Abstract
Phosphorylation is a major post-translational modification widely used in the regulation of many cellular processes. Cyclin-dependent kinase 5 (Cdk5) is a proline-directed serine/threonine kinase activated by activation subunit p35. Cdk5-p35 regulates various neuronal activities such as neuronal migration, spine formation, synaptic activity, and cell death. The kinase activity of Cdk5 is regulated by proteolysis of p35: proteasomal degradation causes down-regulation of Cdk5, whereas cleavage of p35 by calpain causes overactivation of Cdk5. Phosphorylation of p35 determines the proteolytic pathway. We have previously identified Ser8 and Thr138 as major phosphorylation sites using metabolic labeling of cultured cells followed by two-dimensional phosphopeptide mapping and phosphospecific antibodies. However, these approaches cannot determine the extent of p35 phosphorylation in vivo. Here we report the use of Phos-tag SDS-PAGE to reveal the phosphorylation states of p35 in neuronal culture and brain. Using Phos-tag acrylamide, the electrophoretic mobility of phosphorylated p35 was delayed because it is trapped at Phos-tag sites. We found a novel phosphorylation site at Ser91, which was phosphorylated by Ca2+-calmodulin-dependent protein kinase II in vitro. We constructed phosphorylation-dependent banding profiles of p35 and Ala substitution mutants at phosphorylation sites co-expressed with Cdk5 in COS-7 cells. Using the standard banding profiles, we assigned respective bands of endogenous p35 with combinations of phosphorylation states and quantified Ser8, Ser91, and Thr138 phosphorylation. The highest level of p35 phosphorylation was observed in embryonic brain; Ser8 was phosphorylated in all p35 molecules, whereas Ser91 was phosphorylated in 60% and Thr138 was phosphorylated in ∼12% of p35 molecules. These are the first quantitative and site-specific measurements of phosphorylation of p35, demonstrating the usefulness of Phos-tag SDS-PAGE for analysis of phosphorylation states of in vivo proteins.
Phosphorylation is a major post-translational modification of proteins, modulating a variety of cellular functions (1, 2). Because most phosphorylation occurs in a highly site-specific manner, identification of phosphorylation sites has been a subject of intense investigation. Several analytical methods have been utilized to identify phosphorylation sites, including mass spectrometry, amino acid sequencing, and radioisotope phosphate labeling of proteins with mutation(s) at putative phosphorylation site(s) (3, 4). Phosphorylation site-specific antibodies are frequently used to detect phosphorylation at target sites (5, 6). Many phosphospecific antibodies are now commercially available. These phosphospecific antibodies are convenient and useful tools for examining site-specific phosphorylation both in vivo and in vitro. However, they are not appropriate for estimating quantitative ratios of phosphorylation states. Electrophoretic mobility shift on SDS-PAGE is also often used to observe phosphorylation (7–10), but this method is not always applied to site-specific phosphorylation.
Phos-tag is a newly developed dinuclear metal complex that can be used to provide phosphate-binding sites when conjugated to analytical materials such as acrylamide and biotin (11). In SDS-PAGE using Phos-tag acrylamide, phosphorylated proteins are trapped by the Phos-tag sites, delaying their migration and thus separating them from unphosphorylated proteins. Subsequent immunoblot analysis with phosphorylation-independent antibodies reveals both the phosphorylated and unphosphorylated bands. Because the migration of the phosphorylated proteins is greatly delayed compared with migration in Laemmli SDS-PAGE, it is easy to identify the phosphorylated proteins from observed positions on blots. In the past 3 years, this method has been used to detect phosphorylation states for many proteins such as ERK1/2, cdc37, myosin light chain, eIF2α, protein kinase D, β-casein, SIRT7, and dysbindin-1 (12–21).
Cyclin-dependent kinase 5 (Cdk5)1 is a proline-directed serine/threonine kinase that is expressed predominantly in postmitotic neurons and regulates various neuronal events such as neuronal migration, spine formation, synaptic activity, and cell death (22–24). Cdk5 is activated by binding to activation subunit p35 and inactivated by proteasomal degradation of p35 (25). In addition, Cdk5 activity is deregulated by cleavage of p35 to p25 with calpain, resulting in abnormal activation and ultimately causing neuronal cell death (26–29). Proteolysis of p35, either by proteasomal degradation or cleavage by calpain, is regulated by phosphorylation of p35 by Cdk5 (30–33). Therefore, phosphorylation of p35 is essential for proper regulation of Cdk5 activity and function. We previously identified Ser8 and Thr138 as major p35 phosphorylation sites (33). We also showed that phosphorylation of p35 decreased during brain development and proposed its relationship to age-dependent vulnerability of neurons to stress stimuli (32). Thus, to understand the in vivo regulation of Cdk5 activity, it is critical to analyze the phosphorylation states of p35 in brain. However, there is no convenient method to analyze the precise in vivo phosphorylation status of the endogenous proteins.
In this study, we applied the Phos-tag SDS-PAGE method to analyze the phosphorylation states of p35 in vivo and in cultured neurons. We constructed standard band profiles of phosphorylated p35 by Phos-tag SDS-PAGE using Ala mutants at Ser8 and/or Thr138. From these experiments, we observed an unidentified in vivo phosphorylation site at Ser91. We quantified the phosphorylation at each site in cultured neurons and brain, providing the first quantitative estimate of the in vivo phosphorylation states of p35. We discuss the usefulness of Phos-tag SDS-PAGE to analyze the in vivo phosphorylation states of proteins.
EXPERIMENTAL PROCEDURES
Chemicals, Antibodies, and Expression Plasmids
Phos-tag acrylamide was purchased from NARD Institute Ltd. (Amagasaki, Japan). Microcystin LR and bacterial alkaline phosphatase (BAP) were obtained from Wako (Osaka, Japan). N-Methyl-d-aspartate (NMDA) was obtained from Sigma. Pefabloc SC was obtained from Merck. Polyfect transfection reagent was purchased from Qiagen (Hilden, Germany). The site-directed mutagenesis kit was purchased from Stratagene (La Jolla, CA). [γ-32P]ATP was obtained from PerkinElmer Life Sciences. Anti-p35 (C19) and anti-Cdk5 (DC17) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-Thr(P)286 Ca2+-calmodulin-dependent protein kinase II (CaMKII) has been described (34). Anti-phospho-Ser91 p35 (Ser(P)91) was produced in this study. Briefly, unphosphorylated and phosphorylated peptides, KKSLSCANLSC and KKSL(p)SCANLSC, were chemically synthesized, and the phosphorylated peptide was conjugated to keyhole limpet hemocyanin via the C-terminal Cys residue (Invitrogen). Phospho-Ser91 peptide was used to immunize rabbits, and Ser(P)91 antibody was both negatively and positively affinity-purified using unphosphorylated and phosphorylated peptide columns, respectively. Columns were prepared using SulfoLink according to the manufacturer's protocol (Pierce).
S8A, T138A, and 2A mutants of human p35 have been described (33). p35 S91A mutant was generated by the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions using pCMV5-p35 as template and the forward and reverse primers 5′-GAAGAAGTCGCTGGCCTGTGCCAACCTG-3′ and 5′-CAGGTTGGCACAGGCCAGCGACTTCTTC-3′.
Cell and Brain Extracts
COS-7 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 0.1 mg/ml streptomycin. COS-7 cells were transfected for 24 h with p35 and/or Cdk5 expression plasmids by Polyfect transfection reagent and lysed in RIPA buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, 0.1% SDS, 0.5% sodium deoxycholate, 1 mm DTT) containing protease inhibitors (0.4 mm Pefabloc SC and 10 μg/ml leupeptin) and phosphatase inhibitors (10 mm β-glycerophosphate, 5 mm NaF, 1 mm Na3VO4, and 1 μm Microcystin LR). After centrifugation at 10,000 × g for 15 min, the supernatant was collected as COS-7 cell extracts. For dephosphorylation experiments, cells were homogenized in 100 mm Tris-HCl, pH 8.0, containing protease inhibitors, and the cell extracts were incubated with 2 units of BAP at 37 °C for 2 h.
Mice (ICR) or rats (Wistar S/T) were obtained from Sankyo Labo (Tokyo, Japan). Brain cortex of rats at postnatal day 1 (P1) was homogenized in homogenization buffer (25 mm HEPES, pH 7.5, 5 mm KCl, 1.5 mm MgCl2, 1 mm EGTA, 1 mm DTT) with a Teflon pestle homogenizer. After centrifugation at 10,000 × g for 15 min, the supernatant was collected as the rat brain extract. Brain cortices of embryonic day 17 (E17) or adult (10-week-old) mice were homogenized in RIPA buffer containing protease inhibitors and phosphatase inhibitors with a Teflon pestle homogenizer. After centrifugation at 10,000 × g for 15 min, the supernatant was collected as fetal or adult mouse brain extract. Rat and mouse brain extracts were boiled in Laemmli sample buffer and subjected to SDS-PAGE or Phos-tag SDS-PAGE.
