Abstract
Kidney transplant recipients that develop signs of renal dysfunction or proteinuria one or more years after transplantation are at considerable risk for progression to renal failure. To assess the kidney at this time, a “for-cause” biopsy is performed, but this provides little indication as to which recipients will go on to organ failure. In an attempt to identify molecules that could provide this information, we used micorarrays to analyze gene expression in 105 for-cause biopsies taken between 1 and 31 years after transplantation. Using supervised principal components analysis, we derived a molecular classifier to predict graft loss. The genes associated with graft failure were related to tissue injury, epithelial dedifferentiation, matrix remodeling, and TGF-β effects and showed little overlap with rejection-associated genes. We assigned a prognostic molecular risk score to each patient, identifying those at high or low risk for graft loss. The molecular risk score was correlated with interstitial fibrosis, tubular atrophy, tubulitis, interstitial inflammation, proteinuria, and glomerular filtration rate. In multivariate analysis, molecular risk score, peritubular capillary basement membrane multilayering, arteriolar hyalinosis, and proteinuria were independent predictors of graft loss. In an independent validation set, the molecular risk score was the only predictor of graft loss. Thus, the molecular risk score reflects active injury and is superior to either scarring or function in predicting graft failure.
Introduction
Kidney transplants that develop dysfunction or proteinuria after one year following transplantation are at considerable risk for progression to renal failure (1). Certain histopathologic features, particularly interstitial fibrosis and tubular atrophy (IFTA), correlate with graft dysfunction, treatment response, and risk of progression to failure in transplants as well as in native kidneys (2–8). This has led to the belief that late failure of kidney transplants is due to progressive nonspecific scarring, possibly related to calcineurin inhibitor toxicity. However, IFTA is common in kidney transplants, reflecting the burden of injury including donor death, organ harvest, and the transplantation process, and mostly develops in the first year (9). Recent analyses (1, 10) indicate that the main cause of late graft loss is not unexplained scarring or calcineurin inhibitor toxicity but specific disease entities, particularly late antibody-mediated injury and recurrent disease.
Identifying the molecules associated with graft failure could potentially lead to interventions that would slow the progression of organ failure. Some of the individual molecules that predict risk of failure in native proteinuric kidney disease include VEGF and molecules associated with activation of intracellular hypoxia response (11). In kidney transplant biopsies, many molecules show altered expression related to rejection or injury (12–16). However, no comprehensive analysis of the relationship between the transcriptome and allograft survival has been performed.
The emergence of microarrays permits a genome-wide survey of the transcripts associated with future failure in renal allografts presenting with clinical indications for a biopsy, i.e., biopsies for cause (BFCs). The present study analyzed the relationship between gene expression in late BFCs in human kidney transplants and subsequent graft loss and assessed the predictive value of gene expression alone and in combination with histologic lesions and clinical variables. We evaluated the performance of these genes in an independent validation set and in a population of early biopsies that have a very low risk of subsequent graft failure.
Results
Patient demographics and graft survival in the BFC population
Because almost all failures occurred in patients who presented for a BFC after 1 year following transplantation (1), this group was selected for the analysis of risk prediction. The study population included 105 consecutive consenting patients who underwent BFCs between 1 and 31 years after transplantation (median, 57 months). Where more than 1 biopsy was available per patient, only the first biopsy was used for analyses. Median time to graft loss was 14 months, and the median follow-up after biopsy for patients without death or graft loss was 32 months.
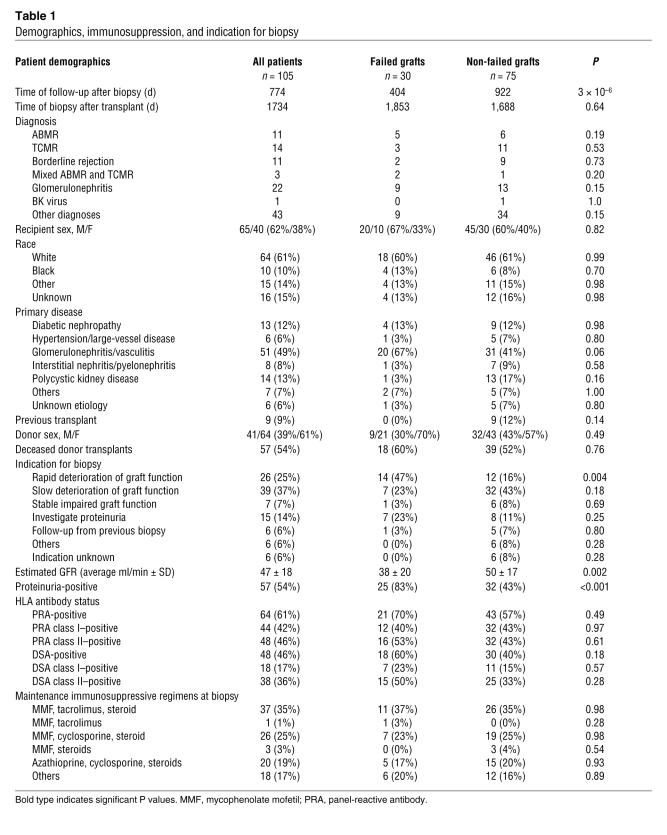
We observed 30 graft failures during follow-up, and 4 patients died with a functioning graft. Demographics and clinical characteristics of all patients are outlined in Table 1. Grafts that subsequently failed had higher incidence of proteinuria and rapid deterioration in function before biopsy and lower glomerular filtration rate (GFR) at time of biopsy. There were no differences in primary disease, time after transplantation, maintenance immunosuppression, or incidence of anti-HLA antibodies between grafts that subsequently failed after biopsy and those that did not. As previously reported, the main disease diagnoses in biopsies from grafts that failed were antibody-mediated rejection (ABMR) (either C4d-positive or C4d-negative) and glomerulonephritis (1).
Table 1 .
Demographics, immunosuppression, and indication for biopsy
Gene expression signature of future graft loss
To identify all genes associated with graft loss, we performed a Cox regression on the entire dataset of 105 late biopsies, using all probesets that passed the interquartile range (IQR) filter (n = 11,500) (see Methods). 886 genes represented by 1,312 probesets were significantly associated with graft failure (598 positively and 288 negatively) at the 0.0001 level (uncorrected P value) (Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI41789DS1).
