Abstract
Recent advances in the proteomics field have allowed a series of high throughput experiments to be conducted on chloroplast samples, and the data are available in several public databases. However, the accurate localization of many chloroplast proteins often remains hypothetical. This is especially true for envelope proteins. We went a step further into the knowledge of the chloroplast proteome by focusing, in the same set of experiments, on the localization of proteins in the stroma, the thylakoids, and envelope membranes. LC-MS/MS-based analyses first allowed building the AT_CHLORO database (http://www.grenoble.prabi.fr/protehome/grenoble-plant-proteomics/), a comprehensive repertoire of the 1323 proteins, identified by 10,654 unique peptide sequences, present in highly purified chloroplasts and their subfractions prepared from Arabidopsis thaliana leaves. This database also provides extensive proteomics information (peptide sequences and molecular weight, chromatographic retention times, MS/MS spectra, and spectral count) for a unique chloroplast protein accurate mass and time tag database gathering identified peptides with their respective and precise analytical coordinates, molecular weight, and retention time. We assessed the partitioning of each protein in the three chloroplast compartments by using a semiquantitative proteomics approach (spectral count). These data together with an in-depth investigation of the literature were compiled to provide accurate subplastidial localization of previously known and newly identified proteins. A unique knowledge base containing extensive information on the proteins identified in envelope fractions was thus obtained, allowing new insights into this membrane system to be revealed. Altogether, the data we obtained provide unexpected information about plastidial or subplastidial localization of some proteins that were not suspected to be associated to this membrane system. The spectral counting-based strategy was further validated as the compartmentation of well known pathways (for instance, photosynthesis and amino acid, fatty acid, or glycerolipid biosynthesis) within chloroplasts could be dissected. It also allowed revisiting the compartmentation of the chloroplast metabolism and functions.
Plastids are semiautonomous organelles that are ubiquitously found in plant cells. They are derived from an endosymbiotic event and are thought to have evolved from an ancient photosynthetic prokaryote related to present-day cyanobacteria. Following endosymbiosis, the plastid genome has been reduced to ∼100 genes, mainly coding for housekeeping functions (translation and transcription of the plastid genome), proteins required for primary photosynthetic reactions, and a few, yet poorly characterized, gene products (1). The most conspicuous plastid type is the chloroplast, found in leaves and carrying out photosynthesis as its main function. Photosynthesis is an integrated biological process involving the coordinated functioning of chloroplast compartments: (a) the thylakoids, a highly organized internal membrane network formed of flat compressed and connected vesicles where solar energy is collected and converted into stored chemical energy (ATP and NADPH) while oxygen, a by-product of the reactions, is evolved; (b) the stroma, an amorphous background rich in soluble proteins that is the site for the reduction of carbon dioxide and its conversion into carbohydrates; and (c) the envelope, a pair of membranes surrounding the chloroplast, that tightly controls the metabolic dialogue between the organelle and the rest of the cell. Among chloroplast subfractions, the envelope membranes are rather unique as they represent a minor chloroplast component (1–2% of the chloroplast proteins) playing a key role in chloroplast metabolism and biogenesis (2). However, the details of chloroplast functions and the compartmentation of chloroplast proteins are not yet fully understood, and there is a major interest in analyzing them for understanding regulation of whole plant cell metabolism. Furthermore, more proteins, new pathways, and their precise localization remain to be discovered.
A step toward this knowledge was provided by proteomics as stroma, thylakoids, their lumen and associated plastoglobules, and envelope membranes have been analyzed in Arabidopsis and in several other plant species and algae. Indeed, subcellular proteomics has attracted considerable interest within the last few years (3–6), and databases such as the Plant Protein Database (PPDB)1 (7), the subcellular proteomics database SUBA (8), or the Plastid Protein Database (plprot) (9) gather plastid proteomics data that help and induce further targeted studies. However, the main limit to these targeted studies resides in the actual cross-contamination of the compartment of interest by major components from other cell compartments. Therefore, the accurate localization of many proteins often remains hypothetical.
A possible strategy to assess protein subcellular localization is quantitative proteomics approaches that are usually used to compare two or more physiological states of a biological system. Quantitative proteomics involves either labeling strategies (metabolic, enzymatic, or chemical) or label-free approaches. For instance, the quantitative MS-based localization of organelle proteins by isotope tagging strategy was used to investigating subcellular localization (3). MS analysis combined with statistical data treatment allowed the assessment of the subcellular distribution of proteins in different plant cell organelles such as the vacuole, ER, or Golgi (10). A similar approach, based on label-free quantification, was also used to set up the organellar map of mouse liver from different density gradients fractions (11). This approach termed “protein correlation profiling” used the peptide MS signals as quantitative measurements. Bergeron and co-workers (12) assessed the localization of proteins within enriched fractions of different endomembrane compartments using spectral counting. The spectral counting strategy is based on the premise that the MS/MS sampling rate of peptides corresponding to a particular protein is related to the abundance of this protein in the mixture being analyzed: the more abundant a protein is, the higher the number of corresponding MS/MS spectra acquired. Spectral counting is a straightforward approach but presents several drawbacks related to the fact that it is a label-free approach and more specifically that it is directly related to protein and peptide identification for which errors in assignment cannot be totally controlled. Consequently, spectral counting is referred to as a semiquantitative approach, and significance thresholds related to the reliability of the results must be carefully indicated. Nevertheless, this approach has been successfully used to assess the subcellular localization of proteins in different systems (12, 13). The accurate mass and time (AMT) method, one of these label-free approaches, combines identification and quantification issues in the context of high throughput quantitative experiments and allows the use of a spectral counting strategy. The AMT method was first elaborated for the study of the Deinococcus radiodurans proteome (14) and has been applied to various types of organisms (15, 16). In the first stage, standard shotgun proteomics approaches are undertaken on extensively fractionated proteins to yield tentative peptide identification. Those experiments yield a database containing the calculated masses based on putative peptide sequences and their corresponding measured chromatographic retention times. This database is subsequently validated in an LC-FTMS experiment measuring the accurate masses of the detected peptides at the normalized retention times observed for their initial identification. Accurate mass and time tags can subsequently be used, in the course of “simple” nano-LC-FTMS measurements, as biomarkers of the presence of a given protein without resorting systematically to MS/MS for identification. Consequently, it becomes possible to identify hundreds of proteins in a single MS spectrum in all subsequent nano-LC-FTMS analyses.
The present work is the first study aimed to specifically address the accurate proteomics-based localization of proteins, based on spectral counting, in the three major chloroplast compartments: stroma, thylakoids, and envelope membranes. Our main goal was to provide reliable data for a better understanding of the respective role of these compartments in the multitude of chloroplast functions. The first objective of the present work was to obtain the most comprehensive repertoire of proteins present in highly purified chloroplasts and their subfractions prepared from Arabidopsis thaliana leaves and analyzed by LC-MS/MS-based approaches. The second objective was to set up a chloroplast protein AMT tag database (AT_CHLORO) gathering identified peptides with their respective and precise analytical coordinates, molecular weight, and retention time. The database we describe here is therefore not only a repository of 1323 chloroplast proteins but also provides detailed proteome information (peptide sequences and molecular weight, chromatographic retention times, and MS/MS identification statistics). These coordinates are essential for further label-free experiments aimed at comparing chloroplast proteomes from various mutants and WT Arabidopsis plants (17, 18). The third objective was to use spectral counting to assess, for the first time in the same set of experiments, protein localization in envelope, thylakoids, and stroma, thus allowing direct comparison of the protein equipment of each compartment. This was made possible because we prepared chloroplast subfractions with a low level of cross-contamination as shown by Western blot experiments and further confirmed by proteomics. As most available chloroplast proteomics data provide more accurate information about proteomes from thylakoids and stroma compared with the envelope, we paid a specific attention on analyzing the proteome of envelope membranes that are a minor chloroplast compartment with essential biological roles (19, 20). The envelope proteome was analyzed at an unprecedented level of sensitivity using a combination of envelope membrane fractionations and LC-MS/MS-based identification approaches, thus providing an improvement in the dynamic range detection of chloroplast envelope trace proteins. To explore these data, manual annotation (subcellular localization and functional annotation) was performed for all the proteins identified. Combined experimental data were then provided to assign subplastidial localization to these proteins, validating genuine envelope components and excluding cross-contaminants. These data provide unexpected information about plastidial or subplastidial localization of some proteins that were not suspected to be associated to this membrane system and allow revisiting the compartmentation of the chloroplast metabolism and functions.
EXPERIMENTAL PROCEDURES
Plant Material and Growth Conditions
Arabidopsis plants, Wassilewskija background (Ws), were grown in culture chambers at 23 °C (12-h light cycle) with a light intensity of 150 μmol·m−2·s−1 in standard conditions (21).
Purification of Chloroplasts and Chloroplast Envelope, Stroma, and Thylakoids from Arabidopsis Leaves
All operations were carried out at 0–5 °C. Percoll-purified chloroplasts were obtained from 100–200 g of A. thaliana leaves with a yield approaching 2%. Several replicates of chloroplast preparation were obtained. Purified intact chloroplasts were then lysed in hypotonic medium, and envelope, thylakoid, and stroma subfractions were purified on a sucrose gradient and stored as described previously (21). Analyses of the purity of chloroplast envelope preparations were performed as described previously (22). They were found to be slightly contaminated by other cell membrane systems (mitochondria, plasma membranes, and tonoplast). Analyses of the cross-contamination of the chloroplast envelope, thylakoids, and stroma preparations with markers from other plastid subfractions were performed using antibodies directed against specific marker enzymes associated to the respective subfractions (see supplemental Data 1).
Western Blot Analyses
For Western blot analyses, gels were transferred to a nitrocellulose membrane (BA85, Schleicher & Schuell, Whatman). The proteins were detected using the polyclonal antibodies using either alkaline phosphatase or ECL detection. To validate purity of the purified envelope membrane fractions, we used rabbit polyclonal antisera raised against the chloroplast envelope ceQORH protein (23) at a 1:5000 dilution, the plasma membrane H+-ATPase (24) at a 1:1000 dilution, the mitochondria TOM40 protein (25) at a 1:1000 dilution, and the thylakoid light-harvesting complex proteins (LHCPs) at a 1:25,000 dilution (a gift from O. Vallon, Institut de Biologie Physico-chimique, Paris, France; see Ref. 26). Chlorophyll content was also determined by spectrophotometry in acetone solutions (27). To quantify the cross-contamination of the purified envelope membrane fractions with proteins derived from other chloroplast subfractions (and the opposite), we used rabbit polyclonal antisera raised against the envelope ceQORH protein (23), the envelope HMA1 ATPase (22), the thylakoid LHCPs (26), the stroma KARI protein (28), and the BCCP1 protein (29) (see supplemental Data 1). To quantify the natural abundance of three identified envelope proteins, HMA1, ceQORH, and P56-4 (At4g00370, ANTR2, or PHt4-4), antibodies used were directed against the envelope HMA1 ATPase (22), ceQORH (23), and the His-308/342 P56-4 recombinant protein (this work) (see supplemental Data 2). To validate the ie18 Arabidopsis mutant (supplemental Data 3), antibodies used were directed against the inner envelope membrane IE18 protein (30) at a 1:1000 dilution, the ceQORH protein (23) at a 1:5000 dilution, and the IE30 protein (31) at a 1:10,000 dilution. To validate the subplastidial localization of the P56-2 (At2g29650, ANTR1, or PHt4-1) protein, a rabbit polyclonal antiserum raised against the His-278/313 P56-2 recombinant protein (this work) (see supplemental Data 4) was used.
Sample Preparation for Mass Spectrometry Analyses
Prior to mass spectrometry analysis, different sample preparation procedures were used for whole chloroplast, stroma, thylakoids, and envelope fractions. Before in-gel or in-solution digestion, specific treatments were undertaken. Some thylakoid fractions were treated using chloroform/methanol (5:4, v/v) extraction or membrane washing with 0.1 m NaOH (see Fig. 1). The stromal fraction, rich in Rubisco protein, was submitted to ammonium acetate precipitation to precipitate this highly abundant protein with or without a Strataclean (Stratagene) purification step for protein concentration (see Fig. 1). Some chloroplast envelope proteins were extracted using alkaline conditions (0.5 m NaOH) as already described (32). To solubilize membrane proteins present in both the outer and the inner surfaces of the envelope vesicles, sonication of the membrane preparations was also performed during this treatment. The resulting mixtures were stored for 15 min on ice before centrifugation (4 °C, 20 min, 12,000 × g). Insoluble proteins were recovered as white pellets and further analyzed (Fig. 1). Following these purification steps, most of the samples were separated by SDS-PAGE either in the conventional separating mode or in the stacking mode (26). SDS-PAGE analyses were performed as described (33). After Coomassie Blue staining to reveal proteins, the gel was cut into discrete bands. In-gel digestion with trypsin (sequencing grade; Promega, Madison, WI) was carried out as described previously (21, 26) with the following modification: after washing and drying, gel pieces were rehydrated in 100 μl of 7% H2O2 at room temperature for 15 min in the dark. This step led to cysteine oxidation and conversion of the methionine residues into sulfone (34). Gel pieces were then extracted with 5% (v/v) formic acid solution. Additionally, some samples were also digested in solution without prior SDS-PAGE separation as described (34).
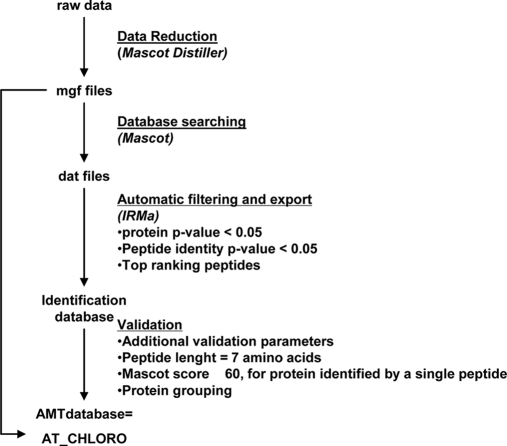
Fig. 1.