Primary Neuronal Culture and Glutamate or NMDA Treatment
Cerebral cortical neurons were prepared from E17-E18 rat brains as described (31). Neurons were treated with 100 μm glutamate or NMDA for 1 or 5 min at 10 days in vitro (DIV 10) and were immediately lysed in RIPA buffer containing protease inhibitors and phosphatase inhibitors.
Phosphorylation of p35 by CaMKII
p35-kinase-negative Cdk5 D144N (knCdk5) complex was purified from Sf9 cells as described (35). p35 was phosphorylated by 0.5 μg/ml CaMKII in the presence of 50 μg/ml calmodulin and 0.5 mm CaCl2 in MOPS buffer (20 mm MOPS, pH 7.2, 0.1 mm EDTA, 0.1 mm EGTA, 1 mm MgCl2, 1 mm ATP) at 37 °C for 60 min. Phosphorylation was detected by electrophoretic mobility shift following SDS-PAGE or by autoradiography after SDS-PAGE when [γ-32P]ATP was used for phosphorylation.
Laemmli SDS-PAGE, Phos-tag SDS-PAGE, and Immunoblotting
Laemmli SDS-PAGE was carried out with 10% polyacrylamide gels. Proteins were transferred to PVDF (Millipore, Bedford, MA) membranes using a semidry blotting apparatus. Phos-tag SDS-PAGE was performed with 7.5% polyacrylamide gels containing 50–100 μm Phos-tag acrylamide and 100–200 μm MnCl2. After electrophoresis, Phos-tag acrylamide gels were washed with transfer buffer (50 mm Tris, 384 mm glycine, 0.1% SDS, 20% methanol) containing 1 mm EDTA for 10 min with gentle shaking and then with transfer buffer without EDTA for 10 min according to the manufacturer's protocol. Proteins were transferred to PVDF membranes using a submarine blotting apparatus. Membranes were probed with anti-p35, anti-Ser(P)8, anti-Ser(P)91, anti-Thr(P)138, or anti-Thr(P)286 CaMKII followed by horseradish peroxidase-conjugated secondary antibody. Immunodetection was carried out with an ECL system (GE Healthcare) or Millipore Immobilon Western chemiluminescent horseradish peroxidase substrate (Millipore).
Quantitative Measurement and Statistical Analysis
Immunoreaction was captured as digital images (eight-bit gray scale) using a Chemidoc XRS apparatus (Bio-Rad) or by scanning x-ray film, and band intensities were measured by Image J software. The digital images were within linear range, which was confirmed by Quantity One software (Bio-Rad) or different intensities of x-ray films. Means and standard errors were calculated statistically from four different samples for COS-7 cells, 10 samples for primary cultured neurons, six samples for E18 mouse brains, and four samples for 10-week-old mouse brains.
RESULTS
Banding Profiles of Phosphorylated p35 in Phos-tag SDS-PAGE
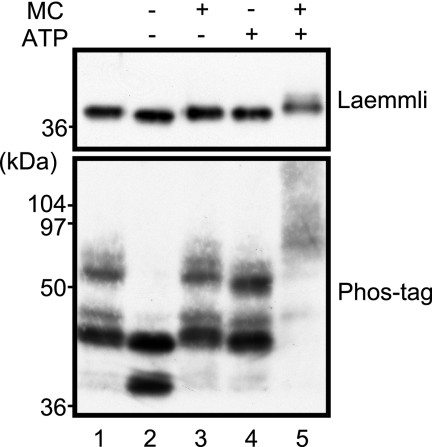
We first examined whether Phos-tag SDS-PAGE can be used to analyze the phosphorylation states of p35. p35 is phosphorylated at Ser8 by Cdk5 in adult rat brain (33). p35 in P1 rat brain extract was compared between Laemmli and Phos-tag SDS-PAGE after incubation with ATP and/or the phosphatase inhibitor Microcystin LR. The dephosphorylated and phosphorylated forms of p35 were then detected by immunoblotting using anti-p35 antibody. After incubation, this analysis showed a slight mobility shift on Laemmli SDS-PAGE (Fig. 1, upper panel). In contrast, using Phos-tag SDS-PAGE (Fig. 1, lower panel), rat brain p35 was separated into several bands with large mobility differences. Incubation in the absence of ATP and Microcystin LR shifted the bands downward to produce a new band (Fig. 1, lane 2), probably due to dephosphorylation. Incubation with Microcystin LR (Fig. 1, lane 3) or ATP (Fig. 1, lane 4) inhibited this downward shift. Thus, phosphorylation states before incubation might be retained under these conditions. Incubation of rat brain with both ATP and Microcystin LR produced considerably upward shifted smear bands (Fig. 1, lane 5), probably due to further phosphorylation. These results indicate that Phos-tag SDS-PAGE can be used to analyze phosphorylation states of p35.
Fig. 1.
Comparison of electrophoretic mobility shift of p35 using Laemmli and Phos-tag SDS-PAGE. Brain extracts prepared from rat at P1 were incubated at 37 °C for 60 min in the presence or absence of 1 mm ATP and/or 10 μm Microcystin LR (MC) as indicated. The extracts were subjected to Laemmli SDS-PAGE (upper panel) or Phos-tag SDS-PAGE (lower panel), and p35 was detected by immunoblotting with anti-p35. Lane 1 is the brain extract before incubation.
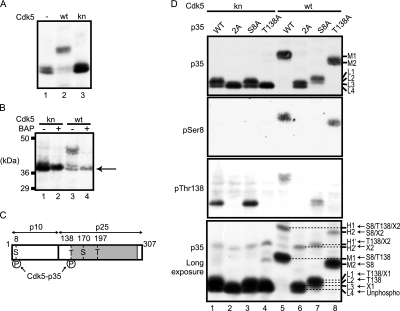
To see the mobility shift by phosphorylation with Cdk5, p35 was co-expressed in COS-7 cells with Cdk5 or kinase-negative Cdk5 D144N (knCdk5). When p35 was expressed alone (Fig. 2A, lane 1) or with knCdk5 (Fig. 2A, lane 3), p35 appeared as two close but distinct bands at about 37 kDa, approximately the same mobility as that of p35 in Laemmli SDS-PAGE. However, co-expression with Cdk5 shifted the p35 mobility remarkably upward (Fig. 2A, lane 2). To confirm that these mobility shifts are indeed caused by phosphorylation, we treated the COS-7 cell extract with BAP. Only a single band was detected after BAP treatment (Fig. 2B, lanes 2 and 4). Dephosphorylation with BAP shifted the upper bands to the lowest position, suggesting that the lowest band corresponds to unphosphorylated p35.
Fig. 2.
Phosphorylation site-specific banding profiles of p35 on Phos-tag SDS-PAGE. A, electrophoretic mobility of p35 co-expressed with Cdk5 or knCdk5 in COS-7 cells by Phos-tag SDS-PAGE. Extracts of COS-7 cells expressing p35 alone (−) or with Cdk5 (wt) or knCdk5 (kn) were subjected to Phos-tag SDS-PAGE followed by immunoblotting with anti-p35. B, effect of BAP treatment on the mobility of p35 by Phos-tag SDS-PAGE. COS-7 cell extracts expressing p35 with Cdk5 (wt) or knCdk5 (kn) were incubated with (+) or without (−) BAP at 37 °C for 2 h and subjected to Phos-tag SDS-PAGE and immunoblotting with anti-p35. The arrow indicates the lowest band. C, a schematic representation of p35 and Cdk5 consensus (S/T)P phosphorylation sequences. p35 is composed of an N-terminal p10 region and a C-terminal p25 Cdk5 activation domain (shaded). Among four possible phosphorylation sites at Ser8, Thr138, Ser170, and Thr197, Ser8 and Thr138 have been shown to be phosphorylated (33). D, mobility shifts in mutants of p35 phosphorylation sites. p35 and its Ala mutants at Ser8/Thr138 (2A), Ser8 (S8A), or Thr138 (T138A), expressed in COS-7 cells with Cdk5 (lanes 5–8) or knCdk5 (lanes 1–4), were subjected to Phos-tag SDS-PAGE and immunoblotting using anti-p35 (top panel), anti-Ser(P)8 (second panel), or anti-Thr(P)138 (third panel). The bottom panel was exposed for a longer time to show minor bands in the top panel. Respective p35 bands are named H1, H2, H1′, H2′, M1, M2, L1, L2, L3, and L4 according to their relative mobility from slow to fast (right side of top and bottom panels). Two unidentified phosphorylation sites are designated here as X1 and X2. Based on the mobility shift of Ala mutants, the phosphorylation site(s) of each band is assigned as described in the right side of bottom panels: H1, Ser8/Thr138/X2; H2, Ser8/X2; M1, Ser8/Thr138; M2, Ser8; H1′, Thr138/X2; H2′, X2; L1, Thr138/X1; L2, Thr138; L3, X1; L4, unphosphorylated (Unphospho).
