Abstract
Although metastasis is the leading cause of cancer-related death, it is not clear why some patients with localized cancer develop metastatic disease after complete resection of their primary tumor. Such relapses have been attributed to tumor cells that disseminate early and remain dormant for prolonged periods of time; however, little is known about the control of these disseminated tumor cells. Here, we have used a spontaneous mouse model of melanoma to investigate tumor cell dissemination and immune control of metastatic outgrowth. Tumor cells were found to disseminate throughout the body early in development of the primary tumor, even before it became clinically detectable. The disseminated tumor cells remained dormant for varying periods of time depending on the tissue, resulting in staggered metastatic outgrowth. Dormancy in the lung was associated with reduced proliferation of the disseminated tumor cells relative to the primary tumor. This was mediated, at least in part, by cytostatic CD8+ T cells, since depletion of these cells resulted in faster outgrowth of visceral metastases. Our findings predict that immune responses favoring dormancy of disseminated tumor cells, which we propose to be the seed of subsequent macroscopic metastases, are essential for prolonging the survival of early stage cancer patients and suggest that therapeutic strategies designed to reinforce such immune responses may produce marked benefits in these patients.
Introduction
Metastatic disease is the major cause of death by cancer (1, 2). Metastasis is a complex multistage process that requires cancer cells within the primary tumor to invade the local tissue and enter the blood or lymphatic vessels. Tumor cells need to survive in the circulation and migrate across vessel walls in order to colonize new sites and grow to form secondary tumors (3). The traditional view has been that tumor cell dissemination occurs late in cancer development (4–6); however, this notion has recently been challenged. Several expression profiling studies (7–10) suggest that the propensity of cancer cells to metastasize is acquired relatively early during tumor progression (reviewed in ref. 11). In addition, examination of bone marrow from early stage cancer patients without overt metastases (reviewed in refs. 12 and 13) and tumor-bearing mice (14) revealed that disseminated tumor cells (DTCs) are present at much earlier time points than expected. We now need to understand the significance of these DTCs. Specifically, we must determine how early DTCs contribute to clinically relevant macrometastases and identify the mechanisms involved in the development, maintenance, and breakdown of dormancy.
Transplanted tumor models in rodents are often used to study metastasis, with most of our current knowledge of cancer cell dissemination being drawn from xenograft models. However, these models often fail to recapitulate the gradual process of tumorigenesis that is observed in humans, and, in the case of immunocompromised recipients, the role of the immune system is masked. Such models are also usually unable to address the issue of metastatic latency, the prolonged interval between cancer cell dissemination and metastatic outgrowth (3). These shortcomings mean that the contributions of DTCs to metastases and of the immune system to metastatic latency remain unclear. Importantly, knowledge of the precise kinetics of metastatic progression is required before we can identify the stages of metastasis that are most amenable to therapeutic intervention.
During the past decade, several studies have demonstrated a critical role for the immune system in preventing cancer initiation (15). However, its role in cancer progression remains unclear. The limited success of current cancer immunotherapy, especially of therapeutic cancer vaccines (16), may be largely attributed to our ignorance of the complex relationships between cancer cells and the immune system. We therefore decided to examine tumor cell dissemination and metastasis in RET.AAD mice, a recently characterized model of spontaneous melanoma (17). RET.AAD mice are transgenic for the human RET oncogene and the chimeric mouse/human MHC antigen AAD. In RET.AAD mice, the RET oncogene is expressed by melanocytes (18), driving enhanced melanogenesis and leading to melanosis and oncogenic transformation. The expression of AAD enables monitoring of CD8+ T cell responses directed against immunodominant epitopes previously identified in melanoma patients.
In the present study, we established that the RET.AAD mouse melanoma model is indeed a model of metastasis, and that tumor cell dissemination to distant organs occurs early during the development of the primary tumor. DTCs remained dormant for varying periods of time depending on the tissue, indicative of organ-specific regulation of metastatic outgrowth. In a previous study, we found that tumor-bearing RET.AAD mice develop a strong and broad anti-melanoma CD8+ T cell response and that depletion of CD8+ T cells reduces survival of these mice (17). This led us to speculate that CD8+ T cells may limit metastatic progression. We now demonstrate that maintenance of dormancy in visceral organs required CD8+ T cells and that depletion of these cells significantly accelerated visceral tumor outgrowth. These findings suggest that immune therapies aimed at prolonging DTC dormancy may be effective in preventing subsequent development of metastasis in early stage cancer patients.
Results
Kinetics of tumor development.
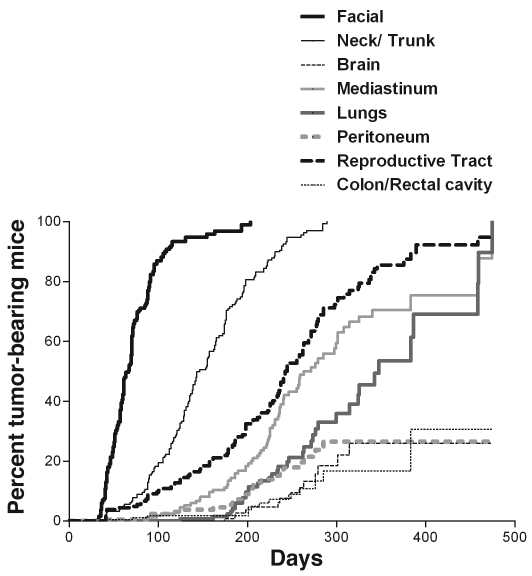
The RET.AAD model of melanoma has been previously described (17). RET.AAD mice, like RET mice, develop spontaneous melanoma, but at a much faster pace because of an increase in C57BL/6 genetic background (19). The melanomas are uveal, and the first lesion always affects one of the eyes, in the choroid or the ciliary body. This is rapidly followed by other facial tumors, including tumors in the second eye within 2 weeks. Tumors then spread to other sites on the trunk, limbs, and tail. When aged mice are necropsied, additional tumors are found on the internal surface of the skin, on the muscles, in the mediastinum, and in various internal organs, including the reproductive and gastrointestinal tracts, the lungs, and the brain. Figure 1 shows the kinetics of tumor development in 180 mice, as assessed by regular clinical examination and necropsy. The median time for onset of facial tumors was 66 days, whereas trunk tumors were detected around 80 days later. Visceral organs were colonized much later and with unique kinetics: the reproductive tract at a median age of 242 days, the mediastinum at a median age of 263 days, and the lungs at a median age of 347 days. The median age for tumor onset in the peritoneum, colorectal cavity, and brain was not reached when mice were 474 days old.
Figure 1. Unique kinetics of tumor onset in various tissues and organs.
The onset of superficial tumors was determined by biweekly clinical examination. Superficial tumors rarely affected mouse viability; therefore, mice bearing such tumors could be analyzed by necropsy at early and late time points for the presence of internal tumors. Kaplan-Meier curves were generated from the analysis of 180 untreated RET.AAD mice.
Most tumors share a common clonal origin.
Primary melanomas in humans have been reported not only in the skin, but also in many of the internal organs. Given that up to 30 tumors were observed in older RET.AAD mice, we examined whether these tumors represented independent transformation events or rather shared a common clonal origin. For this purpose, we performed genome-wide SNP (GW-SNP) profiling to compare somatic mutations detected in different tumors from the same mouse.
