean: molecular cloning and characterization of O-acetylserine (thiol) lyase (cysteine synthase) Crop Science. 2003;43:1819–1827. [Google Scholar]
- Della Mea M, De Filippis F, Genovesi V, Serafini Fracassini D, Del Duca S. The acropetal wave of developmental cell death of Nicotiana tabacum corolla is preceded by activation of transglutaminase in different cell compartments. Plant Physiology. 2007;144:1211–1222. doi: 10.1104/pp.106.092072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos ALW, Wietholter N, EI Gueddari NE, Moerschbacher BM. Protein expression during seed development in Araucaria angustifolia: transient accumulation of class IV chitinases and arabinogalactan proteins. Physiologia Plantarum. 2006;127:138–148. [Google Scholar]
- Filonova LH, Bozhkov PV, Brukhin VB, Daniel G, Zhivotovsky B, von Arnold S. Two waves of programmed cell death occur during formation and development of somatic embryos in the gymnosperm, Norway spruce. Journal of Cell Science. 2000;113:4399–4411. doi: 10.1242/jcs.113.24.4399. [DOI] [PubMed] [Google Scholar]
- Filonova LH, von Arnold S, Daniel G, Bozhkov PV. Programmed cell death eliminates all but one embryo in a polyembryonic plant seed. Cell Death and Differentiation. 2002;9:1057–1062. doi: 10.1038/sj.cdd.4401068. [DOI] [PubMed] [Google Scholar]
- Gallardo K, Firnhaber C, Zuber H, Hericher D, Belghazi M, Henry C, Kuster H, Thompson R. A combined proteome and transcriptome analysis of developing Medicago truncatula seeds. Molecular and Cellular Proteomics. 2007;6:2165–2179. doi: 10.1074/mcp.M700171-MCP200. [DOI] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SP, Puype M, Demol H, Vandekerckhove J, Job D. Proteomic analysis of Arabidopsis seed germination and priming. Plant Physiology. 2001;126:835–848. doi: 10.1104/pp.126.2.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo K, Signor CL, Vandekerckhove J, Thompson RD, Burstin J. Proteomics of Medicago truncatula seed development establishes the time frame of diverse metabolic processes related to reserve accumulation. Plant Physiology. 2003;133:664–682. doi: 10.1104/pp.103.025254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden TA, Schauer SE, Lang JD, Pien S, Mushegian AR, Grossniklaus U, Meinke DW, Ray A. SHORT INTEGUMENTS1/SUSPENSOR1/CARPEL FACTORY, a dicer homolog, is a maternal effect gene required for embryo development in Arabidopsis. Plant Physiology. 2002;130:808–822. doi: 10.1104/pp.003491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimanelli D, Perotti E, Ramirez J, Leblanca O. Timing of the maternal-to-zygotic transition during early seed development in maize. The Plant Cell. 2005;17:1061–1072. doi: 10.1105/tpc.104.029819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbach T, Scheer N, Werr W. Transcriptional activation by the PHD finger is inhibited through an adjacent leucine zipper that binds 14-3-3 proteins. Nucleic Acids Research. 2000;28:3542–3550. doi: 10.1093/nar/28.18.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZQ, Xu Z, Boyd S, Williams D. Chemical composition of decomposing stumps in successive rotation of Chinese fir (Cunninghamia lanceolata (Lamb.) Hook.) plantations. Chinese Science Bulletin. 2005;50:2581–2586. [Google Scholar]
- Huh JH, Bauer MJ, Hsieh TF, Fischer R. Endosperm gene imprinting and seed development. Current Opinion in Genetics and Development. 2007;17:480–485. doi: 10.1016/j.gde.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juge N, Nøhr J, Le Gal-Coëffet MF, Kramhøft B, Furniss CS, Planchot V, Archer DB, Williamson G, Svensson B. The activity of barley α-amylase on starch granules is enhanced by fusion of a starch binding domain from Aspergillus niger glucoamylase. Biochimica et Biophysica Acta. 2006;1764:275–284. doi: 10.1016/j.bbapap.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Klimaszewska K, Morencya F, Jones-Overton C, Cooke J. Accumulation pattern and identification of seed storage proteins in zygotic embryos of Pinus strobus and in somatic embryos from different maturation treatments. Physiologia Plantarum. 2004;121:682–690. [Google Scholar]