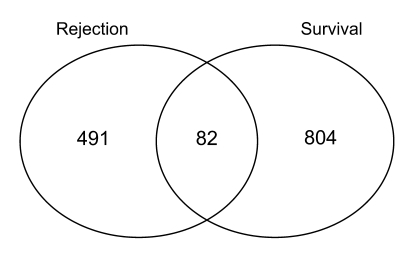
We compared the genes associated with subsequent graft loss with those associated with rejection at the time of biopsy in the same dataset. Rejection-associated genes had been identified in a previous analysis (14) using the BioConductor software package limma (17). Rejection-associated genes and genes associated with future graft loss were derived from the same biopsies. Of the 886 genes associated with graft loss, only 82 (9%) overlapped with the gene list associated with rejection at the same P value cutoff (Figure 1). The transcripts associated with future graft loss were primarily those that had been annotated as associated with tissue injury, matrix remodeling, and epithelial dedifferentiation (Table 2), while the rejection-associated genes had been annotated as reflecting inflammation, i.e., IFN-γ effects, infiltrating T cells, and macrophages (18).
Figure 1. Overlap of genes associated with graft loss and those associated with rejection.
We identified the genes associated with graft loss in a Cox regression model (P < 0.0001) and compared them with those genes whose association with rejection versus nonrejection was significant (t test) at P < 0.0001. The number of genes that were unique to each list as well as the number of overlapping genes are shown.
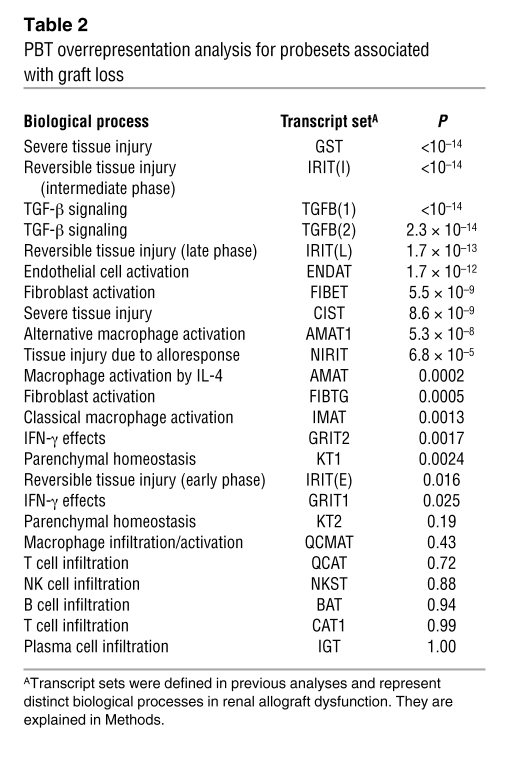
Table 2 .
PBT overrepresentation analysis for probesets associated with graft loss
Developing a gene expression risk score for predicting graft loss
Because we aimed to develop a biopsy-based risk prediction method, we built a gene-based classifier to predict graft loss. Classifier results were obtained using a multiple 10-fold cross-validation method (19). Details of the analysis are described in Methods. A diagram illustrating the process of building the classifier and validation steps is shown in Supplemental Figure 1.
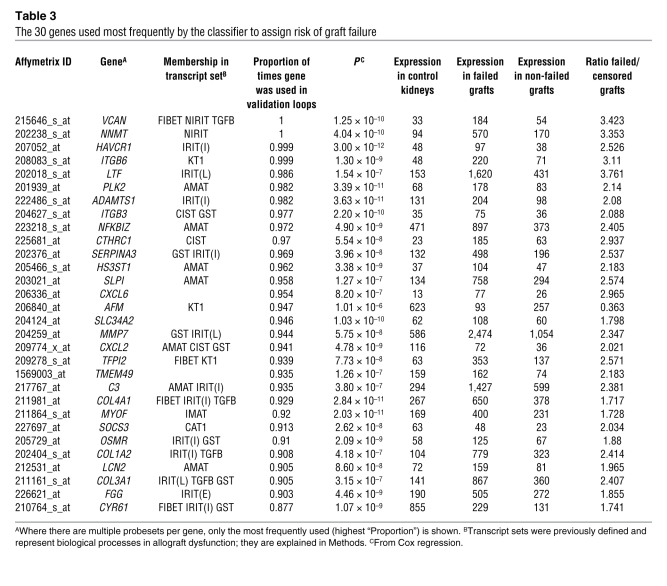
The classifier used 2,748 probesets on at least one occasion, and 117 of the probesets were used in at least 50% of the classifiers. The 30 annotated genes used most frequently by the classifier to assign the risk of graft loss are shown in Table 3, ordered by the proportion of times each gene was used in all the cross-validation/resampling loops. Twenty-seven of the top 30 genes (90%) had previously been annotated in experimental transplantation systems as members of pathogenesis-based transcript sets (PBTs) reflecting tissue injury and matrix remodelling (see Methods and Supplemental Table 2).
Table 3 .
The 30 genes used most frequently by the classifier to assign risk of graft failure
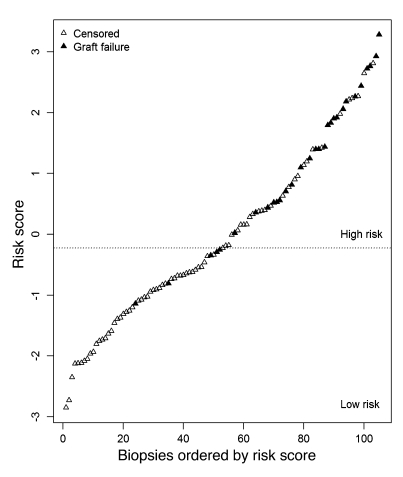
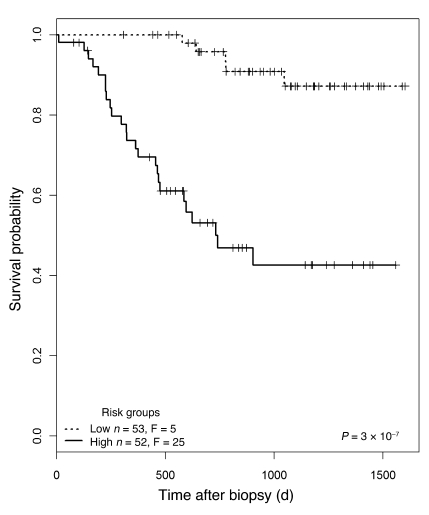
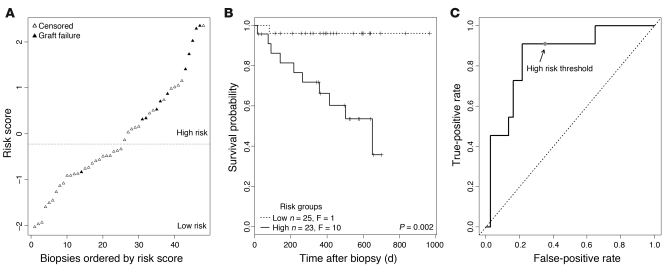
We used the molecular classifier to assign a prognostic molecular risk score to each biopsy using supervised principal components analysis (PCA) (20). Based on the average risk score across the test sets in the 100 validation loops, we assigned patients into either the high- or low-risk group (n = 52 high-risk, n = 53 low-risk patients) (Figure 2). The range of risk scores for each biopsy across the validation loops is shown in Supplemental Figure 2. The mean risk score in those kidney grafts surviving to 1 year after biopsy was –0.31 versus 1.85 in those grafts that failed by 1 year (t test, P = 9.3 × 10–8). In the high-risk group, 25 of 52 patients progressed to graft loss after biopsy, compared with only 5 losses in the low-risk group (log-rank test, P = 3 × 10–7). Kaplan-Meier survival curves for the 2 risk groups are shown in Figure 3.