Analysis of chloroplast fractions: overview of complete strategy. Chloroplasts from Arabidopsis leaves were purified on Percoll density gradients. The chloroplast envelope and other plastid subfractions (thylakoids and stroma) were purified on a sucrose gradient. To get a better view of the chloroplast envelope proteome, we complemented proteomics studies performed on envelope fractions extracted from WT plants with an additional proteomics analysis performed on chloroplast envelope fractions from the iep18 mutant (gray area). Red arrows, analyses corresponding to the proteins included in the initial set used for statistical measurements. Insol., insoluble.
Mass Spectrometry Analyses
For all experiments, tryptic peptides were resuspended in 0.5% aqueous trifluoroacetic acid. Approximately 500 ng of digested sample was injected for each analysis.
For experiments carried out using a Q-TOF type instrument, the samples were injected into a CapLC (Waters) nano-LC system and first preconcentrated on a 300-μm × 5-mm PepMap C18 precolumn. The peptides were then eluted onto a C18 column (75 μm × 150 mm). The chromatographic separation used a gradient from solution A (2% acetonitrile, 98% water, 0.1% formic acid) to solution B (80% acetonitrile, 20% water, 0.08% formic acid) over 60 min at a flow rate of 200 nl/min. The LC system was directly coupled to a Q-TOF Ultima mass spectrometer (Waters). MS and MS/MS data were acquired automatically using the MassLynx 4.0 software (Waters). The MS/MS data were automatically processed using the Mascot Distiller software (supplemental Data 5) to generate mgf files. For each set of samples (WT and iep18 mutant), the Q-TOF mgf files were concatenated with Mascot Daemon (Matrix Science) for further database searching.
Other experiments were performed on a 7-tesla hybrid linear ion trap Fourier transform mass spectrometer (LTQ-FT, Thermo, Bremen, Germany). The experimental sequence consisted of one high resolution MS acquisition in the ICR cell and three MS/MS scans in the linear ion trap in parallel with the MS acquisition. Dynamic exclusion was activated for ions within 5 ppm of a selected peak and eluting in a 3-min window, and one repeat scan was allowed within 30 s. An “Ultimate 3000” nano-HPLC system (LC Packings, Amsterdam, The Netherlands) equipped with a dynamic flow control and a PepMap (LC Packings) column (15 cm, 75-μm diameter, 3-μm C18 particles, 100-Å pore size) was coupled to the FT-ICR instrument. Mobile phases A and B (water, acetonitrile, and formic acid in proportions 97.9:2:0.1 and 19.92:80:0.08, respectively). The gradient was 4–50% B over 60 min followed by a 5-min ramp to 90% B. MS data were acquired in the FTMS detection mode of operation (reduced profile) on a 450–1600 m/z range. The automated gain control target was set to 5 × 105 accumulated charges, and the maximum allowable accumulation time was set to 500 ms. The LTQ-FT raw data were converted into mgf files using Mascot Distiller and Mascot Daemon (Matrix Science) for further database searching (supplemental Data 6).
Database Searching
Database searching was carried out using the Mascot 2.1 program (Matrix Science) available by intranet. Two databases were compiled: a home-made list of well known contaminants (keratins, trypsin, and BSA; 21 entries) and an updated compilation of the A. thaliana protein database provided by TAIR (nuclear, mitochondrial, and plastid genome; TAIR v6.0; July 9, 2006; 30,899 entries). The variable modifications allowed were acetyl (protein), methionine oxidation, methionine sulfone, and cysteic acid. One missed trypsin cleavage was allowed, and trypsin/P was used as the enzyme. The mass tolerances were 10 ppm for precursor ions and 0.8 Da for fragment ions.
For the LTQ-FT data, to assess false positive rate, the corresponding reverse sequences of all entries were added to the compiled database as described by Peng et al. (35). A target-decoy database search was then performed. Validation and false positive rate assessment were performed using the IRMa software (36). Automatic data validation was carried out using the following parameters. (i) The number of report hits was fixed automatically to retrieve proteins with a p value, as defined by Mascot, such as p < 0.05. (ii) Only peptides ranked first and with an identity threshold such as the p value, as defined by Mascot, corresponding to p < 0.05, were kept. Considering those validation parameters, the false discovery rate (FDR), as described by Peng et al. (35), was estimated to be 5.5% for the overall identification database. Identifications were automatically consolidated to a mass spectral identification database (MSIdb) using a software developed in our laboratory (IRMa). Additionally, short peptides (length ≤7 amino acids) were removed. The filtering based on peptide length resulted in a drop in FDR to 4.07%. We subsequently removed peptides pointing to the reverse database. Using a home-made script (37), protein redundancy was eliminated on the basis of proteins being identified by a same set or a subset of peptides. From the so-generated protein groups, an additional validation step was added. Protein groups represented by more than one peptide were automatically kept, and protein groups identified by a single peptide were kept only if the corresponding peptide had a Mascot score above 60. MS/MS queries characterizing single-hit proteins and the corresponding identifications are available as supplementary material (supplemental proteins). An AMT database was created by extracting each unique sequence-calculated molecular weight pair in the identification database and computing the average retention time observed over all occurrences of the corresponding peptide. The maximum score with which a peptide was identified in all analyses was also included in the AMT database as a measure of confidence in this identification.
For Q-TOF data, validation was carried out using the following parameters. (i) The number of report hits was fixed automatically to retrieve proteins with a p value, as defined by Mascot, such as p < 0.05. (ii) A peptide score cutoff was applied so that the FDR was about 1%. (iii) Proteins identified with at least two peptides were automatically validated. (iv) MS/MS spectra corresponding to proteins identified by one peptide were validated manually.
Data Mining: Other Softwares
Protein names and/or associated functions were retrieved from NCBI, PPDB (38), or TAIR. GRAVY, molecular weight, pI, and TargetP annotations were retrieved from PPDB (38). Predictions of membrane-spanning regions (i.e. transmembrane domains) were collected from the ARAMEMNON database (39)). The HMMTOP 2.0 program (40) was also used for transmembrane domain predictions. Chloroplast transit peptides were predicted using the ChloroP 1.1 algorithm (41).
Statistical Model for Subplastidial Localization Determination Based on Spectral Count Data
Spectral counting was used to compare protein amounts in subplastidial fractions. To assess the significance of spectral counts for localization assignment, a logistic regression model was developed. A training set (216 proteins) was defined by selecting proteins whose function (MapManBin description (42)) allowed confident assignment to a specific subcompartment. We selected as representative of the envelope compartment 71 proteins whose functions were determined to be “transporter but not electron transporter” or “protein targeting or (Tic or Toc).” For the stroma, 89 proteins annotated as taking part in the “Calvin cycle” or in “amino acid and metabolism” were selected. For the thylakoids, 56 proteins with the keyword “photosystem” in their annotation were chosen.
A mixture model multinomial logistic regression was used to predict localization based on spectral count data for each subcompartment. All calculations were performed using JMP v.7.0.1 (SAS). A model was built based on the training set described above. For each protein, the model evaluated the probabilities that it belonged to each subcompartment,
 |
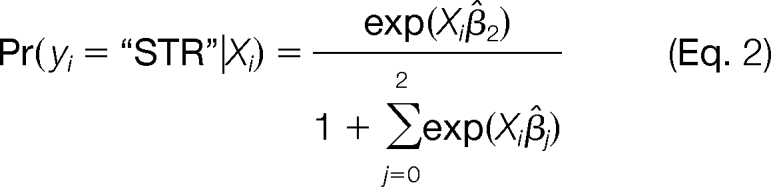 |
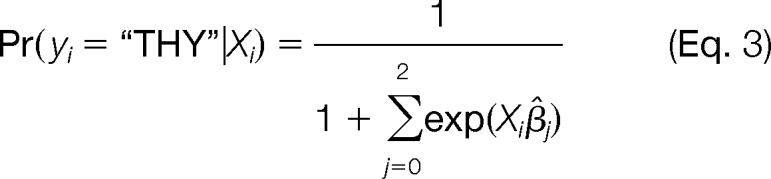 |
where Xi is the spectral count value from each subcompartment for protein i and β̂j is the estimate of the model coefficient. Based on these three probabilities, the model then assigned a most likely localization to each protein.
To test the model, another subset of 339 proteins distinct from the training set was selected based on their PPDB-curated localization. In total, 37 envelope proteins were selected based on the terms “envelope inner,” “envelope inner integral,” “envelope intermembrane space,” “envelope outer,” or “envelope outer-integral” in their PPDB localization. For the stroma, 304 proteins having the terms “plastid stroma” in their PPDB localization were chosen. For the thylakoids, 98 proteins having the terms “thylakoid,” “thylakoid integral,” or “thylakoid peripheral lumenal side” in their PPDB localization were picked. Logistic fits of protein localization versus spectral count values for proteins in the test set also indicated a predictive value of the spectral count data.
Normalized Spectral Counting for Subplastidial Localization Determination
To fully exploit the wealth of information provided by our identification database, we decided to include all experimental data to carry out the localization prediction, not only those from the analyses of the one-dimensional gels of each fraction. Using SQL queries, the number of spectra corresponding to each identified protein was extracted from each series of analyses obtained on chloroplast subfractions: envelope (59 LC-MS/MS analyses), thylakoids (129 LC-MS/MS analyses), and stroma (260 LC-MS/MS analyses). For each series of LC-MS/MS experiments, the spectral count for protein i in subcompartment n was normalized based on the total number of spectra identified in this subcompartment (43).
 |
Finally, the percentage of occurrence for each protein i in a given subcompartment n was estimated by the normalized spectral counts in subcompartment n divided by the sum of normalized spectral counts in all three compartments (envelope, thylakoid, and stroma).
 |
RESULTS AND DISCUSSION
Optimized Chloroplast Fractionation and Generation of AT_CHLORO Database: Overview of Complete Strategy
Chloroplasts from A. thaliana leaves were purified on Percoll density gradients. The plastid subfractions (envelope, thylakoids, and stroma) were purified on a sucrose gradient as described previously (21, 26). As each compartment represents complex fractions with respect to protein content and dynamic range, the three purified subfractions were fractionated by different procedures (Fig. 1). With the objective to obtain the most comprehensive chloroplast protein database, fractionation was set up so that proteins from different subplastidial compartments and present in different copy numbers could be identified. The whole envelope, stroma, or thylakoid samples were first separated by SDS-PAGE or directly digested in solution before analyses.
Rubisco represents about 40–50% of the stroma protein content, making very difficult the analysis of minor proteins. To partially remove this protein from stroma fractions, we precipitated proteins using different concentrations of ammonium sulfate (21). Rubisco was essentially recovered in the supernatant obtained by precipitation with 60% ammonium sulfate, whereas the corresponding pellet was partially devoid of Rubisco. A total of 126 different samples derived from the stroma were obtained; each of them was analyzed at least twice, leading to 260 LC-MS/MS analyses (Fig. 1).
Thylakoids proteins were also separated by SDS-PAGE or directly digested in solution. The purified thylakoid membranes were also treated using alkaline extraction or organic solvent extractions using previously described protocols (26). These different washing and extraction procedures allowed membrane proteins with a wide range of hydrophobic properties to be recovered (26). Altogether, 63 different samples were obtained from thylakoids, leading to 129 LC-MS/MS analyses (Fig. 1).
The purified envelope fractions were also separated by SDS-PAGE or directly digested in solution before analyses. LTQ-FT data were obtained from envelope membrane preparations that were not pretreated (with salts, detergents, or alkaline or hydrophobic extractions), thus increasing the risk of detecting more soluble contaminants from the stroma but potentially allowing identification of genuine peripheral proteins that interact with the inner side of the inner envelope membrane or with the plastid outer surface. Fifty-nine LC-MS/MS analyses were performed on corresponding envelope samples (Fig. 1). Q-TOF data were also obtained from NaOH-treated envelope membranes to enrich the preparations with hydrophobic proteins. Still with the same aim, we performed analyses on envelope membrane extracted from the iep18 mutant (Fig. 1). The IE18 protein (At5g62720) is a component of the inner chloroplast envelope membrane (30). Its function remains to be identified. Interestingly, this protein was one of the few envelope proteins to be under-represented in leaves and roots of Arabidopsis when plants were grown in a medium containing a limiting phosphate concentration (44). Analyzing this iep18 Arabidopsis mutant in the laboratory, we observed a reproducible under-representation of a major protein (IE30 or TPT, the chloroplast envelope phosphate/triose phosphate transporter) in envelope fractions extracted from this mutant (data not shown). We had the idea that lowering the level of a major component of the envelope fraction may give access to minor proteins present in the analyzed fraction, and thus we chose to also analyze envelope fractions from this mutant. Altogether, 40 LC-MS/MS analyses were performed on envelope samples extracted from both plants (WT plants and the iep18 mutant).
Finally, a whole chloroplast fraction was also analyzed by SDS-PAGE. Thus, a total of 241 samples were obtained from separating and stacking gels and from in-solution digestions. Because each of these samples was analyzed at least twice, 494 LC-MS/MS analyses were thus performed from which database searching results were stored in the AT_CHLORO database. From all these analyses, 1323 non-redundant proteins could be identified (see below).
Western Blot Evaluation of Cross-contaminations between Purified Chloroplast Subcompartments
As our objective was to get precise data about subplastidial localization of the identified proteins, great caution was taken to assess purity of the Arabidopsis chloroplast subfractions. Cross-contamination was checked using specific antibodies (supplemental Data 1). Western blot data indicated a low level of cross-contamination (Table I). These data demonstrate that thylakoid fractions contained less than 1% stromal proteins and about 3% envelope proteins. Stroma fractions contained less than 1% envelope and 1% thylakoid proteins. Envelope membranes from Arabidopsis chloroplasts were somewhat less pure as they contained about 3 and 10% proteins derived from the thylakoids or the stroma, respectively. However, as stroma and thylakoids were highly purified, it was straightforward to discriminate whether a protein is actually a stroma, a thylakoid, or an envelope component or presents dual localization. Furthermore, proteomics investigations (semiquantitative information derived from spectral counting) also provided further evidence for protein distribution among chloroplast compartments (see below).
Table I. Assessment of cross-contamination of purified subplastidial fractions using Western blot analyses.
