Abstract
Novel constitutive gene promoters are essential components of crop biotechnology. Our analysis of five such promoters, APX, SCP1, PGD1, R1G1B, and EIF5, in transgenic rice plants is reported here. The five promoter regions were linked to the gfp reporter gene and transformed into rice. Using fluorescent microscopy and q-RT-PCR, promoter activities were analysed in comparison with OsCc1, Act1, and ZmUbi1, previously characterized as strong constitutive promoters. The APX and PGD1 promoters direct high levels of gene expression in all tissues and stages, producing GFP at levels of up to 1.3% of the total soluble protein. PGD1 is particularly active in flowers and mature roots. The R1G1B is active in the whole grain including the embryo, endosperm, and aleurone layer, and thus represents a constitutive promoter with activity in whole seeds that has not been described previously. The ZmUbi1 and R1G1B promoters are markedly less active in young roots and mature leaves whilst the APX, PGD1, OsCc1, and Act1 promoters are highly active in both vegetative and reproductive tissues. Overall, our results demonstrate that APX, PGD1, and R1G1B are novel gene promoters that are highly active at all stages of plant growth with distinct levels of activity.
Keywords: APX, constitutive gene promoter, GFP, PGD1, R1G1B, transgenic rice, vegetative and reproductive tissues
Introduction
Gene promoters that direct high levels of constitutive gene expression are important for crop biotechnology applications. In recent years, several well-characterized gene promoters have become available for transgene expression in plants. The cauliflower mosaic virus (CaMV) 35S gene promoter is the most commonly used to drive transgene expression in a constitutive manner (Odell et al., 1985). However, although the 35S promoter and its derivatives can drive high levels of transgene expression in dicotyledonous plants (Battraw and Hall, 1990; Benfey et al., 1990), their activities are substantially lower in monocotyledonous plants (Christensen et al., 1992; Gupta et al., 2001; Weeks et al., 1993). Conversely, the activity of monocot-derived promoters is higher in monocots than in dicots (Cornejo et al., 1993), necessitating the development of both monocot and dicot promoters.
Several gene promoters have been evaluated for their ability to drive the constitutively high expression of transgenes in monocots including ZmUbi1 (Christensen et al., 1992; Cornejo et al., 1993) from maize (Zea mays), and Act1 (McElroy et al., 1990, 1991), OsTubA1 (Jeon et al., 2000), OsCc1 (Jang et al., 2002), RUBQ1 and 2 (Wang and Oard, 2003), rubi3 (Lu et al., 2008), and OsAct2 (He et al., 2009) from rice (Oryza sativa). These promoters are highly active in monocot crops including rice but are also distinct in various ways. The ZmUbi1 promoter is widely used in monocot crops due to its ability to direct high levels of gene expression in virtually all tissues. This promoter is active in many, although not all, cell types and drives strong expression in young roots and leaves, but these expression levels decrease markedly as these organs mature (Cornejo et al., 1993). The OsCc1 and Act1 promoters are active in both the vegetative and reproductive tissues of transgenic rice plants (McElroy et al., 1990; Jang et al., 2002). The activity profile of the OsAct2 promoter is similar to that of Act1 in leaves and roots of transgenic rice plants, but is slightly stronger (He et al., 2009). The rice ubiquitin gene promoters, RUBQ1 and RUBQ2, have been shown to drive higher GUS expression in transgenic rice plants by 8–35-fold, respectively, when compared with the 35S promoter (Wang and Oard, 2003). The rice polyubiquitin gene promoter, rubi3, has been tested in all tissues and at all growth stages of transgenic rice plants and found to drive a higher level of constitutive expression of reporter genes than the maize ZmUbi1 promoter (Lu et al., 2008). Further, polyubiquitin gene promoters have been isolated and characterized from soybean (Chiera et al., 2007; Hernandez-Garcia et al., 2009), potato (Garbarino et al.,1995; Rockhold et al., 2008), and sugarcane (Wei et al., 2003). Despite such efforts to characterize constitutive gene promoters functionally, few of these have been analysed over the whole spectrum of transgenic plant growth. There is currently therefore a shortage of efficient promoters for high-level constitutive gene expression, particularly in monocot crops. In addition, stacking of several transgenes in a single transgenic plant requires a battery of different promoters, which may otherwise undergo homology-dependent gene silencing, a common occurrence in transgenic plants with multiple copies of the same promoter (Lessard et al., 2002; Potenza et al., 2004).
To characterize novel gene promoters that confer high levels of gene expression in rice, five genes, APX, SCP1, PGD1, R1G1B, and EIF5, were identified by microarray that are expressed at high levels throughout the whole plant. The activity of the corresponding constitutive gene promoters was then quantitatively analysed in transgenic rice plants using the gfp reporter. These promoter activities were analysed in different tissues and at various growth stages in comparison with those of the previously characterized strong constitutive promoters OsCc1, Act1, and ZmUbi1.
Materials and methods
Plant materials
The rice (Oryza sativa L. var. Japonica) plants cv. Nipponbare and cv. Nakdong were used to isolate gene promoters and perform plant transformations, respectively. Rice genomic DNA was prepared from the leaves of 14-d-old Nipponbare plants using the DNAzol plant genomic DNA isolation reagent (Molecular Research Center Inc., Cincinnati, OH) according to the manufacturer's instructions for the isolation of promoter regions by genomic PCR. The T3 generation of three to six independent lines for each of the promoters was used for our subsequent detailed analysis.