Figure 2. Molecular risk scores in individual biopsies.
We built a molecular classifier based on 105 kidney transplant biopsies taken for clinical indications 1 year or more after transplantation and used the result to assign a molecular risk score to each biopsy. Biopsies were sorted by risk score; each biopsy is represented by 1 triangle. Biopsies from patients with subsequent graft loss are indicated in black. Biopsies were assigned into high- or low-risk groups, with the threshold determined by the median risk score.
Figure 3. Kaplan-Meier plots for the two risk groups.
Biopsies were separated into high- and low-risk groups, as shown in Figure 2. F, failed.
Relationship between risk score and clinico-pathological features
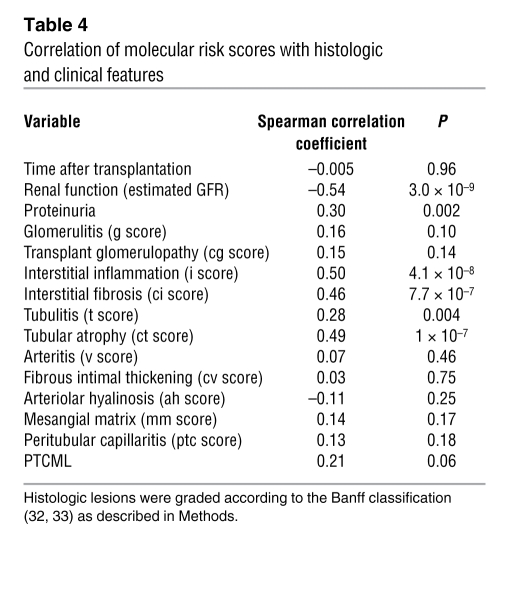
Table 4 shows the correlation between the molecular risk score and histologic and clinical variables. The risk score was negatively correlated with renal function (estimated GFR) at the time of biopsy and positively correlated with proteinuria, interstitial inflammation, atrophy, and fibrosis and less strongly with tubulitis. It was not correlated (P > 0.05) with time of the biopsy after transplantation, glomerular or arterial changes, or arteriolar hyalinosis.
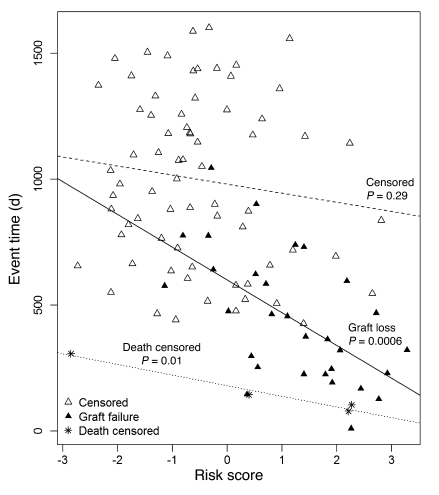
We examined the relationship between risk score and time to failure (Figure 4). Among the patients with graft loss, a higher risk score was associated with shorter time to failure (P = 0.0006). Among patients censored only because of end of study, there was no relationship between risk score and time to censoring. Among patients censored because of death, all 4 of whom died fewer than 400 days after biopsy, there was a significant correlation between risk score and time to death (P = 0.01). This may reflect the effect of a failing kidney transplant on the risk of death, but the small number of observations precludes firm conclusions. Thus, we included deaths with functioning grafts as censored data in the classifier and Cox regression analyses. We conclude that the risk score predicts not only whether failure will occur but also time to failure.
Figure 4. Relationship between risk score and failure/censoring time.
Time to event (graft failure, patient death, or end of follow-up) is plotted against the molecular risk score for each biopsy. Each biopsy is represented by one symbol. Biopsies from patients with subsequent graft loss are represented as black triangles; biopsies from patients who died with a functioning graft are represented by asterisks. All other biopsies are represented by white triangles. Regression lines were drawn separately for patients censored for end of study, those censored for patient death with a functioning graft, and those with graft loss.
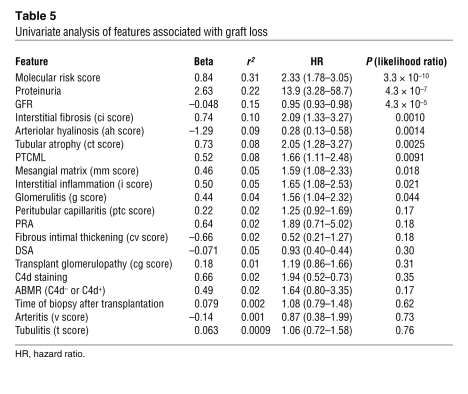
The association of molecular, clinical, and histologic features with graft loss was assessed in a univariate Cox regression analysis (Table 5). In this analysis, the features significantly associated with graft loss were molecular risk score, proteinuria, interstitial fibrosis, tubular atrophy, peritubular capillary basement membrane multilayering (PTCML), mesangial matrix score, interstitial inflammation, glomerulitis, low GFR, and absence of arteriolar hyalinosis. Many variables were not associated with outcome, including time of biopsy after transplantation, the presence of HLA antibody, C4d staining, transplant glomerulopathy, capillaritis, and arterial changes.
Table 5 .
Univariate analysis of features associated with graft loss
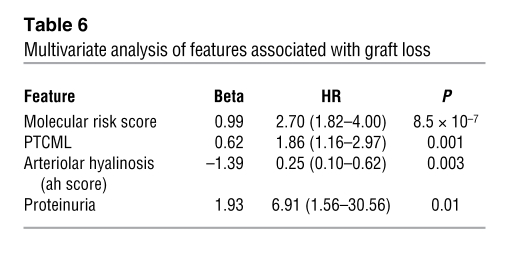
To assess the independent associations of these variables, we performed a forward stepwise multivariate regression (Table 6) of all features that reached significance (P < 0.05) in the univariate analysis. This analysis identified the independent predictors of graft loss as the molecular risk score, peritubular basement membrane multilayering, proteinuria, and arteriolar hyalinosis. Surprisingly, neither GFR nor IFTA was significantly associated with graft loss when the risk score was included in the multivariate analysis.