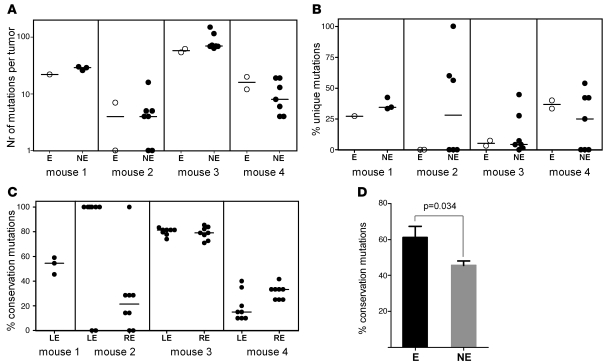
For each sample, the copy number and allelic frequency of 1,415 SNP probes were determined. Genetic alterations were identified by comparing the SNP genotypes of each tumor against the tail sample (germline sequence) from the same mouse. The profiles of different tumors were then compared to assess clonality. If all the tumors in 1 mouse were derived from a single primary lesion, there should be similar patterns of genetic alterations shared among them. On the contrary, if each tumor represented a new primary lesion, genetic alterations in a given tumor should be unrelated to the other lesions from the same mouse. We collected a total of 102 tumors and 5 tail samples from 5 mice aged 198–268 days. SNP profiles were successfully determined for 44 lesions (ranging 4–13 per mouse; Supplemental Table 2; supplemental material available online with this article; doi: 10.1172/JCI42002DS1) and 5 tail samples (to obtain the germline sequence for each mouse). Of the 1,415 probes examined, 274 were mutated in at least 1 tumor compared with the germline sequence, with each tumor displaying on average 23 mutations (range, 0–150; 95% CI, 13–33; Figure 2A).
Figure 2. Pattern of genetic alterations detected by GW-SNP profiling.
(A) Eye tumors (E) and non-eye tumors (NE) displayed similar numbers of total mutations. (B) Eye tumors and non-eye tumors displayed a similar percentage of unique mutations. (C) Percentage of eye mutations that were conserved in the other tumors. The reference tumor is either the left eye (LE) or the right eye (RE) tumor. (A–C) Horizontal bars denote medians. (D) Overall percentage of mutations in the reference tumor that were conserved in the other tumors from the same mouse (n = 4). Reference tumors were either eye or non-eye. Data represent mean ± SEM.
On average, only 22.4% of the total mutations were unique (Figure 2B), indicating that the vast majority of the mutations in 1 particular tumor (77.6%) were also detected in at least 1 other tumor (Figure 3C and Supplemental Figures 3 and 4). For example, in mouse 3, 33 mutations were shared by all 9 tumors, indicating a single clonal origin. Importantly, the vast majority of genetic alterations occurred at distant sites from coding regions (data not shown), which suggests that convergent evolution is unlikely to explain the genetic similarity among tumors from the same mouse.
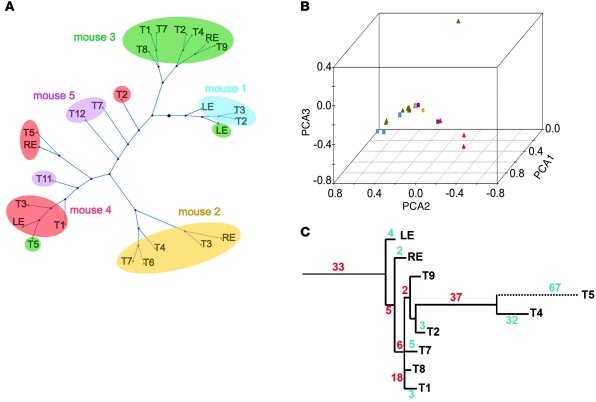
Figure 3. Tumors cluster according to mice.
Tumors (n = 26) taken from 5 mice were clustered according to their pattern of mutations detected by GW-SNP profiling (see Supplemental Figure 4). Distance matrix (A) or PCA (B) showed that tumors from the same mouse tended to cluster together. Colored symbols in B correspond with mice color-coded as in A. (C) Phylogram of the 9 tumors from mouse 3. The number of mutations conserved along each horizontal branch is indicated in red. The number of mutations unique to each tumor is shown in blue. The tissue origin of other tumors is given in Supplemental Table 2.
We further tested the similarity between tumors isolated from the same mouse by comparing all tumors pairwise, either within 1 mouse or between mice. For this purpose, only SNP positions for which all mice shared the same germline genotype were examined (Supplemental Figure 4). When paired tumors were derived from the same mouse, tumors shared more common mutations (on average, 29.6%; 95% CI, 25.3%–33.9%) than when tumors were from 2 different mice (4.8%; 95% CI, 3.6%–5.9%; P = 8 × 10–21; Supplemental Table 1). Tumors from different mice had few mutations in common. Distance matrix clustering of the SNP data showed that in 3 of 5 mice, the vast majority of tumors from the same mouse clustered together, further implying a common clonal relationship (Figure 3A). The same pattern was also observed using principal component analysis (PCA) of the SNP data (Figure 3B). An interesting case was mouse 4, which yielded 3 distinct clusters, suggesting that most of its tumors were derived from 3 independent transformation events.
The results of these GW-SNP analyses showed that tumors developing in a particular mouse were highly related and therefore derived from a small number of independent transformation events. As a consequence, most lesions (including cutaneous tumors) corresponded to metastases and not to independent primary melanomas.
Genetic evidence of early metastatic dissemination.
In cancer patients, the size of the primary tumor correlates with the risk of developing metastases (20), with bigger tumors leading to more metastases. Therefore, metastatic dissemination was usually considered a late event in cancer development (5). It has been difficult to assess whether this is really the case, particularly in humans, because of the scarcity of clinical samples from very early stage patients. We reevaluated this issue in the RET.AAD mouse model, in which the complete process of disease progression can be studied.
Given that the first detectable lesion affects the eyes of all RET.AAD mice, most metastases should be derived from eye tumors. If metastatic dissemination is indeed a late event, most mutations observed in the eye tumor should also be found in the subsequent metastases. These metastases may then accumulate additional mutations. Alternatively, metastases may derive from an early tumoral clone before the primary tumor reaches its full size. In this case, both the primary tumor and the metastases would continue to accumulate mutations independently, so that a far smaller fraction of the genetic alterations would be shared between the eye tumor and the metastases.
Using SNP data, we were able to measure these parameters in 4 mice. Our data indicated that, on average, the number of mutations in the eye tumors was similar to that in the other tumors of the same mouse (Figure 2A). The average 25 mutations detected in eye tumors (95% CI, 8–43) was not statistically different from the average 33 mutations in non-eye tumors (95% CI, 18–51; P = 0.51). Additionally, in the 4 mice analyzed, unique mutations were found in tumors from both the eyes and the other organs, albeit less frequently in eye tumors (eye, 15.9%; non-eye, 25.2%; P = 0.3; Figure 2B). These observations are not compatible with models in which metastases derive from late-disseminating tumor cells.
The average percentage of eye tumor mutations that were also detected at the other sites in the same mouse was 61.1% (Figure 2, C and D). In non-eye tumors, the percentage of mutations conserved in the other sites, including the eyes, was significantly lower (46.5%; P = 0.034), a finding consistent with the clinical observation that the eyes are the site of the first visible tumors. Importantly, a substantial 38.9% of mutations detected in the eye tumors were absent from the other tumors, again showing parallel accumulation of mutations in the eye tumors and metastases. Finally, phylogenic trees of all tumors from the same animals suggested early divergence (Figure 3C and Supplemental Figure 3). These observations can only be explained if metastatic dissemination occurs before the primary tumor reaches its maximal size.
In summary, the patterns of genetic alterations observed in RET.AAD mice were not consistent with the conventional idea of late metastatic dissemination. Rather, they showed that metastases derive from tumor cells that disseminate from the primary tumor at an early time point.
Dissemination of tumor cells begins during the first stages of growth of the primary tumor.