See also supplemental Data 1. <1% means no detected cross-contamination using Western blot analyses.
| Contamination by | Contamination of |
||
|---|---|---|---|
| Envelope | Stroma | Thylakoids | |
| % | |||
| Envelope | <1 | 3 | |
| Stroma | 10 | <1 | |
| Thylakoids | 3 | <1 | |
Estimation of Depth of Analysis
To estimate the depth of the present proteomics analysis, we used a combination of recombinant proteins and antibodies to quantify three independent envelope proteins: HMA1 (22), ceQORH (45), and P56-4 (this work). As presented in supplemental Data 7, we were able to identify proteins representing less than of the chloroplast envelope proteins. When considering the enrichment obtained with chloroplast envelope purification, such minor proteins represent of the chloroplast proteins (i.e. about of the whole cell proteins). It is thus expected that some of the minor proteins that we identified here would not have been identified in a proteomics study performed from crude chloroplasts or crude chloroplast membranes. This was actually the case because many envelope proteins we detected here were far below the detection level when starting from crude chloroplast.
Validation of Protein Identified from FT-ICR Experiments
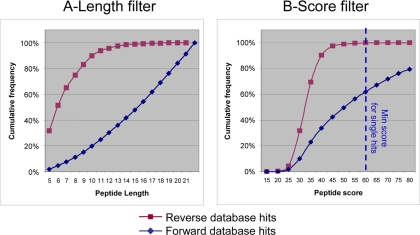
As indicated above, all gel fractions and in-solution digests were analyzed at last twice using an FT-ICR instrument (Fig. 1). Several criteria were applied to filter the Mascot database searching results. First, automatic filtering parameters were applied based on the Mascot p values (0.05) for both proteins and peptides. Results obtained from this automatic filtering process were stored in an identification database (Fig. 2). To refine the results, an additional validation step was performed from results stored in the identification database. While manually inspecting MS/MS spectra, we noticed that some of the spectra, correlated by Mascot to peptides bearing less than 7 amino acids, were not sufficiently informative to guarantee protein identification. Moreover, some peptide sequences of less than 7 amino acids were shown to match both a reverse and a forward protein. This suggested that MS/MS spectra correlated to small peptides (length <7 amino acids) are more prone to random identification than longer peptides (Fig. 3). Thus, we decided to use the length of identified peptides as an additional validation criterion. When rejecting peptides with less than 7 amino acids, about 60% of “reverse peptides” were rejected, whereas most of the “forward peptides” were kept. Eventually, peptides satisfying two criteria, Mascot p values <0.05 and peptide length ≥7 amino acids, were kept (46). This filtering step allowed us to identify proteins with an FDR of 4% at the peptide level. Then, a protein grouping procedure, ensuring the clustering of proteins sharing a subset or a same set of peptides, was applied. In most cases, protein groups are characterized by at least one peptide sequence, named “unique peptide,” that cannot be found in another protein group. However, six protein groups (0.45% of the total number of protein groups) do not have any unique peptide but cannot be classified in another protein group as they share peptides with different protein groups (supplemental Data 7). From the so-generated protein groups, protein groups identified by at least two peptides were automatically validated. For proteins identified by a single peptide sequence, we set a Mascot score threshold above which no reverse protein could be identified. Considering our data set, this threshold was fixed to 60. Overall, the FDR for proteins was below 0.2%. The final results were stored in the AT_CHLORO database (supplemental Data 8).
Fig. 2.
Processing pipeline used to generate AMT tag database AT_CHLORO.
Fig. 3.
Cumulative frequencies of reverse and forward database hits as a function of peptide length (A) are Mascot score (B).
Rationale for Quantification by Spectral Counting: Description of Procedure
Early MS/MS studies indicated that the total number of peptide hits for a specific protein semiquantitatively reflects its abundance in the analyzed protein mixture. These results have been corroborated by many reports to date, and it is commonly accepted that semiquantitative information can be extracted from spectral count data (10–13). In the present study, as we used extensive prefractionation to access the most exhaustive chloroplast proteome, we generated a tremendous amount of MS/MS data, which could be associated to each subchloroplastic compartment. On the basis that a protein should exhibit higher spectral count (i.e. concentration) in analyses of its original subcompartment, we decided to use spectral count data to determine protein subchloroplastic localization.
To assess the relevance of spectral counts for predicting protein localization, we first elaborated a statistical model. For this purpose, we selected the subset of analyses corresponding to one-dimensional gel separations of the three subchloroplastic fractions without any prior treatments (Fig. 1, red arrows). In selecting only this subset of the whole experimental design, we ensured that all three samples were comparable as they were treated in the same way. In each subcompartment, the spectral counts for all the detected proteins were retrieved. In total, a set of 1093 proteins (initial set) identified in duplicate analyses of the three raw chloroplast subfractions (envelope, stroma, and thylakoids) was considered. Results from replicate experiments showed that spectral counts were repeatable (supplemental Data 9, sheet “replicate”). Spectral counts for proteins in the initial set were thus averaged across duplicate analyses for each subcompartment (supplemental Data 9, sheet “initial set”). A training set of 216 proteins, with well characterized localization, was used to set up the logistic model (supplemental Data 9, sheet “initial set,” column “LOC-Training”). Altogether, 204 (94%) of the proteins in the training set were correctly assigned to their subcompartment by the model, indicating that spectral counts indeed contain information about protein localization. To test the model, another subset of 339 proteins, distinct from the training set, was selected based on their PPDB-curated localization (supplemental Data 9, sheet “initial set,” column “LOC-Test”). The logistic model was applied to this test set, and in total, 286 (84%) of the 339 proteins were assigned to the same localization as in PPDB. All envelope proteins were correctly assigned, and contradictory assignments were obtained for only 36 stroma and 17 thylakoid proteins. It is worth noting that some localizations indicated in PPDB could be questionable, and the error rate of the model could be overestimated based on this test set. The statistical analysis thus demonstrated the predictive power of spectral count data for the determination of the subplastidial localization of chloroplast proteins.
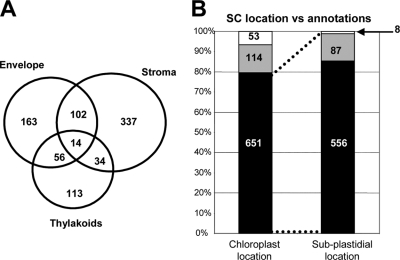
To fully take into account the wealth of information contained in the database, we collected spectral counts from all analyses regardless of the sample treatment. Moreover, because spectral counting is a semiquantitative approach, significant ratios and thresholds are generally high (43, 47). We therefore decided to take into account only proteins identified with at least 10 spectral counts. For each protein and each chloroplast subfraction (envelope, stroma, and thylakoids), the number of associated spectra was retrieved from the identification database (Fig. 2). As each subfraction was characterized by a different number of MS/MS spectra, spectral counts were normalized with respect to the number of assigned MS/MS spectra in each fraction. Based on relative spectral counts across the three chloroplast subfractions, a percentage of occurrence in each subfraction was calculated for all proteins. Taking cross-contamination into consideration based on the Western blot data, proteins were attributed a single, dual, or mixed subplastidial localization. A single localization was thus assigned to proteins for which the percentages of occurrence in the two other subfractions were below a threshold level we arbitrarily fixed at 15% (i.e. higher than the upper level of actual cross-contamination; see Table I). Dual localization was assigned to proteins with a major localization (occurrence >50%) and a secondary localization (occurrence >15%). The remaining proteins were considered to be localized in any of the three subplastidial compartments (mixed localization) (Fig. 4). Eventually, 819 proteins were considered with respect to their subplastidial localization based on spectral count measurements (supplemental Data 9, sheet “SubCellLocalization”). We subsequently verified that the localization given by normalized spectral counts agreed with the most likely localization estimated by the logistic regression model. Only 10 proteins had a contradictory localization, and the localization given by normalized spectral counts was always the one most in agreement with the literature. To further assess the reliability of our spectral count-derived localizations, we compared them with annotations found in PPDB and in TAIR. Fig. 4 shows that about 80% of the selected 819 proteins were actually annotated to be plastidial, whereas only 6% were annotated as being localized in other cellular compartments. Among proteins annotated as being plastidial, the subplastidial proteomic localization of 85% (556 of 651) of them was in agreement with the information found in PPDB and/or TAIR. Overall, these data indicate that about 70% (556 of 819) of the 819-protein data set were properly localized when compared with their annotated subplastidial localization. Only 7% (8 + 53) of these proteins showed a spectral counting-based localization that did not correspond to annotations.
Fig. 4.
Subplastidial localization according to spectral count data (819 proteins). A, distribution of protein subplastidial localization as determined using spectral count (see supplemental Data 9, column “Loc_SC”). B, correlation between spectral count-based localizations and PPDB and/or TAIR annotations. Chloroplast location, the localization according to PPDB and/or TAIR (see supplemental Data 9, column “Annotated as plastidial”): black box, proteins annotated as plastidial; gray box, proteins with no localization annotation; white box, proteins annotated as localized in other subcellular compartments. Subplastidial location, for proteins annotated as plastidial, correlation between the annotation at the subplastidial level (PPDB and/or TAIR) and localization based on spectral count (see supplemental Data 9, column “Correlation loc. SC and annotation at the subplastidial level”): black box, proteins whose subplastidial annotation (PPDB and/or TAIR) matches spectral count-based subplastidial localization; gray box, proteins with no subplastidial localization annotation; white box, proteins annotated as localized in another subplastidial compartment compared with spectral count-based subplastidial localization.
In summary, both statistics and comparisons with literature data indicated that the spectral counting approach used in the present study proved to be efficient in assigning subplastidial localization. This information is essential to further investigate the function and localization of the 201 proteins (114 + 87) for which no subplastidial localization information could be found in either PPDB or TAIR at the time when our data were compiled.
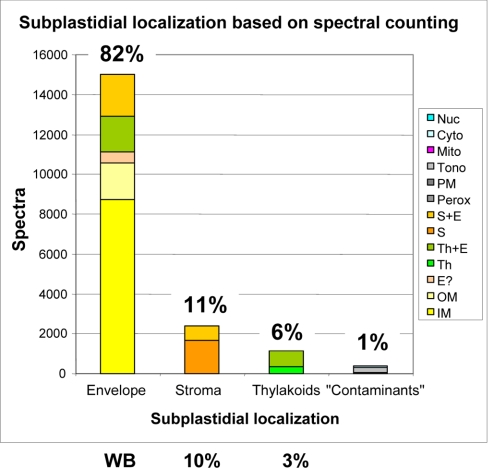
Contamination Measured by Spectral Count Correlates with Western Blot Data
Cross-contamination evaluated using spectral counting was in good agreement with Western blot data (see Table I and supplemental Data 1). For instance, spectral counting estimated the contamination of purified envelope fractions by thylakoid proteins to be about 6% (Fig. 5), whereas it was estimated to be about 3% by Western blot experiments (supplemental Data 1A). Contamination of purified envelope fraction by stroma proteins was estimated to be about 11% by spectral counting (Fig. 5) in good agreement with the 10% found by Western blot (see supplemental Data 1B). However, such quantifications might be biased if the purified thylakoid or stroma fractions were significantly contaminated with envelope membrane proteins. As deduced from reverse experiments (supplemental Data 1, C and D), this was not the case as envelope markers were poorly detected in the stroma (<1%) or the thylakoid (3%), respectively.
Fig. 5.
Spectral count-based subplastidial localization of proteins retrieved from envelope fractions. In good agreement with the levels of cross-contaminations measured using Western blot (WB) experiments (see supplemental Data 1), envelope membranes appear to contain up to 10% stroma proteins and 6% thylakoid membranes proteins. Contamination thresholds were selected accordingly to evaluate subplastidial localization of the proteins identified within the purified envelope fraction (see supplemental Data 10). Nuc, nucleus; Cyto, cytoplasm; Mito, mitochondrion; Tono, tonoplast; PM, plasma membrane; Perox, peroxisome; S+E, stroma and envelope; S, stroma; Th+E, thylakoid and envelope; Th, thylakoid; E?, envelope?, OM, outer membrane; IM, inner membrane.
Although mitochondria are generally expected to be a major source of contamination of the purified chloroplast, we only detected five genuine mitochondrial proteins that were not previously demonstrated to be dually targeted to both the plastid and the mitochondria (supplemental Data 10, lanes 667–670). Indeed, some proteins (51 of 700) identified during chloroplast envelope proteomics analyses were also detected during proteomics analyses of mitochondrial preparations (see supplemental Data 10, column “Expected localization 1”). Some of these proteins are already known to be dually targeted to plastids and mitochondria. This phenomenon was recently reviewed, and 56 proteins were shown to be targeted to both mitochondria and plastids (48) of which 32 were present in our AT_CHLORO database (supplemental Data 11). Considering envelope samples, contamination with mitochondria represents only 0.05% when considering spectral counting. This value is in good agreement with previous demonstration of the low, if any, contamination of the envelope fraction with mitochondrial membranes (22). This is also true for other cell compartments like peroxisomes, plasma membrane, tonoplast, or cytosol. Some contaminants stem from the cytosol and belong to machineries or macromolecular complexes reported to be associated to the outer surface of mitochondria such as the 80 S ribosome in yeast, human, and algae (49–54). By similarity, we cannot exclude that the cytosolic translation machinery is interacting with the outer surface of the chloroplast. However, it is important to note that the number of spectra corresponding to these cytosolic ribosomal subunits is very low, representing altogether less than 0.05% of the value obtained for all proteins detected in purified envelope fractions. The same is true for cytoskeleton proteins (actin isoforms, actin-binding proteins, kinesins, etc.) that were also detected in the purified envelope fraction. Recently, Oikawa et al. (55) isolated an A. thaliana mutant in which light-induced relocalization of chloroplasts was defective and pointed out the CHUP1 protein (for chloroplast unusual positioning 1). This protein contains an actin-binding domain and is localized in the outer envelope of chloroplasts, thus supporting the idea that chloroplasts can interact with actin filaments. Other data have also recently provided evidence for the interaction of members of these protein families with the outer surface of plastids (56). Accordingly, these proteins were classified as proteins potentially interacting with the outer surface of the chloroplast rather than as cytosolic contaminants (supplemental Data 10).
Another possible source of contamination concerns the ER. However, the chloroplast is known to interact with the ER for chloroplast protein and lipid trafficking (1). In support of this, we found a few proteins (eight proteins; 1%) that are known or suspected to interact with or colocalize with the ER (supplemental Data 10, column “Expected localization 1”) and therefore could also interact with the outer surface of the chloroplast. Finally, the estimate of contamination with non-plastid proteins was about 8.5% when considering the number of proteins but less than 2% when considering their actual amount as estimated by spectral counting (Fig. 5).