Next, we evaluated the phosphorylation state of each band using p35 mutants with Ala substitutions at known phosphorylation sites (Fig. 2C): Ser8 (mutant S8A), Thr138 (mutant T138A), and both Ser8 and Thr138 (mutant 2A). These mutants were co-transfected with either Cdk5 or knCdk5 in COS-7 cells, and the cell extracts were subjected to Phos-tag SDS-PAGE and immunoblotting with anti-p35 (Fig. 2D, top panel). When co-expressed with knCdk5, 2A and T138A mutants exhibited a single band with the fastest mobility (Fig. 2D, lanes 2 and 4), whereas wild-type (WT) p35 and the S8A mutant displayed two closely migrating bands with a slightly stronger signal in the upper band (Fig. 2D, lanes 1 and 3), indicating that the upper band was phosphorylated at Thr138. Thus, Thr138 can be phosphorylated by another kinase. When p35 was co-transfected with Cdk5, p35 apparently shifted to a single band with a much slower mobility than that of p35 co-expressed with knCdk5 (Fig. 2D, lane 5). This band pattern is different from that of p35 in lane 3 of Fig. 2B, presumably due to the amount (i.e. kinase activity) of the expressed Cdk5. Upon increasing the expression of Cdk5 in COS-7 cells, the faster migrating bands of p35 shifted to the upper band (supplemental Fig. S1). In contrast, the 2A mutant migrated as two bands at a position close to the unphosphorylated band (Fig. 2D, lane 6), confirming that Ser8 and Thr138 are the major phosphorylation sites on p35 in COS-7 cells. The S8A mutant shifted p35 slightly upward (Fig. 2D, lane 7), and the T138A mutant shifted the band substantially upward (Fig. 2D, lane 8). The mobility change of WT p35 from the position of 2A mutant was almost equal to the combined changes of S8A and T138A mutants. We confirmed phosphorylation at Ser8 and Thr138 using phosphospecific antibodies (Fig. 2D, second and third panels). Ser(P)8 was detected in the major band of WT p35 and T138A, and Thr(P)138 was detected in the major band of WT p35 and S8A, indicating that the mobility shift of p35 is caused by phosphorylation at Ser8 and Thr138 by Cdk5 in COS-7 cells. We labeled the bands for WT p35 and T138A, both with phosphorylation at Ser8, as M1 and M2, respectively (Fig. 2D, top panel).
We observed two other bands that were reproducibly detected throughout these experiments, although they showed some variability based on the method of Phos-tag SDS-PAGE. The upper band associated with the 2A mutant appeared when 2A was co-expressed with Cdk5 (Fig. 2D, lane 6). The phosphorylation site producing this band might correspond to Ser170 or Thr197 in (S/T)P Cdk5 consensus sequences of p35 because the phosphorylation occurred by co-expression with Cdk5. We tentatively identified this phosphorylation site as “X1.” This upward shift was similar to but slightly smaller than that of Thr138 phosphorylation (Fig. 2D, lane 4). Therefore, three slightly separated bands were detected just above the unphosphorylated band. These four bands, including the unphosphorylated p35, were named L1 to L4 from the upper to the lower band, respectively (Fig. 2D, top panel).
Minor phosphorylation bands were detected by exposing blots for a longer time (Fig. 2D, bottom panel). A band with even slower migration was found in WT p35 (lane 5), and corresponding bands with similar upward displacement were seen in the S8A, T138A, and 2A mutants (lanes 6–8), suggesting phosphorylation at a site other than Ser8 and Thr138. We hypothesized that these upper bands are phosphorylated at one site, and we tentatively termed the unidentified phosphorylation site as “X2.” The most highly shifted bands, observed with wild-type p35 and T138A, were designated as H1 and H2, respectively. The corresponding bands observed with S8A and 2A were named H1′ and H2′, respectively. These results enabled us to identify phosphorylation sites in each band of p35. The deduced phosphorylation site(s) at each band are indicated at the right side of the bottom panel of Fig. 2D.
Phosphorylation States of p35 in Cultured Neurons and Mouse Brain
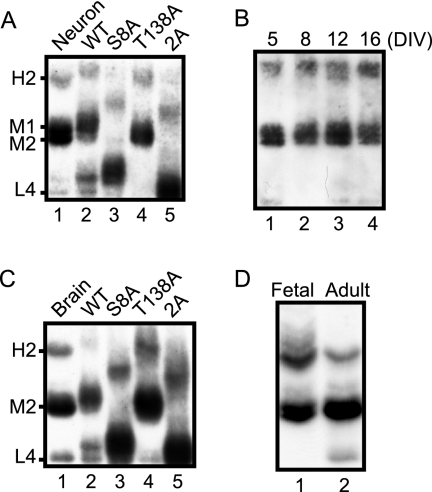
We previously reported that Ser8 was a major phosphorylation site in cultured rat cortical neurons and that Thr138 became phosphorylated in the presence of the protein phosphatase inhibitor Microcystin LR (33). We therefore applied Phos-tag SDS-PAGE to p35 in extracts from rat brain primary neuronal cultures (Fig. 3A and supplemental Fig. S2A). p35 in cultured cortical neurons at DIV 10 was compared with standard phosphorylation-dependent banding profiles of p35 by immunoblotting with anti-p35 after Phos-tag SDS-PAGE. p35 in neuronal cultures displayed two major bands, M1 and M2, and a minor band, H2. We also studied whether the phosphorylation states of p35 change during culture (Fig. 3B). The p35 banding patterns were similar in neurons cultured from 5 to 16 days, although the H2 band appeared slightly stronger in neurons cultured over a longer period. These results indicate that p35 in cultured neurons is phosphorylated mainly at Ser8, moderately at Thr138, and partially at the unidentified X2 site.
Fig. 3.
Phosphorylation states of endogenous p35 in primary neuron culture and in brain. A, electrophoretic mobility of p35 in mouse primary cortical neurons at DIV 10 (lane 1) was compared with that of p35 or its Ala mutants co-expressed with Cdk5 in COS-7 cells (lanes 2–5) using Phos-tag SDS-PAGE followed by immunoblotting with anti-p35. B, time course of phosphorylation states of p35 in a cortical neuron culture from DIV 5 to DIV 16. C, electrophoretic mobility of p35 prepared from mouse brain (10 weeks old; lane 1) was compared with that of p35 mutants co-expressed with Cdk5 in COS-7 cells (lanes 2–5). D, phosphorylation states of p35 in fetal (E18) versus adult (10-week-old) mouse brains.
Next we examined the phosphorylation states of p35 in mouse brain. Adult mouse brain extract was immunoblotted with anti-p35 after Phos-tag SDS-PAGE and compared with the standard phosphorylation banding profiles of p35 expressed in COS-7 cells. Adult brain p35 was composed of H2, M2, and L4 bands, showing a banding pattern similar to p35 in cortical neuron cultures (Fig. 3C and supplemental Fig. S2B). However, the M1 band was weak in brain p35, suggesting that Thr138 is phosphorylated to a lesser degree. We previously showed by electrophoretic mobility shift on Tris-Tricine SDS-PAGE that p35 is more highly phosphorylated in fetal brain than in adult brain (32). Thus, we compared phosphorylation states of p35 between fetal and adult brain using Phos-tag SDS-PAGE (Fig. 3D). The banding profile of fetal brain p35 shifted upward as a whole relative to adult brain p35, despite the similar banding profile. The H2 band was stronger in fetal brain than in adult. There was no unphosphorylated p35 in fetal brain, whereas some p35 remained unphosphorylated in adult brain. These results confirm our previous findings that p35 is more highly phosphorylated in fetal brain than in adult brain (32).