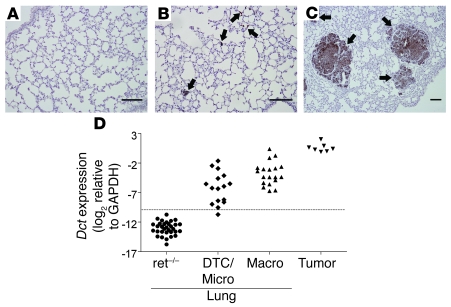
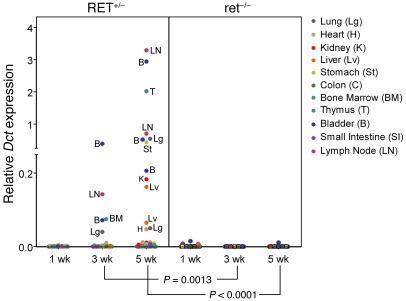
To confirm that tumor cell dissemination is an early event in disease progression, we used immunohistochemistry (IHC) and quantitative real-time PCR (qRT-PCR) to compare the visceral organs of RET.AAD mice with those of their nontransgenic littermates (Figure 4). S100B, a marker of melanocytes, was used to identify tumor cells by IHC. Individual DTCs and clusters of 10–100 S100B+ cells were already detectable in the lungs of 6- to 7-week-old RET.AAD mice (Figure 4, B and C), whereas no S100B+ cells were found in the lungs of 3-week-old RET.AAD mice or nontransgenic littermates (Figure 4A). Dopachrome tautomerase (DCT; also known as TRP-2) is a melanogenic enzyme present exclusively in normal melanocytes and melanomas. Dct gene expression in tumor cells was on average 30 times higher than RET, as assessed by qRT-PCR, which suggests the former is a more sensitive marker for DTCs. The use of Dct as a tumor cell marker was validated by correlating its expression in one lung with detection of S100B+ cells in the contralateral lung (Figure 4D). We then measured Dct and RET expression in tissues collected from young RET.AAD mice. As shown in Figure 5, Dct mRNA was already detectable in lymph nodes and visceral organs 3 weeks after birth, and expression increased further at 5 weeks of age. As determined by Dct expression, organs containing tumor cells included the lungs, heart, kidneys, liver, stomach, colon, bone marrow, thymus, and bladder. In most Dct-positive samples, RET expression was also detected (Supplemental Figure 1). Remarkably, no significant expression of Dct or RET was detected in the corresponding organs of nontransgenic littermates (Figure 5). Tumor dissemination to the brain could not be reliably assessed because of the normal presence of melanocytic cells, which would be expected to express Dct. The direct detection of tumor cells by IHC and qRT-PCR confirmed that tumor cell dissemination is an early event in RET.AAD mice, taking place as soon as 3 weeks after birth and affecting most organs and tissues.
Figure 4. Dct expression in lungs correlates with the presence of S100B+ cells.
Lungs of RET.AAD mice (n = 34) or ret–/–AAD littermates (n = 32) were analyzed for the presence of S100B+ cells by IHC. Dct expression was measured in the contralateral lung by qRT-PCR. (A–C) Representative examples of S100B staining of lungs from ret–/–AAD mice (A) or from RET.AAD mice, either with individual DTCs or micrometastases only (B) or with macroscopic nodules (C). Arrows indicate DTCs or nodules. Scale bars: 100 μm. (D) Dct expression was measured by qRT-PCR in lungs of RET.AAD mice either with individual DTCs and/or micrometastases (DTC/Micro) or with macrometastases (Macro). Nontransgenic littermates (ret–/–) were used as controls. Dct expression is shown as the log2 ratio of Dct over GAPDH. Dotted line represents mean expression in ret–/– lungs + 3 SD. Expression in excised macroscopic nodule (Tumor) is also shown for comparison.
Figure 5. Tumor cell dissemination to internal organs starts at 3 weeks of age.
Dct expression was measured by qRT-PCR in the lymph nodes, lungs, heart, kidney, liver, stomach, colon, bone marrow, thymus, bladder, and small intestine (n = 11 organs per mouse) collected from RET.AAD (RET+/–) mice aged 1 week (n = 3), 3 weeks (n = 3), and 5 weeks (n = 4). As controls, 11 age-matched nontransgenic littermates (ret–/–) were used.
Histopathological evidence of early tumor cell dissemination.
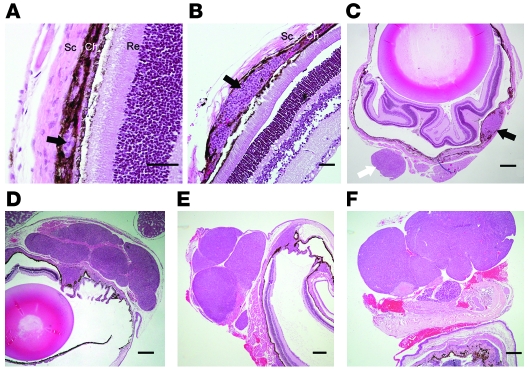
During the course of our studies, we found that in the RET.AAD model, primary eye lesions and local metastases were readily distinguishable by histopathologic examination. We next sought to confirm that the acquisition of a migratory phenotype by tumor cells is concomitant with the growth of the primary tumor, using H&E staining and IHC. At 2 weeks after birth, small hyperplastic lesions were detected in the choroid (Figure 6A) or the ciliary body (data not shown). At this age, hyperplastic cells were still confined within the choroid layer, and the Bruch’s membrane was intact. At 4 weeks of age, in addition to the hyperplastic lesions of the choroids (Figure 6B), small, round nodules were found on the outer surface of the sclera (Figure 6C). This sequence of events indicates that tumor cells had migrated from the primary lesion of the choroid to the sclera, which is normally devoid of melanocytes. Interestingly, these local metastases on the sclera developed without latency, further demonstrating that metastatic latency varies with the target organ. As tumor cells seem to acquire a migratory phenotype concomitantly with the growth of the primary tumor, one may expect this to affect the general morphology of the primary tumor at later time points. Indeed, as shown in Figure 6F, late eye tumors usually displayed a multinodular structure, strongly suggestive of individual nodules originating from tumor cells and migrating short distances. Interestingly, even eye tumors from younger 5- or 6-week-old mice displayed this multinodular structure (Figure 6, D and E), further supporting the notion that acquisition of the migratory phenotype by tumor cells is an early event, possibly even concomitant with the hyperplastic stage.
Figure 6. Morphology of eye tumors at various ages.
Eye sections from 2- (A), 4- (B and C), 5- (D), 6- (E), and 50-week-old (F) mice. (A–C) Black arrows indicate hyperplastic lesions within the choroid (Ch). The white arrow marks a round metastatic nodule attached to the sclera (Sc). Re, retina. (D–F) Note the multinodular morphology of the tumors. Images are representative of more than 20 analyzed eye tumors. Scale bars: 50 μm (A and B); 300 μm (C–F).
Dormancy of DTCs in the lungs.
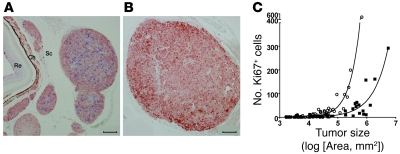
Whereas cancer cells disseminated early, development of overt metastases was delayed in visceral organs, with a median age of onset of 233 days (95% CI, 75–386). Such prolonged dormancy could result from lack of proliferation or from increased cell death. We therefore measured apoptosis and cell proliferation in local (sclera) and distant (lung) metastases. Very few apoptotic tumor cells were detected by TUNEL in lung tumors (data not shown). However, when eye or lung metastases of similar size were stained for the proliferation marker Ki-67, higher mitotic indices were observed in eye tumors (on average, 4.4% versus 1.9%; P = 0.0004; Figure 7 and Supplemental Figure 2). This demonstrates that tumor latency in the lung results from reduced proliferation of tumor cells.
