Abstract
Calmodulin is an essential regulator of intracellular processes in response to extracellular stimuli mediated by a rise in Ca2+ ion concentration. To profile protein-protein interactions of calmodulin in human brain, we probed a high content human protein array with fluorophore-labeled calmodulin in the presence of Ca2+. This protein array contains 37,200 redundant proteins, incorporating over 10,000 unique human neural proteins from a human brain cDNA library. We designed a screen to find high affinity (KD ≤ 1 μm) binding partners of calmodulin and identified 76 human proteins from all intracellular compartments of which 72 are novel. We measured the binding kinetics of 74 targets with calmodulin using a high throughput surface plasmon resonance assay. Most of the novel calmodulin-target complexes identified have low dissociation rates (koff ≤ 10−3 s−1) and high affinity (KD ≤ 1 μm), consistent with the design of the screen. Many of the identified proteins are known to assemble in neural tissue, forming assemblies such as the spectrin scaffold and the postsynaptic density. We developed a microarray of the identified target proteins with which we can characterize the biochemistry of calmodulin for all targets in parallel. Four novel targets were verified in neural cells by co-immunoprecipitation, and four were selected for exploration of the calmodulin-binding regions. Using synthetic peptides and isothermal titration calorimetry, calmodulin binding motifs were identified in the potassium voltage-gated channel Kv6.1 (residues 474–493), calmodulin kinase-like vesicle-associated protein (residues 302–316), EF-hand domain family member A2 (residues 202–216), and phosphatidylinositol-4-phosphate 5-kinase, type I, γ (residues 400–415).
High content protein arrays allow for identification of putative binding partners over all cellular compartments. The technique may be especially valuable for identifying targets of central signaling proteins that are known to regulate a large number of proteins, for example calmodulin. To our knowledge, this method has not been applied to human calmodulin. Array screening has several advantages over other methods, for example affinity chromatography (1). First, affinity chromatography may lead to identification of the more abundant proteins and the capture of secondary proteins that bind to primary calmodulin targets. On the protein arrays, the proteins are presented in distinct locations, and secondary targets are not likely to be identified. Second, array screening is effective in identifying interactions with transmembrane proteins, including receptors and ion channels, which are typically not available in tissue homogenate used for identification through affinity chromatography (2). A further advantage of this array system is the ability to return to the protein-expressing clone of an identified target protein and express it for further characterization.
Calmodulin is present in all eukaryotic cells and constitutes at least 0.1% of the total cellular protein. It is expressed at higher levels in brain, testes, and rapidly growing cells. It participates in signaling pathways that regulate processes such as cell proliferation, learning and memory, growth, and movement (3–5). Regulation of these events is exerted via direct interactions of calmodulin with a large number of cytosolic proteins, including kinases, phosphatases, and cytoskeletal proteins, in response to a rise in intracellular Ca2+ concentration. In the nucleus, calmodulin is also known to transmit Ca2+ signals to a number of transcription factors (5–7). Following an extracellular stimulus, Ca2+ moves into the cytosol either from the outside of the cell via plasma membrane Ca2+ channels or from intracellular stores. Recently, we identified calmodulin as playing a role in the homeostasis of intracellular Ca2+ through binding to the endoplasmic reticulum transmembrane proteins STIM11 and STIM2 (8). Upon binding of Ca2+, calmodulin undergoes a conformational change, and its binding affinity for a number of target proteins increases. The biological action of calmodulin is governed by its biophysical properties, including cooperative Ca2+ binding (9) and structural autonomy of two globular domains (10) that cooperate in target binding (11–14). Previous studies have identified many putative calmodulin-binding proteins in higher and lower organisms (1, 15–19).
In this study, we probed a high content protein array derived from human brain (20) with fluorophore-labeled calmodulin. The fluorophore was attached to an engineered cysteine residue at a surface-exposed position. By this method, we identified 76 high affinity interaction partners of calmodulin. The targets were validated using a high throughput surface plasmon resonance approach. We identified high affinity calmodulin binding to a subset of proteins known to be engaged in the remodeling of the molecular architecture of cells such as the dendritic spine, which is involved in Ca2+-dependent mechanisms of synaptic function and plasticity (21). In addition to four human proteins previously known to bind calmodulin, we identified 72 novel human calmodulin-binding proteins, including receptor tyrosine kinases, cytoplasmic scaffold proteins, signaling enzymes, and cytoskeletal elements that may be regulated in a Ca2+-dependent manner through the concerted action of calmodulin. We demonstrated that such high affinity networks can be characterized using a calmodulin target microarray by examining the Ca2+ sensitivity of calmodulin binding to all interacting proteins in parallel. Four putative binding sites were produced as synthetic peptides, and their binding to calmodulin was verified using isothermal titration calorimetry. We further confirmed the binding of calmodulin to four targets from the array screen in neural cells using immunoprecipitation of endogenous calmodulin with a monoclonal antibody to the C terminus of the protein in a mouse hippocampal cell line.
EXPERIMENTAL PROCEDURES
Calmodulin Mutant Design
Through inspection of all available high resolution structures (x-ray and NMR structures) of calmodulin in its Ca2+-free, Ca2+-bound, and target-bound forms, Ser-17 was identified as a small uncharged hydrophilic residue that is solvent-exposed in all structures. The mutant Ser-17 → Cys was designed to allow for minimal perturbation site-specific fluorophore labeling for array screening and site-specific immobilization for surface plasmon resonance (SPR) studies.
Expression and Purification of Recombinant Human Calmodulin
Full-length human calmodulin was expressed from a modified Pet3a vector (“PetSac” with NdeI and SacI cloning sites) containing a synthetic calmodulin gene that was built with the codons optimal for expression in Escherichia coli (22). The calmodulin gene with mutation Ser-17 → Cys was amplified by PCR from this vector using primers containing the desired base change in two steps using standard procedures. The PCR product was digested by NdeI and SacI and cloned into the PetSac vector. Following transformation, the wild-type and mutant proteins were expressed in E. coli strain BL21 Des3 pLysS star, and the wild-type protein was purified as described (22). The mutant was purified using the same protocol except that 1 mm DTT was included until the final desalting step to keep the protein monomeric. Briefly, the purification procedure involved sonication of the cells, centrifugation, pouring the supernatant into boiling buffer and heating to 85 °C to precipitate E. coli proteins, centrifugation, ion exchange chromatography on DEAE-cellulose in buffer containing CaCl2, phenyl-Sepharose chromatography (loading and washing in CaCl2 followed by elution with EDTA), size exclusion chromatography, and ion exchange chromatography on DEAE-Sephacel in buffer with EDTA. Ca2+-free (apo) calmodulin was prepared by chelating Ca2+ with EDTA and its subsequent removal on a gel filtration column. Purity was confirmed by agarose gel electrophoresis in EDTA and Ca2+, SDS-polyacrylamide gel electrophoresis, and 1H NMR spectroscopy. Titrations in the presence of Quin2 (23) confirmed that the apoprotein was free from Ca2+ (residual Ca2+ less than 0.04 molar eq, which is less than 1% of full saturation).
Alexa Fluor 488 and 546 Labeling of Calmodulin S17C
Apocalmodulin with the S17C (or Ser-17 → Cys) mutation was dissolved in 20 mm sodium phosphate buffer, pH 8 at a concentration of 10 mg/ml. Alexa Fluor 488 or 546 (Invitrogen) (1.2 molar eq) was added from a 5 mg/ml stock in DMSO, and the sample was allowed to react for 1 h in the dark at room temperature. The labeled protein (now called CaM-Alexa488 or CaM-Alexa546) was then separated from excess label in water on a Sephadex G25 size exclusion column, which had been prewashed with EDTA to remove trace metal. The collected protein was divided into aliquots for single use and stored frozen at −20 °C and subsequently diluted for the binding experiments on the arrays. The control proteins calbindin D9k and calbindin D28k were labeled in the same fashion via coupling to an engineered cysteine, whereas secretagogin and anti-His tag antibody were labeled using amine-reactive Alexa Fluor.
Calmodulin-binding Protein Profiling on High Density Protein Arrays
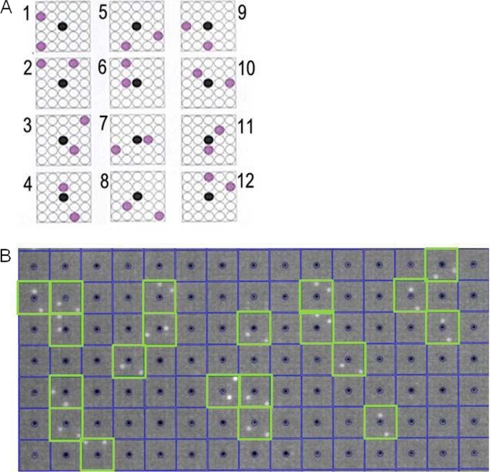
High density protein arrays of the human brain library (hEx1) were obtained from the German Resource Centre for Genome Research (RZPD). The PVDF arrays were soaked in 95% ethanol, rinsed in deionized water, and washed clean of residual bacterial colonies with 20 mm Tris/HCl, 500 mm NaCl, 0.05% (v/v) Tween 20, pH 7.4 (TBST) with 0.5% (v/v) Triton X-100. For calmodulin-binding protein profiling, the protein arrays were blocked in 2% (w/v) nonfat dry milk powder in 20 mm Tris/HCl, 150 mm NaCl, pH 7.4 (TBS) for 2 h; washed twice in TBST; and subsequently incubated with CaM-Alexa488 at a concentration of 1 μm in TBS with 1 mm CaCl2. The protein arrays were washed in TBST with 1 mm CaCl2 six times for 10 min each. The arrays were illuminated with long wave UV light, and the images were taken using a high resolution charge-coupled device detection system (Fuji). Image analysis was performed with VisualGrid (GPC Biotech). Hits were counted as positive if both spots of a clone (in a characteristic pattern around a guiding point; see Fig. 1) were significantly brighter than background.
Fig. 1.
Scoring of human protein (hEx1) arrays with VisualGrid software. A, scoring pattern for each of 12 clones spotted in duplicate around a central guide dot. B, 1 μm CaM-Alexa488 binding to a field of the human protein array with positive clones highlighted in green squares on the blue grid that identifies each block of 24 spots.