Phosphorylation of p35 by CaMKII
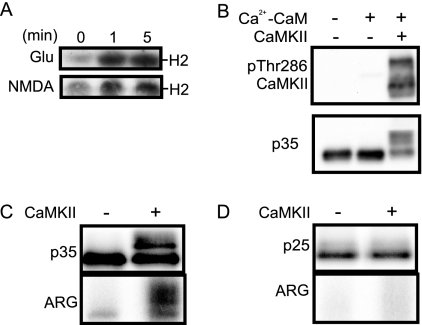
Because the unknown X2 site was phosphorylated in vivo, particularly in fetal brain, we turned our attention to characterizing the X2 site. Because the intensity of band H increased shortly after treatment of primary neurons with NMDA or glutamate (Fig. 4A), we hypothesized that CaMKII is a protein kinase that phosphorylates p35 at the X2 site. First, we phosphorylated p35 bound to knCdk5 in vitro by purified CaMKII in the presence of calcium and calmodulin (Ca2+-CaM). We used p35-knCdk5 in this experiment instead of p35-Cdk5 to avoid phosphorylation of p35 by Cdk5. We found that p35 was shifted substantially upward even in Laemmli SDS-PAGE by incubation with CaMKII in the presence of Ca2+-CaM (Fig. 4B). Autophosphorylation of CaMKII is shown to represent the activation of CaMKII (Fig. 4B, upper panel). The upward shift of p35 was also detected in lysates of HEK293 cells expressing p35 (supplemental Fig. S3A) and in p35 immunoprecipitates of primary neurons (supplemental Fig. S3B) when they were incubated with CaMKII in the presence of Ca2+-CaM. Phosphorylation of shifted p35 was confirmed using [γ-32P]ATP. Shifted p35 was indeed labeled with 32P after incubation with CaMKII in the presence of Ca2+-CaM (Fig. 4C, ARG).
Fig. 4.
In vitro phosphorylation of p35 by CaMKII. A, band H2 of p35 in primary neurons treated with glutamate (Glu) or NMDA. Rat brain cortical neurons at DIV 10 were treated with 100 μm glutamate or NMDA for 1 or 5 min, and the resulting extracts were subjected to Phos-tag SDS-PAGE followed by immunoblotting with anti-p35. B, phosphorylation of p35 by CaMKII in vitro. knCdk5-p35 purified from Sf9 cells was incubated with purified CaMKII in the presence of 0.5 mm Ca2+, 50 μg/ml calmodulin (Ca2+-CaM), and 1 mm ATP. Activation of CaMKII is shown by immunoblotting with anti-Thr(P)286 CaMKII (upper panel). C and D, immunoblots and autoradiography of p35 or p25 incubated with CaMKII. The extracts of COS-7 cells expressing p35 (C) or p25 (D) were immunoprecipitated by anti-p35 and incubated with CaMKII (+) and [γ-32P]ATP in the presence of Ca2+-CaM. Laemmli SDS-PAGE followed by immunoblotting is shown in the upper panel, and the autoradiograph (ARG) is shown in the lower panel.
To determine the CaMKII phosphorylation site in p35, we first asked whether CaMKII phosphorylates p25, the C-terminal fragment of p35 generated by calpain cleavage (26, 27). When p25 was expressed in HEK293 cells and incubated in vitro with CaMKII, phosphorylation of p25 was not seen, whether assayed by electrophoretic mobility shift or by 32P incorporation (Fig. 4D). These results suggested that the phosphorylation site should be present in the N-terminal p10 region.
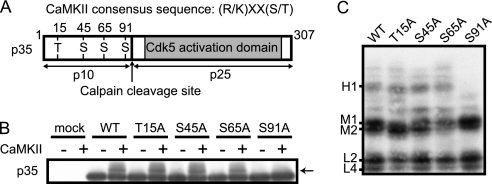
The phosphorylation consensus site for CaMKII is Ser or Thr in (R/K)XX(S/T) (36). There are four such sites at Thr15, Ser45, Ser65, and Ser91 in the p10 region (Fig. 5A). To determine which site is phosphorylated by CaMKII, we constructed an Ala mutant of each site in p35, expressed the mutants in HEK293 cells, and phosphorylated them by CaMKII after immunoprecipitation (Fig. 5B). The S91A mutant was the only one that failed to be shifted upward by incubation with CaMKII, suggesting that Ser91 is a CaMKII phosphorylation site. To see whether Ser91 phosphorylation shifts p35 upward to the position of band H1 on Phos-tag SDS-PAGE, each Ala mutant was expressed with Cdk5 in COS-7 cells, and the cell extracts were subjected to Phos-tag SDS-PAGE followed by immunoblotting with anti-p35 (Fig. 5C). Band H1 was not present in the S91A mutant in contrast to WT and other Ala mutants of p35 in which band H1 was present. Similar results were obtained by co-expression with knCdk5 (supplemental Fig. S3C), suggesting that Ser91 phosphorylation is independent of the kinase activity of Cdk5.
Fig. 5.
Identification of Ser91 in p35 as a CaMKII phosphorylation site. A, schematic representation of p35 and CaMKII phosphorylation consensus (R/K)XX(S/T) sites in the N-terminal p10 region. There are four such sequences (RKAT15, KRHS45, KKNS65, and KSLS91) in the p10 region. The shaded region represents the Cdk5 activation domain. The calpain cleavage site is between Phe98 and Ala99. B, immunoblot of p35 and its Ala mutants phosphorylated by CaMKII, detected with anti-p35. Ala mutants of p35 at the CaMKII consensus phosphorylation sites (T15A, S45A, S65A, and S91A) were expressed in COS-7 cells. Extracts were incubated with CaMKII in the presence of Ca2+-CaM at 37 °C for 60 min and subjected to Laemmli SDS-PAGE followed by immunoblotting with anti-p35. C, Phos-tag SDS-PAGE analysis of p35 Ala mutants phosphorylated by CaMKII. Ala mutants of p35 at CaMKII consensus sites were co-expressed with Cdk5 and subjected to Phos-tag SDS-PAGE followed by immunoblotting with anti-p35.
Phosphorylation of p35 at Ser91 in Primary Neurons and Brains
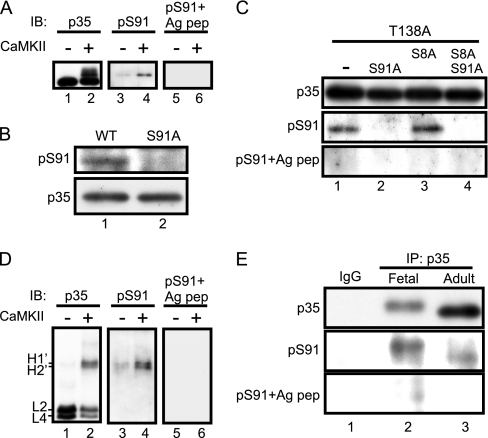
To confirm the phosphorylation of Ser91 in endogenous p35, we generated an anti-phospho-Ser91-specific antibody (anti-Ser(P)91) as described under “Experimental Procedures.” Anti-Ser(P)91 reacted specifically with p35 phosphorylated by CaMKII (Fig. 6A, middle panel). Preabsorption with the antigenic peptide eliminated the reaction (Fig. 6A, right panel). Anti-Ser(P)91 also recognized WT p35 (Fig. 6B, lane 1) but not the S91A mutant (Fig. 6B, lane 2) expressed in COS-7 cells. Furthermore, anti-Ser(P)91 recognized T138A and T138A/S8A mutants but not in combination with the mutation at Ser91 (Fig. 6C). Preabsorption of anti-Ser(P)91 with the phospho-Ser91 antigenic peptide eliminated the immunologic reaction with T138A and T138A/S8A mutants. Band H′ was detected when p35-knCdk5 purified from Sf9 cells was phosphorylated in vitro by CaMKII. Anti-Ser(P)91 reacted specifically with band H′ on Phos-tag SDS-PAGE, although anti-Ser(P)91 preabsorbed with the phospho-Ser91 antigenic peptide did not react, indicating that bands H1′ and H2′, and probably bands H1 and H2, contain phospho-Ser91 (Fig. 6D). Taken together, these data support the specific reaction of anti-Ser(P)91 to phosphorylated Ser91.
Fig. 6.
In vitro phosphorylation of Ser91 shown using phospho-Ser91-specific antibody. A, reaction of anti-Ser(P)91 with CaMKII-phosphorylated p35. p35 purified from Sf9 cells was incubated with purified CaMKII in the presence of Ca2+-CaM and immunoblotted (IB) with anti-p35 (lanes 1 and 2) and anti-Ser(P)91 (lanes 3 and 4). Immunoblotting with anti-Ser(P)91 preabsorbed with the phospho-Ser91 antigenic peptide (Ag pep) is shown in lanes 5 and 6. B, specificity of anti-Ser(P)91. p35 (WT) and its S91A mutant were expressed in COS-7 cells, and the extracts were immunoblotted with anti-p35 and anti-Ser(P)91. C, confirmation of the specificity of anti-Ser(P)91. p35-T138A with Ala mutation at Ser91 or Ser8 was expressed in COS-7 cells, and the cell extracts were immunoblotted with anti-p35, anti-Ser(P)91, and anti-Ser(P)91 preabsorbed with phospho-Ser91 antigenic peptide. D, immunoblotting of CaMKII-phosphorylated p35 separated by Phos-tag SDS-PAGE. p35-knCdk5 purified from Sf9 cells was phosphorylated by purified CaMKII in vitro and subjected to Phos-tag SDS-PAGE followed by immunoblotting with anti-p35 (lanes 1 and 2), anti-Ser(P)91 (lanes 3 and 4), and anti-Ser(P)91 preabsorbed with the phospho-Ser91 antigenic peptide (lanes 5 and 6). E, phosphorylation of p35 at Ser91 in mouse brain. Extracts prepared from fetal (lane 2) or adult (lane 3) mouse brains were immunoprecipitated (IP) with anti-p35. Immunoprecipitates were immunoblotted with anti-p35, anti-Ser(P)91, and anti-Ser(P)91 preabsorbed with the phospho-Ser91 antigenic peptide (lower panel). Immunoprecipitation with control IgG from adult mouse brain is shown in lane 1.