Figure 7. Eye metastases have a higher mitotic index than do lung tumors of similar size.
Eye (A) and lung (B) sections were stained for tumor cells (S100B; red) and the proliferation marker Ki-67 (blue). Scale bars: 100 μm. (C) Number of proliferating tumor cells (S100B+Ki-67+) as a function of tumor size in eye (circles) and lung (squares) tumors. Data are from 12 eyes and 17 lungs from 22 mice, representing a total of 103 tumors.
CD8+ T cells control the growth of disseminated tumor cells in visceral organs.
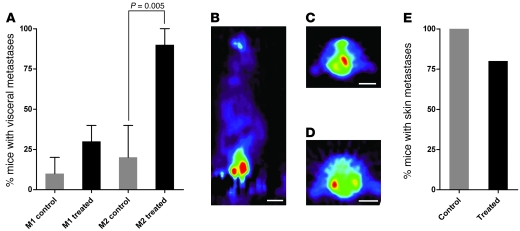
We previously showed that RET.AAD mice develop strong and broad melanoma-specific CD8+ T cell responses, which significantly prolong their lifespan (17). This led us to speculate that CD8+ T cells could be involved in DTC dormancy, thus preventing the development of overt metastases in visceral organs and limiting disease progression. To directly test this hypothesis, young tumor-bearing mice were depleted of CD8+ T cells by weekly injections of a CD8-specific antibody and the development of visceral metastases was monitored using [F18]fluorodeoxyglucose PET (18FDG-PET). 18FDG is a radiolabeled glucose analog that accumulates preferentially in metabolically active tumors. We focused on 2 common sites of metastases in RET.AAD mice, the lung and reproductive tract. Importantly, metastases within either of these organs can be readily detected by 18FDG-PET, which enabled us to determine tumor onset in vivo. The depletion started as soon as the first symptoms were detected in the eye, a time point at which tumor cell dissemination is likely to have already occurred. Flow cytometry analysis indicated a significant 82%–99% reduction in the absolute number of circulating CD8+ T cells throughout the entire experimental period, with minimal effect on monocytes, CD4+ T cells, or granulocytes (data not shown). As shown in Figure 8, CD8-depleted mice rapidly developed internal metastases in the lungs and/or reproductive tract. At 2 months after the initiation of CD8 depletion, visceral metastases were already detected in 9 of 10 depleted mice, whereas 2 control mice were positive (P < 0.005). In contrast, depletion of CD8+ T cells did not affect the onset of cutaneous metastases (Figure 8E).
Figure 8. CD8 depletion accelerates the development of visceral tumors, but has no effect on cutaneous tumors.
(A) Development of visceral metastases was followed by PET scan 1 month (M1) and 2 months (M2) after initiation of the depletion of CD8+ T cells (n = 10). Control mice (n = 9) were injected with rat IgG control. Data represent mean ± SEM. (B–D) Typical images of genital (B) or lung tumors (C and D). Scale bars: ~1 cm. (E) Skin tumors were detected at necropsy. Results are pooled from 2 independent experiments.
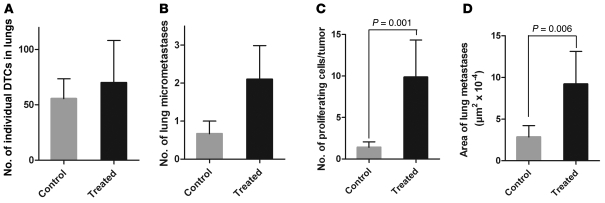
After necropsy, lungs of CD8-depleted and control mice were analyzed for the presence of DTCs and micrometastases. Consistent with our finding that tumor cell dissemination was an early event, no difference was observed in the number of individual DTCs (Figure 9A). However, we found a trend for more micrometastases in the lungs of CD8-depleted mice (Figure 9B). Moreover, the number of Ki-67+ cells per tumor and the size of lung metastases were significantly greater in CD8-depleted mice than in control mice (Figure 9, C and D). Collectively, these results clearly showed that CD8+ T cells play a crucial role in maintaining DTC dormancy in visceral organs.
Figure 9. CD8+ T cells control tumor cell proliferation.
Lungs of CD8-depleted (Treated) or control mice were analyzed by IHC for the presence of individual (A) or clustered (B) S100B+ cells. (C) The number of proliferating cells in each tumor was measured by 2-color IHC using Ki-67– and S100B-specific antibodies. (D) The area of lung tumors was measured using Image-Pro. Results are from 9 control and 11 treated mice in 2 independent experiments, representing a total of 81 and 64 tumors, respectively. Data represent mean ± SEM.
Discussion
The present study in RET.AAD mice demonstrated that metastases were derived from tumor cells that disseminated during the growth of the primary tumor, possibly even during the hyperplastic stage. Furthermore, DTCs in visceral organs remained dormant for prolonged periods of time, in a process requiring CD8+ T cells.
In humans, primary melanomas occur in the skin and other melanocyte-containing sites (eyes, meninges, anus, and vulva), or in sites where primordial melanocytes migrate (2, 21–28). It was therefore conceivable that multiple lesions within the same RET.AAD mouse correspond to independent transformation events. Using GW-SNP profiling, we found that the vast majority of genetic alterations in 1 tumor were shared by at least 1 other tumor from the same mouse. There are 2 possible explanations for this observation: first, that tumors share a common origin; second, that genetic similarity results from convergent evolution of independent tumors. In the latter hypothesis, tumors from 2 different mice should display the same average similarity as would different tumors from the same mouse. In contrast, we found that tumors from the same mouse were overwhelmingly more similar to each other than were those derived from different mice, demonstrating a common clonal origin. This conclusion is further supported by distance matrix clustering and PCA, which showed that tumors from the same mouse usually clustered together.
Until recently, it was widely accepted that metastatic potential was acquired late in a long series of genetic and epigenetic alterations required for full-fledged carcinogenesis (5). This tenet was based on the correlation between primary tumor size and risk of metastatic disease that underlies the TNM tumor staging system. However, recent work has cast doubt on this model (reviewed in ref. 29). Among our most important findings in RET.AAD mice were that tumor cell dissemination occurred early and that metastases derived from these early DTCs. In fact, DTCs were detected in most tissues by qRT-PCR 3 weeks after birth, and by IHC 6 weeks after birth, concurrent with the first clinical detection of the primary eye tumor. Although the results of our SNP analysis suggest these cells are likely to be the precursors of metastases, we cannot fully exclude the possibility that they represent dispersed melanocytic cells that become activated in the transgenic mice. Examination of the primary lesion revealed a multinodular morphology and the presence of small, locally metastatic nodules attached to the sclera at 4 weeks of age. This further implies that the migratory phenotype is acquired early during tumorigenesis.
A recent study in a mouse model of breast cancer also reported early tumor cell dissemination and provided evidence that DTCs can grow into metastases (14). It remains possible that metastases derive from late disseminating tumor cells. Importantly, however, our SNP data were not compatible with this scenario. If metastases were derived from the primary tumor at a late stage, all genetic alterations in the primary tumor would be conserved in the metastases, and additional mutations should only occur in the latter. In contrast, we found similar numbers of mutations in the eye (primary) and non-eye tumors, with only 61% of primary tumor mutations conserved in subsequent metastases. Moreover, tumor phylograms for individual mice showed early genetic divergence between tumors. These observations strongly suggest that early DTCs are indeed the precursors of metastases.