Expression of endogenous genes in rice
Rice 3′-Tiling Microarray analysis was performed as reported previously (Kim et al., 2009). RNA samples from drought-treated and untreated leaf tissues of 14-d-old rice (O. sativa L. cv. Nakdong) plants were used to generate Cy3-labelled complementary DNA probes, which were then hybridized to the microarray. Each dataset was obtained from two biological repeats. Comparisons of the datasets from various tissues identified five genes for promoter analysis out of more than 20 000 on the microarray (see Supplementary Table S1 available at JXB online). For RNA extraction (Oh et al., 2008), rice seeds were germinated in half-strength Murashige-Skoog (MS) (Murahige and Skoog, 1962) solid medium in a growth chamber in the dark at 28 °C for 3 d, followed by light exposure at 28 °C for 1 d, transplanted into soil, and grown in a greenhouse (16/8 h light/dark cycle) at 28–30 °C. Each plant was grown in a pot filled with rice nursery soil (Bio-media, Co., Kyoungju, Korea) for the indicated number of days after germination (DAG) and in the flowering stage when the panicles were at <10 cm in size. Total RNA was extracted from dry seeds, leaves, roots, and flowers using the Qiagen RNeasy plant mini kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. For amplification of the corresponding genes, cDNA was synthesized with random primers using a First-strand cDNA synthesis kit (Fermentas, Burlington, ON) and subsequent RT-PCR was carried out with 40 ng of cDNA template and gene-specific primer pairs designed with Primer Designer 4 software v4.20 (Sci-Ed Software, Durham, NC). These primer pairs are listed in Supplementary Table S2 at JXB online. RT-PCR was performed at 95 °C for 10 min, followed by 28 cycles at 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min, with a final cycle at 4 °C for 10 min. The OsUbi1 (AK121590) gene was used to verify RNA loading. The amplified PCR products were sequenced to ensure fidelity.
Plasmid construction and transformation of rice
The various promoter regions were amplified and isolated by PCR using promoter-specific primer pairs, APX (AK068430), SCP1 (AK101133), PGD1 (AK065920), R1G1B (AF503583), EIF5 (AK060387), OsCc1 (AK060267), Act1 (AK100267), and ZmUbi1 (DQ141598), and the 2× EF-Taq DNA polymerase pre-mix (SolGent, Seoul). Genomic PCR was performed at 95 °C for 10 min, followed by 30 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min, with a final cycle at 4 °C for 10 min using 50 ng of gDNA. The amplified DNAs were re-amplified using the adaptor primer pairs. All primer pairs are listed in Supplementary Table S2 at JXB online. The isolated promoters with adaptor sequences were individually inserted into the rice transformation vector using site-specific recombination. PCR reactions and in vitro recombination reactions were carried out according to the manufacturer's instructions (Gateway® Cloning Technology, Invitrogen, Carlsbad, CA). The rice transformation vector contained the bar gene under the control of the CaMV 35S promoter for use with herbicide-based plant selection. Vectors were introduced into Agrobacterium tumefaciens LBA4404 by tri-parental mating. Embryonic calli from the mature seeds of rice (O. sativa L. cv. Nakdong) were transformed by co-cultivation (Hiei et al., 1994), selected with 7 mg l−1 phosphinothricin, and used to regenerate transgenic plants as previously described by Jang et al. (2002).
Detection of GFP fluorescence
Rice plant leaves, roots, dry seeds, and flowers were observed using a research stereomicroscope (SZX9-3122, Olympus, Tokyo) equipped with an attachment for fluorescence observations. Images were captured using a C5060-ZOOM digital camera (Olympus, Tokyo). Observations under blue light were carried out using a specific filter set (460–480 nm excitation filters, dichroic mirrors of 485 nm and a 495–540 nm barrier filter). The detection of GFP fluorescence was carried out in transgenic plants and also non-transgenic controls.
Real-time PCR analysis
Total RNA was extracted with leaves, roots, dry seeds, and flowers of transgenic and nontransgenic (NT) rice plants using the TRI REAGENT® (Molecular Research Center, Inc., Cincinnati, OH). Total RNAs from dry seeds and flowers were cleaned up by the Qiagen RNeasy mini kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. The First-strand cDNA synthesis kit with random primers for RT-PCR (Fermentas, Burlington, ON) was used for cDNA synthesis. Real-time PCR analysis was carried out using 2× real-time PCR Pre-mix with Evagreen (SolGent, Seoul). The reaction was performed at 95 °C for 15 min followed by 40 cycles of 95 °C for 20 s, 58 °C for 40 s, 72 °C for 20 s in a 20 μl volume mix containing 1 μl of 20× EvaGreen™, 0.25 μM primers and 40 ng cDNA. Thermocycling and fluorescence detection were performed using a Stratagene Mx3000p Real-Time PCR machine and Mx3000P software v2.02 (Stratagene, La Jolla, CA). Melting curve analysis (55–95 °C at a heating rate of 0.1 °C s−1) was performed to ensure that only the required PCR product at a specific melting temperature was measured (Ririe et al., 1997). Real-time PCR reactions were performed in triplicate for each cDNA sample. Following amplification, the experiment was converted to a comparative quantification (calibrator) experiment type and analysed with the Mx3000P software v2.02 (Stratagene, La Jolla, CA). The OsUbi1 (AK121590) gene was used as a reference in the real-time PCR reactions. All primer pairs are listed in Supplementary Table S2 at JXB online.
Immunoblotting analysis
Total soluble proteins were extracted from 30-d-old leaves, roots, and flowers. The extraction buffer consisted of 20 mM TRIS-Cl, pH 8.0, 10 mM EDTA pH 8.0, 30 mM NaCl, and 100 μM phenylmethylsulphonylfluoride (PMSF). The extracts were centrifuged at 9000 g at 4 °C for 30 min and protein concentrations were determined using the Bradford method (Bio-Rad, Hercules, CA). Protein extracts were then separated on 12% SDS polyacrylamide gels and blotted onto a polyvinylidenedifluoride (PVDF) membrane (Immobilon-P, Millipore Co., Billerica, MA) using a semi-dry transfer apparatus (Hoefer, Inc., San Francisco, CA). The immunoreactive proteins were detected using primary antibodies against GFP (Nacalai Tesque, Inc., Kyoto). The chemiluminescence signals generated by the bound antibodies were detected using the Pierce Super Signal Substrate (Pierce, Rockford, IL) according to the manufacturer's protocol. Recombinant GFP proteins were used as a positive control (Abcam, Cambridge, UK).