Building AT_CHLORO Database, a Subchloroplast Proteome Gathering Curated Information about Proteins Identified in Envelope Fractions
Extensive Chloroplast Protein Repertoire with Detailed Proteomic Information about Identified Peptides
The current validated AT_CHLORO database gathers 1323 proteins (supplemental Data 8) identified by 10,654 unique peptide sequences. These figures come very close to results obtained by Zybailov et al. (57) who identified 1325 proteins in high throughput analyses of soluble and membrane chloroplast fractions. Our data, taken together with those results, show that almost 1700 chloroplast proteins or proteins associated to the chloroplast are amenable to proteomics investigations.
The AT_CHLORO database also provides detailed proteomics information (peptide sequences and molecular weight, chromatographic retention times, and MS/MS identification statistics) as we gathered all the information about the 10,654 identified unique peptide sequences. An Access database is available containing all protein identifications (peptide sequences and associated Mascot score) and analytical coordinates (molecular weight and retention time) to be used with the chloroplast protein AMT database http://www.grenoble.prabi.fr/protehome/grenoble-plant-proteomics/ and protein subplastidial localization deduced from spectral counting.
Although the information on chloroplast proteins presently available in public databases (PPDB and SUBA) is highly reliable for proteins from thylakoids and stroma, these databases contain less information about the envelope proteome. We therefore decided to pay special attention to this unique membrane system by combining our proteomics results together with a thorough survey of the literature.
Knowledge Base about Chloroplast Envelope Proteins
Half of the AT_CHLORO proteins were actually identified from an envelope fraction. These data together with additional identifications (Fig. 1; five additional proteins) and manual validation (33 proteins identified with one peptide whose Mascot score was <60) allowed us to identify about 700 proteins in envelope fractions (supplemental Data 10). Such a number is quite remarkable as the envelope represents about 1% of the total protein content (in weight) of the chloroplast. In addition to the present proteomics data, we present in supplemental Data 10 all the publicly available information retrieved from the literature about the 700 proteins identified in purified envelope fractions. We therefore compiled (i) information about the present proteomics data, (ii) results of predictions by bioinformatics tools (transmembrane domains, cell localization, etc.), (iii) information (function, localization, etc.) retrieved from different protein databanks (TAIR, NCBI, UniProt, and PPDB), (iv) appropriate literature, and (v) other proteomics studies targeted to the chloroplast but also to other cell compartments whose proteins are likely to be contaminants. All this information was used and evaluated to provide curated descriptions, functions, and localizations for all proteins identified from proteomics investigations of the chloroplast envelope. The present protein repertoire is organized in several categories according to the expected (supplemental Data 10, column “Expected localization 1”) or experimental (supplemental Data 10, column “Experimental localization 2”) subcellular localization of the proteins in various compartments, i.e.“inner envelope membrane,” “outer envelope membrane,” “envelope?” (i.e. with no more precise envelope subcompartmentation), “stroma,” “thylakoids,” and any combinations of the three subplastidial compartments (envelope, stroma, and thylakoids) when the proteins where recovered in more than one plastidial compartment.
Taking into consideration the present study and previously published proteomes targeted to the chloroplast envelope (26, 58, 59), a total number of 762 proteins has been detected in purified envelope fractions (Fig. 6A) among which 360 (47%) were only identified in this work and 111 (15%) were only identified in the work by Froehlich et al. (58). Compared with the data published by Zybailov et al. (57), 474 of the proteins identified in this work in purified envelope fractions were previously detected in the chloroplast (Fig. 6B). From the 360 proteins identified for the first time in purified envelope fractions, 224 proteins were previously identified in the chloroplast (57), whereas 136 proteins were not. With respect to envelope-specific analyses (26, 58, 59), this work allowed us to identify 170 proteins absent from the chloroplast proteome from Zybailov et al. (57). This suggests that these proteins could not be detected in complex chloroplast subfractions. Alternatively, some of these proteins might also be non-plastid contaminants specifically enriched in chloroplast envelope preparations (e.g. cytosolic proteins interacting with the outer surface of the envelope, major component of other membrane systems, etc.) and that were too poorly represented in more complex chloroplast fractions. However, of the 136 new proteins identified during this work, almost 90 likely reside at the envelope: 66 could be classified as inner envelope membrane components, six could be classified as outer envelope components, and 15 could be classified as envelope proteins shared with the stroma or thylakoid membrane (supplemental Data 10). On the contrary, one-third of these 136 proteins correspond to minor proteins mostly of unknown function and lacking a predictable plastid targeting peptide (supplemental Data 10). These proteins might thus be so far unknown outer envelope components, proteins interacting with the outer surface of the chloroplast, inner envelope proteins with erroneous primary sequences lacking correct N termini, or finally proteins lacking canonical and thus predictable targeting sequences (see below).
Fig. 6.
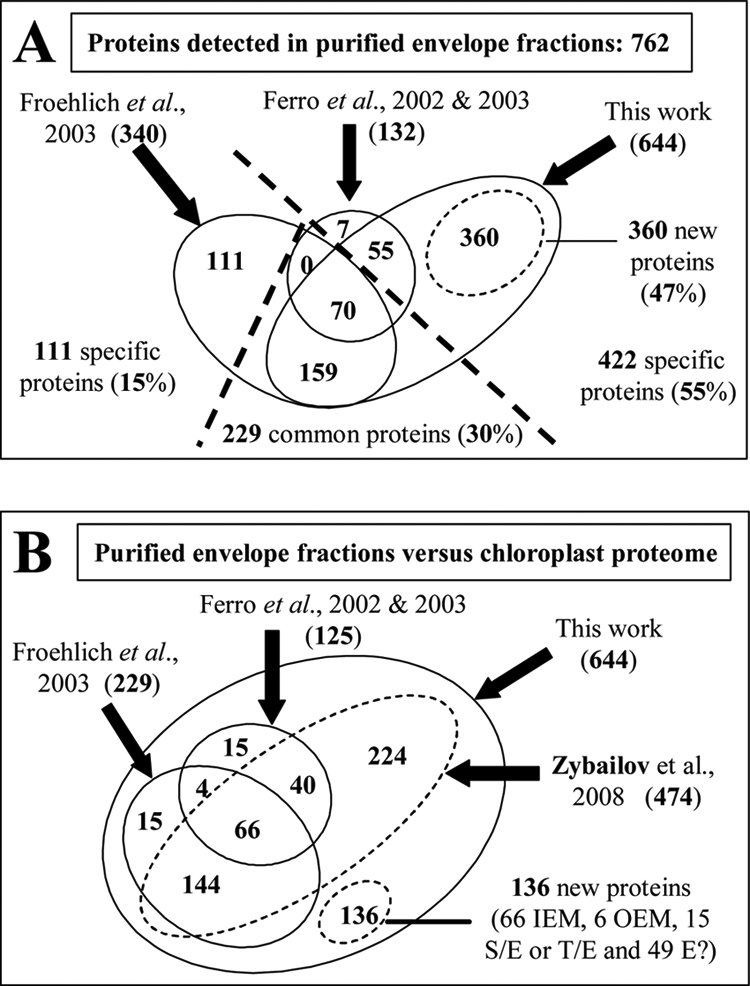
Evaluation of coverage of chloroplast envelope proteome when combining present data with earlier analyses targeted to same membrane system. A, Venn diagram indicating the weight of protein identified during this work when compared with previous data obtained by Ferro et al. (26, 59) or Froehlich et al. (58). Numbers in parentheses are the numbers of proteins identified throughout the diverse studies. For the present study, only 644 of the 700 proteins identified in the purified envelope fraction were considered (proteins classified as “contaminants” and suspected to be derived from non-plastid subcompartments were excluded; see supplemental Data 10). When combining all proteomics studies performed on the chloroplast envelope, a total of 762 proteins was identified. Note that 360 proteins (47%) were only identified during this work. B, overlap of the studies targeted to the chloroplast envelope with the recent and extensive study (more than 1300 identified proteins) performed at the whole chloroplast level (59). In this case, numbers in parentheses are the numbers of proteins identified during this work that were also identified in previous studies. According to these data, of the 360 new envelope proteins identified during this work, 224 proteins were previously detected in the chloroplast (Zybailov et al. (57)), and 136 proteins were only identified during this work. Note that targeting the envelope membrane system to perform the proteomics analyses allowed identification of proteins that could not be detected in more complex chloroplast subfractions. IEM, inner envelope membrane; OEM, outer envelope membrane; E?, envelope?; S/E, stroma/envelope; T/E, thylakoid/envelope.
Functional Annotation
Supplemental Data 10 also describes previously known or expected (when predictable) functional categories assigned to the 700 proteins detected in the purified envelope fractions in the present work. Importantly, clear-cut data from the literature were first considered as the primary criterion to classify the identified proteins within functional categories. When available, functional classification was also deduced from MapManBin (42) data or Pfam predictions (supplemental Data 10). Proteins were classified as “unknown protein” when no functional category could be assigned. These analyses are summarized in the “Curated function” column of supplemental Data 10.
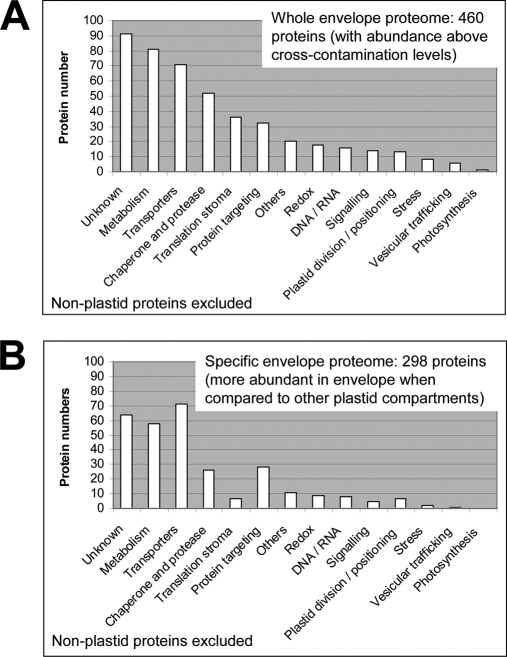
This functional classification provided an overview of the main functions carried out by envelope membranes (Fig. 7). When considering the whole envelope proteome, i.e. the 460 proteins that were detected in purified envelope fractions (with abundance above cross-contamination levels), the three main functional categories were “unknown” (20%), “metabolism” (18%), and “transporters” (15%) (Fig. 7A). As expected, numerous envelope proteins still require functional characterization, and for 90 of them (the 20% unknown proteins), no functional information could be deduced or even predicted from the analysis of their primary sequence (supplemental Data 10). When considering the more specific envelope proteome, i.e. the 298 proteins that were more abundant in purified envelope fractions when compared with levels in other plastid subcompartments, these three main categories still represent 21, 19, and 24%, respectively (Fig. 7B). It is also clear that the two categories “chaperone and protease” (11–8%) and “translation stroma” (8–2%) are reduced within this group of 298 proteins, suggesting that members of these two categories are mostly shared between envelope membranes and the two other plastid subcompartments. Within these 298 envelope proteins, “chaperone and protease” and “protein targeting” stand for 8 and 10%, respectively. In other words, the five above cited main categories (unknown, metabolism, transporters, translation stroma, and chaperone and protease) comprise more than 80% of the chloroplast envelope proteins.
Fig. 7.
Envelope composition: curated annotation and functional categories. Functional annotations were retrieved from supplemental Data 10. Two sets of proteins were analyzed: the “whole envelope proteome” (460 proteins) corresponding to proteins with abundance in purified envelope fraction above measured cross-contamination levels (first 460 proteins in supplemental Data 10) (A) and the more specific envelope proteome (298 proteins), which only contains proteins displaying increased abundance in envelope fractions compared with other plastid compartments (first 298 proteins in supplemental Data 10) (B). In both cases, non-plastid proteins were excluded.
Identification and Subplastidial Localization of Novel Envelope Proteins
In the present work, chloroplast envelope proteins were identified at an unprecedented level of sensitivity. Several parameters explain this progress: (i) the improved sensitivity of recent MS-based technologies, (ii) the lack of physical or chemical treatments of purified membrane fractions, which interestingly remove most soluble contaminants derived from other cell compartments but also eliminate peripheral membrane proteins loosely bound to the inner or outer surface of the envelope membrane system, and (iii) the simultaneous analysis of the three chloroplast subcompartments, which allows identification of genuine envelope proteins that were previously suspected to reside at the stroma or the thylakoid membrane. Finally, of the 700 proteins identified in envelope fractions, 460 proteins were shown to actually reside at the envelope (with abundance above the threshold level of 15%; see above), and 298 of these 460 proteins were demonstrated to be only or mostly detected in purified envelope fractions when compared with other plastid subcompartments (Fig. 7 and supplemental Data 10).
Prior to the present study, the subplastidial localization of most envelope proteins remained unknown (undefined), unclear (chloroplast), or erroneous (wrong subplastidial compartment) as can be seen from supplemental Data 10 (columns I, J, and K, lanes 1–299). This was especially true for the 298 envelope proteins that were only or mostly detected in purified envelope fractions. To evaluate the progress provided by our experimental data, we compared our information with three independent studies or reference databases (see supplemental Data 10, columns I, J, and K). Fewer than 120 of these 298 proteins were described to be associated with the envelope in at least one study or database, and fewer than 50 were assigned to envelope localization by at least two of these three studies. More than 80% of the 86 genuine envelope proteins that are shared with the stroma or the thylakoid fractions (see supplemental Data 10, lanes 375–460) were previously identified, and their precise subplastidial localization was determined. However, it is important to note that for 51 of the 62 envelope proteins shared with the stroma and for 22 of the 24 envelope proteins shared with the thylakoids the alternative localization (stroma or thylakoids) was the only proposed localization (supplemental Data 10, columns I, J, and K, lanes 376–461). Some databases like PPDB contain rigorous information about subcellular localization of proteins that is not based on prediction and only provide it if sufficient information is available. Consequently, our experimental data will be helpful to implement these databases.