Purification of Putative Target Proteins
The individual protein-expressing clones that express putative calmodulin targets were cultured for 20 h in 10 ml of Overnight Express culture medium (24). This medium contains a ratio of the sugars glucose and lactose that supports growth of the bacteria sequentially. Once the glucose is exhausted, the bacteria begin to metabolize the lactose, thereby depressing the T7lac promoter resulting in expression of the recombinant protein from the vector cassette. The recombinant proteins were purified from the cell pellet using metal affinity chromatography under denaturing conditions (100 mm NaH2PO4, 10 mm Tris/HCl, 6 m guanidine hydrochloride, pH 8.0) utilizing the polyhistidine tag for binding to Ni2+-NTA resin. Proteins were eluted in 1 ml of elution buffer (100 mm NaH2PO4, 10 mm Tris/HCl, 8 m urea, pH 4.5), desalted and concentrated in spin columns, and washed three times with PBS (9.5 mm phosphate buffer with 153 mm Na+, 4.1 mm K+, 140 mm Cl−, pH 7.4). In cases where there was more than one expressing clone for the same protein, we set up protein expression and purification from several clones. Phosphorylase kinase, used as a positive control, was purchased from Sigma.
Surface Plasmon Resonance Studies with Immobilized Calmodulin
All SPR experiments were carried out using a Biacore 3000 instrument. S17C calmodulin was immobilized using ligand thiol disulfide exchange coupling. The dextran matrix of a CM5 chip was activated by injecting 25 μl of a fresh mixture of 0.05 m N-hydroxysuccinimide and 0.2 m 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide. A reactive disulfide group was then introduced on the sensor chip surface by injecting 20 μl of 100 μm 2-(2-pyridinyldithioethaneamine), 0.1 m sodium borate, pH 8.5. Calmodulin was then immobilized by injecting 100 μl of 10 μg/ml calmodulin S17C in 10 mm HCO2Na (sodium formate), pH 4.3. Finally, residual 2-(2-pyridinyldithioethaneamine) groups were deactivated by injecting 40 μl of 50 mm l-cysteine, 1 m NaCl, 100 mm HCO2Na, pH 4.3. Blank channels for negative control were prepared by omitting calmodulin in the coupling step. Binding of targets was surveyed by injecting 150 μl of target protein solutions in 10 mm Tris/HCl, 150 mm KCl, 1 mm CaCl2, 0.005% (v/v) Tween 20, pH 7.5. The same buffer was used as running buffer. Dissociation of target protein from calmodulin was followed for 90 min during buffer flow. The chip was then regenerated by injecting 100 μl of 350 mm EDTA, pH 8. The flow rate was 10 μl/min throughout the experiment.
Surface Plasmon Resonance studies with Immobilized Targets
The purified calmodulin target proteins were immobilized on NTA sensor chips. The chip was activated by injecting 20 μl of 0.5 mm NiCl2, and then each protein was immobilized by injecting 150 μl of target protein solution. Calmodulin association was followed during an injection of 150 μl of 700 nm calmodulin in 10 mm Tris/HCl, 150 mm KCl, 1 mm CaCl2, 0.005% (v/v) Tween 20, pH 7.5. The same buffer was used as running buffer, and the flow rate was 10 μl/min. The dissociation of calmodulin was monitored for at least 90 min during buffer flow, and the chip was then regenerated by injecting first 50 μl of 350 mm EDTA, pH 8 and then 100 μl of 0.5% (w/v) SDS followed by a final 50-μl injection of 350 mm EDTA.
Analysis of Surface Plasmon Resonance Data
Curve fitting to all SPR data was performed using the BIAeval software. Dissociation phase data were analyzed by fitting a single exponential decay function to the data,
where R is the response, A is the amplitude, and koff is the dissociation rate constant. The variable parameters were koff and A.
Association phase data were analyzed by fitting the following function to the data with a fixed value of koff as obtained above,
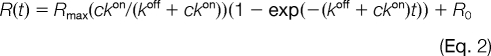 |
where R is the response, c is the protein concentration in the flow, kon is the association rate constant, Rmax is the signal that would have been obtained at full saturation of the immobilized targets, and R0 is the response resulting from injection of the protein. The variable parameters were kon, Rmax, and R0. An estimate of KD was obtained as follows.
In some cases, the data could not be satisfactorily fitted without invoking two processes with separate rates, both for the dissociation phase (Equation 4) and association phase (Equation 5).
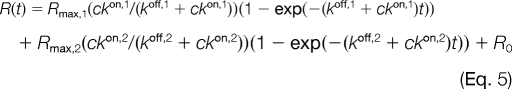 |
Endogenous Calmodulin Co-immunoprecipitation
Confluent monolayers of mouse hippocampal Hpl 3-4 cells (25) from 10-cm2 tissue culture plates were lysed in 0.7 ml of lysis buffer (50 mm Tris/HCl, 150 mm NaCl, 1% Triton X-100, 2 mm CaCl2 with protease inhibitors (0.068 units/ml aprotinin and 2 mm PMSF)), and cells were disrupted with repeated aspiration through a 21-gauge needle. After centrifugation, the supernatant was precleared of nonspecific binders to IgG and/or agarose beads by addition of 1.0 μg of mouse monoclonal anti-M13 IgG (27-9420-01, GE Healthcare), and 20 μl Protein A/G PLUS-agarose (sc-2003, Santa Cruz Biotechnology). After incubation at 4 °C for 30 min, the suspension was centrifuged at 2,500 rpm for 5 min at 4 °C to remove the agarose beads with captured antibody. The supernatant (∼500 μg of total cellular protein) was transferred to a fresh microcentrifuge tube on ice to which 2 μg of mouse anti-calmodulin (ab38841, Abcam) or 2 μg of mouse anti-calbindin D9k (sc-74462, Santa Cruz Biotechnology) antibody was added and incubated for 1 h at 4 °C. Protein A/G PLUS-agarose (20 μl) was added, and the suspension was incubated for 2 h at 4 °C. The suspension was centrifuged at 2,500 rpm for 5 min at 4 °C, and the pellet was washed our times with lysis buffer and once with PBS with 2 mm CaCl2. The pellet (immunoprecipitates) was resuspended in 0.125 m Tris/HCl, 20% glycerol, 10% 2-mercaptoethanol, 4% SDS, 0.004% bromphenol and subjected to SDS-PAGE on either 7.5 or 12% polyacrylamide gels followed by transfer to PVDF membrane (Amersham Biosciences). The membranes were incubated in 2% nonfat milk in TBST buffer containing rabbit polyclonal anti-potassium voltage-gated channel Kv6.1 (anti-KCNG1; ab49063, Abcam), anti-glutamate (NMDA) receptor subunit ζ1 (anti-NMDAR1; G8913, Sigma), or anti-CaM kinase-like vesicle-associated protein (anti-CaMKV; ab69564, Abcam) antibody, or mouse monoclonal anti-spectrin α chain (ab11755, Abcam) antibody followed by 3 × 5-min washes. The membranes were then incubated with horseradish peroxidase-conjugated secondary antibodies in TBST buffer, and the enhanced chemiluminescence was measured 1 min after adding luminol ECL reagent (Pierce).
Generation of Protein Microarrays
FAST slides (Schleicher & Schuell) were placed in a QArray System (Genetix, New Milton, UK; humidity controlled at 60–65%) with 16 blunt-ended stainless steel print tips with a tip diameter of 150 μm that were used to generate the protein arrays. Each identified calmodulin-binding protein was spotted on these FAST slides eight times in separate spots from a stock solution with a concentration of 0.1 mg/ml. For most of the proteins, spotting of the undiluted purified proteins led to immobilization of 15 fmol of protein/spot on the microarray. The amount of protein was estimated based on gel band intensity on SDS-PAGE of the spotted solutions. Each protein microarray contained several control spots, including mouse monoclonal anti-calmodulin IgG (Sigma-Aldrich) at 0.1 mg/ml, phosphorylase kinase, and myo-inositol-1(or 4)-monophosphatase. A stock solution of 200 nm CaM-Alexa546 was spotted as an internal control for signal intensity. The microarrays were blocked in 2% milk, TBST for 1 h and washed twice for 10 min in TBS with 1 mm EDTA and twice in TBS with 1 mm Ca2+.
CaM-Alexa Screening of Protein Microarrays
The microarrays were incubated with 1 μm CaM-Alexa546 in TBS with either 1 mm CaCl2 or 1 mm EDTA for 1 h at room temperature in the dark. The microarrays were washed in the same buffer as used in the binding experiment with 0.05% (v/v) Tween 20 3 × 5 min, dried, and imaged using a Genepix scanner (4000B, Axon Instruments). The same conditions were used for all control proteins tested, including secretagogin (purified in our laboratory; Ref. 26), calbindin D28k (purified in our laboratory; Ref. 27), and anti-His tag antibody (28). All experiments were conducted at least twice.
Peptide Synthesis and Purification
The four peptides corresponding to the putative calmodulin-binding sites from the CaM kinase-like vesicle-associated protein (AQIEKNFARAKWKKA; CaMKV(302–316)); EF-hand domain family member A2 (VWKGSSKLFRNLKEKG; EFHA2(202–216)); phosphatidylinositol-4-phosphate 5-kinase, type I, γ (LQSYRFIKKLEHTWKA; PIP5K1C(400–415)), and the potassium voltage-gated channel Kv6.1 (QERVMFRRAQFLIKTKSQLS; KVGCh(474–493)) were made by solid-phase methodology using a standard Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry protocol utilizing an Applied Biosystems 433A or a PS3 Protein Technologies Peptide Synthesizer. For all peptides, the PAL resin was used to furnish a C-terminal carboxamide. A mixture of O-benzotriazole-N,N,N′,N′-tetramethyluronium hexafluorophosphate and N-hydroxybenzotriazole was used in the activation of the free carboxylic acids (4–10 molar eq). β-Branched amino acids and residues immediately following a β-branched residue were coupled twice. Prior to cleavage of the peptide from the resin, the peptides were N-acetylated with a solution containing 0.5 m pyridine and 0.5 m acetic anhydride in N,N-dimethylformamide. The cleavage of the peptide from the resin was achieved with TFA/thioanisole/1,2-ethanedithiol/anisole in a volume ratio of 9.0:0.5:0.3:0.2 for 2 h at room temperature. The solution was then concentrated with a stream of N2 gas, and the crude peptide was precipitated with ice-cold diethyl ether, collected by filtration, dissolved in a mixture of water and acetonitrile, and lyophilized. The crude peptide was purified by HPLC using a C4 preparative column (Vydac) and a linear gradient of water containing 0.1% TFA by volume (solvent A) and acetonitrile/water 9:1 containing 0.1% TFA (solvent B) with a gradient of 0.5% increase in solvent B/min. The identity of each peptide was verified by ESI MS performed by Texas A&M University, College Station, TX and found to be within 0.1% of the expected molecular weight.