Results
Identification of novel constitutively expressed genes in rice
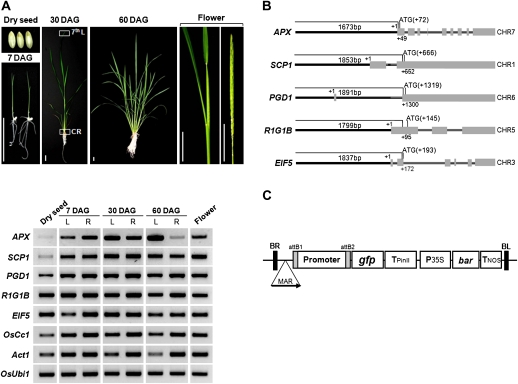
To screen for genes that are constitutively expressed in rice, expression profiling was performed using RNAs from leaves, roots, and flowers and the Rice 3'-Tiling Microarray (Oh et al., 2009). These analyses identified the genes APX, SCP1, PGD1, R1G1B, and EIF5 as being expressed in all tissues at high levels (see Supplementary Table S1 at JXB online). Expression of these five genes was further analysed by RT-PCR using total RNAs from eight different tissues and/or stages of rice plant growth. These included dry seeds, leaves, and roots at the indicated days after germination (DAG), and also the flowers (Fig. 1A). The transcript levels of APX and SCP1 were high in all tissues throughout the developmental stages except dry seeds and 60 DAG roots. The transcript levels of PGD1, R1G1B, and EIF5 were comparable to those of the positive control genes, OsCc1 (Jang et al., 2002) and Act1 (McElroy et al., 1990) in all tissues and stages of plant growth in rice. It was found that expression levels of R1G1B in flower was the lowest in microarray data (see Supplementary Table S1 at JXB online) but that the gfp mRNA levels (Fig. 1A) were comparable to other promoter:gfp plants. The discrepancy could be due to the difference in stages of flowers used for microarray and RT-PCR.
Fig. 1.
Expression and genomic structures of five constitutively expressed rice genes. (A) The expression of five constitutive rice genes in eight different tissues and/or three stages of plant growth was analysed by RT-PCR. These include dry seeds, leaves (L), and roots (R) at the indicated days after germination (DAG), and also the panicles (flower; <10 cm in size). Bars, 3 cm. The genes OsCc1 (Jang et al., 2002), Act1 (McElroy et al., 1990), and OsUbi1 (AK121590) were used as constitutive controls. 7th L, 7th leaf blade; CR, crown root. (B) Genomic structures of the five newly identified constitutive genes. Bold lines, 5′ upstream region; filled boxes, exons; lines between boxes, introns. All boxes were drawn to scale and chromosome numbers (CHR) are shown. The transcriptional start site and the translational start codon are indicated by +1 and ATG, respectively. (C) Schematic representation the T-DNA of the rice transformation vector used in this study. Rice promoters were linked to the gfp reporter gene by site-specific recombination. Expression of the gfp reporter gene is then regulated by the promoter and the potato Pin II 3′ region (TpinII). The bacterial phosphinothricin acetyl transferase gene (bar), a selectable marker for rice transformation, was fused between the CaMV 35S promoter (P35S) and the nopaline synthase polyadenylation region (Tnos). MAR, the 5′-matrix attachment region from the chicken lysozyme gene (Oh et al., 2005), was inserted to produce stable expression of the transgene.
Approximately 2.0 kb of the region immediately upstream of a translation initiation codon (ATG) was predicted to be the promoter region in each case (Fig. 1B). The predicted promoter regions of the five constitutive genes were amplified by PCR using rice (Oryza sativa cv. Nipponbare) genomic DNA. The resulting PCR products were then inserted into a rice transformation vector via site-specific recombination (Fig. 1C), generating constructs with each promoter sequence linked to the green fluorescent protein gene gfp. Several independent transgenic lines were obtained for each gene promoter using the Agrobacterium-mediated transformation method (Hiei et al., 1994) and were grown to maturity in a greenhouse. Through the self-pollination and selection of germinating transgenic seeds on phosphinothricin-containing MS media, T3 homozygous lines were obtained. Six independent lines for each construct were chosen for further analysis.