- Lippert D, Zhuang J, Ralph S, Ellis DE, Gilbert M, Olafson R, Ritland K, Ellis B, Douglas CJ, Bohlmann J. Proteome analysis of early somatic embryogenesis in Picea glauca. Proteomics. 2005;5:461–473. doi: 10.1002/pmic.200400986. [DOI] [PubMed] [Google Scholar]
- Lombardi L, Ceccarelli N, Picciarelli P, Lorenzi R. Caspase-like proteases involvement in programmed cell death of Phaseolus coccineus suspensor. Plant Science. 2007;172:573–578. doi: 10.1016/j.plaphy.2007.01.014. [DOI] [PubMed] [Google Scholar]
- Massicotte L, Coenen K, Mourot M, Sirard MA. Maternal housekeeping proteins translated during bovine oocyte maturation and early embryo development. Proteomics. 2006;6:3811–3820. doi: 10.1002/pmic.200500803. [DOI] [PubMed] [Google Scholar]
- Nagmani R, Diner S, Garton S, Zipf AE. Anatomical comparison of somatic and zygotic embryogeny in confers. In: Jain SM, Gupta PK, Newton RJ, editors. Somatic embryogenesis in woody plants. Vol. I. Dordrecht: Kluwer Academic Press; 2000. pp. 23–48. [Google Scholar]
- Pawłowski TA. Proteomics of European beech (Fagus sylvatica L.) seed dormancy breaking: influence of abscisic and gibberellic acids. Proteomics. 2007;7:2246–2257. doi: 10.1002/pmic.200600912. [DOI] [PubMed] [Google Scholar]
- Ravanel S, Gakière B, Job D, Douce R. The specific features of methionine biosynthesis and metabolism in plants. Proceedings of the National Academy of Sciences, USA. 1998;95:7805–7812. doi: 10.1073/pnas.95.13.7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallandrouze A, Faurobert M, EI Maâtaoui M. Characterization of the developmental stages of cypress zygotic embryos by two-dimensional electrophoresis and by cytochemistry. Physiologia Plantarum. 2002;114:608–618. doi: 10.1034/j.1399-3054.2002.1140415.x. [DOI] [PubMed] [Google Scholar]
- Serafini-Fracassini D, Del Duca S, Monti F, Poli F, Sacchetti G, Bregoli AM, Biondi S, Della Mea M. Transglutaminase activity during senescence and programmed cell death in the corolla of tobacco (Nicotiana tabacum) flowers. Cell Death and Differentiation. 2002;9:309–321. doi: 10.1038/sj.cdd.4400954. [DOI] [PubMed] [Google Scholar]
- Silveira V, Santa-Catarina C, Balbuena TS, Moraes FMS, Ricart CAO, Sousa MV, Guerra MP, Handro WF, Floh EIS. Endogenous abscisic acid and protein contents during seed development of Araucaria angustifolia. Biologia Plantarum. 2008;52:101–104. [Google Scholar]
- Solari F, Bateman A, Ahringer J. The Caenorhabditis elegans genes egl-27 and egr-1 are similar to MTA1, a member of a chromatin regulatory complex, and are redundantly required for embryonic patterning. Development. 1999;126:2483–2494. doi: 10.1242/dev.126.11.2483. [DOI] [PubMed] [Google Scholar]
- Tian RH, Zhang GY, Yan CH, Dai YR. Involvement of poly (ADP-ribose) polymerase and activation of caspase-3-like protease in heat shock-induced apoptosis in tobacco suspension cells. FEBS letters. 2000;474:11–15. doi: 10.1016/s0014-5793(00)01561-1. [DOI] [PubMed] [Google Scholar]
- Turner MA, Yang X, Yin D, Kuczera K, Borchardt R, Howell PL. Structure and function of S-adenosylhomocysteine hydrolase. Biochimica et Biophysica Acta. 2000;33:101–125. doi: 10.1385/CBB:33:2:101. [DOI] [PubMed] [Google Scholar]
- von Arnold S, Sabala I, Bozhkov P, Dyachok J, Filonova L. Developmental pathways of somatic embryogenesis. Plant Cell Tissue and Organ Culture. 2002;69:233–249. [Google Scholar]
- Wan L, Xia Q, Qiu X, Selvaraj G. Early stages of seed development in Brassica napus: a seed coat-specific cysteine proteinase associated with programmed cell death of the inner integument. The Plant Journal. 2002;30:1–10. doi: 10.1046/j.1365-313x.2002.01262.x. [DOI] [PubMed] [Google Scholar]
- Weretilnyk EA, Alexander KJ, Drebenstedt M, Snider JD, Summers PS, Moffatt BA. Maintaining methylation activities during salt stress. The involvement of adenosine kinase. Plant Physiology. 2001;125:856–865. doi: 10.1104/pp.125.2.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Mediation of poly (ADP-Ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]