Using this antibody, we analyzed the phosphorylation of endogenous p35 in mouse brain. Because the antibody could not detect phosphorylated Ser91 in brain extracts, immunoprecipitated p35 was examined in this experiment. Anti-Ser(P)91 reacted more strongly to p35 in immunoprecipitates prepared from fetal brain than those prepared from adult brain (Fig. 6E). This indicate that Ser91 is phosphorylated in fetal brains more than in adult brains, consistent with the result of Fig. 3D, if the X2 site is indeed Ser91.
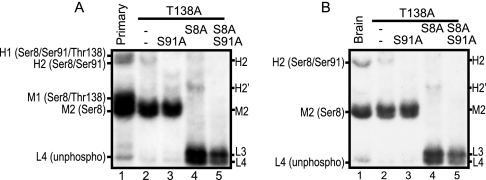
Mutating Ser91 to Ala in T138A or S8A/T138A mutants, we further examined whether band H2 of primary cultured neurons (Fig. 7A) or adult mouse brain (Fig. 7B) includes phosphorylated Ser91. T138A mutants were used in this experiment because they showed a similar p35 banding patterns in brain and primary cultured neurons (Fig. 3, A and C). The S91A mutation abolished band H2 from the T138A mutant (Fig. 7, A and B, lane 3) and band H2′ from the T138A/S8A mutant (Fig. 7, A and B, lane 5), indicating that band H2 is phosphorylated at least at Ser8 and Ser91 and that band H2′ is phosphorylated at Ser91. All phosphorylated bands at the H and M regions disappeared with triple mutations at Thr138, Ser8, and Ser91. In the L region, L3 was still detected in the triple mutant S8A/S91A/T138A due to phosphorylation at the X1 site in COS-7 cells. These results indicate that p35 is primarily phosphorylated at Ser8 and partially at Ser91 in cultured cortical neurons and mouse brain.
Fig. 7.
Assignment of phosphorylation states of endogenous p35. Comparison of p35 prepared from primary cortical neurons (A) and mouse brain (B) with p35 mutants expressed in COS-7 cells by Phos-tag SDS-PAGE. Extracts of primary cortical neurons (lane 1 in A), adult mouse brain (lane 1 in B), and COS-7 cells expressing p35-T138A (lane 2), p35-T138A/S91A (lane 3), p35-T138A/S8A (lane 4), and p35-T138A/S8A/S91A (lane 5) were separated by Phos-tag SDS-PAGE and immunoblotted with anti-p35. Five different phosphorylation states were observed for p35 in primary neurons: H1, phosphorylation at Ser8, Ser91, and Thr138; H2, phosphorylation at Ser8 and Ser91; M1, phosphorylation at Ser8 and Thr138; M2, phosphorylation at Ser8; and L4, unphosphorylated. Three different phosphorylation states were observed in adult brain: H2, phosphorylation at Ser8 and Ser91; M2, phosphorylation at Ser8; and L4, unphosphorylated (unphospho).
Quantitative Analysis of Phosphorylated p35 in Cultured Neurons and Mouse Brain
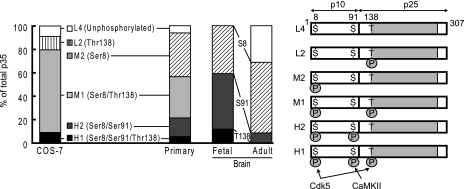
Because one phosphorylation-independent antibody (C19) is sufficient for detection of different phosphorylation states of p35 by immunoblotting after Phos-tag SDS-PAGE, we can estimate the phosphorylation states of p35 quantitatively by densitometric measurement of the respective p35 bands. Tables I and II show the results of p35 in primary cultured neurons at DIV 10–12 and mouse brain at E17 and 10 weeks as well as p35 in COS-7 cells in which high Cdk5 activity was expressed. Table I shows the percent ratio of each band, and Table II shows the percent ratio of phosphorylation at each site. Fig. 8 shows a graphic representation of these calculations (left panel) and schematic representations of p35 with corresponding phosphorylation states and their responsible kinases (right panel). Phosphorylation states differed between COS-7 cells and cultured neurons. Although Ser8 was phosphorylated strongly in both types of cells, Thr138 was more highly phosphorylated in COS-7 cells than in cultured neurons, and Ser91 was more highly phosphorylated in cultured neurons than in COS-7 cells.
Table I. Percent ratio of p35 in respective phosphorylation states.
p35 prepared from COS-7 cells, primary neurons, or mouse brains was separated into three to five bands, H1, H2, M1, M2, L2, or L4, depending on their phosphorylation states in Phos-tag SDS-PAGE. The amount of each band was estimated by densitometric scanning, and their ratios were expressed as percentage of the total amount of p35. Data are represented as mean ± S.E. from four independent preparations of COS-7 cells transfected with Cdk5-p35, 10 preparations of primary cultured cortical neurons at DIV 10–12, six preparations of fetal mouse brains, and four preparations of adult mouse brains.
| COS-7 Cdk5-p35 (n = 4) | Primary (n = 10) | Fetal (n = 6) | Adult (n = 4) | |
|---|---|---|---|---|
| H1 (Ser8/Ser91/Thr138) | 9.06 ± 2.9 | 5.30 ± 1.4 | 11.7 ± 2.6 | N.D.a |
| H2 (Ser8/Ser91) | N.D. | 16.0 ± 3.0 | 47.3 ± 1.7 | 8.40 ± 0.4 |
| M1 (Ser8/Thr138) | 70.6 ± 10.1 | 35.2 ± 2.3 | N.D. | N.D. |
| M2 (Ser8) | N.D. | 37.4 ± 3.0 | 41.0 ± 3.3 | 60.3 ± 0.5 |
| L2 (Thr138) | 11.5 ± 5.6 | N.D. | N.D. | N.D. |
| L4 (unphosphorylated) | 8.94 ± 3.7 | 6.15 ± 0.9 | N.D. | 31.3 ± 0.5 |
a N.D., not detected.
Table II. Ratio of phosphorylation at each site in p35.
Ratios of p35 phosphorylation at Ser8, Ser91, and Thr138 were calculated in COS-7 cells, primary neurons, and brains from the values shown in Table I. Values are represented as mean ± S.E.
| COS-7 | Primary | Fetal | Adult | |
|---|---|---|---|---|
| Ser8 | 79.6 ± 9.1 | 93.9 ± 0.9 | 100 | 68.7 ± 0.5 |
| Ser91 | 9.06 ± 2.9 | 21.3 ± 3.7 | 59.0 ± 3.3 | 8.42 ± 0.4 |
| Thr138 | 91.1 ± 3.7 | 40.5 ± 2.2 | 11.7 ± 2.6 | N.D.a |
a Not detected.
Fig. 8.
Graphic and schematic representation of quantitative ratio for each phosphorylation band of p35 in COS-7 cells, primary neurons, and mouse brain. The graph on the left side was made using the data shown in Table I. L4 (white), unphosphorylated p35; L2 (vertical lines), p35 phosphorylated at Thr138; M2 (sloped lines), p35 phosphorylated at Ser8; M1 (light gray), p35 phosphorylated at Ser8/Thr138; H2 (dark gray), p35 phosphorylated at Ser8/Ser91; H1 (black), p35 phosphorylated at Ser8/Ser91/Thr138. In brain p35, the ratio of phosphorylation at Ser8, Ser91, and Thr138 is represented by Ser(P)8, Ser(P)91, and Thr(P)138, respectively, in the middle between fetal and adult brain. p35 and its phosphorylation sites and responsible kinases are shown schematically in the right panel.
Phosphorylation of p35 also changed in mouse brain based on age. In fetal brain, almost all p35 molecules were phosphorylated at Ser8, 59% of p35 molecules were phosphorylated at Ser91, and ∼12% of p35 molecules were phosphorylated at Thr138. Phosphorylation at all three sites decreased in adult brains, down to 69% for Ser8, down to 8% for Ser91, and to undetectable levels for Thr138. As a result of decreased phosphorylation, the unphosphorylated form of p35 increased to 31% in adult brains from an undetectable amount in fetal brain.