Activity of the promoter:gfp constructs in various tissues of transgenic rice plants
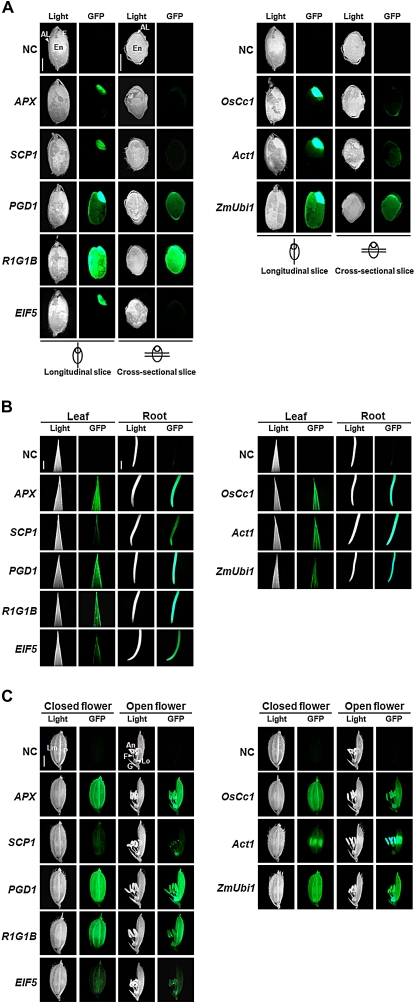
To investigate promoter activity in the whole rice plant, GFP fluorescence in the vegetative and reproductive tissues of the promoter:gfp transgenic plants was visually detected with a fluorescence microscope. Transgenic plants harbouring the APX, SCP1, PGD1, R1G1B, and EIF5 promoter constructs were analysed in comparison with those containing the OsCc1 (Jang et al., 2002), Act1 (McElroy et al., 1990), and ZmUbi1 (Cornejo et al., 1993), three known constitutive promoters. Images of non-transgenic (NT) tissues were used as negative controls. For the observation of GFP fluorescence, one transgenic line for each construct was chosen whose expression pattern was closest to that of the endogenous gene. To detect GFP fluorescence separately in embryos, endosperms, and aleurones of dry seeds, hand-cut vertical and horizontal sections of dry seeds were used for analysis by fluorescence microscopy (Fig. 2A). The APX, SCP1, and EIF5 promoters were found to be active only in embryos, as were the OsCc1 and Act1 promoters. GFP fluorescence level of the PGD1:gfp seeds was similar to that of the ZmUbi1:gfp seeds, being positive in the aleurones as well as the embryos. Interestingly, the R1G1B promoter directed high levels of GFP fluorescence in the whole grain including the embryo, endosperm, and aleurone layer. GFP fluorescence was examined further in the leaves and roots of 30 DAG plants by microscopy (Fig. 2B). A tip of 7th leaf blade and a crown root around the node (denoted the 7th L and CR in Fig. 1A, respectively) were used for the detection of GFP fluorescence. In vegetative tissues, the APX, PGD1, and R1G1B promoters produced high levels of GFP fluorescence in a similar manner to the constitutive control promoters. The levels of GFP fluorescence in the leaves, roots and flowers (Fig. 2B, C) driven by the SCP1:gfp and EIF5:gfp plants were markedly lower. GFP fluorescence was also examined in flowers (spikelets) at the meiosis stage (Fig. 2C). As observed in the vegetative tissues, the transgenic flowers all showed a high level of GFP fluorescence except for those harbouring SCP1:gfp and EIF5:gfp. More specifically, the APX, PGD1, and R1G1B promoters directed high levels of gfp expression in all floral organs including the lemma, palea, lodicule, anther, and filament, which is a similar pattern to that in the OsCc1:gfp and ZmUbi1:gfp flowers.
Fig. 2.
Promoter activity in various tissues of the promoter:gfp transgenic plants. Transgenic plants harbouring the constitutive gene promoters, APX, SCP1, PGD1, R1G1B, and EIF5, were analysed in comparison with the transgenic plants containing the known constitutive gene promoters, OsCc1 (Jang et al., 2002), Act1 (McElroy et al., 1990), and ZmUbi1 (Cornejo et al., 1993). Light and GFP indicate images captured using light and fluorescence microscopy, respectively. Bars, 2 cm. (A) GFP fluorescence in hand-cut vertical and horizontal sections of dry seeds. AL, aleurone layer; E, embryo; En, endosperm. (B) GFP fluorescence in the leaves and roots of 30 DAG plants. (C) GFP fluorescence in flowers. An, anther; G, glume; Lm; lemma; Lo, lodicule; F, filament; P, palea.
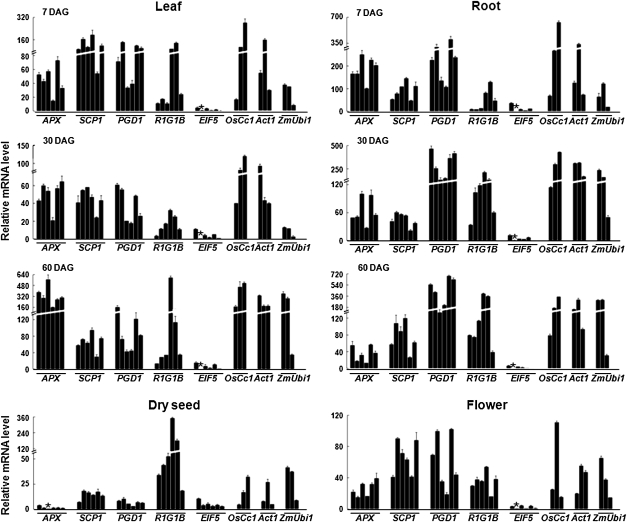
Quantitative analysis of promoter activity
To compare promoter activity levels, the gfp mRNA levels of six independent T3 promoter:gfp transgenic lines was measured by qPCR (Fig. 3). Total RNA was extracted from dry seeds, leaves, and roots at the indicated days after germination (DAG), and also from the flowers. The levels of promoter-driven gfp transcripts were calculated relative to the gfp mRNA concentration in the ZmUbi1:gfp transgenic lines (Table 1). Overall, promoter-driven gfp mRNA was found to be abundant at various levels in all of the vegetative tissues and flowers in most promoter:gfp transgenic lines. In particular, the gfp mRNA levels driven by the APX, SCP1, PGD1, and R1G1B promoters were found to be comparable to those produced by the OsCc1, Act1, and ZmUbi1 constitutive control promoters. By contrast, the activity of the EIF5 promoter was markedly lower in all vegetative tissues and flowers, although was still widely constitutive. Interestingly, the PGD1 and R1G1B promoters were far more active than the others in 60 DAG roots and dry seeds, respectively. Conversely, the R1G1B and ZmUbi1 promoters were found to have significantly lower activity than all others in 7 DAG roots and 30 DAG leaves.
Fig. 3.
Quantitative comparison of promoter activity in six independent lines of each of the promoter:gfp transgenic plants. Total RNA was extracted from the leaves and roots at the indicated days after germination (DAG), from dry seeds, and from the flowers of five independent T3 lines of transgenic plants harbouring the APX, SCP1, PGD1, R1G1B, and EIF5 gene promoters. Levels of gfp mRNAs were measured using real-time PCR, normalized with respect to that of the internal control gene OsUbi1 (AK121590), and plotted relative to the level of gfp mRNAs in the lowest-expressing transgenic plants (indicated by the asterisk). Three independent lines from transgenic plants harbouring the OsCc1 (Jang et al., 2002), Act1 (McElroy et al., 1990), and ZmUbi1 (Cornejo et al., 1993) gene promoters, were included as constitutive controls. Data bars represent the mean ±SD of triplicate measurements. Note that the y-axis is discontinuous.