In humans, melanoma is one of the most frequent metastatic cancers of unknown primary origin, which likely reflects the propensity of melanoma cells to disseminate early from small, indolent primary lesions (30). Recently, circulating tumor cells (CTCs) were reported in a large proportion of uveal melanomas (31–34). CTC frequencies were low but independent of disease stage, consistent with early dissemination. Moreover, CTCs were detected in patients years after eye enucleation, which indicates that CTCs can remain dormant for prolonged periods of time. DTCs and CTCs have also been detected in patients with early stage cutaneous melanomas (35, 36) and epithelial cancers (37–41). Interestingly, tumor cell dissemination was even observed in stage 0 melanomas (36) and dysplastic lesions of the cervix (42). Therefore, there is extensive evidence to support the notion that cancer cell dissemination is an early event in human cancers.
Indirect evidence suggests that the immune system plays a role in prolonging melanoma dormancy. For example, donor-derived melanomas have developed in immunosuppressed graft recipients (43–45). Tumor infiltration by T cells, especially CD8+, correlates with survival in early stage cancer patients (46, 47). Moreover, CXCR3 expression by peripheral CD8+ T cells is associated with survival (48), whereas expression of the corresponding chemokines — CXCL9 and CXCL10 — by metastases is associated with CD8+ T cell recruitment (49). Importantly, expression of the T cell activation marker CD69 correlates positively with survival and negatively with metastasis (50). Conversely, T cell death, or decreased expression of TCRζ or the downstream proteins P56lck and ZAP-70, correlates with shorter survival (51). There is also clear evidence in uveal melanoma that patients with functional CD8+ T cells have a better prognostic, whereas absence of TCRζ expression correlates with a worse outcome (52).
The finding that tumor cells display a migratory phenotype early in tumorigenesis challenges the traditional view of a late acquisition of metastatic potential (11). What could be the selective advantages of this early migratory phenotype if, upon migration to distant organs, DTCs remain dormant for months or years? How is a migratory phenotype selected for if additional alterations, such as the capacity to evade the immune response, are required to thrive? Cancer cells compete with each other for space and nutrients (53). A plausible explanation for our observations is that tumor cells acquire a migratory phenotype to escape this competition. Activation of genes that promote such local invasion might also facilitate more distant dissemination. In support of this interpretation, breast ductal adenocarcinomas display a higher fractal dimension than does normal breast tissue (54, 55). In fact, primary breast tumors can be considered as conglomerates of many small, fast-growing nodules. Interestingly, pathological examination of RET.AAD tumors, including primary eye tumors, also revealed a multinodular structure, suggesting further similarities to human cancers. We propose that early acquisition of a migratory phenotype is selected in the context of the primary tumor because it facilitates its expansion. Primary tumors do not metastasize as a consequence of being large, but rather in order to become large (54).
Upon migration to a distant site, cancer cells are subjected to site-specific selective pressures. The permissiveness of the immune environment may vary widely in different tissues. Our data showed that tumor cells disseminated to various organs early, but developed into overt macrometastases with different kinetics, which implies that metastatic latency varies among organs. Indeed, metastatic eye tumors display a higher mitotic index than do lung nodules of similar size. It took less than 2 weeks for the primary eye tumor to metastasize to the contralateral eye, 2 months to metastasize to another cutaneous site, and more than 5 or 6 months to grow in visceral sites. Therefore, in our model, early disseminating tumor cells were not particularly “immature” or intrinsically unable to develop into macrometastases, as discussed for human breast cancer (12). Rather, prolonged DTC latency in visceral organs reflects the need for additional site-specific adaptation. Interestingly, unlike human uveal melanoma, in which the liver is the major site of metastasis, liver metastases in RET.AAD mice were rarely observed, despite the presence of early DTCs in this organ. This provided further evidence for organ-specific control of DTCs. Our results clearly showed that one major barrier that DTC must overcome is of immune origin, since CD8+ T cell depletion accelerated the growth of visceral tumors. Besides having well-known cytotoxic activity, CD8+ T cells can produce several antiproliferative cytokines, including IFN-γ, TNF-α, and lymphotoxin. The best example is IFN-γ (56), which reduces tumor cell proliferation directly via STAT1-mediated activation of cell cycle inhibitors, such as p21WAF1/CIP1 and p27Kip1 (57), and indirectly by inhibiting angiogenesis or activating antitumor immunity (reviewed in ref. 58). Th1 cells also produce cytokines that inhibit tumor proliferation (59). In a mouse model of chemical carcinogenesis, both CD4+ and CD8+ T cells controlled late metastatic outgrowth (60). Further studies are necessary to investigate the precise mechanisms by which CD8+ T cells prevent metastatic progression in RET.AAD mice.
From a therapeutic perspective, preventing tumor cell dissemination is probably not realistic if, as shown here, metastases derive from early DTCs, sometimes even before the primary tumor is clinically detectable. Indeed, whether extensive lymphadenectomy improves breast cancer patient survival is still debated (61–63). If DTCs are dormant, standard chemotherapy, which targets rapidly dividing cells, is unlikely to eliminate these cells (64). Relevant preclinical models, such as RET.AAD, which recapitulate early tumor cell dissemination and metastatic latency, are invaluable tools for identifying mechanisms that may control metastatic outgrowth in cancer patients. Although the concept of cancer immunosurveillance has received considerable experimental support, cancer immune therapies remain disappointing (65). The present study uncovers what we believe to be a novel facet of immunosurveillance by showing that CD8+ T cells controlled the metastatic outgrowth in visceral organs. Monitoring the quality of T cell responses in cancer patients with minimal residual disease may help predict disease progression, and reinforcing T cell–mediated control of DTCs could become a realistic objective of cancer immunotherapy. A better understanding of the role of the immune system in metastatic dormancy may lead to new immunotherapeutic approaches that convert aggressive cancers into stable chronic conditions.
Methods
Mice.
Animal care and experimental procedures were approved by the IACUC of the Biological Resource Center and of Singhealth. The generation of RET.AAD mice was previously described (17). In this particular transgenic line (304/B6 mice), RET expression is driven by the metallothionein gene promoter and is expressed by melanocytes (66).
SNP genotyping.
DNA was extracted from tumor and tail samples collected from 5 RET.AAD tumor-bearing mice using the Qiagen DNeasy kit and RNAse A (Qiagen) treatment. Genotyping was performed by Origen Labs using the Affymetrix GeneChip Mouse Mapping 5K SNP Kit according to the manufacturer’s protocol.
SNP array data normalization and genotyping.
Raw signal intensities were normalized and genotyped using the Affymetrix GeneChip Targeted Genotype System 1.5. Normalization consisted of background subtraction, spectral overlap correction, and allele balancing, and genotyping was based on an expectation-maximization clustering algorithm of the transformed probe allele signals, as described previously (67). We selected the 1,415 probes with genotype calls for tail samples that correspond to known SNP alleles for BALB/c and C57BL/6. Known SNP alleles were obtained from dbSNP Build 128.
Mutation classification.
Mutations were classified by comparing the genotype of the tumors to that of the tail (germline). To identify regions of copy number variation, the ratio of signal sum of SNP probe alleles of the tumor samples to the tail sample was calculated. Intensity ratios of all SNP probes and the corresponding chromosome number and position were used for DNA copy number analysis by the circular binary segmentation method (68, 69). Prior to segmentation into regions, intensity ratios were smoothed. Based on the mean intensity ratio of each copy number segment, mutation classes of SNPs in the tumors were then classified as copy number amplifications, copy number reductions, or no copy number variation.
Clustering by distance matrix, phylograms, and PCA.
Only SNPs for which all 5 mice displayed identical genotypes were used for this analysis. The distance between any 2 tumors was determined based on a weighted pairwise score with a modification of the weighting factor (70). Scores were calculated for SNP array–determined genetic alterations by summing scores for genotype mutations and copy number variation at a particular SNP. The score was negative for different genetic alterations and positive for similar genetic alterations. In contrast to Waldman (70), the weight of the score was calculated based on the log probability for not having the observed mutation(s) at that SNP for all analyzed tumors. The distance matrix for all tumors was clustered based on the Fitch and Margoliash method implemented by Phylodraw (71, 72). For clustering by PCA, mutation class was converted to a numerical matrix to which singular value decomposition was applied.