Isothermal Titration Calorimetry
Isothermal titration calorimetry (ITC) was carried out using a VP-ITC microcalorimeter (MicroCal, Piscataway, NJ). All samples were prepared by dissolving desalted lyophilized protein or peptide in 10 mm Tris, 150 mm KCl, pH 7.50 with either 1 mm CaCl2 or 1 mm EDTA and adjustment of pH to 7.50 if needed. Each of four synthetic peptides was titrated from a 200 μm stock solution into 10 μm wild-type human calmodulin at 25 °C. An initial injection of 5 μl was followed by 29 injections of 10 μl of peptide solution with 5-min equilibration time between injections. The concentrations of peptide stocks were determined by amino acid analysis after acid hydrolysis (analysis purchased from BMC, Uppsala, Sweden), and the calmodulin concentration was determined by absorbance using ε = 3200 liters mol−1 cm−1. Data were fitted using the Orgin 7 software (MicroCal) and a 1:1 binding model.
RESULTS
Identification of Putative Calmodulin Targets on Human Protein Array
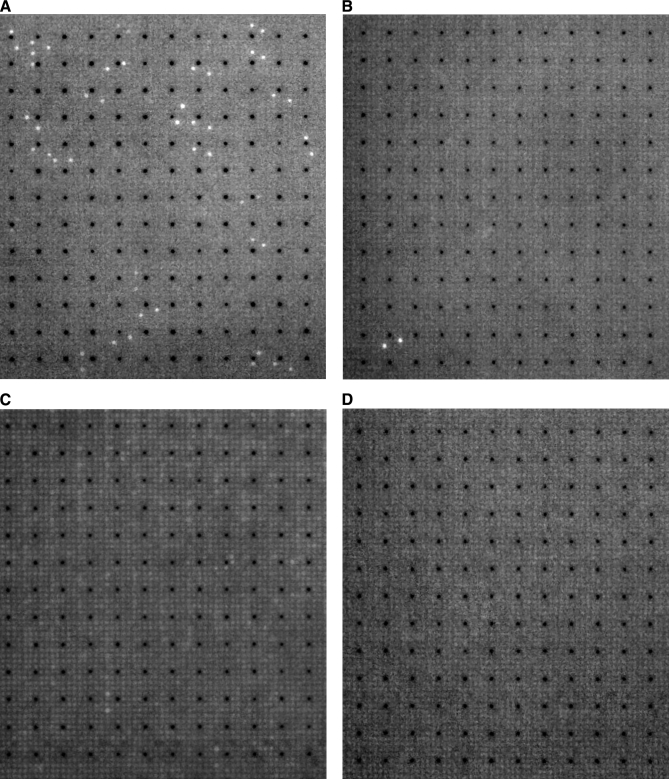
In this work, we used high content human protein arrays on 22.2 × 22.2-cm PVDF membranes (20). Each protein array was robotically spotted in a standard 5 × 5 spotting pattern with each array containing 27,648 protein spots in duplicate, making a total of 55,296 protein spots per array. The protein collection of 37,200 clones was spotted as follows: one membrane with 27,648 individual clones spotted in duplicate and one membrane with 9,552 individual clones spotted in duplicate multiple times. Together, the two membranes held 37,200 redundant proteins of which over 10,000 were estimated to be unique (non-redundant) human proteins based on sequencing of a subset of the clones (29). Twelve proteins were arrayed in duplicate in the 5 × 5 spotting pattern around a central ink guiding point (Fig. 1). Before use, the membranes were washed clean of residual bacterial colonies, leaving the expressed proteins on the membrane. The array was screened for calmodulin-binding proteins in the presence of 1 mm Ca2+ using calmodulin labeled at a cysteine residue with the fluorescent probe Alexa Flour 488 (CaM-Alexa488; Fig. 2A). The cysteine was site-specifically substituted for serine 17, which is solvent-exposed in all known structures of calmodulin, causing minimum perturbation of the binding interactions.
Fig. 2.
Equivalent fields of high content protein array incubated overnight with 1 μm protein labeled with Alexa Fluor 488 in TBS buffer with 1 mm CaCl2 followed by 6 × 10-min washes in TBST buffer for total of 60 min. A, calmodulin; B, secretagogin; C, calbindin D28k; D, calbindin D9k.
We initially identified 1,200 clones as binding CaM-Alexa488 on the high content redundant array after a 10-min wash in 20 mm Tris/HCl, 150 mm NaCl, 1 mm CaCl2, pH 7.5. This set may include a number of lower affinity target proteins. Following a series of more stringent washes (6 × 10 min with 20 mm Tris/HCl, 500 mm NaCl, 1 mm CaCl2, 0.05% Tween 20, pH 7.5) this number was reduced to 480 positives clones, representing a set of high affinity binding proteins. DNA sequencing of the clones led to the identification of a non-redundant set of 76 high affinity partners of human calmodulin, 72 of which were not previously identified as calmodulin binding (Table I and supplemental Table S1). For 13 of these 72 proteins, either a mouse or nematode (Caenorhabditis elegans) homologue has been reported to bind to calmodulin (1, 19). A subset of these proteins was identified as proteins involved in the organization of the postsynaptic density consisting of four known and eight novel calmodulin-binding proteins (supplemental Table S2).
Table I. Calmodulin target proteins with accession number (Swiss-Prot), KD, and whether previously identified as a calmodulin-binding protein indicated (italics).
The affinity estimated from fitting to SPR data is given as KD with values estimated within 1 order of magnitude (±0.5 log10 units). Human proteins that are previously known as calmodulin targets are highlighted in italics. References to earlier work are given for mouse (1), rat (59) and C. elegans (19) homologues. ND, not determined. CTD, C-terminal domain.
| Protein | Accession no. | KD |
|---|---|---|
| Membrane proteins | ||
| Solute carrier family 16, member 8/MCT3 | O95907 | 1 nm |
| Solute carrier family 7, member 5 | Q01650 | 100 nm |
| Neuron-specific protein family member 2 | Q9Y328 | 1 nm |
| Plasticity-related protein 2 (1) | Q6T4P5 | 100 pm |
| Potassium voltage-gated channel Kv6.1 | Q9UIX4 | 100 nm |
| Glutamate (NMDA) receptor subunit ζ1 precursor | Q05586 | 10 nm |
| Tetraspanin-7 | P41732 | 1 nm |
| Lysophospholipid acyltransferase 7 | Q96N66 | 100 pm |
| Semaphorin 3A (1) | P51805 | 1 nm |
| Transmembrane protein 9B precursor | Q9NQ34 | 10 nm |
| Semaphorin 4C precursor | Q9C0C4 | 10 nm |
| Cleft lip- and palate-associated transmembrane protein 1 | O96005 | 1 nm |
| Receptor accessory protein 2 | Q9BRK0 | 10 nm |
| Fibroblast growth factor receptor 3 precursor | P22607 | 100 pm |
| Yip1-interacting factor homologue B isoform 2 | Q5BJH7 | 10 nm |
| STIM1 | Q13586 | 100 nm |
| Similar to double C2-like domain-containing protein β (1) | Q14184 | 1 μm |
| Ras-related protein Rab-11B (1) | Q15907 | 10 nm |
| Syntaxin-18 | Q9P2W9 | 10 nm |
| Sterol-regulatory element-binding protein cleavage-activating protein | Q12770 | 1 nm |
| β-Sarcoglycan | Q16585 | 1 nm |
| Regulator of G-protein signaling 19 | P49795 | 10 nm |
| EF-hand domain family, member A2 | Q86XE3 | 10 nm |
| CaM kinase-like vesicle-associated (59) | Q8NCB2 | 1 nm |
| Synaptosomal associated protein 29 | O95721 | 1 nm |
| Serine incorporator 2 | Q96SA4 | 1 μm |
| Receptor-type tyrosine-protein phosphatase ζ precursor | P23471 | 100 nm |
| Coiled-coil-helix-coiled-coil-helix domain containing protein 2 | Q9Y6H1 | 10 nm |
| CaM-kinase II α chain | Q9UQM7 | 100 nm |
| APLP1 | P51693 | 10 nm |
| Coiled-coil domain-containing 88C | Q9P219 | 10 pm |
| Nuclear proteins | ||
| ZNF527 | Q8NB42 | 100 nm |
| ZNF238 | Q99592 | 10 nm |
| ZNF358 | Q9NW07 | 10 nm |
| ZNF330 | Q9Y3S2 | 10 nm |
| Transcription factor IIIA | Q92664 | 100 nm |
| Heterogeneous nuclear ribonucleoprotein D-like | O14979 | 1 μm |
| Zinc fingers and homeoboxes 2 | Q9Y6X8 | 1 nm |
| Bromodomain and PHD finger-containing protein 3 | Q9ULD4 | 1 nm |
| RNA binding motif protein 4 | Q9BWF3 | 10 nm |
| RNA-binding protein 4B | Q9BQ04 | 10 nm |
| Decapping enzyme, scavenger | Q96C86 | 10 nm |
| RNA-binding protein 5 | P52756 | 1 nm |
| G1 to S phase transition 1 (1) | P15170 | 10 nm |
| Inhibitor of growth family, member 4 (p29ING4) | Q9UNL4 | 10 nm |
| DnaJ homologue subfamily C member 2/zuotin-related factor 1 | Q99543 | 10 nm |
| Ran-binding protein 3 | Q9H6Z4 | 100 pm |
| Glioma tumor suppressor candidate region gene 2/p60 | Q9NZM5 | 1 nm |
| TP53I11 | O14683 | 10 nm |
| Phosphorylated CTD-interacting factor 1 | Q9H4Z3 | ND |
| Cytoskeletal proteins | ||
| Tubulin folding cofactor B | Q99426 | 10 nm |
| Dynein, cytoplasmic, heavy polypeptide 1 | Q14204 | 1 μm |
| Actin, β | P60709 | 1 nm |
| Actin, γ1 | P63261 | 1 nm |
| Centromere protein B, 80 kDa | P07199 | 1 nm |
| Kinesin heavy chain isoform 5C (1) | O60282 | 1 nm |
| Spectrin α chain | Q13813 | 10 nm |
| Cytoplasmic proteins | ||
| NLR family member X1 | Q86UT6 | 1 nm |
| Elongation factor 2 | P13639 | 1 nm |
| SRC-like adapter | Q13239 | 1 nm |
| STIP1 homology and U-box-containing protein 1 | Q9UNE7 | 1 nm |
| Pyruvate kinase isozymes M1/M2 (1) | P14618 | 1 nm |
| Phosphatidylinositol-4-phosphate 5-kinase, type I, γ (19) | O60331 | 1 nm |
| Triose-phosphate isomerase 1 variant (1) | P60174 | 10 nm |
| Diphosphomevalonate decarboxylase | P53602 | 100 nm |
| PDZ domain-containing 4 | Q76G19 | 10 nm |
| Stathmin 1/oncoprotein 18 (1) | P16949 | ND |
| Ubiquitin-like protein FUBI | P35544 | 10 nm |
| Kelch-like protein 21 | Q9UJP4 | 1 nm |
| Mitochondrial proteins | ||
| NADH dehydrogenase (ubiquinone) Fe-S protein 7, 20 kDa (19) | O75251 | 1 nm |
| Protein from Golgi | ||
| Proprotein convertase subtilisin/kexin type 1 inhibitor (1) | Q9UHG2 | 100 nm |
| Ribosomal proteins | ||
| 40 S ribosomal protein S2 (1) | P15880 | 1 nm |
| 60 S ribosomal protein L9 | P32969 | 1 nm |
| 60 S ribosomal protein L14 | P50914 | 1 μm |
| Uncategorized proteins | ||
| hCG1805852 | EAW73369 | 1 nm |
| Unnamed protein product | BAC85305 | 1 nm |
The inbuilt redundancy of arrayed proteins serves as a useful internal control in determining protein-protein interactions. This is manifested by binding of calmodulin to many spots that correspond to the same protein. For example, the ribosomal protein S2 was expressed by approximately half of the clones whose protein product was bound by calmodulin on the array. On the other hand, not all existing proteins are represented on the array. The lack of identification of some known targets may be due to factors such as lack of full-length protein, structural constraints, high off rates, etc.