Table 1.
Relative promoter activities in various tissues and stages of growth in rice plants
| Promoter | Dry seed | 7 DAG |
30 DAG |
60 DAG |
Flower | |||
| Leaf | Root | Leaf | Root | Leaf | Root | |||
| APX | 0.1±0.0 | 1.7±0.7 | 2.6±0.7 | 5.3±1.7 | 0.4±0.2 | 1.4±0.6 | 0.2±0.1 | 0.7±0.2 |
| SCP1 | 0.5±0.1 | 4.5±1.9 | 1.3±0.5 | 4.7±1.3 | 0.3±0.1 | 0.3±0.1 | 0.4±0.2 | 1.7±0.6 |
| PGD1 | 0.2±0.1 | 3.1±1.6 | 3.3±1.5 | 4.0±2.0 | 2.0±0.9 | 0.4±0.2 | 2.1±1.1 | 1.6±0.9 |
| R1G1B | 3.9±4.5 | 1.8±2.0 | 0.7±0.7 | 1.8±1.1 | 0.7±0.4 | 0.6±1.0 | 0.8±0.8 | 0.9±0.3 |
| EIF5 | 0.2±0.1 | 0.1±0.1 | 0.2±0.2 | 0.4±0.4 | 0.03±0.0 | 0.03±0.0 | 0.01±0.0 | 0.1±0.0 |
| OsCc1 | 0.6±0.5 | 5.1±5.0 | 4.6±4.0 | 8.3±4.1 | 1.9±1.0 | 1.7±0.8 | 1.0±0.7 | 1.3±1.3 |
| Act1 | 0.5±4 | 3.1±2.6 | 2.5±1.9 | 6.1±3.0 | 1.8±0.6 | 1.0±0.4 | 0.9±0.5 | 1.0±0.5 |
| ZmUbi1 | 1.0±0.6 | 1.0±0.6 | 1.0±0.7 | 1.0±0.6 | 1.0±0.6 | 1.0±0.7 | 1.0±0.7 | 1.0±0.6 |
Levels of promoter-driven gfp mRNA in different transgenic lines were calculated relative to those of gfp mRNAs in ZmUbi1:gfp transgenic lines. Data are the means ±SD of the gfp mRNA levels in six independent transgenic lines of APX, SCP1, PGD1, R1G1B, and EIF5 or three independent transgenic lines of OsCc1, Act1, and ZmUbi1.
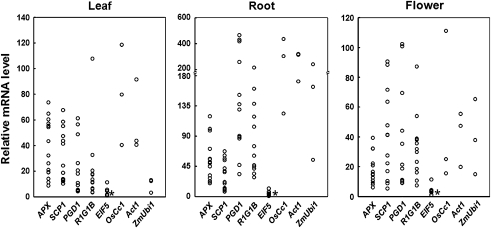
Promoter activities are known to vary greatly among transgenic lines. To assess this variation, the gfp mRNA levels in the leaves, roots, and flowers of 9–15 transgenic lines were compared for each promoter. The transcript levels were measured by q-RT-PCR and plotted relative to those in the EIF5 transgenic plants. As shown in Fig. 4, the promoter activity levels ranged from 6.0–13.7-fold in leaves and roots and from 2.2–18.2-fold in flowers in different promoter:gfp plants.
Fig. 4.
Range of variation in the promoter activity in different transgenic lines. A total of 9–15 independent transgenic lines per promoter were analysed. Total RNA was isolated from the leaf and root tissues of 30 DAG plants and from flowers prior to the pollination stage. The levels of gfp mRNA were measured using real-time PCR and plotted relative to the gfp mRNA levels in EIF5 transgenic plants (indicated by the asterisk). All mRNA levels were normalized with respect to that of the internal control gene OsUbi1 (AK121590). Three independent lines of the transgenic plants harbouring the OsCc1 (Jang et al., 2002), Act1 (McElroy et al., 1990), and ZmUbi1 (Cornejo et al., 1993) gene promoters were used as constitutive controls. The data points represent the mean of triplicate measurements.
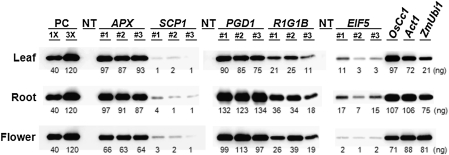
It was observed that the levels of GFP fluorescence do not always reflect the mRNA levels in promoter:gfp plants. It was found that the GFP fluorescence in SCP1:gfp plants was by far the lowest but that the gfp mRNA levels were comparable to other promoter:gfp plants. Therefore, the GFP protein levels among the promoter:gfp plants were determined by immunoblotting. As shown in Fig. 5, these levels were strikingly low in SCP1:gfp plants (0.02–0.06% of total soluble proteins) given the high levels of gfp mRNA. This observation prompted us to investigate the 5′ UTR sequence of SCP1. As shown in Supplementary Fig. S1 at JXB online, SCP1 contains a 489 bp 5′-UTR that contains three upstream ATGs (uATG) and ORFs (uORFs), which may have a negative impact upon the translation efficiency of the SCP1:gfp mRNA (Fig. 5). The GFP protein levels of APX:gfp and PGD1:gfp plants were high and comparable to those of OsCc1:gfp, Act1:gfp, and ZmUbi1:gfp plants (0.6–1.3% of the total soluble proteins; Fig. 5). The GFP protein levels of R1G1B:gfp and EIF5:gfp plants ranged from 0.11% to 0.34% and from 0.01% to 0.17% of the total soluble proteins, respectively, depending on the specific transgenic lines and/or tissues. Overall, our current results demonstrate that at least two out of our five gene promoters are constitutively active in a whole plant body. The other three have distinct levels of activity in different tissues and/or at different stages of plant growth.