Dual Localization of Proteins within Chloroplast
Proteins Detected in Both Envelope and Thylakoids
The presence of low, but significant, levels of thylakoid proteins in purified envelope fractions raises the question of the relevance of such a dual localization. In fact, dual localization in envelope and thylakoid membranes of some enzymes can result from either the actual compartmentation of chloroplast metabolism (see below) or contamination. Many proteins belonging to major protein complexes involved in the light-dependent reactions of photosynthesis (i.e. PSI, PSII including OEE subunits, b6/f, LHCI, LHCII, and ATP synthase) were identified during this study. We analyzed the localization of their subunits that were identified by more than 10 spectral counts (63 proteins). As expected, most of these proteins were localized in the thylakoids (supplemental Data 8 and 10). With a closer look at the localization data, noticeable differences appeared. Indeed, most subunits of the PSI (except LHCI) and of the thylakoid ATPase complex were detected in the envelope fraction in significant proportion (15–25%; i.e. far above the 3% cross-contamination of envelope by thylakoids; see Table I and supplemental Data 1), but it was not the case for b6/f, PSII subunits, or LHC proteins (supplemental Data 12). Questions about the half-life of synthesis and probability of accumulation within the envelope (during import) of nucleus-encoded precursors were excluded because this enrichment also concerns chloroplast-encoded subunits of both the ATPase and PSI. Another option was the specific contamination of purified envelope fractions with specific thylakoid vesicles during the subplastidial fractionation process. The lateral distribution of the main chlorophyll-protein complexes between appressed and non-appressed thylakoid membranes is a well described process (60) that participates in adaptation of photosynthesis to changeable environmental conditions. PSI and thylakoid ATP synthase complexes are almost exclusively localized in unstacked regions, whereas PSII is mostly present in the stacked regions of thylakoid membranes (for a review, see Ref. 61). Thus, the subplastidial localization of PSI and ATPase synthase partly in the envelope proteins could be explained by two phenomena: either (i) the unstacked regions of thylakoids, being lighter than grana, are co-purified with envelope membranes or (ii) the unstacked regions of the thylakoids really interact with the envelope. In any case, the partial localization of thylakoid proteins in the envelope could indicate an actual localization in the unstacked region of thylakoids.
Proteins Detected in Both Envelope and Stroma
According to spectral count data, many proteins appear to be shared by the envelope and the stroma (supplemental Data 10). This represents 38 of the 298 proteins found to be more abundant in purified envelope fractions and 111 of the 460 envelope proteins shown to actually reside at the envelope. Some specific classes of proteins seem to be specifically affected by this phenomenon.
Of the 54 ribosomal or ribosome-binding proteins identified in the present study, 36 were shared by envelope and stroma fractions. Seven proteins were mainly recovered in the envelope fraction (supplemental Data 10, lanes 648–669), and 29 were mainly recovered in the stroma (supplemental Data 10, lanes 406–434) with an average envelope representation of 30%, thus far above the expected representation of stroma contaminants in the envelope fraction (see supplemental Data 1 and Table I). On the other hand, 18 proteins were only detected in the stroma (supplemental Data 10, lanes 544–561), and one nucleus-encoded protein (RPS30) was even found to be more abundant in the thylakoid (68%) than in the stroma. This might indicate that the plastidial ribosomal complexes are dynamic structures that are highly mobile within the chloroplast to be recruited at a precise localization for protein synthesis.
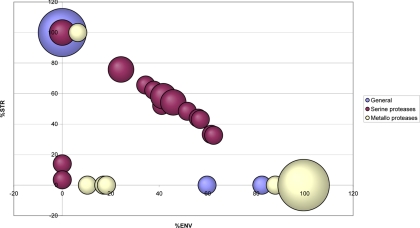
Proteases are critical regulatory factors for many metabolic and degradation processes within the cell but also within the plastids (62). According to their localization in the chloroplast, proteases have different roles. We extracted from the AT_CHLORO database proteins classified as proteases by MapManBin (42) and examined the spectral count-based suborganellar localization. Strikingly, major protease classes have specific subplastidial localization (Fig. 8). It is currently acknowledged that Clp proteases are localized in the stroma (63). However, some of the Clp proteases identified here were more abundant in the envelope (supplemental Data 10, lanes 2–7 and 377–386). Some Ftsh proteins have been shown to be anchored to the stromal surface of the thylakoid membrane and are involved in degradation of unassembled proteins and in the turnover of the D1 protein of the PSII reaction center (64). Among the 12 identified Ftsh proteins, seven were only detected in the envelope. This finding addresses the question of the specific role of Ftsh proteins in the envelope.
Fig. 8.
Relative spectral counts and subplastidial localization of proteases. Subplastidial localization of the different classes of proteases as defined by MapManBin (42) is shown. The size of the spots is proportional to the number of proteins having the same relative spectral count distribution between envelope (ENV), stroma (STR), and thylakoids localization.
Two subunits, α and β, of ACCase were previously shown to strongly interact with the inner membrane of the chloroplast envelope (65). In good agreement with these data, ACCDβ and the chloroplast-encoded ACCDα subunits were only (i.e. 95%) detected in the envelope (supplemental Data 10, lanes 51 and 52). On the contrary, the two other members (BCCP1 and BCCP2) of the same ACCase complex were found to be more enriched in the stroma (supplemental Data 10, lanes 395 and 396) in good agreement with other previously published data localizing the ACCase complex within the stroma (66). One explanation for the different localization of members of the same protein complex would be that, during chloroplast breakage, some subunits are released in the stroma because of a weak interaction within the complex. Another explanation would be related to the regulation of the ACCase complex function through interaction. This last hypothesis strengthens previous data demonstrating that (i) the chloroplast ACCase complex can dissociate and reassociate (29, 67), (ii) ACCase is at least partially associated to the inner membrane of the chloroplast envelope (68), and (iii) the biotin carrier subunit is more susceptible than the other subunits to solubilization during extraction (69).
Cases Studies: New Perspectives for Already Described Proteins
Ycf Proteins
The chloroplast genomes of most higher plants contain two giant open reading frames designated as ycf1 and ycf2. Although their function is unknown, these chloroplast genes are essential for cell life as homoplasmic mutant plants could not be obtained from attempts to disrupt these genes (70). Surprisingly, the two chloroplast gene products, Ycf1.2 and Ycf2.1, were almost exclusively found (>95%) in chloroplast envelope fractions. Ycf1.2 contains several predicted α helices and was thus suspected to be associated to the thylakoid (see public database information; supplemental Data 10, column I, lane 272); however, it was never detected within thylakoids by proteomics. Ycf2.1 protein does not contain predicted transmembrane helices and was previously suspected to be associated to the stroma (cf. public database information; supplemental Data 10, column I, lane 273), but it was not detected in the stroma (71). Therefore, the present data clearly demonstrate that these two proteins reside at the chloroplast envelope. These data allow unambiguously upgrading the list of chloroplast-encoded and envelope-localized proteins. The lack of detectable Ycf10 protein, which was previously associated to the chloroplast envelope (72), in any of the chloroplast subfractions suggests that this protein might either be too poorly expressed to be detected or only transiently expressed.
Proteins of Outer Chloroplast Envelope Membranes: Why So Few?
Only 25 proteins could be unambiguously associated to the outer envelope membrane due to availability of previously published data. These proteins could be classified into four categories. The first category included a series of five proteins (OEP21, OEP24, OEP37, and isoforms), one (OEP21-like) identified for the first time, one identified previously (73), and two more identified recently (74, 75), were demonstrated to act as solute or ion channels. The second category included six members of the OEP16 protein family with four of them being previously known envelope proteins (26, 58). There is still some controversy about the exact role of these proteins (76, 77), which have been proposed as either amino acid or specific preprotein transporters. Surprisingly, OEP16-3, which was not previously detected in envelope fractions but was shown to be only targeted to mitochondria (78), was also identified during this work. However, because this protein is one of the rare proteins detected in the envelope fraction with only one peptide, this result should be considered with caution. The third category included seven components of the TOC translocon. Again, the almost certainly less abundant protein (Toc132) was the only protein not to have been identified during earlier proteomics works. It is interesting to note that only one isoform of the Toc64 family (Toc64-III) was identified here in good agreement with the cytosolic and mitochondrial localization of Toc64-I and Toc64-V, respectively (1). Finally, if one excludes proteins recently associated to the outer envelope membrane through targeted biochemical studies, e.g. PVD2 (79), CRL (80), or SFR2 (81), very few identified proteins could be associated to this membrane system during this work. This limit directly results from the fact that about 90% of these proteins lack a predicted chloroplast transit peptide (supplemental Data 10, column I, lanes 275–299). As a consequence, it is expected that several proteins identified during this work and classified in the envelope? category (see supplemental Data 10, lanes 300–374, column “Expected localization 1”) might be new components of the outer envelope membrane (the fourth category).
Chloroplast Proteins Lacking “Canonical” Transit Peptides
About 400 proteins present in the AT_CHLORO database have no ChloroP-predictable transit peptides and therefore could have various origins. Some of them can be encoded by the chloroplast genome: indeed, this was the case for 52 of them, i.e. 4% of the total proteome. Other possibilities exist for the remaining 342 proteins (25%). They can reside at the outer envelope as most of them are expected not to bear a predictable chloroplast transit peptide (1). They can also be derived from extraplastidial contamination. However, in several cases, the ChloroP prediction could be wrong. For instance, ChloroP does not predict any transit peptide for the IEP30 phosphate/triose phosphate translocator (At5g46110), a major envelope protein known to be cleaved to its mature form during import to the inner envelope membrane (see supplemental Data 10). Furthermore, some genes are not properly predicted, and this can lead to erroneous ChloroP predictions for the actual targeting of the deduced protein. In addition, some chloroplast proteins do not have any canonical transit peptide and utilize alternatives routes for protein import into the chloroplast (23, 45, 82–87). We therefore performed complementary experiments to address these questions for some of the proteins identified here.
We first wanted to validate the subcellular localization of proteins recently associated to the chloroplast but lacking a predictable transit peptide. Two proteins, SFR2 and AtTSP9, were fused to cyan fluorescent protein, and their subcellular localization was analyzed in a transgenic Arabidopsis plant by fluorescence microscopy (supplemental Data 13). In good agreement with recently published data, we obtained evidence for plastid targeting of both SFR2, which behaves as an outer envelope membrane protein (81), and AtTSP9, which was previously demonstrated through subcellular fractionation and antibody detection to behave as a thylakoid membrane protein (88). Altogether, these data demonstrate that these two proteins are genuine chloroplast proteins.
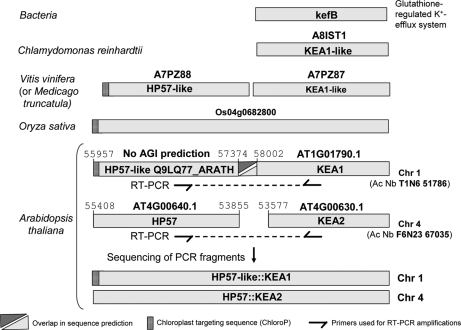
The conclusion was very different for the two proteins, KEA1 and KEA2, we detected in envelope fractions. Having been predicted as potassium transporters, one would expect that they contain a classical transit peptide like most inner envelope membrane proteins and especially transporters. Searching for experimental information about their N termini, we could not find any ESTs in Arabidopsis databases. These two proteins were of similar size to orthologues in bacteria, algae, and other plants (Fig. 9). However, we detected rice orthologues containing a huge additional N terminus (Fig. 9). Searching for sequences similar to this additional N terminus within the Arabidopsis genome, we detected two unknown Arabidopsis proteins (supplemental Data 14). The N terminus of KEA2 was identified as HP57 that was also detected in the envelope during this work (supplemental Data 14). The N terminus of KEA1 was identified as an HP57-like protein (no AGI accession number) (supplemental Data 14). Performing PCR amplification of cDNAs using primers selected within the HP57 and KEA sequences, we demonstrated that these two predicted proteins are in fact two parts of the same large envelope protein (Fig. 9). In other words, the N-terminal regions of KEA proteins are not predicted to contain plastid targeting peptides because these predicted N termini are in fact central parts of a genuine chloroplast envelope protein. Because of the huge size of the expected cDNAs (above 4 kbp), the N terminus of the HP57 proteins was not covered by existing EST sequences. However, the N terminus of the HP57-like protein is predicted to contain a classical plastid targeting peptide (Fig. 9) in good agreement with its localization within the inner envelope membrane. Furthermore, 5′ rapid amplification of cDNA end PCR experiments are required to validate these predicted N-terminal sequences, and it is expected that the correct N terminus of HP57 should also contain such a classical plastid targeting peptide.
Fig. 9.
KEA1 and KEA2 were detected in chloroplast envelope proteome. These two proteins are of similar size when compared with their homologues in bacteria, algae, and some plants. However, some predicted rice proteins contain a huge additional N terminus. The N terminus of KEA2 (AT4G00630.1) was identified as the HP57 protein (AT4G00640.1) that was also detected in the envelope fraction during this work (supplemental Data 14). The N terminus of KEA1 (AT1G01790.1) was identified as an HP57-like protein (no AGI accession number; Q9LQ77_ARATH) (supplemental Data 14) for which peptides were also detected during this work. PCR amplification demonstrated that these predicted proteins are two parts of the same large envelope protein. The N terminus of one of the two HP57 proteins is predicted to contain a classical plastid targeting peptide in good agreement with the genuine localization of this protein within the inner membrane of the chloroplast envelope. Chr, chromosome; Ac Nb, accession number.
We also detected the carbonic anhydrases CA1 and CA2 in the envelope and the stroma (see supplemental Data 10, lanes 40 and 41). However, if CA1 is effectively expected to be localized within the stroma, CA2 was recently demonstrated to be exclusively localized in the cytosol (89). As discussed above, the envelope is poorly contaminated with cytosolic proteins (only 0.3% when considering spectral count). Trying to understand the presence of the cytosolic CA2 in plastids, we found that the TAIR database contains five predicted structures of the CA2 protein (At5g14740.1 to At5g14740.5). One model (At5g14740.2) has a short N terminus that is not predicted to contain a plastid targeting sequence. The four other models contain an identical additional N-terminal sequence that is predicted as a chloroplast transit peptide. Checking the sequence of the primer used previously (89) to amplify the 5′-end of the CA2 cDNA before expression of CA2-green fluorescent protein fusions in planta confirmed that the shortest model (At5g14740.2) was chosen. In other words, previous experiments performed to associate CA2 to the cytosol might have led to this conclusion due to the use of a truncated version of the predicted CA2 protein. Alternative splicing might explain the dual localization of the protein in the cytosol and the chloroplast. However, if the present data clearly demonstrate that CA2 is at least associated to plastids, alternative splicing leading to the cytosolic localization of CA2 remains to be demonstrated.