DISCUSSION
In this study, we analyzed the in vivo phosphorylation states of a p35 Cdk5 activator quantitatively using Phos-tag SDS-PAGE followed by immunoblotting with a phosphorylation-independent antibody to p35. p35 displayed phosphorylation site-dependent upward shifts in electrophoretic mobility on Phos-tag SDS-PAGE that enabled us to identify the phosphorylation states of p35 without using radioisotope or phosphospecific antibodies. After establishing the standard phosphorylation profile of p35, we evaluated the in vivo phosphorylation states of p35 in cultured neurons and mouse brain. In fetal mouse brain, for example, Ser8 was phosphorylated in all p35 molecules, and Thr138 was phosphorylated in 20% of p35 molecules. In addition, we identified Ser91 as a novel in vivo phosphorylation site that was phosphorylated by CaMKII in vitro and was phosphorylated in about 60% of p35 molecules in fetal mouse brain. The overall phosphorylation at each of these residues was reduced in adult mouse brain. Because Cdk5 activity is regulated by phosphorylation-dependent proteolysis of p35, the quantitative analysis of in vivo phosphorylation states of p35 will be useful for exploring in vivo regulation of Cdk5 activity. Our analysis also demonstrates the general usefulness of Phos-tag SDS-PAGE as a method for in vivo phosphorylation analysis of proteins.
Phos-tag SDS-PAGE is a recently developed method that is capable of separating phosphorylated proteins on SDS-PAGE (12) and has been used to show phosphorylation of proteins (12–21). When this technique was initially reported, Kinoshita et al. (12) predicted its use in quantitative estimation of in vivo phosphorylation. However, most previous reports used this technique qualitatively as one of several methods indicating phosphorylation of particular proteins. A quantitative application of Phos-tag SDS-PAGE was reported with myosin light chain, but site-specific quantification was not addressed (15). We report here that the upward mobility shifts differed among the phosphorylation sites in p35 where the largest upward shift of phosphorylation was at Ser91 followed by Ser8 and finally Thr138, which was a small but distinct shift. Although the chemical basis for phosphorylation site-specific mobility variation is not known, we were able to use this method successfully to identify site-specific phosphorylation-dependent shifts.
Using two-dimensional phosphopeptide mapping and anti-phosphoantibody analysis, we previously reported that p35 is phosphorylated at Ser8 and Thr138 in COS-7 cells and at Ser8 in cultured neurons (33). Furthermore, we showed that phosphorylation decreases in developing rat brain (32). However, we did not have a method to address quantitative phosphorylation of endogenous p35 in cultured neurons or brain at that time. The quantitative measurement obtained here supports the previous qualitative results and further demonstrates that there are differences in the extent of phosphorylation between transfected cells, primary cultured neurons, and brain cells in vivo. Ser8 was phosphorylated in most p35 molecules whether they were exogenously expressed in transfected COS-7 cells or endogenously expressed in primary cultured neurons and embryonic brain. The phosphorylation of Thr138 was detected most extensively in COS-7 cells (91% of p35 molecules) followed by primary neurons (40%) and embryonic brain (<12%). In contrast, the phosphorylation of Ser91 was least extensive in COS-7 cells (9%) followed by primary neurons (21%) and embryonic brain (59%). These results suggest that phosphorylation of ectopically expressed proteins is not always identical to that of proteins in endogenous tissues.
The phosphorylation of Ser8 was found in p35 co-expressed with Cdk5 but not with knCdk5. Thus, Ser8 was phosphorylated by Cdk5. In fetal brain, Ser8 was phosphorylated in all p35 molecules. This result was somewhat surprising even though Ser8 is autophosphorylated. This high phosphorylation level is not due to its inability to dephosphorylate after it has been once phosphorylated. Ser8 was dephosphorylated by incubation of brain extract in the absence of phosphatase inhibitors. Ser8 was strongly phosphorylated over a short time during metabolic labeling experiments of cultured neurons (33). These results suggest that phosphorylation of Ser8 is maintained by continuous phosphorylation even after it is dephosphorylated. Continuous phosphorylation may be accomplished by intramolecular phosphorylation in the active Cdk5-p35 complex. In contrast, phosphorylation of Thr138 was not the case. We hypothesized that Thr138 could be the phosphorylation site for Cdk5 because Thr138 is in the TPKR sequence, the most preferred consensus sequence for Cdk5 (37). This possibility is supported by the observation that co-expression of p35 with Cdk5 enhanced phosphorylation of Thr138. However, this site was also phosphorylated when p35 was co-expressed with knCdk5 in COS-7 cells. Cycling Cdks such as Cdk1 and Cdk2, whose substrate specificity is similar to Cdk5 (38), are potential candidates for phosphorylating Thr138 in COS-7 cells. These results indicate that Thr138 can be phosphorylated by other proline-directed kinases and that, as opposed to Ser8, Thr138 phosphorylation would be intermolecular even when the site is phosphorylated by Cdk5.
In addition to Ser8 and Thr138, several unidentified minor phosphorylation sites were detected in p35 expressed in COS-7 cells using Phos-tag SDS-PAGE analysis. For the site designated X1, phosphorylation shifted the 2A mutant to the L3 position by Phos-tag SDS-PAGE when co-expressed with Cdk5. Because this phosphorylation was observed in the 2A mutant by co-expression with Cdk5 but not with knCdk5, this site could be one of the other p35 (S/T)P sites, Ser170 and Thr197 (Fig. 2C). This is consistent with the recent report by He et al. (39) that the 2A mutant co-expressed with Cdk5 in HEK293 cells is still phosphorylated to some extent. However, this phosphorylation appeared only to a small extent, if at all, in neurons of mouse brain because the bands that could possibly include phosphorylated X1 were not clearly detectable in p35 prepared from brain. Therefore, in this study, we did not attempt to determine whether Ser170 or Thr197 corresponds to the X1 site. Another novel phosphorylation site named X2 became obvious when the immunoblot using antibody C19 (anti-p35) was overexposed. This band was detected even in the 2A mutant co-expressed with knCdk5, indicating that the site was phosphorylated at an amino acid other than the (S/T)P sites by a different kinase. The finding that phosphorylation at this site was detected in endogenous p35 in neurons prompted us to identify the X2 site (see below). These two phosphorylation sites were not evident in our previous analysis by two-dimensional phosphopeptide mapping following metabolic labeling of neurons or COS-7 cells with [32P]phosphate (33). Lower levels of phosphorylation and/or a slower turnover rate may explain why the X1 and X2 sites were not identified in earlier studies. In either case, Phos-tag SDS-PAGE can be applied to detect minor phosphorylation sites as well as those with a slow turnover rate.
Because phosphorylation of X2 increased upon NMDA treatment, which activates CaMKII (40), we hypothesized that the X2 site could be a CaMKII phosphorylation site. Using Ala mutants at CaMKII consensus (R/K)XX(S/T) phosphorylation sites, the X2 site was identified as Ser91. We confirmed the phosphorylation of Ser91 in fetal and adult brains using phosphospecific antibody (Ser(P)91) generated in this study. Mass spectrometric analysis may be an alternative way to confirm this phosphorylation, although this might not be easy for p35 because only a small percentage of p35, that is a small amount of unstable protein, was phosphorylated at Ser91 in adult brains. In general, however, combinatory use of mass spectroscopy to map phosphorylation sites after identifying phosphorylation using Phos-tag SDS-PAGE will be a powerful biochemical analytic tool.
Although we cannot conclude that Ser91 is phosphorylated exclusively by CaMKII in vivo, based on published results that a population of p35 localizes in the postsynaptic region (41, 42) and associates with CaMKII (43), it is likely that CaMKII phosphorylates p35 at Ser91 in neurons. Indeed, the phosphorylation of Ser91 was lower in COS-7 cells than in primary neurons and fetal brain, reflecting expression levels of CaMKII, which are abundant in neuronal cells compared with rat kidney cells (44). Ser91 phosphorylation was reduced considerably with development, dropping from 59% in embryonic brain to 8% in adult brain. This result may simply reflect the decreased activation frequency of CaMKII in adult mouse brain. In fact, it is reported that active CaMKII decreases during brain development (45, 46). On the other hand, we reported previously that Cdk5 activity suppresses CaMKII activation (41). Thus, the interaction between Cdk5 and CaMKII is bidirectional. CaMKII is a well known mediator of long term potentiation (47, 48), and Cdk5 has also been shown to be involved in memory formation (42, 49–52). The cross-talk between these molecules would be an important regulatory mechanism in synaptic plasticity.