Fig. 5.
Levels of GFP protein in the promoter:gfp transgenic rice plants. Protein extracts were prepared from the leaf and root tissues at 30 DAG and from flowers just before pollination. Three independent promoter:gfp transgenic plants and non-transgenic (NT) plants were analysed. One line of each of the transgenic plants harbouring the OsCc1 (Jang et al., 2002), Act1 (McElroy et al., 1990), and ZmUbi1 (Cornejo et al., 1993) gene promoters were included as constitutive controls. Ten μg of total soluble proteins were separated on a SDS-polyacrylamide gel, transferred to a membrane, and then immunoblotted with an anti-GFP antibody. The amount of GFP protein in promoter:gfp transgenic plants were calculated by comparing band intensities with those of known amounts of recombinant GFP protein used as positive controls (PC).
Discussion
In our current study, the activity of five different gene promoters throughout the entire growth cycle of transgenic rice plants was quantitatively analysed. All of these promoters were found to be active in various tissues and/or stages of plant growth with different patterns of relative activity. The APX and PGD1 promoters direct significantly higher levels of gene expression than any of the others tested in all the tissues and at all growth stages. In addition, PGD1 was found to have outstanding levels of activity in flowers and roots at 60 DAG. Overall, the levels of APX, and PGD1 promoter activity were comparable to those of OsCc1, Act1 and ZmUbi1, which are previously characterized strong constitutive promoters. Hence, the APX and PGD1 promoters may provide useful alternatives that will facilitate strong constitutive expression of a transgene in the whole rice plant. R1G1B is another constitutive promoter found to have high levels of activity in all the tissues and stages except for 7 DAG roots and 30 DAG leaves (Table 1). It was interesting to learn also that the R1G1B promoter is very active in the whole grain including the embryo, endosperm, and aleurone layer. To our knowledge, a constitutive gene promoter that is also active in the whole grain has not been described previously. In addition, it was found that the activity of the ZmUbi1 and R1G1B promoters is markedly reduced in 7 DAG roots and 30 DAG leaves whilst the APX, PGD1, OsCc1, and Act1 promoters are active in both vegetative and reproductive tissues of transgenic rice plants (Fig. 3; Table 1). Interestingly, the APX, PGD1, R1G1B, OsCc1, and ZmUbi1 promoters showed similar activity levels in all of the floral organs whereas the activity of the Act1 promoter was observed to be predominant in the anthers. The PGD1and ZmUbi1 promoters were similarly active in the embryos and aleurones yet distinct from the other promoters (Fig. 2A). EIF5 was found to be a constitutive promoter but to have low levels of activity in all tissues and at all growth stages. EIF5 exhibited the lowest activity among the five novel promoters analysed despite the high expression levels of its endogenous gene on the microarray (see Supplementary Table S1 at JXB online) and evidenced by RT-PCR (Fig. 1A). It thus appears that cis-acting elements located at regions other than the promoter are important for high levels of endogenous gene expression. There is a demand for such weak constitutive promoters, for example, for the expression of transgenes that are toxic or otherwise inhibitory in abundance, and for which a very strong promoter would not be suitable.
It was found that the GFP protein levels of SCP1:gfp plants are much lower than those of the other promoter:gfp plants although their gfp mRNA levels are high. The presence of three upstream ATGs in the 5′-UTR of the SCP1 promoter may have a negative impact upon the translation efficiency of the SCP1:gfp mRNA (Fig. 5). Consistent with our current results, the Arabidopsis ABSCISIC ACID-INSENSITIVE3 promoter was previously reported to contain three uORFs within its 405 bp 5′-UTR and the excision of this region dramatically increased GUS expression (Ng et al., 2004). The UTRs within transcripts often determine gene expression by influencing mRNA stability and translation efficiency. About 30% of the genes in rice and Arabidopsis are likely to be influenced at the translational level by signals present in their 5′-UTR as these motifs possess uAUG/uORFs with sequence similarities to regulatory consensus sequences (Shashikanth et al., 2008). A potential mechanism for uORFs to affect translational efficiency is therefore to act as a decoy from the principal ORF, and thereby potentially to cause premature termination, the triggering of mRNA decay via NMD (Nonsense-Mediated mRNA Decay), or a delay in the attachment of the ribosomal complex and thus the initiation of translation at the correct AUG start codon (Lawless et al., 2009).
Using the PlantCare program (Lescot et al., 2002), several cis-elements were identified in the promoter regions of the APX, SCP1, PGD1, R1G1B, and EIF5 as well as OsCc1, Act1, and ZmUbi1 genes (see Supplementary Fig. S2 at JXB online). A putative TATA box sequence was located at a region approximately between –23 and –62 relative to the transcription initiation site, and two to three CAAT boxes and common cis-acting elements were found in all of these promoter regions. The G-box motif, a cis-acting element involved in light responsiveness, is also located in all of these promoter regions. The G-box element has been found to be essential for transcriptional activity in the Arabidopsis rbcS-1A (Donald and Cashmore, 1990), the spinach Rubisco activase (Rca) (Orozco and Ogren, 1993), and in the rice OsAct2 (He et al., 2009) genes. The TGACG motif, which is important for high-level gene expression of Agrobacterium tumefaciens T-DNA octopine synthase (ocs) (Ellis et al., 1987), nopaline synthase (nos) (Mitra and An, 1989), CaMV 35S (Fang et al., 1989), and rice GOS2 (De Pater et al., 1992), was also found to be present within the APX, SCP1, PGD1, R1G1B, EIF5, and Act1 promoter regions. The rice endosperm-specific cis-acting elements, AACA and ACGT, were also identified (Wu et al., 2000; Qu et al., 2008) in the PGD1 and R1G1B promoters that are active in dry seeds.