Chloroplast envelope fractions were found to be contaminated with abundant markers from the tonoplast. Indeed, 12 subunits of the same complex, the vacuolar ATPase, were detected in the purified envelope fraction (supplemental Data 10, lanes 688–699). These abundant tonoplast proteins were only seen in envelope membranes and never detected in the stroma or the thylakoid membranes, thus suggesting that envelope is the only plastidial compartment contaminated with abundant tonoplast proteins. Surprisingly, two TIP-type aquaporins were specifically detected in the thylakoid fraction; one of them was even enriched in the thylakoid when compared with its content in the envelope (supplemental Data 10, lanes 685 and 686). As none of the abundant tonoplast markers could be detected in purified envelope fractions, this suggests that the association of these TIP proteins with the thylakoid membrane might be real. The presence of aquaporins in the chloroplast has recently been suggested (90). In good agreement with these data, we also detected in envelope preparations (but not in thylakoids) four members of the PIP family during this study. Again, these four proteins were classified as putative contaminants (supplemental Data 10, lanes 679–682), but the genuine envelope localization of one or several isoforms of the PIP family cannot be excluded.
Controversy about Envelope Localization of Some Proteins
The present work produced some data that are in conflict with previously published studies. Recent data (91) described the identification, expression, and functional analyses of TAAC, a novel thylakoid ATP/ADP carrier from Arabidopsis. In fact, proteomics provides a strikingly different view as this protein was detected in chloroplast envelope membranes (26) but never in extensive proteome analyses targeted to thylakoid membranes (38, 92, 93). Therefore, a number of proteomics studies favor the hypothesis that TAAC is a genuine envelope protein.
Another recent study (94) described the identification, expression, and functional expression in Escherichia coli of ANTR1, a thylakoid Na+-dependent phosphate transporter from Arabidopsis. In this report, ANTR1 was only detected in the thylakoids and not detected in purified envelope membranes (see Fig. 2 of Ref. 94). Again, in our study, ANTR1 was only detected in the envelope (supplemental Data 10, lane 205) and not detected during extensive proteomics analyses targeted to the thylakoid membranes (38, 92, 93). We thus raised specific polyclonal antibodies against the ANTR1 protein. In good agreement with the proteomics data, the ANTR1 protein was only detected within the purified envelope membrane and not detected in the thylakoid membranes (supplemental Data 4). As detection of this signal in the envelope membranes might have resulted from cross-hybridization of the anti-ANTR1 antibodies with another envelope protein, the ANTR1 protein was overexpressed in Arabidopsis, and envelope and thylakoid membranes from two independent overexpressing plants were purified (supplemental Data 4). Altogether, these data demonstrate that ANTR1 is a genuine envelope protein and does not reside at the thylakoids.
Envelope Membranes and Compartmentation of Chloroplast Metabolism
Chloroplast metabolism is highly compartmentalized, and spectral counting provided an interesting view of the interacting roles of envelope, stroma, and thylakoids. As expected, protein localization and compartmentation of well known chloroplast functions (photosynthesis, transport, amino acid metabolism, protein synthesis, etc.) were strongly correlated (Fig. 10). For instance, transport functions are mainly localized in the envelope (Fig. 10B). The case of proteins involved in amino acid metabolism is also remarkable as most of the proteins of this functional class were exclusively identified in stroma fractions (Fig. 10C). Among the numerous chloroplast pathways essential for carbon fixation, lipid metabolism, and starch and amino acid biosynthesis, we will emphasize here on only two examples, lipid and terpenoid metabolism, because they involve the three main chloroplast compartments. About a hundred proteins involved in lipid metabolism (fatty acid biosynthesis, export, and metabolism; desaturation and oxylipin metabolism; and the synthesis of chloroplast-specific glycerolipids) have been identified in chloroplasts by proteomics (see supplemental Data 8 and 10), and we know now which genes are actually expressed in mature chloroplasts. For instance, several key proteins are clearly missing from the chloroplast protein repertoire: a conspicuous example concerns several galactolipid-synthesizing enzymes as only the major MGD1 was identified. In fact, this is not so surprising as the “missing” MGD2 and MGD3 are mostly expressed in non-green tissues (95), and therefore only very minor amounts should be present in leaves, whereas expression of the genes encoding MGD2, MGD3, DGD1, and DGD2 is induced when plants are subjected to phosphate deprivation (44). Spectral counting provides further molecular evidence for two main chloroplast compartments playing a major role in lipid biosynthesis. The initial steps of fatty acid biosynthesis take place in the stroma (up to the formation of acyl-acyl carrier protein) with some interaction with the envelope at key steps (such as the formation of acetyl-CoA, the precursor for fatty acids, or the export of fatty acids for phospholipid biosynthesis in the endomembrane system). Then, the envelope membranes concentrate most of the proteins involved in chloroplast glycerolipid metabolism, fatty acid desaturation, and formation of fatty acid-derived signaling molecules (for a review, see Ref. 96). Furthermore, proteomics also provides some clues on the complex dialogue, which is essential to produce prokaryotic and eukaryotic chloroplast lipids, that takes place between the envelope membranes and the endomembranes as the whole set of TGD proteins (96) was identified by proteomics in envelope membranes.
Fig. 10.
Relative spectral counts and subplastidial localization for selected functional classes. The size of the spots is proportional to the number of proteins having the same relative spectral count distribution between envelope, stroma, and thylakoids fractions. A, how to read the diagrams. x axis, percentage of spectral count corresponding to envelope fractions; y axis, percentage of spectral count corresponding to stroma fractions; yellow box (ENV), proteins that are mainly localized in the envelope; orange box (STR), proteins that are mainly localized in the stroma; green box (THY), proteins that are mainly localized in the thylakoids. B–H, diagrams showing the subplastidial localization, based on spectral count, for some functional classes as defined by MapManBin (42) (see supplemental Data 9, column “MapManBin”). The functional classes considered are transport (B), amino acid metabolism (C), photosystems (D), not assigned (E), protein degradation (F), protein synthesis (G), and Calvin cycle (H).
Targeted proteomics also helped draw a picture of the tight metabolic network for terpenoid metabolism involving about a hundred of proteins residing within all chloroplast compartments. Supplemental Data 8 demonstrates that almost all enzymes involved in the biosynthesis of soluble precursors for chlorophyll, carotenoids, or prenylquinones were identified unambiguously in the stroma fraction. Then protoporphyrin IX is formed within the chloroplast membranes and is channeled into different pathways: the ferrochelatase involved in heme biosynthesis is tightly linked to thylakoids, whereas chlorophyll biosynthesis takes place in both thylakoids and envelope membranes. In contrast, proteomics revealed that the biosynthesis of hydrophobic carotenoids and prenylquinones is almost restricted to envelope membranes. Although our understanding of the carotenoid biosynthetic pathway in plants has been advanced greatly in the past decade primarily as a result of molecular genetic and biochemical genomics-based approaches (97), very little was known about its precise localization within chloroplasts mostly because these enzymes have historically proven to be extremely difficult to purify and analyze. Indeed, the present data allowed careful examination of the subplastidial localization of proteins of carotenoid metabolism (supplemental Data 15) and led to the conclusion that enzymes involved in phytoene desaturation and cyclization of lycopene reside tat the envelope membranes that are responsible for generating carotenoid diversity as these pathways mark a branch point to two major cyclic carotenoid groups: the β,β-carotenoids (such as β-carotene, zeaxanthin, and violaxanthin) and β,ε-carotenoids (such as lutein). Zeaxanthin is then converted to the epoxycarotenoid violaxanthin by an epoxidase, whereas the reverse reaction is catalyzed by a de-epoxidase. Here, the enzymes involved show a unique distribution among the chloroplast membranes that is related to the function of xanthophylls within these two membrane systems. Whereas the epoxidase is present in both envelope membranes and thylakoids, the de-epoxidase is restricted to thylakoids. The envelope membranes, which are devoid of de-epoxidase activity, are therefore involved in violaxanthin synthesis, whereas in thylakoids, both epoxidase and de-epoxidase catalyze the reactions of the so-called xanthophyll cycle responsible for non-photochemical quenching of singlet-excited chlorophyll during photosynthesis. This suggests that envelope membranes, because of carotenoids and prenylquinones, have a safeguarding function in chloroplasts, especially against oxidative stress.
Conclusion
This investigation concerns chloroplasts, the organelles of the plant cell that harbor the chemical process by which green plants synthesize organic compounds from carbon dioxide and water in the presence of sunlight. Analyzing the chloroplast proteome not only helps understanding how plants efficiently harvest light energy but will also lead to a better understanding of the multitude of functions of the chloroplast as the chloroplast is essential for carbon fixation, lipid metabolism, and starch and amino acid biosynthesis. This constitutes an important economic issue not only from the plant biotechnology standpoint but also for the production of highly energetic compounds via cultivation of microalgae, such as Chlamydomonas reinhardtii. The present work can also provide a strong basis to carry out differential proteomics in other physiologically and economically important plastids such as amyloplasts and chromoplasts.
The present work is the first study designed to address the accurate proteomics-based localization of chloroplast proteins with respect to the three major chloroplast compartments: stroma, thylakoids, and envelope. We identified 1323 proteins, thus matching previous results obtained in a comparable high throughput experiment (57). As a whole, the data currently available indicate that almost 1,700 chloroplast proteins are amenable to proteomics investigations with current MS technologies and conventional fractionation techniques. This only represents one-third to one-half of the total proteins that are estimated to be localized within the chloroplast (1). As mentioned above, several key proteins are still clearly missing from the chloroplast protein repertoire. To get a more exhaustive repertoire of the Arabidopsis chloroplast, forthcoming generations of mass spectrometers will probably be more sensitive and will help to increase this repertoire. Nevertheless, getting a better overview of the chloroplast proteome will mainly rely on more targeted strategies. First, sample preparation and fractionation of plastid subfractions (outer and inner envelope, stroma, thylakoids, lumen, and plastoglobules) could be improved to gain access to minor chloroplast proteins. Those types of fractions will be worth revisiting with current MS instrumentations and bioinformatics tools. Also, other types of plastids (amyloplasts, chromoplasts, etc.) could be analyzed to identify proteins that are more abundant in those types of plastids (98). Unfortunately, not all plants species can be used for purifying specialized plastids. Second, MS-targeted approaches can be used to check whether proteins that are suspected to be chloroplast-localized (e.g. by prediction tools) are actually localized in the chloroplast. To do so, selected reaction monitoring could be used to detect proteins of interest. With the use of internal standards (peptides or proteins), an assessment of the amount of the targeted proteins, within the a given fraction, could also be given (99). Another interesting extension of our work would be through the use of a “top-down” analytical strategy (100, 101) to study full-length envelope proteins from their native environment to access information about protein processing and post-translational modifications.
The present report also represents a unique resource for plastid envelope proteins with respect to both functional and localization issues. Highly curated information on proteins identified in the envelope fraction revealed new insight and ascertained some hypotheses about the different metabolisms associated to the envelope. These data provide unexpected localization information for many chloroplast proteins that were often either not known to reside within the envelope membranes or incorrectly associated to the wrong subplastidial compartment (see for instance supplemental Data 10). Forthcoming review articles, targeted to specific chloroplast metabolisms, are required to delve deeper into the wealth of information contained in our experimental data and to compare these results with previously known or suspected subplastidial localization of the corresponding proteins (see for instance Refs. 102 and 103). The information derived from the AT-CHLORO database will thus be of valuable interest for the plant community. This is particularly true for the envelope compartment as the envelope membranes are the only membrane structure common to all types of plastids and present at all stages of plastid differentiation (19).
Finally, the present work also provides the first AMT database dedicated to plant and usable for label-free quantification experiments. Thus, it now becomes possible to identify and quantify thousands of chloroplast peptides via high throughput nano-LC-MS experiments (17, 18). Such experiments are currently being carried out to screen the impact of different environmental conditions or of knock-out mutations at the chloroplast level. Moreover, such an AT_CHLORO database is usable by any laboratory having a high resolution mass spectrometer (FT-ICR or Orbitrap) provided that standardization of the retention times is achieved (104) and that tools dedicated to the AMT strategy are used.
Acknowledgments
We acknowledge Renaud Dumas, Claude Alban, and Olivier Vallon for the generous gift of the KARI, BCCP1, and LHCPs antibodies, respectively. Véronique Dupierris and Yves Vandenbrouck are acknowledged for having made available on line the AT_CHLORO database (Microsoft Access database).
* This work was supported in part by the CNRS Institut des Sciences Biologiques, CEA, INSERM, INRA, and Grenoble University and by Fondation Rhône-Alpes Futur (to S. B.).
 This article contains supplemental Data 1–15 and validated proteins.
This article contains supplemental Data 1–15 and validated proteins.
1 The abbreviations used are:
- PPDB
- Plant Protein Database
- AMT
- accurate mass and time
- PS
- photosystem
- ER
- endoplasmic reticulum
- WT
- wild type
- LHCP
- light-harvesting complex protein
- TAIR
- The Arabidopsis Information Resource
- FDR
- false discovery rate
- NCBI
- National Center for Biotechnology Information
- LHC
- light-harvesting complex
- EST
- expressed sequence tag
- AGI
- Arabidopsis Genome Initiative
- ceQORH
- chloroplast envelope quinone oxydo-reductase homolog
- KARI
- keto-acid reducto liomerase
- OEE
- oxygen-evolving element
- ACCase
- acetyl-coa carboxylase
- ACCD
- acetyl-coa carboxylase
- Toc
- translocon at the outer membrane of the chloroplast envelope
- Tic
- translocon at the inner membrane of the chloroplast envelope.