Control experiments were performed with three Alexa Fluor 488-labeled proteins from the calmodulin superfamily of EF-hand proteins, which all are negatively charged at neutral pH. For the hydrophilic protein calbindin D9k, we found no putative targets on the arrays (Fig. 2D), indicating that the hits found for calmodulin are neither mediated by a negative EF-hand protein surface nor the Alexa Fluor 488 label. For each of the hydrophobic EF-hand proteins, calbindin D28k (Fig. 2C) and secretagogin (Fig. 2B), we observed a distinct set of putative targets. None of the 13 targets for secretagogin are shared with calmodulin, whereas one of 48 targets for calbindin D28k is shared with calmodulin (pyruvate kinase isozymes M1/M2). This clearly shows that the putative calmodulin targets are not binding because of an unspecific hydrophobic interaction or a higher abundance of the target on the arrays. In total, 75 of the 76 putative targets are unique to calmodulin.
Validation of Binding Partners by SPR
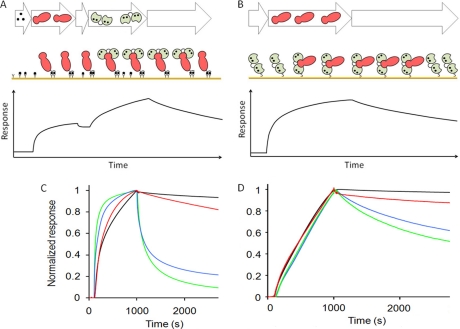
Using SPR assays with either immobilized calmodulin (thiol-linked to CM5 sensor chip) or linked target protein (His tag to Ni2+-NTA sensor chip), 74 of the 76 interactions were successfully validated. Two proteins were not validated because they failed to express in liquid culture. A range of binding affinities and rate constants was observed for the different classes of binding proteins as apparent from variable shapes of the binding curves (Fig. 3 and supplemental Fig. S2). For the proteins that were validated using thiol-linked calmodulin, the dissociation rate constant, koff, was estimated from fitting Equation 1 to the dissociation phase data and found to be in the range of 10−3–10−6 s−1 (supplemental Table S3). The concentration of each target was estimated from the intensity of gel bands on SDS-PAGE (supplemental Fig. S1), providing association rate constants by fitting Equation 2 to the association phase data. The kon values were found to be in the range of 102–105 m−1 s−1 (supplemental Table S3) and are accurate to within 1 order of magnitude (±0.5 log10 units). The equilibrium dissociation constant, KD, was calculated from the estimated association and dissociation rate constants according to Equation 3 (Table I). All targets were studied at least twice using either one or both SPR approaches. The precision of the SPR measurements was evaluated by using the known calmodulin-binding protein, phosphorylase kinase. Using the same method as for the novel targets, we determined a KD of 0.6 nm for the interaction between calmodulin and phosphorylase kinase (supplemental Fig. S3), whereas previously published values for peptide fragments derived from phosphorylase kinase are 1 nm or higher, i.e. within approximately 0.2 log10 units (30). To check the reproducibility of the reported rate and equilibrium constants, six randomly chosen proteins (coiled-coil-helix-coiled-coil-helix domain-containing protein 2, lysophospholipid acyltransferase 7, bromodomain and PHD finger-containing protein 3, NADH dehydrogenase, and Znf358) were analyzed three times on different SPR chips. Both association and dissociation rates showed little difference between experiments, and the standard deviation for the equilibrium dissociation constants was 0.3 log10 K units. Together with the precision of the protein concentration determination, which is needed to estimate the association rate constant, we thus found it appropriate to report rate constants and dissociation constants with a precision of 1 order of magnitude (±0.5 log10 units). The complementary approach using immobilization of the target proteins to nickel (Ni2+)-coated NTA sensor chips may be preferred because the high precision in the injected calmodulin concentration (700 nm) permitted better estimation of the association rate constant and thereby equilibrium constant for 17 proteins. For several proteins, however, calmodulin binding after His tag immobilization was ineffective, possibly because of steric hindrance of the calmodulin binding segment.
Fig. 3.
Surface plasmon resonance studies. A and B, schematics outlining the two SPR approaches with target proteins immobilized via His tag to Ni2+-NTA sensor chips (A) or calmodulin immobilized via a thiol linker to CM5 sensor chips (B). Shown are representative sensorgrams from SPR studies of calmodulin-target interactions in different kinetic ranges for calmodulin binding to His tag-immobilized ribosomal protein S2 (black), APLP1 (red), dynein (blue), and transcription factor IIIA (green) (C) and target protein binding to immobilized calmodulin for ZHX2 (black), elongation factor 2 (red), solute carrier family 16, member 8/MCT3 (blue), and semaphorin 4C (green) (D). All data were obtained in 10 mm Tris/HCl, 150 mm KCl, 1 mm CaCl2, 0.005% (v/v) Tween 20, pH 7.5.
Endogenous Calmodulin Co-immunoprecipitation
Mouse hippocampal Hpl 3-4 cell lysates were subjected to co-immunoprecipitation using anti-calmodulin or mouse anti-calbindin D9k antibody as the precipitating antibody followed by Western blotting with rabbit polyclonal antibodies against glutamate (NMDA) receptor subunit ζ1 (NMDAR1; Fig. 4A), KVGCh (Fig. 4B), and CaMKV (Fig. 4C) and a mouse monoclonal antibody against spectrin α chain (Fig. 4D). All four targets were detected in complex with calmodulin in the cell lysates, whereas no target was found in complex with the negative control protein calbindin D9k. All four proteins chosen for this study are novel human calmodulin targets, and the potassium voltage-gated channel Kv6.1 has not previously been found in any organism.
Fig. 4.
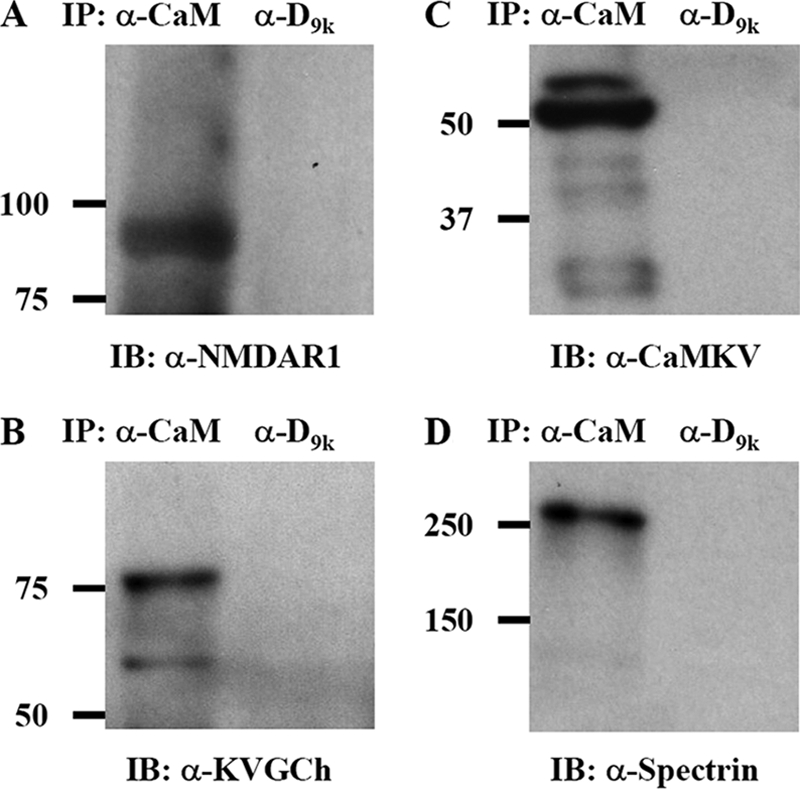
Western blots of immunoprecipitates from hippocampal cell lysates using either anti-calmodulin IgG (left lane in each panel) or anti-calbindin D9k IgG (right lane in each panel) in immunoprecipitation (IP) step and anti-glutamate (NMDA) receptor subunit ζ1 (A), anti-potassium voltage-gated channel Kv6.1 (B), anti-CaMKV (C), or anti-spectrin α chain IgG (D) in the immunoblotting (IB) detection step.