In summary, five different gene promoters in transgenic rice plants were analysed throughout all stages of plant growth. These promoters were each found to be active in the whole rice plant body including flowers and dry seeds with distinct patterns of activity. These elements therefore provide novel putative constitutive gene promoters with great potential for use in crop biotechnology applications. The use of alternative promoters with similar characteristics is essential for the stacking of several transgenes in order to avoid homology-dependent gene silencing, a phenomenon which often occurs in transgenic plants with multiple copies of the same promoter. Our novel rice promoters may also be useful for transgene expression in other monocot crops such as maize, barley, and wheat.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Table S1. Microarray data for seven constitutive rice genes in 14-d-old leaves and roots and early stage flowers.
Supplementary Table S2. Primers used in this study for RT-PCR and promoter isolation.
Supplementary Fig. S1. Structure of the SCP1 5′-UTR.
Supplementary Fig. S2. Potential regulatory elements in the promoter regions of the eight constitutive genes analysed in this study.
Supplementary Material
Acknowledgments
This work was supported by the Ministry of Education, Science, and Technology, Korea, through the Crop Functional Genomics Center (CG2111 to J-KK) and by the Rural Development Administration through the Biogreen21 Program (PJ0007149 to J-KK).
References
- Battraw MJ, Hall TC. Histochemical analysis of CaMV 35S promoter-β-glucuronidase gene expression in transgenic rice plants. Plant Molecular Biology. 1990;15:527–538. doi: 10.1007/BF00017828. [DOI] [PubMed] [Google Scholar]
- Benfey PN, Ren L, Chua NH. Tissue-specific expression from CaMV 35S enhancer subdomains in early stages of plant development. The EMBO Journal. 1990;9:1677–1684. doi: 10.1002/j.1460-2075.1990.tb08291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiera JM, Bouchard RA, Dorsey SL, Park EH, Buenrostro-Nava MT, Ling PP, Finer JJ. Isolation of two highly active soybean (Glycine max (L.) Merr.) promoters and their characterization using a new automated image collection and analysis system. Plant Cell Reports. 2007;26:1501–1509. doi: 10.1007/s00299-007-0359-y. [DOI] [PubMed] [Google Scholar]
- Christensen AH, Sharrock RA, Quail PH. Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Molecular Biology. 1992;18:675–689. doi: 10.1007/BF00020010. [DOI] [PubMed] [Google Scholar]
- Cornejo MJ, Luth D, Blankenship KM, Anderson OD, Blechl AE. Activity of a maize ubiquitin promoter in transgenic rice. Plant Molecular Biology. 1993;23:567–581. doi: 10.1007/BF00019304. [DOI] [PubMed] [Google Scholar]
- De Pater BS, van der Mark F, Rueb S, Katagiri F, Chua N-H, Schilperoort RA, Hensgens LAM. The promoter of the rice gene GOS2 is active in various different monocot tissues and binds rice nuclear factor ASF-1. The Plant Journal. 1992;2:837–844. doi: 10.1111/j.1365-313x.1992.00837.x. [DOI] [PubMed] [Google Scholar]
- Donald RGK, Cashmore AR. Mutation of either G box or I box sequences profoundly effects expression from the Arabidopsis rbcS-1A promoter. EMBO Journal. 1990;9:1717–1726. doi: 10.1002/j.1460-2075.1990.tb08295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JG, Llewellyn DJ, Walker JC, Dennis ES, Peacock WJ. The ocs element: a 16 base pair palindrome essential for activity of the octopine synthase enhancer. EMBO Journal. 1987;6:3203–3208. doi: 10.1002/j.1460-2075.1987.tb02636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang R-X, Nagy F, Sivasubramaniam S, Chua N- H. Multiple cis regulatory elements for maximal expression of the cauliflower mosaic virus 35S promoter in transgenic plants. The Plant Cell. 1989;1:141–150. doi: 10.1105/tpc.1.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbarino JE, Oosumi T, Belknap WR. Isolation of a polyubiquitin promoter and its expression in transgenic potato plants. Plant Physiology. 1995;109:1371–1378. doi: 10.1104/pp.109.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P, Raghuvanshi S, Tyagi AK. Assessment of the efficiency of various gene promoters via biolistics in leaf and regenerating seed callus of millets, Eleusine coracana and Echinochloa crus-galli. Plant Biotechnology. 2001;18:275–282. [Google Scholar]
- He C, Lin Z, McElroy D, Wu R. Identification of a rice Actin2 gene regulatory region for high-level expression of transgenes in monocots. Plant Biotechnology Journal. 2009;7:227–239. doi: 10.1111/j.1467-7652.2008.00393.x. [DOI] [PubMed] [Google Scholar]
- Hernandez-Garcia CM, Adriana P, Martinelli AP, Bouchard RA, Finer JJ. A soybean (Glycine max) polyubiquitin promoter gives strong constitutive expression in transgenic soybean. Plant Cell Reports. 2009;28:837–849. doi: 10.1007/s00299-009-0681-7. [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. The Plant Journal. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- Jang IC, Choi WB, Lee KH, Song SI, Nahm BH, Kim JK. High-level and ubiquitous expression of the rice cytochrome c gene OsCc1 and its promoter activity in transgenic plants provides a useful promoter for transgenesis of monocots. Plant Physiology. 2002;129:1473–1481. doi: 10.1104/pp.002261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J-S, Lee S, Jung K-H, Jun S-H, Kim C, An G. Tissue-preferential expression of a rice α-tubulin gene, OsTubA1, mediated by the first intron. Plant Physiology. 2000;123:1005–1014. doi: 10.1104/pp.123.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EH, Kim YS, Park SH, Koo YJ, Choi YD, Chung YY, Lee IJ, Kim JK. Methyl jasmonate reduces grain yield by mediating stress signals to alter spikelet development in rice. Plant Physiology. 2009;149:1751–1760. doi: 10.1104/pp.108.134684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawless C, Pearson RD, Selley JN, Smirnova JB, Grant CM, Ashe MP, Graham D, Pavitt GD, Hubbard SJ. Upstream sequence elements direct post-transcriptional regulation of gene expression under stress conditions in yeast. BMC Genomics. 2009 doi: 10.1186/1471-2164-10-7. doi:10.1186/1471-2164-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard PA, Kulaveerasingam H, York GM, Strong A, Sinskey AJ. Manipulating gene expression for the metabolic engineering of plants. Metabolic Engineering. 2002;4:67–79. doi: 10.1006/mben.2001.0210. [DOI] [PubMed] [Google Scholar]