REFERENCES
- 1.Jarvis P. (2008) Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol 179, 257–285 [DOI] [PubMed] [Google Scholar]
- 2.Joyard J., Block M., Pineau B., Albrieux C., Douce R. (1990) Envelope membranes from mature spinach chloroplasts contain a NADPH:protochlorophyllide reductase on the cytosolic side of the outer membrane. J. Biol. Chem 265, 21820–21827 [PubMed] [Google Scholar]
- 3.Lilley K. S., Dupree P. (2007) Plant organelle proteomics. Curr. Opin. Plant Biol 10, 594–599 [DOI] [PubMed] [Google Scholar]
- 4.Haynes P. A., Roberts T. H. (2007) Subcellular shotgun proteomics in plants: looking beyond the usual suspects. Proteomics 7, 2963–2975 [DOI] [PubMed] [Google Scholar]
- 5.Rossignol M., Peltier J. B., Mock H. P., Matros A., Maldonado A. M., Jorrín J. V. (2006) Plant proteome analysis: a 2004–2006 update. Proteomics 6, 5529–5548 [DOI] [PubMed] [Google Scholar]
- 6.Ephritikhine G., Ferro M., Rolland N. (2004) Plant membrane proteomics. Plant Physiol. Biochem 42, 943–962 [DOI] [PubMed] [Google Scholar]
- 7.Sun Q., Zybailov B., Majeran W., Friso G., Olinares P. D., van Wijk K. J. (2009) PPDB, the Plant Proteomics Database at Cornell. Nucleic Acids Res 37, D969–D974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heazlewood J. L., Verboom R. E., Tonti-Filippini J., Small I., Millar A. H. (2007) SUBA: the Arabidopsis Subcellular Database. Nucleic Acids Res 35, D213–D218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleffmann T., Hirsch-Hoffmann M., Gruissem W., Baginsky S. (2006) plprot: a comprehensive proteome database for different plastid types. Plant Cell Physiol 47, 432–436 [DOI] [PubMed] [Google Scholar]
- 10.Dunkley T. P., Hester S., Shadforth I. P., Runions J., Weimar T., Hanton S. L., Griffin J. L., Bessant C., Brandizzi F., Hawes C., Watson R. B., Dupree P., Lilley K. S. (2006) Mapping the Arabidopsis organelle proteome. Proc. Natl. Acad. Sci. U.S.A 103, 6518–6523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foster L. J., de Hoog C. L., Zhang Y., Zhang Y., Xie X., Mootha V. K., Mann M. (2006) A mammalian organelle map by protein correlation profiling. Cell 125, 187–199 [DOI] [PubMed] [Google Scholar]
- 12.Gilchrist A., Au C. E., Hiding J., Bell A. W., Fernandez-Rodriguez J., Lesimple S., Nagaya H., Roy L., Gosline S. J., Hallett M., Paiement J., Kearney R. E., Nilsson T., Bergeron J. J. (2006) Quantitative proteomics analysis of the secretory pathway. Cell 127, 1265–1281 [DOI] [PubMed] [Google Scholar]
- 13.Kislinger T., Cox B., Kannan A., Chung C., Hu P., Ignatchenko A., Scott M. S., Gramolini A. O., Morris Q., Hallett M. T., Rossant J., Hughes T. R., Frey B., Emili A. (2006) Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell 125, 173–186 [DOI] [PubMed] [Google Scholar]
- 14.Lipton M. S., Pasa-Tolic L., Anderson G. A., Anderson D. J., Auberry D. L., Battista J. R., Daly M. J., Fredrickson J., Hixson K. K., Kostandarithes H., Masselon C., Markillie L. M., Moore R. J., Romine M. F., Shen Y., Stritmatter E., Tolic' N., Udseth H. R., Venkateswaran A., Wong K. K., Zhao R., Smith R. D. (2002) Global analysis of the Deinococcus radiodurans proteome by using accurate mass tags. Proc. Natl. Acad. Sci. U.S.A 99, 11049–11054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hixson K. K., Adkins J. N., Baker S. E., Moore R. J., Chromy B. A., Smith R. D., McCutchen-Maloney S. L., Lipton M. S. (2006) Biomarker candidate identification in Yersinia pestis using organism-wide semiquantitative proteomics. J. Proteome Res 5, 3008–3017 [DOI] [PubMed] [Google Scholar]
- 16.May D., Fitzgibbon M., Liu Y., Holzman T., Eng J., Kemp C. J., Whiteaker J., Paulovich A., McIntosh M. (2007) A platform for accurate mass and time analyses of mass spectrometry data. J. Proteome Res 6, 2685–2694 [DOI] [PubMed] [Google Scholar]
- 17.Smith R. D., Pasa-Tolic L., Lipton M. S., Jensen P. K., Anderson G. A., Shen Y., Conrads T. P., Udseth H. R., Harkewicz R., Belov M. E., Masselon C., Veenstra T. D. (2001) Rapid quantitative measurements of proteomes by Fourier transform ion cyclotron resonance mass spectrometry. Electrophoresis 22, 1652–1668 [DOI] [PubMed] [Google Scholar]
- 18.Pasa-Tolic L., Lipton M. S., Masselon C. D., Anderson G. A., Shen Y., Toliæ N., Smith R. D. (2002) Gene expression profiling using advanced mass spectrometric approaches. J. Mass Spectrom 37, 1185–1198 [DOI] [PubMed] [Google Scholar]
- 19.Douce R., Joyard J. (1990) Biochemistry and function of the plastid envelope. Annu. Rev. Cell Biol 6, 173–216 [DOI] [PubMed] [Google Scholar]
- 20.Block M. A., Douce R., Joyard J., Rolland N. (2007) Chloroplast envelope membranes: a dynamic interface between plastids and the cytosol. Photosynth. Res 92, 225–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salvi D., Rolland N., Joyard J., Ferro M. (2008) Purification and proteomic analysis of chloroplasts and their sub-organellar compartments. Methods Mol. Biol 432, 19–36 [DOI] [PubMed] [Google Scholar]
- 22.Seigneurin-Berny D., Gravot A., Auroy P., Mazard C., Kraut A., Finazzi G., Grunwald D., Rappaport F., Vavasseur A., Joyard J., Richaud P., Rolland N. (2006) HMA1, a new Cu-ATPase of the chloroplast envelope, is essential for growth under adverse light conditions. J. Biol. Chem 281, 2882–2892 [DOI] [PubMed] [Google Scholar]
- 23.Miras S., Salvi D., Ferro M., Grunwald D., Garin J., Joyard J., Rolland N. (2002) Non-canonical transit peptide for import into the chloroplast. J. Biol. Chem 277, 47770–47778 [DOI] [PubMed] [Google Scholar]
- 24.Morsomme P., Dambly S., Maudoux O., Boutry M. (1998) Single point mutations distributed in 10 soluble and membrane regions of the Nicotiana plumbaginifolia plasma membrane PMA2 H+-ATPase activate the enzyme and modify the structure of the C-terminal region. J. Biol. Chem 273, 34837–34842 [DOI] [PubMed] [Google Scholar]
- 25.Werhahn W., Niemeyer A., Jänsch L., Kruft V., Schmitz U. K., Braun H. (2001) Purification and characterization of the preprotein translocase of the outer mitochondrial membrane from Arabidopsis. Identification of multiple forms of TOM20. Plant Physiol 125, 943–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferro M., Salvi D., Brugière S., Miras S., Kowalski S., Louwagie M., Garin J., Joyard J., Rolland N. (2003) Proteomics of the chloroplast envelope membranes from Arabidopsis thaliana. Mol. Cell. Proteomics 2, 325–345 [DOI] [PubMed] [Google Scholar]
- 27.Bruinsma J. (1961) A comment on the spectrophotometric determination of chlorophyll. Biochim. Biophys. Acta 52, 576–578 [DOI] [PubMed] [Google Scholar]
- 28.Dumas R., Joyard J., Douce R. (1989) Purification and characterization of acetohydroxyacid reductoisomerase from spinach chloroplasts. Biochem. J 262, 971–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alban C., Jullien J., Job D., Douce R. (1995) Isolation and characterization of biotin carboxylase from pea chloroplasts. Plant Physiol 109, 927–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seigneurin-Berny D., Rolland N., Garin J., Joyard J. (1999) Technical Advance: Differential extraction of hydrophobic proteins from chloroplast envelope membranes: a subcellular-specific proteomic approach to identify rare intrinsic membrane proteins. Plant J 19, 217–228 [DOI] [PubMed] [Google Scholar]
- 31.Block M. A., Dorne A. J., Joyard J., Douce R. (1983) Preparation and characterization of membrane fractions enriched in outer and inner envelope membranes from spinach chloroplasts. II. Biochemical characterization. J. Biol. Chem 258, 13281–13286 [PubMed] [Google Scholar]
- 32.Ferro M., Seigneurin-Berny D., Rolland N., Chapel A., Salvi D., Garin J., Joyard J. (2000) Organic solvent extraction to identify hydrophobic chloroplast membrane proteins. Electrophoresis 21, 3517–3526 [DOI] [PubMed] [Google Scholar]
- 33.Chua N. H. (1980) Electrophoresis analysis of chloroplast proteins. Methods Enzymol 69, 434–436 [Google Scholar]
- 34.Jaquinod M., Villiers F., Kieffer-Jaquinod S., Hugouvieux V., Bruley C., Garin J., Bourguignon J. (2007) A proteomics dissection of Arabidopsis thaliana vacuoles isolated from cell culture. Mol. Cell. Proteomics 6, 394–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng J., Elias J. E., Thoreen C. C., Licklider L. J., Gygi S. P. (2003) Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res 2, 43–50 [DOI] [PubMed] [Google Scholar]
- 36.Dupierris V., Masselon C., Court M., Kieffer-Jaquinod S., Bruley C. (2009) A toolbox for validation of mass spectrometry peptides identification and generation of database: IRMa. Bioinformatics 25, 1980–1981 [DOI] [PubMed] [Google Scholar]
- 37.Kearney R. E., Bergeron J. J., Bell A., McPherson P., Blondeau F., Drapeau M., Servant F., De Grandpre S., Gilchrist A., Lesimple S., Au C. (December3, 2007) Reduction of redundant protein identification in high throughput proteomics. CA Patent 2,559,108
- 38.Friso G., Giacomelli L., Ytterberg A. J., Peltier J. B., Rudella A., Sun Q., Wijk K. J. (2004) In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. Plant Cell 16, 478–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwacke R., Schneider A., van der Graaff E., Fischer K., Catoni E., Desimone M., Frommer W. B., Flügge U. I., Kunze R. (2003) ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol 131, 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tusnády G. E., Simon I. (2001) The HMMTOP transmembrane topology prediction server. Bioinformatics 17, 849–850 [DOI] [PubMed] [Google Scholar]
- 41.Emanuelsson O., Nielsen H., von Heijne G. (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8, 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L. A., Rhee S. Y., Stitt M. (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37, 914–939 [DOI] [PubMed] [Google Scholar]
- 43.Usaite R., Wohlschlegel J., Venable J. D., Park S. K., Nielsen J., Olsson L., Yates J. R., 3rd (2008) Characterization of global yeast quantitative proteome data generated from the wild-type and glucose repression Saccharomyces cerevisiae strains: the comparison of two quantitative methods. J. Proteome Res 7, 266–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Misson J., Raghothama K. G., Jain A., Jouhet J., Block M. A., Bligny R., Ortet P., Creff A., Somerville S., Rolland N., Doumas P., Nacry P., Herrerra-Estrella L., Nussaume L., Thibaud M. C. (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc. Natl. Acad. Sci. U.S.A 102, 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miras S., Salvi D., Piette L., Seigneurin-Berny D., Grunwald D., Reinbothe C., Joyard J., Reinbothe S., Rolland N. (2007) Toc159- and Toc75-independent import of a transit sequence-less precursor into the inner envelope of chloroplasts. J. Biol. Chem 282, 29482–29492 [DOI] [PubMed] [Google Scholar]
- 46.Adachi J., Kumar C., Zhang Y., Mann M. (2007) In-depth analysis of the adipocyte proteome by mass spectrometry and bioinformatics. Mol. Cell. Proteomics 6, 1257–1273 [DOI] [PubMed] [Google Scholar]
- 47.Bantscheff M., Schirle M., Sweetman G., Rick J., Kuster B. (2007) Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem 389, 1017–1031 [DOI] [PubMed] [Google Scholar]
- 48.Carrie C., Giraud E., Whelan J. (2009) Protein transport in organelles: Dual targeting of proteins to mitochondria and chloroplasts. FEBS J 276, 1187–1195 [DOI] [PubMed] [Google Scholar]
- 49.Crowley K. S., Payne R. M. (1998) Ribosome binding to mitochondria is regulated by GTP and the transit peptide. J. Biol. Chem 273, 17278–17285 [DOI] [PubMed] [Google Scholar]
- 50.Marc P., Margeot A., Devaux F., Blugeon C., Corral-Debrinski M., Jacq C. (2002) Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep 3, 159–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kellems R. E., Allison V. F., Butow R. A. (1975) Cytoplasmic type 80S ribosomes associated with yeast mitochondria. IV. Attachment of ribosomes to the outer membrane of isolated mitochondria. J. Cell Biol 65, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kellems R. E., Allison V. F., Butow R. A. (1974) Cytoplasmic type 80 S ribosomes associated with yeast mitochondria. II. Evidence for the association of cytoplasmic ribosomes with the outer mitochondrial membrane in situ. J. Biol. Chem 249, 3297–3303 [PubMed] [Google Scholar]
- 53.Atteia A., Adrait A., Brugière S., Tardif M., van Lis R., Deusch O., Dagan T., Kuhn L., Gontero B., Martin W., Garin J., Joyard J., Rolland N. (2009) A proteomic survey of Chlamydomonas reinhardtii mitochondria sheds new light on the metabolic plasticity of the organelle and on the nature of the alpha-proteobacterial mitochondrial ancestor. Mol. Biol. Evol 26, 1533–1548 [DOI] [PubMed] [Google Scholar]
- 54.MacKenzie J. A., Payne R. M. (2004) Ribosomes specifically bind to mammalian mitochondria via protease-sensitive proteins on the outer membrane. J. Biol. Chem 279, 9803–9810 [DOI] [PubMed] [Google Scholar]