Table 6 .
Multivariate analysis of features associated with graft loss
Since the presence of proteinuria and GFR were known before the biopsy, we repeated the multivariate analysis excluding these factors to determine which information arising from the biopsy (histology, microarray results) predicted failure. When GFR and proteinuria were excluded, the independent risk factors were the molecular risk score, PTCML, and absence of arteriolar hyalinosis.
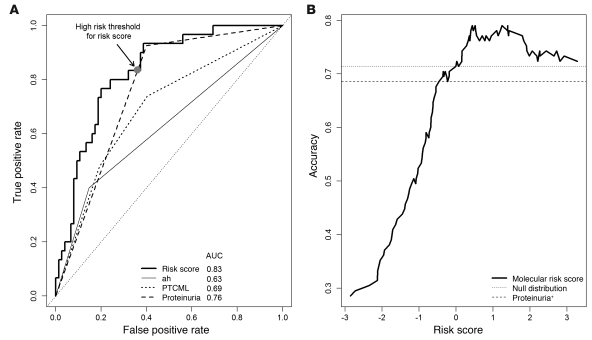
When the receiver operating characteristic (ROC) curves for each of the features independently associated with graft loss in the multivariate analysis were compared (Figure 5A), the molecular risk score showed a greater area under the curve (AUC = 0.83) than the clinical or histologic features (AUC = 0.63–0.76). When the P values for the differences in AUCs were calculated by permutation test, they were not significant: P = 0.17 for risk score versus proteinuria, P = 0.06 versus PTCML, and P = 0.002 versus arterial hyalinosis. However, the maximum accuracy obtainable using the risk score was significantly greater than with proteinuria (Figure 5B) (P = 0.001). In fact, using the presence of proteinuria as a threshold led to a lower accuracy (i.e., 0.69) than did the null distribution (accuracy, 0.71) (i.e., guessing that all samples survived). In contrast, all risk score thresholds greater than 0.0 produced higher accuracies than the null distribution.
Figure 5. ROC curves of molecular risk score compared with clinical and histologic features.
(A) To assess the sensitivity and specificity of the molecular classifier compared with that of clinical features, we compared the ROC curve derived from the molecular risk score with variables significantly associated with graft loss in a multivariate analysis. AUC is indicated for each parameter. (B) Accuracy (total correct predictions/total samples) for varying risk score thresholds for predicting graft failure. The dashed line represents the accuracy when the presence of proteinuria was used as the single threshold for predicting failure. The dotted line is the accuracy when all samples are predicted to survive. ah, arteriolar hyalinosis.
Robustness of the classifier results
Single gene classifiers.
We assessed the strength of predictions from simple single gene classifiers, based solely on the expression values of the highest ranked and the 100th, the 500th, and the 5,000th ranked genes in each training set (Supplemental Figure 3). Predictions from single genes, even those that were not near the top of the ranked list of genes, were very similar to those using the full PCA model. Thus, the risk for graft loss can be predicted adequately by any of a great many genes. This is due to the highly coordinated changes in expression found in many thousands of genes.
Application of the classifier to an independent validation set.
We assessed the performance of the classifier in an independent set of biopsies (n = 48) taken for clinical indication more than 1 year after transplantation at the University of Minnesota, with patient characteristics and clinical and histologic features similar to those in the studies described above. This population experienced 11 graft losses and had a median follow-up time of 406 days. Risk scores were calculated by supplying the classifier built from our main dataset with the gene expression values from the Minnesota dataset. The relationship of the molecular risk score to graft loss, the Kaplan-Meier plots of high- versus low-risk groups (defined using the risk threshold in the main dataset), and the ROC curve illustrating the tradeoff between sensitivity and specificity (Figure 6, A–C) showed results similar to those in the original data set. In the univariate analysis in this validation set, only the molecular risk score was significantly associated with subsequent graft loss (P = 0.0004). Thus, the risk score was superior to classic predictors such as IFTA and low GFR.
Figure 6. Performance of the classifier in an independent validation set.
Based on the classifier results derived from the 105 biopsies in our dataset, we calculated risk scores for each sample in an independent set of biopsies (n = 48) with similar clinical and histologic features. (A) Risk scores in relation to graft loss in the validation set, showing the risk threshold derived from the main (n = 105) dataset. (B) Kaplan-Meier plots of the 2 risk groups in the validation set, using the threshold shown in Figure 6A. (C) ROC curves illustrating sensitivity/specificity of the risk scores in the validation set.
Application of the classifier to a patient population at low risk of graft loss.
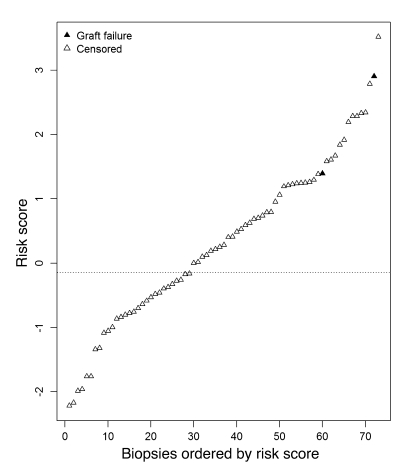
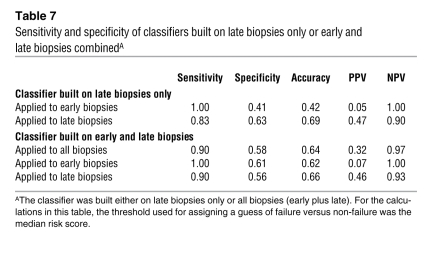
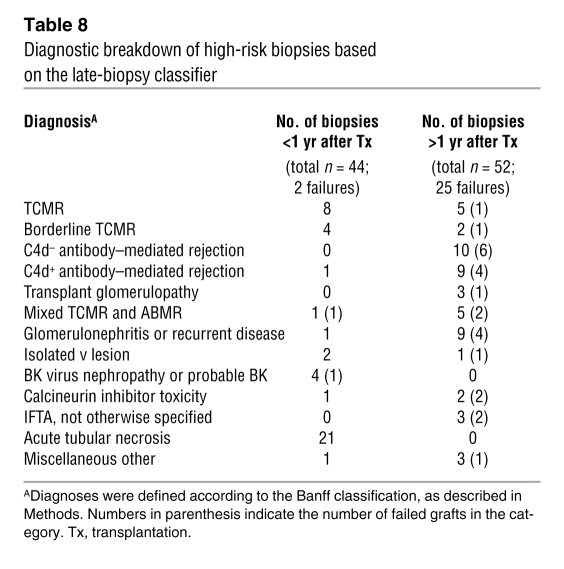
To assess whether high risk scores always predict risk for graft loss, we applied the classifier algorithm (based on late biopsies) to a set of biopsies taken within the first year after transplantation, which had very little risk of graft loss during follow-up in our study (Figure 7). Of 73 early biopsies (<1 year after transplantation), 44 (60%) had a risk score above the high-risk threshold from the late biopsy analysis (sensitivity, 1.00; specificity, 0.41; Table 7). However, only 2 of these grafts subsequently failed. Both of these were actually biopsied late in the first year, at 273 days and 360 days. Most of the remaining 42 early biopsies, which were classified as high risk but did not fail during the follow-up period, were diagnosed as T cell–mediated rejection (TCMR) (n = 8), borderline TCMR (n = 4), polyoma virus nephropathy (n = 3), or acute tubular necrosis (n = 21). When comparing early and late biopsies with high-risk scores, we found differences in diagnostic categories: early biopsies with high-risk scores (but low incidence of failure) had mainly acute tubular necrosis and TCMR, while late biopsies with high-risk scores (and high incidence of failure) were often diagnosed as having ABMR or glomerulonephritis (Table 8).