In summary, we used Phos-tag SDS-PAGE for the first time to estimate site-specific in vivo phosphorylation of p35. Quantification of site-specific phosphorylation can be investigated by other means, for example by stable isotope mass spectrometry using multiple reaction monitoring (53–55). However, the number of studies using this method has been limited. We successfully used the Phos-tag SDS-PAGE method to determine populations of p35 with different combinations of phosphorylation sites. The successful use of the method is due to our previous determination of two major phosphorylation sites in p35 and the limited number of phosphorylation sites. We note that this method has advantages over other methods that have been used to analyze phosphorylation of proteins. This method does not require special equipment other than that used for typical electrophoresis and blotting, nor does it require radioisotope labeling, purification of the proteins, or a catalogue of site-specific phosphorylation-dependent antibodies. Lysates of tissues or cells can be directly subjected to this method followed by immunoblotting with a phosphorylation-independent antibody, although prior identification of phosphorylation sites of target proteins would be preferable to maximally utilize this method. We also have to note the negative aspects of this method. This method takes more time than mass spectroscopy and requires reliable antibody for quantification. Nevertheless, we believe that this method is a simple and convenient way to quantify the in vivo phosphorylation states of proteins.
Acknowledgments
We thank Dr. Masaki Inagaki (Aichi Cancer Center Research Institute, Nagoya, Japan) for helpful advice about generating the phosphospecific antibody. We also thank Dr. Junjiro Horiuchi (Tokyo Metropolitan University, Tokyo, Japan) for reading the manuscript.
* This work was supported by a grant-in-aid from the Japan Society for the Promotion of Science (to T. H.) and a grant-in-aid from the Ministry of Education, Culture, Sports, and Science and Technology of Japan (to S. H.).
 This article contains supplemental Figs. S1–S3.
This article contains supplemental Figs. S1–S3.
1 The abbreviations used are:
- Cdk
- cyclin-dependent kinase
- Phos-tag
- phosphate-binding tag
- NMDA
- N-methyl-d-aspartate
- BAP
- bacterial alkaline phosphatase
- CaMKII
- Ca2+-calmodulin-dependent protein kinase II
- ERK
- extracellular signal-regulated kinase
- P
- postnatal day
- E
- embryonic day
- DIV
- days in vitro
- kn
- kinase-negative
- WT
- wild-type
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- Ca2+-CaM
- calcium and calmodulin.
REFERENCES
- 1.Hunter T. (2000) Signaling—2000 and beyond. Cell 100, 113–127 [DOI] [PubMed] [Google Scholar]
- 2.Cohen P. (2002) Protein kinases—the major drug targets of the twenty-first century? Nat. Rev. Drug. Discov 1, 309–315 [DOI] [PubMed] [Google Scholar]
- 3.Thingholm T. E., Jensen O. N., Larsen M. R. (2009) Analytical strategies for phosphoproteomics. Proteomics 9, 1451–1468 [DOI] [PubMed] [Google Scholar]
- 4.Kalume D. E., Molina H., Pandey A. (2003) Tackling the phosphoproteome: tools and strategies. Curr. Opin. Chem. Biol 7, 64–69 [DOI] [PubMed] [Google Scholar]
- 5.Inagaki M., Inagaki N., Takahashi T., Takai Y. (1997) Phosphorylation-dependent control of structures of intermediate filaments: a novel approach using site- and phosphorylation state-specific antibodies. J. Biochem 121, 407–414 [DOI] [PubMed] [Google Scholar]
- 6.Kaufmann H., Bailey J. E., Fussenegger M. (2001) Use of antibodies for detection of phosphorylated proteins separated by two-dimensional gel electrophoresis. Proteomics 1, 194–199 [DOI] [PubMed] [Google Scholar]
- 7.Wegener A. D., Jones L. R. (1984) Phosphorylation-induced mobility shift in phospholamban in sodium dodecyl sulfate-polyacrylamide gels. Evidence for a protein structure consisting of multiple identical phosphorylatable subunits. J. Biol. Chem 259, 1834–1841 [PubMed] [Google Scholar]
- 8.Baudier J., Cole R. D. (1987) Phosphorylation of tau proteins to a state like that in Alzheimer's brain is catalyzed by a calcium/calmodulin-dependent kinase and modulated by phospholipids. J. Biol. Chem 262, 17577–17583 [PubMed] [Google Scholar]
- 9.Morishima M., Ihara Y. (1994) Posttranslational modifications of tau in paired helical filaments. Dementia 5, 282–288 [DOI] [PubMed] [Google Scholar]
- 10.Hisanaga S., Kusubata M., Okumura E., Kishimoto T. (1991) Phosphorylation of neurofilament H subunit at the tail domain by CDC2 kinase dissociates the association to microtubules. J. Biol. Chem 266, 21798–21803 [PubMed] [Google Scholar]
- 11.Kinoshita E., Kinoshita-Kikuta E., Takiyama K., Koike T. (2006) Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol. Cell. Proteomics 5, 749–757 [DOI] [PubMed] [Google Scholar]
- 12.Kinoshita-Kikuta E., Aoki Y., Kinoshita E., Koike T. (2007) Label-free kinase profiling using phosphate affinity polyacrylamide gel electrophoresis. Mol. Cell. Proteomics 6, 356–366 [DOI] [PubMed] [Google Scholar]
- 13.Miyata Y., Nishida E. (2007) Analysis of the CK2-dependent phosphorylation of serine 13 in Cdc37 using a phospho-specific antibody and phospho-affinity gel electrophoresis. FEBS J 274, 5690–5703 [DOI] [PubMed] [Google Scholar]
- 14.Oh H., Irvine K. D. (2008) In vivo regulation of Yorkie phosphorylation and localization. Development 135, 1081–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeya K., Loutzenhiser K., Shiraishi M., Loutzenhiser R., Walsh M. P. (2008) A highly sensitive technique to measure myosin regulatory light chain phosphorylation: the first quantification in renal arterioles. Am. J. Physiol. Renal Physiol 294, F1487–F1492 [DOI] [PubMed] [Google Scholar]
- 16.Tatematsu K., Yoshimoto N., Okajima T., Tanizawa K., Kuroda S. (2008) Identification of ubiquitin ligase activity of RBCK1 and its inhibition by splice variant RBCK2 and protein kinase Cbeta. J. Biol. Chem 283, 11575–11585 [DOI] [PubMed] [Google Scholar]
- 17.Igarashi J., Murase M., Iizuka A., Pichierri F., Martinkova M., Shimizu T. (2008) Elucidation of the heme binding site of heme-regulated eukaryotic initiation factor 2alpha kinase and the role of the regulatory motif in heme sensing by spectroscopic and catalytic studies of mutant proteins. J. Biol. Chem 283, 18782–18791 [DOI] [PubMed] [Google Scholar]
- 18.Sumara G., Formentini I., Collins S., Sumara I., Windak R., Bodenmiller B., Ramracheya R., Caille D., Jiang H., Platt K. A., Meda P., Aebersold R., Rorsman P., Ricci R. (2009) Regulation of PKD by the MAPK p38delta in insulin secretion and glucose homeostasis. Cell 136, 235–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kinoshita E., Kinoshita-Kikuta E., Matsubara M., Aoki Y., Ohie S., Mouri Y., Koike T. (2009) Two-dimensional phosphate-affinity gel electrophoresis for the analysis of phosphoprotein isotypes. Electrophoresis 30, 550–559 [DOI] [PubMed] [Google Scholar]
- 20.Grob A., Roussel P., Wright J. E., McStay B., Hernandez-Verdun D., Sirri V. (2009) Involvement of SIRT7 in resumption of rDNA transcription at the exit from mitosis. J. Cell Sci 122, 489–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oyama S., Yamakawa H., Sasagawa N., Hosoi Y., Futai E., Ishiura S. (2009) Dysbindin-1, a schizophrenia-related protein, functionally interacts with the DNA-dependent protein kinase complex in an isoform-dependent manner. PLoS One 4, e4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dhavan R., Tsai L. H. (2001) A decade of CDK5. Nat. Rev. Mol. Cell Biol 2, 749–759 [DOI] [PubMed] [Google Scholar]
- 23.Cheung Z. H., Ip N. Y. (2007) The roles of cyclin-dependent kinase 5 in dendrite and synapse development. Biotechnol. J 2, 949–957 [DOI] [PubMed] [Google Scholar]
- 24.Lim A. C., Qi R. Z. (2003) Cyclin-dependent kinases in neural development and degeneration. J. Alzheimers Dis 5, 329–335 [DOI] [PubMed] [Google Scholar]