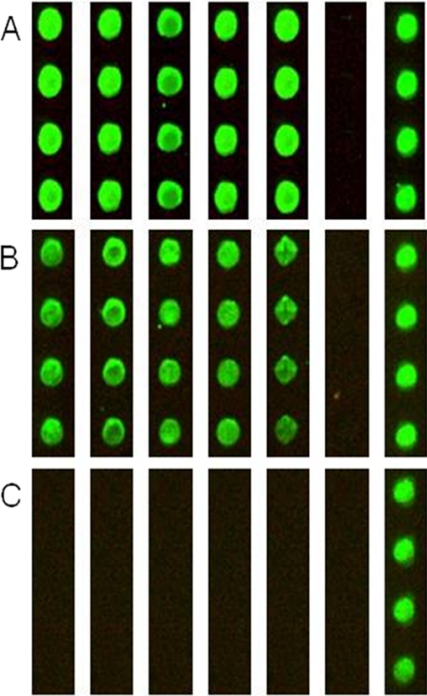
Preparation and Studies of Calmodulin-binding Protein Microarray
Expressed proteins were printed onto FAST slides and incubated with anti-His tag antibody (Fig. 5A), 1 μm CaM-Alexa546 in 1 mm Ca2+ (Fig. 5B), or 1 mm EDTA (Fig. 5C). The slides were washed with buffer at the same Ca2+ or EDTA concentration followed by imaging. In the presence of Ca2+, strong binding was observed to the proteins on the microarray (Fig. 5B), whereas no signal was detected in Ca2+-free buffer (Fig. 5C), highlighting the Ca2+ dependence of the interaction between calmodulin and the arrayed proteins. We also observed a differential intensity of binding of calmodulin to individual members of the microarray that will warrant future studies over a range of Ca2+ concentrations. Control experiments were performed with a 1 μm concentration of the EF-hand proteins secretagogin-Alexa546, calbindin D28k-Alexa546, and calbindin D9k-Alexa546 in the presence of 1 mm Ca2+. No binding of these proteins was observed to the calmodulin target microarray, whereas all microarray proteins were detected with the anti-His tag antibody.
Fig. 5.
Ca2+ sensitivity of binding of calmodulin to calmodulin-interacting proteins on protein microarray. The microarray was incubated with anti-RGSHis6 and Cy3-labeled anti-mouse IgG (A), 1 μm CaM-Alexa546 in 1 mm CaCl2 (B), and 1 μm CaM-Alexa546 in 1 mm EDTA (C). Lane 1, diphosphomevalonate decarboxylase; lane 2, ribosomal protein S2; lane 3, dynein; lane 4, ZNF358; lane 5, CaM kinase II α; lane 6, buffer; lane 7, CaM-Alexa546.
Putative Calmodulin Binding Motifs
The identified calmodulin-binding proteins were investigated for the presence of putative calmodulin binding motifs using the search engine at a web-based database (31). Putative binding sites were found for 67 of 74 proteins, and a list of the highest scoring motif for each protein is presented in supplemental Table S4. Based on this list, synthetic peptides were made corresponding to the putative binding sites of three transmembrane proteins and one cytoplasmic protein. A so-called 1-5-10 motif was predicted at residues 302–316 of CaM kinase-like vesicle-associated protein (AQIEKNFARAKWKKA). A peptide with this sequence, CaMKV(302–316), was synthesized, purified, and used in binding studies. For two other proteins, containing no known motifs, the regions with highest scores were selected (supplemental Table S4). Thus, peptides corresponding to residues 200–216 of EF-hand domain family member A2 (VWKGSSKLFRNLKEKG; EFHA2(202–216)) and residues 400–415 of phosphatidylinositol-4-phosphate 5-kinase, type I, γ (LQSYRFIKKLEHTWKA; PIP5K1C(400–415)) were synthesized, purified, and used in binding studies. For the potassium voltage-gated channel Kv6.1, seven putative binding regions were found by the database search, and four of these (residues 284–303, 308–347, 361–380, and 474–493) are part of the protein expressed by the clone on the array (residues 218–513). The highest score was obtained for residues 361–380 (LGLQTLGLTARRCTREFGLL). The score for the sequence comprising residues 474–493 (QERVMFRRAQFLIKTKSQLS) was 4 times lower but was judged more promising by manual inspection. The 474–493 sequence was therefore chosen, and the corresponding synthetic peptide, KVGCh(474–493), was synthesized, purified, and used in binding studies.
Calmodulin Binding to Motif Peptides by ITC
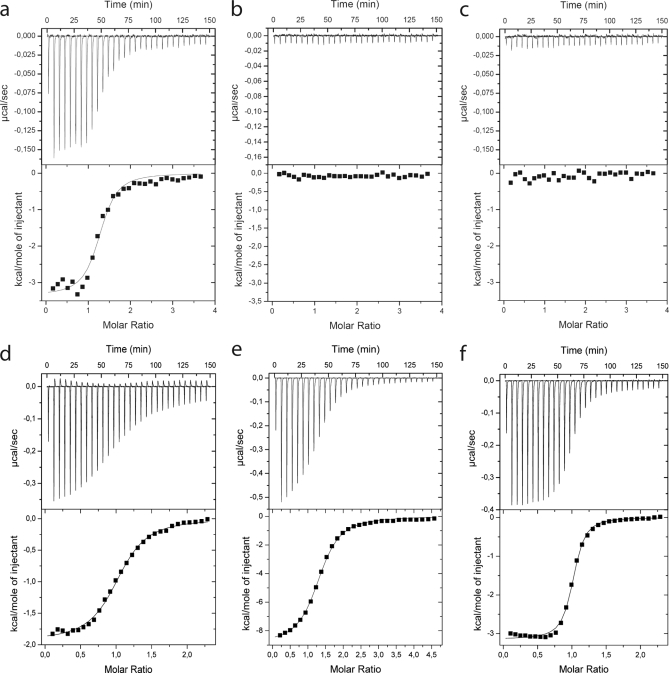
The binding of calmodulin to the four synthetic peptides was studied in the presence and absence of Ca2+ using ITC (Fig. 6). An exothermic process was observed for calmodulin binding to the KVGCh(474–493) peptide in 10 mm Tris/HCl, 150 mm KCl, 1 mm CaCl2, pH 7.5 (Fig. 6a, upper panel). The negative signals obtained indicate that the reaction produces heat, and therefore less heat needs to be added to the sample compared with the reference cell to keep them at constant temperature. Fitting to the integrated data (Fig. 6a, lower panel) using a 1:1 binding model resulted in an estimation of the equilibrium dissociation constant KD of 500 nm. The signals obtained in the presence of 1 mm EDTA (Fig. 6b) were much smaller than in the presence of Ca2+ and of the same magnitude as the friction heat observed when water was injected into water (not shown) or the peptide was injected into buffer (Fig. 6c).
Fig. 6.
Shown is isothermal titration calorimetry at 25 °C for peptides titrated from 200 or 400 μm solutions into 10 μm calmodulin in 10 mm Tris, 150 mm KCl, pH 7. 50 with either 1 mm CaCl2 (a, d, e, and g) or 1 mm EDTA (b) or peptide titrated into buffer (c). An initial injection of 5 μl was followed by 29 injections of 10 μl of peptide solution with 5-min equilibration time between injections. The upper panels show the raw data. The lower panels show integrated heats versus molar ratio of peptide to protein, and the solid lines represent the best fit to the data using a 1:1 binding model. a and b, KVGCh(474–493) titrated into calmodulin in the presence of 1 mm CaCl2 (a) or 1 mm EDTA (b). c, KVGCh(474–493) titrated into buffer with no calmodulin. d, EFHA2(202–216) titrated into calmodulin in the presence of 1 mm CaCl2. e, CaMKV(302–316) titrated into calmodulin in the presence of 1 mm CaCl2. d, PIP5K1C(400–415) titrated into calmodulin in the presence of 1 mm CaCl2.
Exothermic processes were observed in buffer with 1 mm CaCl2 for calmodulin binding to the three peptides EFHA2(202–216) with a KD of 1.7 μm (Fig. 6d), CaMKV(302–316) with a KD of 740 nm (Fig. 6e), and PIP5K1C(400–415) with a KD of 400 nm (Fig. 6f). Weaker binding was observed for all three peptides in the presence of EDTA (data not shown): EFHA2(202–216) with a KD of 3.2 μm, CaMKV(302–316) with a KD of 1.9 μm, and PIP5K1C(400–415) with a KD of 30 μm.
DISCUSSION
Specificity and sensitivity are undoubtedly the most critical issues for high throughput protein-protein interaction screening methods. Validation of the affinities for targets that are identified in high throughput systems using independent quantitative methods must therefore be an integral part of the experimental design. This consideration is of particular relevance in this study where more than 90% of the identified proteins are novel, having not been identified as human calmodulin binding partners in previous studies, and comprise many different protein classes and pathways that may be dependent on Ca2+ signaling in cells.
Through the use of two complementary high throughput SPR approaches, a high degree of confidence in a large number of identified targets was achieved (Table I and supplemental Table S1) with 97% (74 of 76) of the targets confirmed. The high level of validation is particularly powerful considering the large number of novel interacting proteins and in particular for those proteins that have, as yet, no known function. With the microarray, we can further investigate the role of calmodulin in regulating neural processes and study in parallel the differential inhibition of calmodulin targets by for example motif peptides. The human neural calmodulin set of binding proteins described here includes two proteins that have not been characterized in any way beyond their sequence identification and genome location. Their interaction with calmodulin offers a first insight into the function of these uncategorized proteins, which may be a Ca2+-regulated process.