Figure 7. Molecular risk scores in biopsies taken within 1 year after transplantation.
The molecular risk score has high predictive value in patients at risk of graft loss. To assess whether the associated gene expression changes are unique to the population at risk, we applied the classifier results to a set of biopsies taken within the first year after transplantation.
Table 7 .
Sensitivity and specificity of classifiers built on late biopsies only or early and late biopsies combinedA
Table 8 .
Diagnostic breakdown of high-risk biopsies based on the late-biopsy classifier
To exclude that the high rate of false-positive predictions in the early biopsy group was due to a statistical bias (based on the fact that the classifier was built on late biopsies and applied to the clinically different group of early biopsies), we built a separate classifier on the entire dataset (early and late biopsies). In this analysis, the molecular risk score continued to have a high predictive value for graft loss, but the difference between early and late biopsies in terms of positive predictive value remained (Table 7).
Thus reversible/treatable early injury and rejection can induce the same gene expression changes as the disease processes leading to graft loss in the late biopsies, without leading to graft failure.
Discussion
The present study developed a gene expression–based classifier to predict kidney allograft failure after a late biopsy (>1 year after transplantation) for clinical indications. The molecular risk scores were strongly associated with graft loss. Using previously annotated transcript sets reflecting major biological events in renal transplants, we found that the transcripts used by the classifier to predict graft failure were those reflecting tissue injury, dedifferentiation of the epithelium, and tissue remodeling, but not those reflecting inflammation (IFNG effects and T cell or macrophage infiltration). The predictive ability of the classifier was high, with a sensitivity of 0.83 and a specificity of 0.63 at the high/low risk threshold. The gene expression risk score correlated with fibrosis and atrophy, interstitial inflammation, and glomerulitis but independently predicted graft failure in a multivariate analysis. The risk score was validated in an independent set of late BFCs.
The risk score is an indicator of active injury, whose significance depends on the disease state of the patient. Risk predictions were accurate when applied to the late biopsy population, where subsequent progression is considerable, but not when applied to early biopsies, where the diseases operating have a low risk of progression when treated. Early after transplantation the diseases causing injury are self-limited (acute tubular necrosis) or reversible by therapy (e.g., TCMR), but in late biopsies the diseases causing injury are progressive and unresponsive to treatment (ABMR, recurrence of primary disease). Thus, the probability of progression after a biopsy for clinical indications is conferred both by the presence of active injury (the risk score) and by the diseases inducing the active injury response and their potential for spontaneous resolution or response to therapy. The classifier and the individual genes predict risk not in absolute terms but in relation to the diseases or injury mechanisms disturbing the kidney and triggering the biopsy.
The transcripts predicting graft loss reflect an ongoing response to injury, including epithelial distress and dedifferentiation, with reexpression of developmental genes and loss of transcripts associated with differentiated epithelium as well as remodeling of the matrix. Many of the molecules used by the classifier to predict progression to renal failure are already known to be involved in responses to injury and play important roles in kidney development. For example, nicotinamide N-methyltransferase (NNMT) and versican (VCAN), both associated with cell migration in cancer (21–24), may represent dedifferentiation and epithelial-mesenchymal transition. ITGB6 is expressed by renal epithelium during stress and injury (25) and activates TGFB1 (26). HAVCR1 (also known as KIM1) is a well-established feature of the injured kidney (27). Interestingly, the collagen genes were not prominent in the list, suggesting that the genes predicting graft loss represent an active response to injury in the epithelium and matrix but not fibrogenesis per se. In previous analyses, we have performed extensive comparisons between microarray results and RT-PCR, which showed excellent reproducibility of both methods and confirmed the robustness of microarray results (28).
The genes associated with graft loss in late biopsies indicate that an active ongoing tissue response to injury is the final common pathway linking mechanisms of inflammation and noninflammatory disease states to parenchymal loss, dysfunction, and eventual kidney failure. The changes in the expression of these genes represent a stereotyped response of the tissue to injury, a structured program. Many transcripts were also found in mouse isografts, which allowed us to map the time-dependent intrinsic responses of the nephron to a clearly defined and self-limited stress, indicating that these genes represent inherently reversible injury (29, 30), if the injury mechanism is self limited or treatable. The gene list was similar to the transcript changes we recently found correlating with GFR disturbances in BFCs (31), which also reflect perturbations in the parenchyma and are indicators of an ongoing response to injury and thus potentially of a reversible epithelial injury repair process. This was further corroborated by observing high-risk scores in early biopsies from kidneys that did not progress to failure. Thus, the transcript changes reflect a perturbation in kidney biology that indicates ongoing remodeling and repair but not inevitable decline toward failure.
The risk score emerges as an independent predictor in multivariate analysis, indicating that transcriptome changes provide information beyond that derived from histology, demographics, GFR, or proteinuria. A molecularly based test has the potential to be more objective than a biopsy read by a pathologist, despite the standardization of the Banff criteria, because of intraobserver variation. In addition, the features of active injury are not captured by morphology but are reflected by gene expression changes. Thus, the risk score emerges as a more robust predictor of risk in the validation set than the classical factors: atrophy and fibrosis, low GFR, and proteinuria. The fact that not all grafts with a high-risk score failed, resulting in false positives, is not unexpected given the fact that patients were treated after the biopsy, resulting in reversal of the injury process in some cases. Since we did not include protocol biopsies performed after 1 year, we do not know whether there is a molecular signature of patients at high risk for graft failure in those without symptoms/signs of ongoing injury.