- 55.Oikawa K., Kasahara M., Kiyosue T., Kagawa T., Suetsugu N., Takahashi F., Kanegae T., Niwa Y., Kadota A., Wada M. (2003) Chloroplast unusual positioning1 is essential for proper chloroplast positioning. Plant Cell 15, 2805–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumatani T., Sakurai-Ozato N., Miyawaki N., Yokota E., Shimmen T., Terashima I., Takagi S. (2006) Possible association of actin filaments with chloroplasts of spinach mesophyll cells in vivo and in vitro. Protoplasma 229, 45–52 [DOI] [PubMed] [Google Scholar]
- 57.Zybailov B., Rutschow H., Friso G., Rudella A., Emanuelsson O., Sun Q., van Wijk K. J. (2008) Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One 3, e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Froehlich J. E., Wilkerson C. G., Ray W. K., McAndrew R. S., Osteryoung K. W., Gage D. A., Phinney B. S. (2003) Proteomic study of the Arabidopsis thaliana chloroplastic envelope membrane utilizing alternatives to traditional two-dimensional electrophoresis. J. Proteome Res 2, 413–425 [DOI] [PubMed] [Google Scholar]
- 59.Ferro M., Salvi D., Riviere-Rolland H., Vermat T., Seigneurin-Berny D., Grunwald D., Garin J., Joyard J., Rolland N. (2002) Integral membrane proteins of the chloroplast envelope: identification and subcellular localization of new transporters. Proc. Natl. Acad. Sci. U.S.A 99, 11487–11492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andersson B., Anderson J. M. (1980) Lateral heterogeneity in the distribution of chlorophyll-protein complexes of the thylakoid membranes of spinach chloroplasts. Biochim. Biophys. Acta 593, 427–440 [DOI] [PubMed] [Google Scholar]
- 61.Mustárdy L., Buttle K., Steinbach G., Garab G. (2008) The three-dimensional network of the thylakoid membranes in plants: quasihelical model of the granum-stroma assembly. Plant Cell 20, 2552–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van der Hoorn R. A. (2008) Plant proteases: from phenotypes to molecular mechanisms. Annu. Rev. Plant Biol 59, 191–223 [DOI] [PubMed] [Google Scholar]
- 63.Sakamoto W. (2006) Protein degradation machineries in plastids. Annu. Rev. Plant Biol 57, 599–621 [DOI] [PubMed] [Google Scholar]
- 64.Adam Z., Rudella A., van Wijk K. J. (2006) Recent advances in the study of Clp, FtsH and other proteases located in chloroplasts. Curr. Opin. Plant Biol 9, 234–240 [DOI] [PubMed] [Google Scholar]
- 65.Alban C., Job D., Douce R. (2000) Biotin metabolism in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol 51, 17–47 [DOI] [PubMed] [Google Scholar]
- 66.Thelen J. J., Ohlrogge J. B. (2002) Both antisense and sense expression of biotin carboxyl carrier protein isoform 2 inactivates the plastid acetyl-coenzyme A carboxylase in Arabidopsis thaliana. Plant J 32, 419–431 [DOI] [PubMed] [Google Scholar]
- 67.Alban C., Baldet P., Douce R. (1994) Localization and characterization of two structurally different forms of acetyl-CoA carboxylase in young pea leaves, of which one is sensitive to aryloxyphenoxypropionate herbicides. Biochem. J 300, 557–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hirsch S., Soll J. (1995) Import of a new chloroplast inner envelope protein is greatly stimulated by potassium phosphate. Plant Mol. Biol 27, 1173–1181 [DOI] [PubMed] [Google Scholar]
- 69.Shorrosh B. S., Savage L. J., Soll J., Ohlrogge J. B. (1996) The pea chloroplast membrane-associated protein, IEP96, is a subunit of acetyl-CoA carboxylase. Plant J 10, 261–268 [DOI] [PubMed] [Google Scholar]
- 70.Drescher A., Ruf S., Calsa T., Jr., Carrer H., Bock R. (2000) The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J 22, 97–104 [DOI] [PubMed] [Google Scholar]
- 71.Peltier J. B., Cai Y., Sun Q., Zabrouskov V., Giacomelli L., Rudella A., Ytterberg A. J., Rutschow H., van Wijk K. J. (2006) The oligomeric stromal proteome of Arabidopsis thaliana chloroplasts. Mol. Cell. Proteomics 5, 114–133 [DOI] [PubMed] [Google Scholar]
- 72.Rolland N., Dorne A. J., Amoroso G., Sültemeyer D. F., Joyard J., Rochaix J. D. (1997) Disruption of the plastid ycf10 open reading frame affects uptake of inorganic carbon in the chloroplast of Chlamydomonas. EMBO J 16, 6713–6726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pohlmeyer K., Soll J., Grimm R., Hill K., Wagner R. (1998) A high-conductance solute channel in the chloroplastic outer envelope from pea. Plant Cell 10, 1207–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Goetze T. A., Philippar K., Ilkavets I., Soll J., Wagner R. (2006) OEP37 is a new member of the chloroplast outer membrane ion channels. J. Biol. Chem 281, 17989–17998 [DOI] [PubMed] [Google Scholar]
- 75.Hemmler R., Becker T., Schleiff E., Bölter B., Stahl T., Soll J., Götze T. A., Braams S., Wagner R. (2006) Molecular properties of Oep21, an ATP-regulated anion-selective solute channel from the outer chloroplast membrane. J. Biol. Chem 281, 12020–12029 [DOI] [PubMed] [Google Scholar]
- 76.Pollmann S., Springer A., Buhr F., Lahroussi A., Samol I., Bonneville J. M., Tichtinsky G., von Wettstein D., Reinbothe C., Reinbothe S. (2007) A plant porphyria related to defects in plastid import of protochlorophyllide oxidoreductase A. Proc. Natl. Acad. Sci. U.S.A 104, 2019–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Philippar K., Geis T., Ilkavets I., Oster U., Schwenkert S., Meurer J., Soll J. (2007) Chloroplast biogenesis: the use of mutants to study the etioplast-chloroplast transition. Proc. Natl. Acad. Sci. U.S.A 104, 678–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Murcha M. W., Elhafez D., Lister R., Tonti-Filippini J., Baumgartner M., Philippar K., Carrie C., Mokranjac D., Soll J., Whelan J. (2007) Characterization of the preprotein and amino acid transporter gene family in Arabidopsis. Plant Physiol 143, 199–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Miyagishima S. Y., Froehlich J. E., Osteryoung K. W. (2006) PDV1 and PDV2 mediate recruitment of the dynamin-related protein ARC5 to the plastid division site. Plant Cell 18, 2517–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Asano T., Yoshioka Y., Kurei S., Sakamoto W., Machida Y. (2004) A mutation of the CRUMPLED LEAF gene that encodes a protein localized in the outer envelope membrane of plastids affects the pattern of cell division, cell differentiation, and plastid division in Arabidopsis. Plant J 38, 448–459 [DOI] [PubMed] [Google Scholar]
- 81.Fourrier N., Bédard J., Lopez-Juez E., Barbrook A., Bowyer J., Jarvis P., Warren G., Thorlby G. (2008) A role for SENSITIVE TO FREEZING2 in protecting chloroplasts against freeze-induced damage in Arabidopsis. Plant J 55, 734–745 [DOI] [PubMed] [Google Scholar]
- 82.Armbruster U., Hertle A., Makarenko E., Zühlke J., Pribil M., Dietzmann A., Schliebner I., Aseeva E., Fenino E., Scharfenberg M., Voigt C., Leister D. (2009) Chloroplast proteins without cleavable transit peptides: rare exceptions or a major constituent of the chloroplast proteome? Mol. Plant 2, 1325–1335 [DOI] [PubMed] [Google Scholar]
- 83.Kitajima A., Asatsuma S., Okada H., Hamada Y., Kaneko K., Nanjo Y., Kawagoe Y., Toyooka K., Matsuoka K., Takeuchi M., Nakano A., Mitsui T. (2009) The rice alpha-amylase glycoprotein is targeted from the Golgi apparatus through the secretory pathway to the plastids. Plant Cell 21, 2844–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nanjo Y., Oka H., Ikarashi N., Kaneko K., Kitajima A., Mitsui T., Muñoz F. J., Rodríguez-López M., Baroja-Fernández E., Pozueta-Romero J. (2006) Rice plastidial N-glycosylated nucleotide pyrophosphatase/phosphodiesterase is transported from the ER-Golgi to the chloroplast through the secretory pathway. Plant Cell 18, 2582–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Villarejo A., Burén S., Larsson S., Déjardin A., Monné M., Rudhe C., Karlsson J., Jansson S., Lerouge P., Rolland N., von Heijne G., Grebe M., Bako L., Samuelsson G. (2005) Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat. Cell Biol 7, 1224–1231 [DOI] [PubMed] [Google Scholar]
- 86.Turkina M. V., Villarejo A., Vener A. V. (2004) The transit peptide of CP29 thylakoid protein in Chlamydomonas reinhardtii is not removed but undergoes acetylation and phosphorylation. FEBS Lett 564, 104–108 [DOI] [PubMed] [Google Scholar]
- 87.Nada A., Soll J. (2004) Inner envelope protein 32 is imported into chloroplasts by a novel pathway. J. Cell Sci 117, 3975–3982 [DOI] [PubMed] [Google Scholar]
- 88.Hansson M., Dupuis T., Strömquist R., Andersson B., Vener A. V., Carlberg I. (2007) The mobile thylakoid phosphoprotein TSP9 interacts with the light-harvesting complex II and the peripheries of both photosystems. J. Biol. Chem 282, 16214–16222 [DOI] [PubMed] [Google Scholar]
- 89.Fabre N., Reiter I. M., Becuwe-Linka N., Genty B., Rumeau D. (2007) Characterization and expression analysis of genes encoding alpha and beta carbonic anhydrases in Arabidopsis. Plant Cell Environ 30, 617–629 [DOI] [PubMed] [Google Scholar]
- 90.Uehlein N., Otto B., Hanson D. T., Fischer M., McDowell N., Kaldenhoff R. (2008) Function of Nicotiana tabacum aquaporins as chloroplast gas pores challenges the concept of membrane CO2 permeability. Plant Cell 20, 648–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Thuswaldner S., Lagerstedt J. O., Rojas-Stütz M., Bouhidel K., Der C., Leborgne-Castel N., Mishra A., Marty F., Schoefs B., Adamska I., Persson B. L., Spetea C. (2007) Identification, expression, and functional analyses of a thylakoid ATP/ADP carrier from Arabidopsis. J. Biol. Chem 282, 8848–8859 [DOI] [PubMed] [Google Scholar]
- 92.Peltier J. B., Ytterberg A. J., Sun Q., van Wijk K. J. (2004) New functions of the thylakoid membrane proteome of Arabidopsis thaliana revealed by a simple, fast, and versatile fractionation strategy. J. Biol. Chem 279, 49367–49383 [DOI] [PubMed] [Google Scholar]
- 93.Peltier J. B., Emanuelsson O., Kalume D. E., Ytterberg J., Friso G., Rudella A., Liberles D. A., Söderberg L., Roepstorff P., von Heijne G., van Wijk K. J. (2002) Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell 14, 211–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pavón L. R., Lundh F., Lundin B., Mishra A., Persson B. L., Spetea C. (2008) Arabidopsis ANTR1 is a thylakoid Na+-dependent phosphate transporter: functional characterization in Escherichia coli. J. Biol. Chem 283, 13520–13527 [DOI] [PubMed] [Google Scholar]
- 95.Awai K., Maréchal E., Block M. A., Brun D., Masuda T., Shimada H., Takamiya K., Ohta H., Joyard J. (2001) Two types of MGDG synthase genes, found widely in both 16:3 and 18:3 plants, differentially mediate galactolipid syntheses in photosynthetic and nonphotosynthetic tissues in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A 98, 10960–10965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Benning C. (2009) Mechanisms of lipid transport involved in organelle biogenesis in plant cells. Annu. Rev. Cell Dev. Biol 25, 71–91 [DOI] [PubMed] [Google Scholar]
- 97.Breitenbach J., Sandmann G. (2005) zeta-Carotene cis isomers as products and substrates in the plant poly-cis carotenoid biosynthetic pathway to lycopene. Planta 220, 785–793 [DOI] [PubMed] [Google Scholar]
- 98.Baginsky S. (2009) Plant proteomics: concepts, applications, and novel strategies for data interpretation. Mass Spectrom. Rev 28, 93–120 [DOI] [PubMed] [Google Scholar]
- 99.Brun V., Masselon C., Garin J., Dupuis A. (2009) Isotope dilution strategies for absolute quantitative proteomics. J. Proteomics 72, 740–749 [DOI] [PubMed] [Google Scholar]
- 100.Whitelegge J. P., Laganowsky A., Nishio J., Souda P., Zhang H., Cramer W. A. (2006) Sequencing covalent modifications of membrane proteins. J. Exp. Bot 57, 1515–1522 [DOI] [PubMed] [Google Scholar]
- 101.Zabrouskov V., Whitelegge J. P. (2007) Increased coverage in the transmembrane domain with activated-ion electron capture dissociation for top-down Fourier-transform mass spectrometry of integral membrane proteins. J. Proteome Res 6, 2205–2210 [DOI] [PubMed] [Google Scholar]
- 102.Joyard J., Ferro M., Masselon C., Seigneurin-Berny D., Salvi D., Garin J., Rolland N. (2010) Chloroplast proteomics highlights the subcellular compartmentation of lipid metabolism. Prog. Lipid Res 49, 128–158 [DOI] [PubMed] [Google Scholar]
- 103.Joyard J., Ferro M., Masselon C., Seigneurin-Berny D., Salvi D., Garin J., Rolland N. (2009) Chloroplast proteomics and the compartmentation of plastidial isoprenoid biosynthetic pathways. Mol. Plant 2, 1154–1180 [DOI] [PubMed] [Google Scholar]
- 104.Tarasova I. A., Guryca V., Pridatchenko M. L., Gorshkov A. V., Kieffer-Jaquinod S., Evreinov V. V., Masselon C. D., Gorshkov M. V. (2009) Standardization of retention time data for AMT tag proteomics database generation. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci 877, 433–440 [DOI] [PubMed] [Google Scholar]