Screening the large arrays of human proteins with three different control proteins from the same protein family showed that the putative calmodulin targets are specific and were not found due to unspecific electrostatic or hydrophobic interactions or were not due to interaction of array proteins with the Alexa Fluor 488 label on an EF-hand protein surface. In array screening, there is also risk for artifacts due to the higher abundance of some proteins on the array; however, screening with anti-His tag antibody showed that the identified calmodulin targets are of similar abundance as other expressed proteins on the array.
The verified targets displayed low dissociation rate constants (koff ranging from 10−3 to 10−6 s−1) and high affinity (KD ranging from 10 pm to 1 μm), consistent with the design of the screen. The variation in binding kinetics may be related to the biological roles of the targets in the cell. For example, transcription factor IIIA has a KD of 100 nm and is characterized by a high on rate and a high off rate (Fig. 3), whereas the protein RPS2 displayed a much higher affinity for calmodulin with a KD of about 1 nm, a lower on rate, and a significantly lower off rate. These proteins may be regulated differently by calmodulin in response to Ca2+ oscillation in the cell.
The human proteins on the high content arrays are heterologously expressed in E. coli and are therefore not post-translationally modified; however, modifications such as glycosylation are not expected for intracellular binding targets for calmodulin. Refolding of arrayed proteins may not be spontaneous after induced protein expression on PVDF membranes followed by lysis of bacterial colonies, and some proteins may remain partially denatured on the array membrane. For many known targets, however, the binding region has been found to be a contiguous segment that is unfolded when not bound to calmodulin. Previous studies have also shown that short peptides with the target binding sequence often retain high affinity binding to calmodulin (32). Unfolding of the targets on the array may therefore not significantly affect their affinity for calmodulin, which is a key consideration in the success of our screen. For membrane proteins, this feature may be especially important as they are unlikely to be correctly folded on the arrays. However, it is recognized that the binding of calmodulin to discontinuous epitopes is not likely to be identified using this method.
The identified target proteins were investigated for the presence of putative calmodulin binding motifs using a web-based database (31). As summarized in supplemental Table S4, this procedure identified putative binding regions in most of the targets. However, only for a few proteins are there previously known types of target motifs. It is therefore likely that a number of novel motifs will be identified within this set of binding partners. For example, the calmodulin binding domain of STIM1 and -2 is a novel motif containing hydrophobic and positively charged residues (8) not identified by the search engine. In addition, mutational studies have shown retained affinity for calmodulin upon removal of positive charges or substitution of hydrophobic residues in binding motifs (33, 34). This reflects a relatively high level of sequence variability and heterogeneity of calmodulin target motifs, and improved bioinformatics tools may be developed as more motif sequences become identified.
Four proteins were taken all the way from being identified as putative calmodulin targets on the large arrays of human proteins through validation as high affinity targets by SPR and finally to calmodulin binding motif discovery, and one of them, the potassium voltage-gated channel Kv6.1, was also verified as a calmodulin binder in neural cells by co-immunoprecipitation. A peptide corresponding to one of the predicted binding sites for the potassium voltage-gated channel Kv6.1 was thus found to bind calmodulin with high affinity in a Ca2+-dependent manner and with an equilibrium binding constant similar to the one found for the protein by SPR. Likewise, we discovered novel calmodulin-binding sites in EF-hand domain family member A2; phosphatidylinositol-4-phosphate 5-kinase, type I, γ; and a motif of known type (1-5-10 motif) at residues 302–316 of the CaM kinase-like vesicle-associated protein.
Immunoprecipitation of endogenous calmodulin with a monoclonal antibody facilitated the successful detection of the membrane proteins potassium voltage-gated channel Kv6.1 and NMDA receptor subunit 1 in complex with calmodulin, suggesting an expanded role of calmodulin in regulation of these ion channels in hippocampal cells. The co-immunoprecipitation of vesicle-associated calmodulin kinase in these cells is added confirmation of high affinity binding of calmodulin to a protein that can travel to dendritic processes in the vesicular machinery supporting synaptic plasticity. The co-immunoprecipitation of spectrin α chain from these cells with calmodulin may serve as a starting point to study the activity of calmodulin in organizing the spectrin scaffold. The confirmation of biophysical data with in-cell binding assays although non-quantitative provides evidence of the significance of these high affinity networks in neural cells.
Target Classification
We identified proteins located in the cytosol, nucleus, mitochondria, endoplasmic reticulum, Golgi apparatus, cytoskeleton, and intracellular and plasma membranes (supplemental Table S1). These proteins include metabolic enzymes, transcription factors, ion channels, transporters, receptors, and transmembrane proteins. The identification of a protein as a calmodulin target and the measurement of the affinity and exchange rate do not prove any regulatory role of calmodulin. Nevertheless, the data presented here may serve as a starting point to investigate the role of calmodulin in a number of critical processes, and we will, in the following discussion, highlight the roles of some of the identified targets. A significant number of the identified target proteins are known to be involved in neural processes including central nervous system development, synaptic function, and neuroendocrine secretion. For example, four previously known calmodulin-binding proteins, NMDAR, spectrin, actin, and Ca2+- and calmodulin-dependent protein kinase IIα (CaMKIIα), play a fundamental role in the organization of the postsynaptic density (PSD) in dendritic spines (35, 36). In addition, we identified a number of novel partners for calmodulin that are fundamentally important to this process (supplemental Table S2). The PSD dynamically changes its structure and composition during development and in response to synaptic activity and is described as a huge membrane-associated protein complex specialized for postsynaptic signaling and plasticity (37–40). The Ca2+ regulation of these processes in response to NMDA receptor activation is exemplified by the role of CaMKIIα in determining the strength of the synaptic transmission (41) and, ultimately, in storage of information in the brain. The identification here of high affinity calmodulin binding to several new proteins involved in PSD architecture raises important questions as to the greater role of calmodulin in synaptic plasticity.
Transmembrane Proteins
We identified the binding of calmodulin to 29 novel transmembrane proteins and two previously known partners (Table I). The previously known partners, CaMKII and glutamate receptor, are integrally involved in processes underlying learning and memory. The novel high affinity targets may reveal new insights into the Ca2+ regulation of synaptic plasticity. All 31 membrane proteins were validated, and their affinity of binding to calmodulin was estimated by fitting to the SPR data. The affinities for the 31 membrane proteins range from 106 to 1011 m−1 (KD values range from 10 pm to 1 μm) in the presence of Ca2+. A very strong effect of Ca2+ is illustrated by the fact that no binding was observed to the calmodulin target microarrays in the absence of Ca2+. The identification of this set of membrane proteins as calmodulin targets may also provide new insights into the role of calmodulin in regulating transport processes over membranes. For example, we have recently indicated a role for calmodulin in regulating store-operated Ca2+ entry by binding to the endoplasmic reticulum single pass transmembrane protein STIM1, a protein identified using this approach (8). Another single pass transmembrane protein that we identified here as a calmodulin target is the EF-hand domain family member A2 (KD = 10 nm). Using ITC, a calmodulin-binding site was found in the cytosolic portion of this protein at residues 202–216 (VWKGSSKLFRNLKEKG; KD = 1.6 μm). The drop in affinity for the peptide compared with intact protein suggests that either additional intermolecular interactions with the intact target contribute to the affinity or that the binding site is somewhat larger or shifted by a few residues.
Potassium voltage-gated channels are important components of the dendritic spine and PSD (42). We observed high affinity calmodulin binding (KD = 100 nm) to the potassium voltage-gated channel isoform Kv6.1 (Swiss-Prot accession number Q9UIX4) and to a peptide corresponding to residues 474–493 (QERVMFRRAQFLIKTKSQLS; KD = 500 nm). The affinity for the peptide is of the same order of magnitude as for the intact protein, suggesting complete coverage of this binding site. Moreover, strong Ca2+ dependence was found both for the interaction between calmodulin and Kv6.1 protein and for the interaction between calmodulin and KVGCh(474–493) peptide. Residues 474–493 constitute a novel calmodulin-binding site, located in the C-terminal cytoplasmic part of Kv6.1, which is of different length and sequence compared with the previously identified calmodulin-binding Kv7.1–7.5 (43). Thus, calmodulin may regulate voltage-gated K+ currents through several targets.
A number of other important proteins in the dendritic spine and PSD were identified as calmodulin targets, for example semaphorin 4C, semaphorin 3A, and dynein heavy chain. Semaphorins are known to act as chemorepulsive molecules that guide axons during neural development. For example, semaphorin 3A signaling pathways have been shown to play an important role in the regulation of dendritic spine maturation in cerebral cortex neurons (44, 45). Dynein heavy chain is proposed to be a major component of PSD and is present in dendritic spines, raising the possibility that cytoplasmic dynein plays structural and functional roles in the postsynaptic terminal (46).
Vesicle fusion and secretion are important synaptic processes that are regulated by Ca2+. We identified the CaM kinase-like vesicle-associated protein as a high affinity calmodulin target (KD = 1 nm). This protein contains a so called 1-5-10 motif at residues 302–316 (AQIEKNFARAKWKKA), and we showed here that a peptide with this sequence, CaMKV(302–316), binds to calmodulin (KD = 600 nm). The novel calmodulin target monocarboxylate transporter 2 (MCT2) is located on vesicular membranes within the postsynaptic spine in rodent brain (47). Synaptosomal associated protein 29 (SNAP29) was found here to bind to calmodulin with high affinity, whereas SNAP25 was previously identified as a target of another Ca2+-regulatory protein of the calmodulin superfamily, secretagogin (26). The protein similar to double C2-like domain-containing protein β (DOC2-β) belongs to a family of proteins identified in mouse brain to regulate synaptic vesicle docking (48).