The superiority of the risk score to the classical features associated with progression to renal failure after a biopsy (atrophy, fibrosis, low GFR, proteinuria) indicates that these actually predict progression because they are correlated with an active injury response in biopsies for clinical indications. Atrophy, fibrosis, and low GFR indicate nephron loss, but the risk score subsumes this risk because it actually reflects nephrons in distress. This may offer hope that progression to renal failure is not inevitable if we can arrest the diseases causing progression. In other words, we find evidence here not of an overwhelming “point of no return” but of active injury that could be reversed if we find therapies that arrest the cause of the active injury.
Methods
Patient population, specimens, and data collection.
Written informed consent was obtained from all study patients. The study was approved by the institutional review boards of the University of Alberta (issue 5299), the University of Illinois, Chicago (protocol 2006-0544), the University of Minnesota (protocol HSC#0606 M87646), and the Hennepin County Medical Center (protocol HSR#06-2670). All consenting renal transplant patients undergoing BFCs as standard of care between September 2004 and October 2007 at the University of Alberta or between November 2006 and February 2007 at the University of Illinois were included in the analysis. In addition to our cross-validation analysis of the Edmonton dataset, biopsies obtained from Minnesota between September 2006 and September 2007 were used as an independent validation set.
Biopsies were obtained under ultrasound guidance by using spring-loaded needles (ASAP Automatic Biopsy, Microvasive). In addition to the cores required for standard histopathology, we collected one core for gene expression studies. The biopsy sample processing was performed as described in detail in our previous study (14).
Histopathologic diagnosis.
All biopsies were assessed using the updated Banff 07 criteria (32, 33) by a pathologist who was blinded to the results of molecular studies. All biopsies had adequate cortical tissue for analysis by Banff criteria, with the exception of 2 biopsies that had no arteries. Diagnostic groups included TCMR, borderline TCMR, ABMR, IFTA not otherwise specified, glomerulonephritis, acute tubular necrosis, and BK virus nephropathy. In addition to ABMR with positive C4d staining (as specified in the Banff classification), we included the emerging category of C4d-negative ABMR (published in ref. 1), which is defined by the presence of circulating donor-specific HLA antibodies and evidence of microcirculation changes in the biopsy (presence of glomerulitis, transplant glomerulopathy, peritubular capillaritis, PTCML).
PBTs.
PBTs represent biological processes during rejection and other types of injury in renal allografts. They serve as an annotation tool, with which genes identified in our analysis can be assigned to biological processes relevant to transplantation. PBTs were derived previously from mouse transplant models and in vitro human cell lines (15, 29, 30, 34–40). The definitions and algorithms for the PBTs referenced in the tables can be found in Supplemental Table 2. The PBTs included IFN-γ–inducible transcripts: GRIT1 and GRIT2 (34); cytotoxic T cell–associated transcripts: CAT1 (15) and QCAT (35); NK cell–associated transcripts: NKST (36); classical macrophage activation transcripts: IMAT (37); alternative macrophage activation transcripts: AMAT1 (38) and AMAT (IL-4–inducible transcripts); B cell–associated transcripts: BAT (39); immunoglobulin transcripts: IGT (39); endothelial transcripts: ENDAT (40); injury and repair transcripts: IRIT subsets IRIT(E), IRIT(I), and IRIT(L) (29); transcripts associated with severe tissue injury: GST and CISTS (38); kidney parenchymal transcripts: KT1 and KT2 (30); and transcript sets reflecting fibroblast activation (FIBET and FIBTG) or TGF-β activation (TGFB[1] and TGFB[2]).
Data analysis.
Because we aimed to develop a biopsy-based risk prediction method, graft survival was assessed as time between biopsy and graft failure/censoring, not time between transplantation and failure/censoring. Patients were censored for the end of study (July 26, 2009), death with functioning graft, or loss to follow-up. Graft failure was defined as return to dialysis (n = 30).
Microarray data files were preprocessed using robust multichip averaging (RMA) in Bioconductor and subjected to variance-based filtering (41) as described previously (34). Nonspecific IQR filtering was used to eliminate probesets with low variation across the dataset. 11,500 of the original 54,675 probesets passed this filtering step and were retained for further analysis. Expression and phenotype data, as well the CEL files, are available at the Gene Expression Omnibus database ( http://www.ncbi.nlm.nih.gov/geo/; accession number GSE21374).
Classifier results were obtained using a multiple 10-fold cross-validation method (19). In each iteration, the data were divided into 10 roughly equal-sized subsamples. Nine of the subsamples were used to predict the risk scores of the remaining “left-out” subsample. This procedure was repeated 10 times, each time using a different left-out subsample, so that all biopsies received a single predicted risk score. Within each of the 10 folds of this algorithm, the genes used in the classifier were reselected based only on those samples not being left out, i.e., only using the training set for that left-out subsample. The supervised PCA method uses a shrinkage algorithm (20, 42), ordering the selected genes by their Cox regression P values within each training set. The shrinkage procedure itself selects for genes whose expression is most stable within samples from the same phenotypic class (e.g., within failures and within non-failures). One of the desirable consequences of this method is that many genes with similar P values, in terms of their association with graft loss, can be eliminated in such a way as to retain those with the most informative predictive content. No fixed P value cutoff is used. In each training set, the shrinkage threshold (equal to the number of genes used) was chosen by maximizing the predictive accuracy in that particular training set. This entire procedure was repeated 100 times (each time using a different random 10-fold data split, following the method used in ref. 19), and each biopsy’s average risk score over all 100 iterations was recorded. Based on the average risk score, patients were assigned to one of 2 risk groups (high and low), using the median across all biopsies as the cutoff between both groups. In the case of multiple biopsies from one patient, only the first biopsy was used. A diagram illustrating the process of building the classifier and validation steps is shown in Supplemental Figure 1.
Single gene classifiers were assessed by the same cross-validation method. In each training set, the nth ranked gene (n = 1, n = 100, n = 500, or 5,000) was determined, and its expression value in the corresponding test set was used as the risk score. Since a different nth ranked gene might occur in every training/test set split, all expression values were first standardized by subtracting that gene’s mean expression value and dividing by the gene’s standard deviation.
The full classifier derived from the Edmonton dataset was also applied to the gene expression values of an independent validation set from Minnesota, and risk scores were assigned.