- 25.Hisanaga S., Saito T. (2003) The regulation of cyclin-dependent kinase 5 activity through the metabolism of p35 or p39 Cdk5 activator. Neurosignals 12, 221–229 [DOI] [PubMed] [Google Scholar]
- 26.Patrick G. N., Zukerberg L., Nikolic M., de la Monte S., Dikkes P., Tsai L. H. (1999) Conversion of p35 to p25 deregulates Cdk5 activity and promotes neurodegeneration. Nature 402, 615–622 [DOI] [PubMed] [Google Scholar]
- 27.Kusakawa G., Saito T., Onuki R., Ishiguro K., Kishimoto T., Hisanaga S. (2000) Calpain-dependent proteolytic cleavage of the p35 cyclin-dependent kinase 5 activator to p25. J. Biol. Chem 275, 17166–17172 [DOI] [PubMed] [Google Scholar]
- 28.Lee M. S., Kwon Y. T., Li M., Peng J., Friedlander R. M., Tsai L. H. (2000) Neurotoxicity induces cleavage of p35 to p25 by calpain. Nature 405, 360–364 [DOI] [PubMed] [Google Scholar]
- 29.Saito T., Konno T., Hosokawa T., Asada A., Ishiguro K., Hisanaga S. (2007) p25/cyclin-dependent kinase 5 promotes the progression of cell death in nucleus of endoplasmic reticulum-stressed neurons. J. Neurochem 102, 133–140 [DOI] [PubMed] [Google Scholar]
- 30.Patrick G. N., Zhou P., Kwon Y. T., Howley P. M., Tsai L. H. (1998) p35, the neuronal-specific activator of cyclin-dependent kinase 5 (Cdk5) is degraded by the ubiquitin-proteasome pathway. J. Biol. Chem 273, 24057–24064 [DOI] [PubMed] [Google Scholar]
- 31.Saito T., Ishiguro K., Onuki R., Nagai Y., Kishimoto T., Hisanaga S. (1998) Okadaic acid-stimulated degradation of p35, an activator of CDK5, by proteasome in cultured neurons. Biochem. Biophys. Res. Commun 252, 775–778 [DOI] [PubMed] [Google Scholar]
- 32.Saito T., Onuki R., Fujita Y., Kusakawa G., Ishiguro K., Bibb J. A., Kishimoto T., Hisanaga S. (2003) Developmental regulation of the proteolysis of the p35 cyclin-dependent kinase 5 activator by phosphorylation. J. Neurosci 23, 1189–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamei H., Saito T., Ozawa M., Fujita Y., Asada A., Bibb J. A., Saido T. C., Sorimachi H., Hisanaga S. (2007) Suppression of calpain-dependent cleavage of the CDK5 activator p35 to p25 by site-specific phosphorylation. J. Biol. Chem 282, 1687–1694 [DOI] [PubMed] [Google Scholar]
- 34.Liu J., Fukunaga K., Yamamoto H., Nishi K., Miyamoto E. (1999) Differential roles of Ca(2+)/calmodulin-dependent protein kinase II and mitogen-activated protein kinase activation in hippocampal long-term potentiation. J. Neurosci 19, 8292–8299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamada M., Saito T., Sato Y., Kawai Y., Sekigawa A., Hamazumi Y., Asada A., Wada M., Doi H., Hisanaga S. (2007) Cdk5-p39 is a labile complex with the similar substrate specificity to Cdk5-p35. J. Neurochem 102, 1477–1487 [DOI] [PubMed] [Google Scholar]
- 36.Means A. R. (2000) Regulatory cascades involving calmodulin-dependent protein kinases. Mol. Endocrinol 14, 4–13 [DOI] [PubMed] [Google Scholar]
- 37.Kesavapany S., Li B. S., Amin N., Zheng Y. L., Grant P., Pant H. C. (2004) Neuronal cyclin-dependent kinase 5: role in nervous system function and its specific inhibition by the Cdk5 inhibitory peptide. Biochim. Biophys. Acta 1697, 143–153 [DOI] [PubMed] [Google Scholar]
- 38.Hisanaga S., Uchiyama M., Hosoi T., Yamada K., Honma N., Ishiguro K., Uchida T., Dahl D., Ohsumi K., Kishimoto T. (1995) Porcine brain neurofilament-H tail domain kinase: its identification as cdk5/p26 complex and comparison with cdc2/cyclin B kinase. Cell. Motil. Cytoskeleton 31, 283–297 [DOI] [PubMed] [Google Scholar]
- 39.He L., Hou Z., Qi R. Z. (2008) Calmodulin binding and Cdk5 phosphorylation of p35 regulate its effect on microtubules. J. Biol. Chem 283, 13252–13260 [DOI] [PubMed] [Google Scholar]
- 40.Bliss T. V., Collingridge G. L. (1993) A synaptic model of memory: long-term potentiation in the hippocampus. Nature 361, 31–39 [DOI] [PubMed] [Google Scholar]
- 41.Hosokawa T., Saito T., Asada A., Ohshima T., Itakura M., Takahashi M., Fukunaga K., Hisanaga S. (2006) Enhanced activation of Ca2+/calmodulin-dependent protein kinase II upon downregulation of cyclin-dependent kinase 5-p35. J. Neurosci. Res 84, 747–754 [DOI] [PubMed] [Google Scholar]
- 42.Lai K. O., Ip N. Y. (2009) Recent advances in understanding the roles of Cdk5 in synaptic plasticity. Biochim. Biophys. Acta 1792, 741–745 [DOI] [PubMed] [Google Scholar]
- 43.Dhavan R., Greer P. L., Morabito M. A., Orlando L. R., Tsai L. H. (2002) The cyclin-dependent kinase 5 activators p35 and p39 interact with the alpha-subunit of Ca2+/calmodulin-dependent protein kinase II and alpha-actinin-1 in a calcium-dependent manner. J. Neurosci 22, 7879–7891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tobimatsu T., Fujisawa H. (1989) Tissue-specific expression of four types of rat calmodulin-dependent protein kinase II mRNAs. J. Biol. Chem 264, 17907–17912 [PubMed] [Google Scholar]
- 45.Molloy S. S., Kennedy M. B. (1991) Autophosphorylation of type II Ca2+/calmodulin-dependent protein kinase in cultures of postnatal rat hippocampal slices. Proc. Natl. Acad. Sci. U.S.A 88, 4756–4760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamamoto H., Hiragami Y., Murayama M., Ishizuka K., Kawahara M., Takashima A. (2005) Phosphorylation of tau at serine 416 by Ca2+/calmodulin-dependent protein kinase II in neuronal soma in brain. J. Neurochem 94, 1438–1447 [DOI] [PubMed] [Google Scholar]
- 47.Thiel G., Czernik A. J., Gorelick F., Nairn A. C., Greengard P. (1988) Ca2+/calmodulin-dependent protein kinase II: identification of threonine-286 as the autophosphorylation site in the alpha subunit associated with the generation of Ca2+-independent activity. Proc. Natl. Acad. Sci. U.S.A 85, 6337–6341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fukunaga K., Stoppini L., Miyamoto E., Muller D. (1993) Long-term potentiation is associated with an increased activity of Ca2+/calmodulin-dependent protein kinase II. J. Biol. Chem 268, 7863–7867 [PubMed] [Google Scholar]
- 49.Wei F. Y., Tomizawa K., Ohshima T., Asada A., Saito T., Nguyen C., Bibb J. A., Ishiguro K., Kulkarni A. B., Pant H. C., Mikoshiba K., Matsui H., Hisanaga S. (2005) Control of cyclin-dependent kinase 5 (Cdk5) activity by glutamatergic regulation of p35 stability. J. Neurochem 93, 502–512 [DOI] [PubMed] [Google Scholar]
- 50.Ohshima T., Ogura H., Tomizawa K., Hayashi K., Suzuki H., Saito T., Kamei H., Nishi A., Bibb J. A., Hisanaga S., Matsui H., Mikoshiba K. (2005) Impairment of hippocampal long-term depression and defective spatial learning and memory in p35 mice. J. Neurochem 94, 917–925 [DOI] [PubMed] [Google Scholar]
- 51.Hawasli A. H., Benavides D. R., Nguyen C., Kansy J. W., Hayashi K., Chambon P., Greengard P., Powell C. M., Cooper D. C., Bibb J. A. (2007) Cyclin-dependent kinase 5 governs learning and synaptic plasticity via control of NMDAR degradation. Nat. Neurosci 10, 880–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sananbenesi F., Fischer A., Wang X., Schrick C., Neve R., Radulovic J., Tsai L. H. (2007) A hippocampal Cdk5 pathway regulates extinction of contextual fear. Nat. Neurosci 10, 1012–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mayya V., Rezual K., Wu L., Fong M. B., Han D. K. (2006) Absolute quantification of multisite phosphorylation by selective reaction monitoring mass spectrometry: determination of inhibitory phosphorylation status of cyclin-dependent kinases. Mol. Cell. Proteomics 5, 1146–1157 [DOI] [PubMed] [Google Scholar]
- 54.Wolf-Yadlin A., Hautaniemi S., Lauffenburger D. A., White F. M. (2007) Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc. Natl. Acad. Sci. U.S.A 104, 5860–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ciccimaro E., Hanks S. K., Yu K. H., Blair I. A. (2009) Absolute quantification of phosphorylation on the kinase activation loop of cellular focal adhesion kinase by stable isotope dilution liquid chromatography/mass spectrometry. Anal. Chem 81, 3304–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]