Cytoplasmic Proteins
The 12 cytoplasmic proteins are all novel. The binding of calmodulin to PIP5K1C (KD = 1 nm) may be of particular interest as a potential new link between two signaling pathways of central importance, Ca2+ signaling and phosphoinositide signaling (5). PIP5K1C acts to phosphorylate phosphatidylinositol 4-phosphate to generate phosphatidylinositol 4,5-bisphosphate. Phosphatidylinositol 4,5-bisphosphate regulates several cellular processes, including actin assembly and vesicle trafficking, processes that seem to involve calmodulin at several levels. A calmodulin-binding site in human PIP5K1C was identified around residues 400–415 (LQSYRFIKKLEHTWKA; KD = 400 nm). A C. elegans homologue of PIP5K1C has previously been found to bind calmodulin in a screening study (19). Besides the novel interaction between calmodulin and PIP5K1C, the Ca2+ and phosphoinositide signaling pathways are linked at several levels. For example, inositol 1,4,5-trisphosphate is a major trigger of Ca2+ release in the cell though binding to the inositol 1,4,5-trisphosphate receptor, which in turn is regulated by calmodulin (49). Calmodulin also binds to and regulates phosphatidylinositol 3-kinase and inositol-1,4,5-trisphosphate 3-kinase (50). Additional links are provided by calbindin D28k, which activates myo-inositol-1(or 4)-monophosphatase (51), and frequenin (neuronal calcium sensor-1), which modulates the activity of phosphatidylinositol 4-kinase (52). Among the other 11 cytoplasmic proteins found in the screen are two proteins that may play a role in the assembly of macromolecular complexes in PSD in response to Ca2+ signaling, i.e. the PDZ domain-containing protein 4 and SRC-like adapter protein.
Nuclear Proteins
All 19 nuclear proteins are novel binding partners of calmodulin, and 18 were validated by SPR technology, including transcription factor IIIA; zinc finger proteins ZNF330, ZNF527, ZNF358, ZNF238, ZHX2, and bromodomain and PHD finger-containing protein 3; RNA binding motif proteins RBM4 and RBM5; and inhibitor of growth family, member 4 (ING4). The related zinc finger protein 268 has been described in mouse hippocampus as having a role in regulation of structural plasticity (53).
Cytoskeletal Proteins
Two of the seven cytoskeletal proteins, spectrin and β-actin, have previously been characterized as human calmodulin-binding proteins. γ-Actin has also been found to be involved in the assembly of these complexes and binds to human calmodulin with equally high affinity as β-actin. The spectrin scaffold in neurons binds to cytoplasmic domains of NMDA receptor subunits (41) and associates with other cytoskeletal structures, protein kinases, and phosphatases enriched in synapses (35, 36, 54, 55). It is fascinating to speculate that calmodulin may be involved in regulating the postsynaptic machinery by linking members of these assemblies in a Ca2+-dependent fashion.
Ribosomal Proteins
Among the three ribosomal proteins are two novel targets (L9 and L14), whereas the mouse ribosomal protein S2 was previously identified as a calmodulin binder by affinity chromatography (1). Future studies may be designed to find out whether calmodulin binds only to free ribosomal proteins, as verified in the present study, or also to these proteins when present in the intact ribosome. Interestingly, translation machinery components, e.g. polyribosomes, have been detected in dendrite shafts and dendritic spines (56–58).
Mitochondrial and Golgi Apparatus Proteins
The mitochondrial protein NADH dehydrogenase (ubiquinone) Fe-S protein 7 is a component of the mitochondrial respiratory chain. A homologue of the proprotein convertase subtilisin/kexin type I inhibitor was previously identified as a calmodulin target in mouse brain (1). Conversion of prohormones and precursor proteins into biologically active peptides and proteins involves the concerted action of a number of convertases and post-translation modification enzymes. The regulation of these enzymes is not well characterized, but the Ca2+-dependent regulation by calmodulin in neural tissues is a potential mechanism by which these processes are controlled.
Unclassified Proteins
Identification and validation of two unclassified proteins as calmodulin targets is intriguing and motivates further studies of the occurrence and function of these proteins.
Concluding Remarks
The present work provides a large number of validated novel neural calmodulin-interacting proteins. Our results suggest that Ca2+/calmodulin regulate a significantly higher number of proteins involved in structural plasticity than previously identified. This serves as a starting point for biochemical and biological studies toward a deeper understanding of Ca2+ signaling in the brain. Particularly interesting is the very large number of membrane proteins that are found to bind calmodulin in the presence of Ca2+. The set of human neural calmodulin targets provides an exciting new foundation to explore the involvement of calmodulin in regulating macromolecular assemblies such as the postsynaptic density and ultimately synaptic strength and potentiation. The calmodulin target microarray provides a versatile tool for investigating factors that regulate the interactions of calmodulin with a large number of targets in parallel.
Acknowledgments
The help with purification of calmodulin S17C by Eva Thulin is gratefully acknowledged. We are grateful to Professor Takashi Onodera, University of Tokyo for the gift of hippocampal cell line Hpl 3-4.
* This work was supported by the Swedish Research Council (Vetenskapsrådet); the Royal Physiographic Society, Lund, Sweden; Science Foundation Ireland and its Walton Visitor Award; and the Koshland Integrated Science Center at Haverford College.
 This article contains supplemental Tables S1–S4 and Figs. S1–S3.
This article contains supplemental Tables S1–S4 and Figs. S1–S3.
1 The abbreviations used are:
- STIM
- stromal interaction molecule
- DEAE
- N,N′-diethylaminoethyl
- ITC
- isothermal titration calorimetry
- NTA
- nitrilotriacetic acid
- PSD
- postsynaptic density
- SPR
- surface plasmon resonance
- CaM
- calmodulin
- NMDA
- N-methyl-d-aspartate
- NMDAR
- N-methyl-d-aspartate receptor
- CaMVK
- CaM kinase-like vesicle-associated protein
- EFHA2
- EF-hand domain family member A2
- PIP5K1C
- phosphatidylinositol-4-phosphate 5-kinase, type I, γ
- KVGCh
- potassium voltage-gated channel Kv6.1
- PHD
- plant homeodomain
- CaMKII
- Ca2+- and calmodulin-dependent protein kinase II
- RZRD
- imaGenes
- CTD
- C-terminal domain.
REFERENCES
- 1.Berggård T., Arrigoni G, Olsson O., Fex M., Linse S., James P. (2006) 140 mouse brain proteins identified by Ca2+-calmodulin affinity chromatography and tandem mass spectrometry. J. Proteome Res 5, 669–687 [DOI] [PubMed] [Google Scholar]
- 2.Larkin D., Murphy D., Reilly D. F., Cahill M., Sattler E., Harriott P., Cahill D. J., Moran N. (2004) ICln, a novel integrin alpha IIb beta 3 associated protein, functionally regulates platelet activation. J. Biol. Chem 279, 27286–27293 [DOI] [PubMed] [Google Scholar]
- 3.Xia Z., Storm D. R. (2005) The role of calmodulin as a signal integrator for synaptic plasticity. Nat. Rev. Neurosci 6, 267–276 [DOI] [PubMed] [Google Scholar]
- 4.Kahl C. R., Means A. R. (2003) Regulation of cell cycle progression by calcium/calmodulin-dependent pathways. Endocr. Rev 24, 719–736 [DOI] [PubMed] [Google Scholar]
- 5.Clapham D. E. (2007) Calcium signaling. Cell 131, 1047–1058 [DOI] [PubMed] [Google Scholar]
- 6.Ikura M., Osawa M., Ames J. B. (2002) The role of calcium-binding proteins in the control of transcription: structure to function. BioEssays 24, 625–636 [DOI] [PubMed] [Google Scholar]
- 7.West A. E., Chen W. G., Dalva M. B., Dolmetsch R. E., Kornhauser J. M., Shaywitz A. J., Takasu M. A., Tao X., Greenberg M. E. (2001) Calcium regulation of neuronal gene expression. Proc. Natl. Acad. Sci. U.S.A 98, 11024–11031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bauer M. C., O'Connell D., Cahill D. J., Linse S. (2008) Calmodulin binding to the polybasic C-termini of STIM proteins involved in store-operated calcium entry. Biochemistry 47, 6089–6091 [DOI] [PubMed] [Google Scholar]
- 9.Linse S., Helmersson A., Forsén S. (1991) Calcium binding to calmodulin and its globular domains. J. Biol. Chem 266, 8050–8054 [PubMed] [Google Scholar]
- 10.Finn B. E., Evenäs J., Drakenberg T., Waltho J. P., Thulin E., Forsén S. (1995) Calcium-induced structural changes and domain autonomy in calmodulin. Nat. Struct. Biol 2, 777–783 [DOI] [PubMed] [Google Scholar]
- 11.Linse S., Drakenberg T., Forsén S. (1986) Mastoparan binding induces a structural change affecting both the N-terminal and C-terminal domains of calmodulin. A 113Cd-NMR study. FEBS Lett 199, 28–32 [DOI] [PubMed] [Google Scholar]
- 12.Ikura M., Clore G. M., Gronenborn A. M., Zhu G., Klee C. B., Bax A. (1992) Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science 256, 632–638 [DOI] [PubMed] [Google Scholar]
- 13.Meador W. E., Means A. R., Quiocho F. A. (1992) Target enzyme recognition by calmodulin: 2.4 Å structure of a calmodulin-peptide complex. Science 257, 1251–1255 [DOI] [PubMed] [Google Scholar]
- 14.Shuman C. F., Jiji R., Åkerfeldt K. S., Linse S. (2006) Reconstitution of calmodulin from domains and subdomains: influence of target peptide. J. Mol. Biol 358, 870–881 [DOI] [PubMed] [Google Scholar]
- 15.Popescu S. C., Popescu G. V., Bachan S., Zhang Z., Seay M., Gerstein M., Snyder M., Dinesh-Kumar S. P. (2007) Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc. Natl. Acad. Sci. U.S.A 104, 4730–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen X., Valencia C. A., Szostak J. W., Szostak J., Dong B., Liu R. (2005) Scanning the human proteome for calmodulin-binding proteins. Proc. Natl. Acad. Sci. U.S.A 102, 5969–5974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu H., Bilgin M., Bangham R., Hall D., Casamayor A., Bertone P., Lan N., Jansen R., Bidlingmaier S., Houfek T., Mitchell T., Miller P., Dean R. A., Gerstein M., Snyder M. (2001) Global analysis of protein activities using proteome chips. Science 293, 2101–2105 [DOI] [PubMed] [Google Scholar]