Univariate associations between gene expression and graft survival were assessed by Cox regression analysis (P value cutoff = 0.0001). Multivariate regressions using the molecular risk score, pathology lesions, and clinical variables were built using a forward stepwise model using all features that reached significance (P < 0.05) in the univariate analysis. False discovery rates were estimated using the R package fdrtool (43). Unless otherwise specified, a P value of 0.05 was considered statistically significant.
Supplementary Material
Table 4 .
Correlation of molecular risk scores with histologic and clinical features
Acknowledgments
Special thanks go to Zija Jacaj for help with collection of the clinical data and to Vido Ramassar and Anna Hutton for technical support. This research has been supported by funding and/or resources from Genome Canada, Genome Alberta, the University of Alberta, the University of Alberta Hospital Foundation, Roche Molecular Systems, Hoffmann–La Roche Canada Ltd., the Alberta Ministry of Advanced Education and Technology, the Roche Organ Transplant Research Foundation, the Kidney Foundation of Canada, and Astellas Canada. P. Halloran held a Canada Research Chair in Transplant Immunology until 2008 and currently holds the Muttart Chair in Clinical Immunology.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(6):1862–1872. doi:10.1172/JCI41789.
See the related Commentary beginning on page 1803.
References
- 1.Einecke G, et al. Antibody-mediated microcirculation injury is the major cause of late kidney transplant failure. Am J Transplant. 2009;9(11):2520–2531. doi: 10.1111/j.1600-6143.2009.02799.x. [DOI] [PubMed] [Google Scholar]
- 2.Gaber LW, et al. Correlation between Banff classification, acute renal rejection scores and reversal of rejection. Kidney Int. 1996;49(2):481–487. doi: 10.1038/ki.1996.68. [DOI] [PubMed] [Google Scholar]
- 3.Colvin RB, et al. Evaluation of pathologic criteria for acute renal allograft rejection: reproducibility, sensitivity, and clinical correlation. J Am Soc Nephrol. 1997;8(12):1930–1941. doi: 10.1681/ASN.V8121930. [DOI] [PubMed] [Google Scholar]
- 4.Haas M, Kraus ES, Samaniego-Picota M, Racusen LC, Ni W, Eustace JA. Acute renal allograft rejection with intimal arteritis: histologic predictors of response to therapy and graft survival. Kidney Int. 2002;61(4):1516–1526. doi: 10.1046/j.1523-1755.2002.00254.x. [DOI] [PubMed] [Google Scholar]
- 5.Agodoa L, et al. Assessment of structure and function in progressive renal disease. Kidney Int Suppl. 1997;63:S144–S150. [PubMed] [Google Scholar]
- 6.Bohle A, Grund KE, Mackensen S, Tolon M. Correlations between renal interstitium and level of serum creatinine. Morphometric investigations of biopsies in perimembranous glomerulonephritis. Virchows Arch A Pathol Anat Histol. 1977;373(1):15–22. doi: 10.1007/BF00432465. [DOI] [PubMed] [Google Scholar]
- 7.Bohle A, Christ H, Grund KE, Mackensen S. The role of the interstitium of the renal cortex in renal disease. Contrib Nephrol. 1979;16:109–114. doi: 10.1159/000402883. [DOI] [PubMed] [Google Scholar]
- 8.Schainuck LI, Striker GE, Cutler RE, Benditt EP. Structural-functional correlations in renal disease. II. The correlations. Hum Pathol. 1970;1(4):631–641. doi: 10.1016/S0046-8177(70)80061-2. [DOI] [PubMed] [Google Scholar]
- 9.Nankivell BJ, Borrows RJ, Fung CL, O’Connell PJ, Allen RD, Chapman JR. The natural history of chronic allograft nephropathy. N Engl J Med. 2003;349(24):2326–2333. doi: 10.1056/NEJMoa020009. [DOI] [PubMed] [Google Scholar]
- 10.El Zoghby ZM, et al. Identifying specific causes of kidney allograft loss. Am J Transplant. 2009;9(3):527–535. doi: 10.1111/j.1600-6143.2008.02519.x. [DOI] [PubMed] [Google Scholar]
- 11.Rudnicki M, et al. Hypoxia response and VEGF-A expression in human proximal tubular epithelial cells in stable and progressive renal disease. Lab Invest. 2009;89(3):337–346. doi: 10.1038/labinvest.2008.158. [DOI] [PubMed] [Google Scholar]
- 12.Sarwal M, et al. Molecular heterogeneity in acute renal allograft rejection identified by DNA microarray profiling. N Engl J Med. 2003;349(2):125–138. doi: 10.1056/NEJMoa035588. [DOI] [PubMed] [Google Scholar]
- 13.Flechner SM, et al. Kidney transplant rejection and tissue injury by gene profiling of biopsies and peripheral blood lymphocytes. Am J Transplant. 2004;4(9):1475–1489. doi: 10.1111/j.1600-6143.2004.00526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mueller TF, et al. Microarray analysis of rejection in human kidney transplants using pathogenesis-based transcript sets. Am J Transplant. 2007;7(12):2712–2722. doi: 10.1111/j.1600-6143.2007.02005.x. [DOI] [PubMed] [Google Scholar]
- 15.Einecke G, et al. Expression of CTL associated transcripts precedes the development of tubulitis in T-cell mediated kidney graft rejection. Am J Transplant. 2005;5(8):1827–1836. doi: 10.1111/j.1600-6143.2005.00974.x. [DOI] [PubMed] [Google Scholar]
- 16.Strehlau J, et al. Quantitative detection of immune activation transcripts as a diagnostic tool in kidney transplantation. Proc Natl Acad Sci U S A. 1997;94(2):695–700. doi: 10.1073/pnas.94.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Smythe GK. Limma: linear models for microarray data. In Gentlemen R, Carey V, Dudoit S, Irizarry R, Huber W, eds.Bioinformatics and Computational Biology Solutions using R and Bioconductor . New York, New York, USA: Springer Publishing; 2005:398–420. [Google Scholar]
- 18.Reeve J, et al. Diagnosing rejection in renal transplants: a comparison of molecular- and histopathology-based approaches. Am J Transplant. 2009;9(8):1802–1810. doi: 10.1111/j.1600-6143.2009.02694.x. [DOI] [PubMed] [Google Scholar]
- 19.Kim JH.Estimating classification error rate: Repeated cross-validation, repeated hold-out and bootstrap. Comput Stat Data Anal. 200953113735–3745. . 10.1016/j.csda.2009.04.009 [DOI] [Google Scholar]