- Lu J, Sivamani E, Li X, Qu R. Activity of the 5′ regulatory regions of the rice polyubiquitin rubi3 gene in transgenic rice plants as analysed by both GUS and GFP reporter genes. Plant Cell Reports. 2008;27:1587–1600. doi: 10.1007/s00299-008-0577-y. [DOI] [PubMed] [Google Scholar]
- McElroy D, Zhang W, Cao J, Wu R. Isolation of an efficient actin promoter for use in rice transformation. The Plant Cell. 1990;2:163–171. doi: 10.1105/tpc.2.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy D, Blowers AD, Jenes B, Wu R. Construction of expression vectors based on the rice actin1 (Act1) 5′ region for use in monocot transformation. Molecular and General Genetics. 1991;231:150–160. doi: 10.1007/BF00293832. [DOI] [PubMed] [Google Scholar]
- Mitra A, An G. Three distinct regulatory elements comprise the upstream promoter region of the nopaline synthase gene. Molecular and General Genetics. 1989;215:294–299. doi: 10.1007/BF00339731. [DOI] [PubMed] [Google Scholar]
- Murahige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- Ng DW-K, Chandrasekharan MB, Hall TC. The 5′-UTR negatively regulates quantitative and spatial expression from the ABI3 promoter. Plant Molecular Biology. 2004;54:25–38. doi: 10.1023/B:PLAN.0000028767.06820.34. [DOI] [PubMed] [Google Scholar]
- Odell JT, Nagy F, Chua NH. Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature. 1985;313:810–812. doi: 10.1038/313810a0. [DOI] [PubMed] [Google Scholar]
- Oh S-J, Jeong JS, Kim E-H, Yi NR, Yi S-I, Jang I-C, Kim YS, Suh S-C, Nahm BH, Kim J- K. Matrix attachment region from the chicken lysozyme locus reduces variability in transgene expression and confers copy number-dependence in transgenic rice plants. Plant Cell Reports. 2005;24:145–154. doi: 10.1007/s00299-005-0915-2. [DOI] [PubMed] [Google Scholar]
- Oh SJ, Kim SJ, Kim YS, Park SH, Ha SH, Kim JK. Arabidopsis cyclin D2 expressed in rice forms a functional cyclin-dependent kinase complex that enhances seedling growth. Plant Biotechnology Reports. 2008;2:227–231. [Google Scholar]
- Oh SJ, Kim YS, Kwon CW, Park HK, Jeong JS, Kim JK. Overexpression of the transcription factor AP37 in rice improves grain yield under drought conditions. Plant Physiology. 2009;150:1368–1379. doi: 10.1104/pp.109.137554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco BM, Ogren WL. Localization of light-inducible and tissue-specific regions of the spinach ribulose bisphosphate carboxylase/oxygenase (rubisco) activase promoter in transgenic tobacco plants. Plant Molecular Biology. 1993;23:1129–1138. doi: 10.1007/BF00042347. [DOI] [PubMed] [Google Scholar]
- Potenza C, Aleman L, Sengupta-Gopalan C. Targeting transgene expression in research, agricultural, and environmental applications: promoters used in plant transformation. In Vitro Cellular and Developmental Biology. 2004;40:1–22. [Google Scholar]
- Qu LQ, Xing YP, Liu WX, Xu XP, Song YR. Expression pattern and activity of six glutelin gene promoters in transgenic rice. Journal of Experimental Botany. 2008;59:2417–2424. doi: 10.1093/jxb/ern110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ririe KM, Rasmussen RP, Wittwer CT. Product differentitation by analysis of DNA melting curves during the polymerase chain reaction. Analytical Biochemistry. 1997;245:154–160. doi: 10.1006/abio.1996.9916. [DOI] [PubMed] [Google Scholar]
- Rockhold DR, Chang S, Nathan Taylor N, Allen PV, McCue KF, Belknap WR. Structure of two Solanum bulbocastanum polyubiquitin genes and expression of their promotersin transgenic potatoes. American Journal of Potato Research. 2008;85:219–226. [Google Scholar]
- Shashikanth M, Krishna AR, Ramya G, Devi G, Ulaganathan K. Genome-wide comparative analysis of Oryza sativa (japonica) and Arabidopsis thaliana 5′-UTR sequences for translational regulatory signals. Plant Biotechnology. 2008;25:553–563. [Google Scholar]
- Wang J, Oard JH. Rice ubiquitin promoters: deletion analysis and potential usefulness in plant transformation systems. Plant Cell Reports. 2003;22:129–134. doi: 10.1007/s00299-003-0657-y. [DOI] [PubMed] [Google Scholar]
- Weeks JT, Anderson OD, Blechl AE. Rapid production of multiple independent lines of fertile transgenic wheat (Triticum aestivum) Plant Physiology. 1993;102:1077–1084. doi: 10.1104/pp.102.4.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei HR, Wang ML, More PH, Albert HH. Comparative expression analysis of two sugarcane polyubiquitin promoters and flanking sequences in transgenic plants. Journal of Plant Physiology. 2003;160:1241–1251. doi: 10.1078/0176-1617-01086. [DOI] [PubMed] [Google Scholar]
- Wu C-Y, Washida H, Onodera Y, Harada K, Takaiwa F. Quantitative nature of the prolamin-box, ACGT and AACA motifs in a rice glutelin gene promoter: minimal cis-element requirements for endosperm-specific gene expression. The Plant Journal. 2000;23:415–421. doi: 10.1046/j.1365-313x.2000.00797.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.