In vivo CD8+ T cell depletion.
CD8+ T cells were depleted using rat anti-mouse CD8 depleting antibody (ATCC TIB-210; provided by L. Renia, Singapore Immunology Network, A-STAR). Mice were administered intraperitoneally 0.25 mg of anti-CD8 antibody or control rat IgG (Sigma-Aldrich) at 6 weeks of age, and then 0.1 mg antibody weekly for 7 weeks. Efficiency of CD8+ T cell depletion was monitored by flow cytometry analysis of circulating blood. Mice were clinically assessed once per week for palpable tumors and once per month for visceral metastasis by 18FDG-PET scan (see below). On the day of sacrifice, mice were examined for superficial and internal macroscopic tumors. Half of the lungs and reproductive tract were analyzed by IHC, and half were used for qRT-PCR.
IHC.
Formalin-fixed paraffin-embedded sections (5 μm) were immunostained for S100B to identify melanocytic cells. Briefly, sections were heat-treated with Target Retrieval Solution (Dako S1699), blocked with 3% (v/v) hydrogen peroxide in methanol for 30 minutes and 10% (v/v) normal goat serum (Dako) in PBS for 2 hours, and then incubated overnight at 4°C with rabbit polyclonal anti-S100B (Dako Z0331; diluted 1:4,000 in PBS with 10% [v/v] normal goat serum). Anti-S100B binding was revealed with Envision anti-rabbit HRP (Dako K4003) and Sigma-Aldrich Fast DAB with metal enhancer tablet. Nuclear Fast Red Solution (Sigma-Aldrich) was used as a counterstain. To assess proliferation, sections were incubated with rabbit polyclonal anti-S100B and rat anti–Ki-67 mAb (Abcam clone TEC-3; diluted 1:25). Anti–Ki-67 binding was revealed using biotinylated donkey polyclonal anti-rat (Jackson Lab; AffiniPure F(ab′)2 Fragment; diluted 1:300), alkaline phosphatase–conjugated streptavidin (Rockland Inc.; diluted 1:2,000) and Alkaline Phosphatase Substrate Kit III (Vector Laboratories). Anti-S100B binding was revealed using Envision anti-rabbit HRP (Dako K4003) and AEC peroxidase substrate (Vector Laboratories). At least 1 section per sample was stained with H&E. Numbers of DTCs (single S100B+ cells) or metastases (>1 DTC) were counted in 5–10 random fields of view by 2 independent observers. Areas of metastases were calculated using ImagePro Analyzer 6.2 software (Media Cybernetics Inc.).
Tumor cell detection by qRT-PCR.
Tissue and organ samples were collected in RNAlater (Qiagen) and homogenized in TRIzol (Qiagen). RNA was extracted using the Qiagen RNeasy 96-well Universal Tissue Kit. cDNA was reverse transcribed (Roche Applied Biosystems reagents) from 2,000 ng RNA. The presence of melanocytic tumor cells in tissues and organs was determined by qRT-PCR with SYBR green (Bio-rad iTaq SYBR Green Supermix with ROX) and specific primers against the melanocyte-specific gene Dct (primers 5′-CTTCCTGAATGGGACCAATG-3′ and 5′-ACGGCGTAATTGTAGCCAAG-3′). Dct expression was normalized against GAPDH expression (primers 5′-TGCGACTTCAACAGCAACTC-3′ and 5′-ATGTAGGCCATGAGGTCCAC-3′). Real-time PCR cycling conditions were as follows: 95°C for 10 minutes; 40 cycles of 95°C for 30 seconds, 55°C for 1 minute, and 72°C for 30 seconds; 95°C for 1 minute; 55°C for 30 seconds; and 95°C for 30 seconds.
18FDG-PET.
Prior to micro-PET imaging, mice were fasted overnight, with water available, until imaging was completed. On the day of imaging, mice were prewarmed to 37°C before an approximate 5.5-MBq dose of 18FDG (0.6 mM; obtained from the Department of Nuclear Medicine of Singapore General Hospital) was administered intraperitoneally. Body temperatures were maintained at 37°C throughout the uptake period. Micro-PET imaging was performed with a R4 microPET scanner (Concordes Microsystems Inc.) 1 hour after 18FDG injections. The reconstructed resolution was 1.8 mm full width at half maximum in the center of the field of view and 3 mm at 4 cm radial offset. Each mouse was subjected to a 15-minute acquisition. During imaging, mice were placed in an imaging chamber and maintained under 2% (v/v) isoflurane anesthesia in oxygen. For image reconstruction, an energy window of 350–700 keV and a coincidence timing window of 6 ns were used. We obtained 2-dimensional histograms by Fourier rebinning, and image reconstruction by filtered backprojection was used. Image data were corrected for nonuniformity of the scanner response, dead time count losses, and physical decay to the time of injection. No correction was applied for attenuation, scatter, or partial-volume averaging. For quantitative image analysis, regions of interest (ROIs) were manually drawn over tumors. Counting rates in the reconstructed images were converted to standardized uptake values (SUVs) by use of a system calibration factor derived from the imaging of a mouse-sized water-equivalent phantom containing 18F.
Statistics.
For statistical analysis, we used 2-tailed, unpaired t test for comparing mutation number, unicity, or conservation and Mann-Whitney test for Dct expression in the various organs from RET.AAD or ret–/–AAD littermates. Development of metastases after CD8+ T cell depletion was assessed by Fisher exact test. Prism (GraphPad Inc.) and Excel (Microsoft) softwares were used for calculations and graphing. P values less than 0.05 were considered significant.
Supplementary Material
Acknowledgments
The authors thank Vida Ong for help in mouse husbandry and necropsy, Yvonne de Kozak and Jerry Ward for useful comments on the pathological data, Laurent Renia for the gift of the hybridoma TIB-210, and Masafumi Inoue for help in its production. J.-P. Abastado also thanks Lucy Robinson, Alessandra Nardin, Jean-Paul Thiery, and Philippe Kourilsky for critical reading of the manuscript.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(6):2030–2039. doi:10.1172/JCI42002.
See the related Commentary beginning on page 1800.