- 20.Bair E, Tibshirani R. Semi-supervised methods to predict patient survival from gene expression data. PloS Biology. 2004;2(4):E108. doi: 10.1371/journal.pbio.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Igarashi P, Shao X, McNally BT, Hiesberger T. Roles of HNF-1beta in kidney development and congenital cystic diseases. Kidney Int. 2005;68(5):1944–1947. doi: 10.1111/j.1523-1755.2005.00625.x. [DOI] [PubMed] [Google Scholar]
- 22.Wight TN, Merrilees MJ. Proteoglycans in atherosclerosis and restenosis: key roles for versican. Circ Res. 2004;94(9):1158–1167. doi: 10.1161/01.RES.0000126921.29919.51. [DOI] [PubMed] [Google Scholar]
- 23.Kinsella MG, Bressler SL, Wight TN. The regulated synthesis of versican, decorin, and biglycan: extracellular matrix proteoglycans that influence cellular phenotype. Crit Rev Eukaryot Gene Expr. 2004;14(3):203–234. doi: 10.1615/critreveukaryotgeneexpr.v14.i3.40. [DOI] [PubMed] [Google Scholar]
- 24.Wu Y, Siadaty MS, Berens ME, Hampton GM, Theodorescu D. Overlapping gene expression profiles of cell migration and tumor invasion in human bladder cancer identify metallothionein 1E and nicotinamide N-methyltransferase as novel regulators of cell migration. Oncogene. 2008;27(52):6679–6689. doi: 10.1038/onc.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trevillian P, Paul H, Millar E, Hibberd A, Agrez MV. A(v)beta(6) integrin expression in diseased and transplanted kidneys. Kidney Int. 2004;66(4):1423–1433. doi: 10.1111/j.1523-1755.2004.00904.x. [DOI] [PubMed] [Google Scholar]
- 26.Wipff PJ, Hinz B. Integrins and the activation of latent transforming growth factor beta 1 - An intimate relationship. Eur J Cell Biol. 2008;87(8–9):601–615. doi: 10.1016/j.ejcb.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 27.Bonventre JV. Kidney Injury Molecule-1 (KIM-1): a specific and sensitive biomarker of kidney injury. Scand J Clin Lab Invest Suppl. 2008;241:78–83. doi: 10.1080/00365510802145059. [DOI] [PubMed] [Google Scholar]
- 28.Allanach K, et al. Comparing microarray versus RT-PCR assessment of renal allograft biopsies: similar performance despite different dynamic ranges. Am J Transplant. 2008;8(5):1006–1015. doi: 10.1111/j.1600-6143.2008.02199.x. [DOI] [PubMed] [Google Scholar]
- 29.Famulski KS, et al. Transcriptome analysis reveals heterogeneity in the injury response of kidney transplants. Am J Transplant. 2007;7(11):2483–2495. doi: 10.1111/j.1600-6143.2007.01980.x. [DOI] [PubMed] [Google Scholar]
- 30.Einecke G, Broderick G, Sis B, Halloran PF. Early loss of renal transcripts in kidney allografts: relationship to the development of histologic lesions and alloimmune effector mechanisms. Am J Transplant. 2007;7(5):1121–1130. doi: 10.1111/j.1600-6143.2007.01797.x. [DOI] [PubMed] [Google Scholar]
- 31.Bunnag S, et al. Molecular correlates of renal function in human kidney transplant biopsies. J Am Soc Nephrol. 2009;20(5):1149–1160. doi: 10.1681/ASN.2008080863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Racusen LC, et al. The Banff 97 working classification of renal allograft pathology. Kidney Int. 1999;55(2):713–723. doi: 10.1046/j.1523-1755.1999.00299.x. [DOI] [PubMed] [Google Scholar]
- 33.Solez K, et al. Banff 07 classification of renal allograft pathology: updates and future directions. Am J Transplant. 2008;8(4):753–760. doi: 10.1111/j.1600-6143.2008.02159.x. [DOI] [PubMed] [Google Scholar]
- 34.Famulski KS, et al. Changes in the transcriptome in allograft rejection: IFN-γ induced transcripts in mouse kidney allografts. Am J Transplant. 2006;6(6):1342–1354. doi: 10.1111/j.1600-6143.2006.01337.x. [DOI] [PubMed] [Google Scholar]
- 35.Hidalgo LG, et al. The transcriptome of human cytotoxic T cells: measuring the burden of CTL-associated transcripts in human kidney transplants. Am J Transplant. 2008;8(3):637–646. doi: 10.1111/j.1600-6143.2007.02129.x. [DOI] [PubMed] [Google Scholar]
- 36.Hidalgo LG, Einecke G, Allanach K, Halloran PF. The transcriptome of human cytotoxic T cells: similarities and disparities among allostimulated CD4(+) CTL, CD8(+) CTL and NK cells. Am J Transplant. 2008;8(3):627–636. doi: 10.1111/j.1600-6143.2007.02128.x. [DOI] [PubMed] [Google Scholar]
- 37.Einecke G, Mengel M, Hidalgo LG, Allanach K, Famulski KS, Halloran PF. The early course of renal allograft rejection: Defining the time when rejection begins. Am J Transplant. 2009;9(3):483–493. doi: 10.1111/j.1600-6143.2008.02546.x. [DOI] [PubMed] [Google Scholar]
- 38.Famulski KS, Sis B, Billesberger L, Halloran PF. Interferon-gamma and donor MHC class I control alternative macrophage activation and activin expression in rejecting kidney allografts: a shift in the Th1-Th2 paradigm. Am J Transplant. 2008;8(3):547–556. doi: 10.1111/j.1600-6143.2007.02118.x. [DOI] [PubMed] [Google Scholar]
- 39.Einecke G, et al. Expression of B cell and immunoglobulin transcripts is a feature of inflammation in late allografts. Am J Transplant. 2008;8(7):1434–1443. doi: 10.1111/j.1600-6143.2008.02232.x. [DOI] [PubMed] [Google Scholar]
- 40.Sis B, Jhangri G, Bunnag S, Allanach K, Kaplan B, Halloran PF. Endothelial gene expression in kidney transplants with alloantibody indicates antibody-mediated damage despite lack of C4d staining. Am J Transplant. 2009;9(10):2312–2323. doi: 10.1111/j.1600-6143.2009.02761.x. [DOI] [PubMed] [Google Scholar]
- 41. Scholtens D, von Heydebreck A. Analysis of differential gene expression studies. In: Gentleman R, Carey VJ, Huber W, Irizarray R, Dudoit S, eds.Statistics for Biology and Health: Bioinformatics and Computational Biology Solutions Using R and Bioconductor . New York, New York, USA: Springer Publishing; 2005:229–248. [Google Scholar]
- 42.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99(10):6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strimmer K. fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics. 2008;24(12):1461–1462. doi: 10.1093/bioinformatics/btn209. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.