- 18.Calábria L. K., Garcia Hernandez L., Teixeira R. R., Valle de Sousa M., Espindola F. S. (2008) Identification of calmodulin-binding proteins in brain of worker bees. Comp. Biochem. Physiol. B Biochem. Mol. Biol 151, 41–45 [DOI] [PubMed] [Google Scholar]
- 19.Shen X., Valencia C. A., Gao W., Cotten S. W., Dong B., Huang B. C., Liu R. (2008) Ca(2+)/calmodulin binding proteins from the C. elegans proteome. Cell Calcium 43, 444–456 [DOI] [PubMed] [Google Scholar]
- 20.Büssow K., Cahill D., Nietfeld W., Bancroft D., Scherzinger E., Lehrach H., Walter G. (1998) A method for global protein expression and antibody screening on high-density filters of an arrayed cDNA library. Nucleic Acids Res 26, 5007–5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helmchen F., Svoboda K., Denk W., Tank D. W. (1999) In vivo dendritic calcium dynamics in deep-layer cortical pyramidal neurons. Nat. Neurosci 2, 989–996 [DOI] [PubMed] [Google Scholar]
- 22.Waltersson Y., Linse S., Brodin P., Grundström T. (1993) Mutational effects on the cooperativity of Ca2+ binding in calmodulin. Biochemistry 32, 7866–7871 [DOI] [PubMed] [Google Scholar]
- 23.André I., Linse S. (2002) Measurement of Ca2+-binding constants of proteins and presentation of the CaLigator software. Anal. Biochem 305, 195–205 [DOI] [PubMed] [Google Scholar]
- 24.Grabski A, Mehler M., Drott D. (2005) The Overnight Express Autoinduction System: high-density cell growth and protein expression while you sleep. Nat. Methods 2, 233–235 [Google Scholar]
- 25.Kuwahara C., Takeuchi A. M., Nishimura T., Haraguchi K., Kubosaki A., Matsumoto Y., Saeki K., Matsumoto Y., Yokoyama T., Itohara S., Onodera T. (1999) Prions prevent neuronal cell-line death. Nature 400, 225–226 [DOI] [PubMed] [Google Scholar]
- 26.Rogstam A., Linse S., Lindqvist A., James P., Wagner L., Berggård T. (2007) Binding of calcium ions and SNAP-25 to the hexa EF-hand protein secretagogin. Biochem. J 401, 353–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thulin E., Linse S. (1999) Expression and purification of human calbindin D28k. Protein Expr. Purif 15, 265–270 [DOI] [PubMed] [Google Scholar]
- 28.Lueking A., Possling A., Huber O., Beveridge A., Horn M., Eickhoff H., Schuchardt J., Lehrach H., Cahill D. J. (2003) A nonredundant human protein chip for antibody screening and serum profiling. Mol. Cell. Proteomics 2, 1342–1349 [DOI] [PubMed] [Google Scholar]
- 29.Herwig R., Schmitt A. O., Steinfath M., O'Brien J., Seidel H., Meier-Ewert S., Lehrach H., Radelof U. (2000) Information theoretical probe selection for hybridisation experiments. Bioinformatics 16, 890–898 [DOI] [PubMed] [Google Scholar]
- 30.Dasgupta M., Blumenthal D. K. (1995) Characterization of the regulatory domain of the γ-subunit of phosphorylase kinase. J. Biol. Chem 270, 22283–22289 [DOI] [PubMed] [Google Scholar]
- 31.Yap K. L., Kim J., Truong K., Sherman M., Yuan T., Ikura M. (2000) Calmodulin target database. J. Struct. Funct. Genomics 1, 8–14 [DOI] [PubMed] [Google Scholar]
- 32.Kranz J. K., Lee E. K., Nairn A. C., Wand A. J. (2002) A direct test of the reductionist approach to structural studies of calmodulin activity. J. Biol. Chem 277, 16351–16354 [DOI] [PubMed] [Google Scholar]
- 33.André I., Kesvatera T., Jönsson B., Åkerfeldt K. S., Linse S. (2004) The role of electrostatic interactions in calmodulin-peptide complex formation. Biophys. J 87, 1929–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montigiani S., Neri G., Neri P., Neri D. (1996) Alanine substitutions in calmodulin-binding peptides result in unexpected affinity enhancement. J. Mol. Biol 258, 6–13 [DOI] [PubMed] [Google Scholar]
- 35.Wechsler A., Teichberg V. I. (1998) Brain spectrin binding to the NMDA receptor is regulated by phosphorylation, calcium and calmodulin. EMBO J 17, 3931–3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sytnyk V., Leshchyns'ka I., Nikonenko A. G., Schachner M. (2006) NCAM promotes assembly and activity-dependent remodeling of the postsynaptic signaling complex. J. Cell Biol 174, 1071–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kasai H., Matsuzaki M., Noguchi J., Yasumatsu N., Nakahara H. (2003) Structure-stability-function relationships of dendritic spines. Trends Neurosci 26, 360–368 [DOI] [PubMed] [Google Scholar]
- 38.Siekevitz P. (1985) The postsynaptic density: a possible role in long-lasting effects in the central nervous system. Proc. Natl. Acad. Sci. U.S.A 82, 3494–3498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kennedy M. B. (2000) Signal-processing machines at the postsynaptic density. Science 290, 750–754 [DOI] [PubMed] [Google Scholar]
- 40.Sheng M., Kim M. J. (2002) Postsynaptic signaling and plasticity mechanisms. Science 298, 776–780 [DOI] [PubMed] [Google Scholar]
- 41.Asrican B., Lisman J., Otmakhov N. (2007) Synaptic strength of individual spines correlates with bound Ca2+-calmodulin-dependent kinase II. J. Neurosci 27, 14007–14011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brooke R. E., Atkinson L., Batten T. F., Deuchars S. A., Deuchars J. (2004) Association of potassium channel Kv3.4 subunits with pre- and post-synaptic structures in brainstem and spinal cord. Neuroscience 126, 1001–1010 [DOI] [PubMed] [Google Scholar]
- 43.Hautin Haitin Y., Attali B. (2008) The C-terminus of Kv7 channels: a multifunctional module. J. Physiol 586, 1803–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morita A., Yamashita N., Sasaki Y., Uchida Y., Nakajima O., Nakamura F., Yagi T., Taniguchi M., Usui H., Katoh-Semba R., Takei K., Goshima Y. (2006) Regulation of dendritic branching and spine maturation by semaphorin3A-Fyn signalling. J. Neurosci 26, 2971–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamashita N., Morita A., Uchida Y., Nakamura F., Usui H., Ohshima T., Taniguchi M., Honnorat J., Thomasset N., Takei K., Takahashi T., Kolattukudy P., Goshima Y. (2007) Regulation of spine development by semaphorin3A through cyclin-dependent kinase 5 phosphorylation of collapsin response mediator protein 1. J. Neurosci 27, 12546–12554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng H. H., Liu S. H., Lee H. C., Lin Y. S., Huang Z. H., Hsu C. I., Chen Y. C., Chang Y. C. (2006) Heavy chain of cytoplasmic dynein is a major component of the postsynaptic density fraction. J. Neurosci. Res 84, 244–254 [DOI] [PubMed] [Google Scholar]
- 47.Bergersen L. H., Magistretti P. J., Pellerin L. (2005) Selective postsynaptic co-localization of MCT2 with AMPA receptor GluR2/3 subunits at excitatory synapses exhibiting AMPA receptor trafficking. Cereb. Cortex 15, 361–370 [DOI] [PubMed] [Google Scholar]
- 48.Verhage M., de Vries K. J., Røshol H., Burbach J. P., Gispen W. H., Südhof T. C. (1997) DOC2 proteins in rat brain: complementary distribution and proposed function as vesicular adapter proteins in early stages of secretion. Neuron 18, 453–461 [DOI] [PubMed] [Google Scholar]
- 49.Yamada M., Miyawaki A., Saito K., Nakajima T., Yamamoto-Hino M., Ryo Y., Furuichi T., Mikoshiba K. (1995) The calmodulin-binding domain in the mouse type 1 inositol1,4,5-trisphosphate receptor. Biochem. J 308, 83–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Erneux C., Moreau C., Vandermeers A., Takazawa K. (1993) Interaction of calmodulin with a putative calmodulin-binding domain of inositol 1,4,5-triphosphate 3-kinase. Effects of synthetic peptides and site-directed mutagenesis of Trp165. Eur. J. Biochem 214, 497–501 [DOI] [PubMed] [Google Scholar]
- 51.Berggård T., Szczepankiewicz O., Thulin E., Linse S. (2002) myo-Inositol monophosphatase is an activated target of calbindin D28k. J. Biol. Chem 277, 41954–41959 [DOI] [PubMed] [Google Scholar]
- 52.Rajebhosale M., Greenwood S., Vidugiriene J., Jeromin A., Hilfiker S. (2003) Phosphatidylinositol 4-OH kinase is a downstream target of neuronal calcium sensor-1 in enhancing exocytosis in neuroendocrine cells. J. Biol. Chem 278, 6075–6084 [DOI] [PubMed] [Google Scholar]
- 53.Nahm W. K., Noebels J. L. (1998) Nonobligate role of early or sustained expression of immediate-early gene proteins c-fos, c-jun, and Zif/268 in hippocampal mossy fiber sprouting. J. Neurosci 18, 9245–9255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Matteis M. A., Morrow J. S. (2000) Spectrin tethers and mesh in the biosynthetic pathway. J. Cell Sci 113, 2331–2343 [DOI] [PubMed] [Google Scholar]
- 55.Leshchyns'ka I., Sytnyk V., Morrow J. S., Schachner M. (2003) Neural cell adhesion molecule (NCAM) association with PKCβ2 via βI spectrin is implicated in NCAM-mediated neurite outgrowth. J. Cell Biol 161, 625–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gardiol A., Racca C., Triller A. (1999) Dendritic and postsynaptic protein synthetic machinery. J. Neurosci 19, 168–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ostroff L. E., Fiala J. C., Allwardt B., Harris K. M. (2002) Polyribosomes redistribute from dendritic shafts into spines with enlarged synapses during LTP in developing rat. Neuron 35, 535–545 [DOI] [PubMed] [Google Scholar]
- 58.Sheng M., Hoogenraad C. C. (2007) the postsynaptic architecture of excitatory synapses: a more quantitative view. Annu. Rev. Biochem 76, 823–847 [DOI] [PubMed] [Google Scholar]
- 59.Godbout M., Erlander M. G., Hasel K. W., Danielson P. E., Wong K. K., Battenberg E. L., Foye P. E., Bloom F. E., Sutcliffe J. G. (1994) 1G5: a calmodulin-binding, vesicle-associated, protein kinase-like protein enriched in forebrain neurites. J. Neurosci 14, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]