References
- 1.Jemal A, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58(2):71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2. Pazdur R, Wagman L, Camphausen K, Hoskins W.Cancer management: a multidisciplinary approach. Huntington, New York: PRR; 2008. [Google Scholar]
- 3.Nguyen DX, Bos PD, Massague J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9(4):274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 4.Fidler IJ, Kripke ML. Metastasis results from preexisting variant cells within a malignant tumor. Science. 1977;197(4306):893–895. doi: 10.1126/science.887927. [DOI] [PubMed] [Google Scholar]
- 5.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61(5):759–767. doi: 10.1016/0092-8674(90)90186-I. [DOI] [PubMed] [Google Scholar]
- 6.Fidler IJ, Hart IR. Biological diversity in metastatic neoplasms: origins and implications. Science. 1982;217(4564):998–1003. doi: 10.1126/science.7112116. [DOI] [PubMed] [Google Scholar]
- 7.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33(1):49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 8.van‘t Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 9.Sotiriou C, Piccart MJ. Taking gene-expression profiling to the clinic: when will molecular signatures become relevant to patient care? Nat Rev Cancer. 2007;7(7):545–553. doi: 10.1038/nrc2173. [DOI] [PubMed] [Google Scholar]
- 10.Desmedt C, Ruiz-Garcia E, Andre F. Gene expression predictors in breast cancer: current status, limitations and perspectives. Eur J Cancer. 2008;44(18):2714–2720. doi: 10.1016/j.ejca.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 11.Bernards R, Weinberg RA. A progression puzzle. Nature. 2002;418(6900):823. doi: 10.1038/418823a. [DOI] [PubMed] [Google Scholar]
- 12.Pantel K, Brakenhoff RH. Dissecting the metastatic cascade. Nat Rev Cancer. 2004;4(6):448–456. doi: 10.1038/nrc1370. [DOI] [PubMed] [Google Scholar]
- 13.Wikman H, Vessella R, Pantel K. Cancer micrometastasis and tumour dormancy. Apmis. 2008;116(7–8):754–770. doi: 10.1111/j.1600-0463.2008.01033.x. [DOI] [PubMed] [Google Scholar]
- 14.Husemann Y, et al. Systemic spread is an early step in breast cancer. Cancer Cell. 2008;13(1):58–68. doi: 10.1016/j.ccr.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 15.Dunn GP, Old LJ, Schreiber RD. The three Es of cancer immunoediting. Annu Rev Immunol. 2004;22:329–360. doi: 10.1146/annurev.immunol.22.012703.104803. [DOI] [PubMed] [Google Scholar]
- 16.Rosenberg SA, Yang JC, Restifo NP. Cancer immunotherapy: moving beyond current vaccines. Nat Med. 2004;10(9):909–915. doi: 10.1038/nm1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lengagne R, et al. Distinct role for CD8 T cells toward cutaneous tumors and visceral metastases. J Immunol. 2008;180(1):130–137. doi: 10.4049/jimmunol.180.1.130. [DOI] [PubMed] [Google Scholar]
- 18.Iwamoto T, et al. Aberrant melanogenesis and melanocytic tumour development in transgenic mice that carry a metallothionein/ret fusion gene. EMBO J. 1991;10(11):3167–3175. doi: 10.1002/j.1460-2075.1991.tb04878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dragani TA, et al. Mapping of melanoma modifier loci in RET transgenic mice. Jpn J Cancer Res. 2000;91(11):1142–1147. doi: 10.1111/j.1349-7006.2000.tb00897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minn AJ, et al. Lung metastasis genes couple breast tumor size and metastatic spread. Proc Natl Acad Sci U S A. 2007;104(16):6740–6745. doi: 10.1073/pnas.0701138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.North JH, Pack MS. Malignant tumors of the small intestine: a review of 144 cases. Am Surg. 2000;66(1):46–51. [PubMed] [Google Scholar]
- 22.Macak J. Melanoma of the stomach: reality or fiction? Pathologica. 1998;90(4):388–390. [PubMed] [Google Scholar]
- 23.Chalkiadakis G, Wihlm JM, Morand G, Weill-Bousson M, Witz JP. Primary malignant melanoma of the esophagus. Ann Thorac Surg. 1985;39(5):472–475. doi: 10.1016/s0003-4975(10)61963-7. [DOI] [PubMed] [Google Scholar]
- 24.Christova S, Meinhard K, Mihailov I, Alexiev B. Three cases of primary malignant melanoma of the alimentary tract. Gen Diagn Pathol. 1996;142(1):63–67. [PubMed] [Google Scholar]
- 25.Sachs DL, Lowe L, Chang AE, Carson E, Johnson TM. Do primary small intestinal melanomas exist? Report of a case. J Am Acad Dermatol. 1999;41(6):1042–1044. doi: 10.1016/s0190-9622(99)70273-2. [DOI] [PubMed] [Google Scholar]
- 26.Kadivar TF, Vanek VW, Krishnan EU. Primary malignant melanoma of the small bowel: a case study. Am Surg. 1992;58(7):418–422. [PubMed] [Google Scholar]
- 27.Stranks GJ, Mathai JT, Rowe-Jones DC. Primary malignant melanoma of the oesophagus: case report and review of surgical pathology. Gut. 1991;32(7):828–830. doi: 10.1136/gut.32.7.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tarantino L, et al. Primary small-bowel melanoma: color Doppler ultrasonographic, computed tomographic, and radiologic findings with pathologic correlations. J Ultrasound Med. 2007;26(1):121–127. doi: 10.7863/jum.2007.26.1.121. [DOI] [PubMed] [Google Scholar]
- 29.Klein CA. Parallel progression of primary tumours and metastases. Nat Rev Cancer. 2009;9(4):302–312. doi: 10.1038/nrc2627. [DOI] [PubMed] [Google Scholar]
- 30.Chin L, Garraway LA, Fisher DE. Malignant melanoma: genetics and therapeutics in the genomic era. Genes Dev. 2006;20(16):2149–2182. doi: 10.1101/gad.1437206. [DOI] [PubMed] [Google Scholar]
- 31.Ulmer A, et al. Visualization of circulating melanoma cells in peripheral blood of patients with primary uveal melanoma. Clin Cancer Res. 2008;14(14):4469–4474. doi: 10.1158/1078-0432.CCR-08-0012. [DOI] [PubMed] [Google Scholar]
- 32.Schuster R, et al. Circulating tumor cells as prognostic factor for distant metastases and survival in patients with primary uveal melanoma. Clin Cancer Res. 2007;13(4):1171–1178. doi: 10.1158/1078-0432.CCR-06-2329. [DOI] [PubMed] [Google Scholar]
- 33.Callejo SA, Antecka E, Blanco PL, Edelstein C, Burnier MN., Jr Identification of circulating malignant cells and its correlation with prognostic factors and treatment in uveal melanoma. A prospective longitudinal study. Eye (Lond). 2007;21(6):752–759. doi: 10.1038/sj.eye.6702322. [DOI] [PubMed] [Google Scholar]
- 34.Ossowski L, Aguirre-Ghiso JA. Dormancy of metastatic melanoma. Pigment Cell Melanoma Res. 2009;23(1):41–56. doi: 10.1111/j.1755-148X.2009.00647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mocellin S, Hoon D, Ambrosi A, Nitti D, Rossi CR. The prognostic value of circulating tumor cells in patients with melanoma: a systematic review and meta-analysis. Clin Cancer Res. 2006;12(15):4605–4613. doi: 10.1158/1078-0432.CCR-06-0823. [DOI] [PubMed] [Google Scholar]
- 36.Palmieri G, et al. Prognostic value of circulating melanoma cells detected by reverse transcriptase-polymerase chain reaction. J Clin Oncol. 2003;21(5):767–773. doi: 10.1200/JCO.2003.01.128. [DOI] [PubMed] [Google Scholar]
- 37.Allard WJ, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10(20):6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 38.Braun S, et al. Cytokeratin-positive cells in the bone marrow and survival of patients with stage I, II, or III breast cancer. N Engl J Med. 2000;342(8):525–533. doi: 10.1056/NEJM200002243420801. [DOI] [PubMed] [Google Scholar]
- 39.Bidard FC, et al. Disseminated tumor cells of breast cancer patients: a strong prognostic factor for distant and local relapse. Clin Cancer Res. 2008;14(11):3306–3311. doi: 10.1158/1078-0432.CCR-07-4749. [DOI] [PubMed] [Google Scholar]
- 40.Meng S, et al. Circulating tumor cells in patients with breast cancer dormancy. Clin Cancer Res. 2004;10(24):8152–8162. doi: 10.1158/1078-0432.CCR-04-1110. [DOI] [PubMed] [Google Scholar]
- 41.Schlimok G, et al. Micrometastatic cancer cells in bone marrow: in vitro detection with anti-cytokeratin and in vivo labeling with anti-17-1A monoclonal antibodies. Proc Natl Acad Sci U S A. 1987;84(23):8672–8676. doi: 10.1073/pnas.84.23.8672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vinokurova S, et al. Clonal history of papillomavirus-induced dysplasia in the female lower genital tract. J Natl Cancer Inst. 2005;97(24):1816–1821. doi: 10.1093/jnci/dji428. [DOI] [PubMed] [Google Scholar]
- 43.Buell JF, et al. Donor transmitted malignancies. Ann Transplant. 2004;9(1):53–56. [PubMed] [Google Scholar]
- 44.Milton CA, Barbara J, Cooper J, Rao M, Russell C, Russ G. The transmission of donor-derived malignant melanoma to a renal allograft recipient. Clin Transplant. 2006;20(5):547–550. doi: 10.1111/j.1399-0012.2006.00514.x. [DOI] [PubMed] [Google Scholar]
- 45.MacKie RM, Reid R, Junor B. Fatal melanoma transferred in a donated kidney 16 years after melanoma surgery. N Engl J Med. 2003;348(6):567–568. doi: 10.1056/NEJM200302063480620. [DOI] [PubMed] [Google Scholar]
- 46.Mihm MC, Jr, Clemente CG, Cascinelli N. Tumor infiltrating lymphocytes in lymph node melanoma metastases: a histopathologic prognostic indicator and an expression of local immune response. Lab Invest. 1996;74(1):43–47. doi: 10.1111/1523-1747.ep12514601. [DOI] [PubMed] [Google Scholar]
- 47.Piras F, et al. The predictive value of CD8, CD4, CD68, and human leukocyte antigen-D-related cells in the prognosis of cutaneous malignant melanoma with vertical growth phase. Cancer. 2005;104(6):1246–1254. doi: 10.1002/cncr.21283. [DOI] [PubMed] [Google Scholar]
- 48.Mullins IM, et al. CXC chemokine receptor 3 expression by activated CD8+ T cells is associated with survival in melanoma patients with stage III disease. Cancer Res. 2004;64(21):7697–7701. doi: 10.1158/0008-5472.CAN-04-2059. [DOI] [PubMed] [Google Scholar]
- 49.Harlin H, et al. Chemokine expression in melanoma metastases associated with CD8+ T-cell recruitment. Cancer Res. 2009;69(7):3077–3085. doi: 10.1158/0008-5472.CAN-08-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hillen F, et al. Leukocyte infiltration and tumor cell plasticity are parameters of aggressiveness in primary cutaneous melanoma. Cancer Immunol Immunother. 2008;57(1):97–106. doi: 10.1007/s00262-007-0353-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dworacki G, et al. Decreased zeta chain expression and apoptosis in CD3+ peripheral blood T lymphocytes of patients with melanoma. Clin Cancer Res. 2001;7(3 suppl):947s–957s. [PubMed] [Google Scholar]
- 52.Staibano S, et al. Tumor infiltrating lymphocytes in uveal melanoma: a link with clinical behavior? Int J Immunopathol Pharmacol. 2006;19(1):171–179. [PubMed] [Google Scholar]
- 53.Gatenby RA, Gillies RJ. A microenvironmental model of carcinogenesis. Nat Rev Cancer. 2008;8(1):56–61. doi: 10.1038/nrc2255. [DOI] [PubMed] [Google Scholar]
- 54.Norton L, Massague J. Is cancer a disease of self-seeding? Nat Med. 2006;12(8):875–878. doi: 10.1038/nm0806-875. [DOI] [PubMed] [Google Scholar]
- 55.Norton L. Conceptual and practical implications of breast tissue geometry: toward a more effective, less toxic therapy. Oncologist. 2005;10(6):370–381. doi: 10.1634/theoncologist.10-6-370. [DOI] [PubMed] [Google Scholar]
- 56.Wall L, Burke F, Smyth JF, Balkwill F. The anti-proliferative activity of interferon-gamma on ovarian cancer: in vitro and in vivo. Gynecol Oncol. 2003;88(1 pt 2):S149–S151. doi: 10.1006/gyno.2002.6707. [DOI] [PubMed] [Google Scholar]
- 57.Chin YE, Kitagawa M, Su WC, You ZH, Iwamoto Y, Fu XY. Cell growth arrest and induction of cyclin-dependent kinase inhibitor p21 WAF1/CIP1 mediated by STAT1. Science. 1996;272(5262):719–722. doi: 10.1126/science.272.5262.719. [DOI] [PubMed] [Google Scholar]
- 58.Ikeda H, Old LJ, Schreiber RD. The roles of IFN gamma in protection against tumor development and cancer immunoediting. Cytokine Growth Factor Rev. 2002;13(2):95–109. doi: 10.1016/S1359-6101(01)00038-7. [DOI] [PubMed] [Google Scholar]
- 59.Muller-Hermelink N, et al. TNFR1 signaling and IFN-gamma signaling determine whether T cells induce tumor dormancy or promote multistage carcinogenesis. Cancer Cell. 2008;13(6):507–518. doi: 10.1016/j.ccr.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 60.Koebel CM, et al. Adaptive immunity maintains occult cancer in an equilibrium state. Nature. 2007;450(7171):903–907. doi: 10.1038/nature06309. [DOI] [PubMed] [Google Scholar]
- 61.Weir L, Speers C, D’Yachkova Y, Olivotto IA. Prognostic significance of the number of axillary lymph nodes removed in patients with node-negative breast cancer. J Clin Oncol. 2002;20(7):1793–1799. doi: 10.1200/JCO.2002.07.112. [DOI] [PubMed] [Google Scholar]
- 62.Morrow M. A survival benefit from axillary dissection: was Halsted correct? Ann Surg Oncol. 1999;6(1):17–18. doi: 10.1007/s10434-999-0017-4. [DOI] [PubMed] [Google Scholar]
- 63.Veronesi U, Marubini E, Mariani L, Valagussa P, Zucali R. The dissection of internal mammary nodes does not improve the survival of breast cancer patients. 30-year results of a randomised trial. Eur J Cancer. 1999;35(9):1320–1325. doi: 10.1016/S0959-8049(99)00133-1. [DOI] [PubMed] [Google Scholar]
- 64.Braun S, et al. Lack of effect of adjuvant chemotherapy on the elimination of single dormant tumor cells in bone marrow of high-risk breast cancer patients. J Clin Oncol. 2000;18(1):80–86. doi: 10.1200/JCO.2000.18.1.80. [DOI] [PubMed] [Google Scholar]
- 65.Old LJ. Cancer vaccines: an overview. Cancer Immun. 2008;8(suppl 1):1. [PubMed] [Google Scholar]
- 66.Kato M, et al. Transgenic mouse model for skin malignant melanoma. Oncogene. 1998;17(14):1885–1888. doi: 10.1038/sj.onc.1202077. [DOI] [PubMed] [Google Scholar]
- 67.Moorhead M, et al. Optimal genotype determination in highly multiplexed SNP data. Eur J Hum Genet. 2006;14(2):207–215. doi: 10.1038/sj.ejhg.5201528. [DOI] [PubMed] [Google Scholar]
- 68.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5(4):557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 69.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23(6):657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 70.Waldman FM, DeVries S, Chew KL, Moore DH, 2nd, Kerlikowske K, Ljung BM. Chromosomal alterations in ductal carcinomas in situ and their in situ recurrences. J Natl Cancer Inst. 2000;92(4):313–320. doi: 10.1093/jnci/92.4.313. [DOI] [PubMed] [Google Scholar]
- 71.Choi JH, Jung HY, Kim HS, Cho HG. PhyloDraw: a phylogenetic tree drawing system. Bioinformatics. 2000;16(11):1056–1058. doi: 10.1093/bioinformatics/16.11.1056. [DOI] [PubMed] [Google Scholar]
- 72.Felsenstein J. An alternating least squares approach to inferring phylogenies from pairwise distances. Syst Biol. 1997;46(1):101–111. doi: 10.1093/sysbio/46.1.101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.