Abstract
Despite progress in the characterization of their genomes, proteomes of several model organisms are often only poorly characterized. This problem is aggravated by the presence of large numbers of expressed sequence tag clones that lack homologues in other species, which makes it difficult to identify new proteins irrespective of whether such molecules are involved in species-specific biological processes. We have used a pulsed stable isotope labeling with amino acids in cell culture (SILAC)-based mass spectrometry method, which is based on the detection of paired peptides after [13C6]lysine incorporation into proteins in vivo, to greatly increase the confidence of protein identification in cross-species database searches. The method was applied to identify nearly 3000 proteins in regenerating tails of the urodele amphibian Notophthalmus viridescens, which possesses outstanding capabilities in the regeneration of complex tissues. We reason that pulsed in vivo SILAC represents a versatile tool to identify new proteins in species for which only limited sequence information exists.
Lack of sequence information has greatly impeded analysis of biological processes and identification of proteins in several nonstandard model organisms for which no comprehensive genome characterization is available. Even if nucleotide sequence data (i.e. EST libraries) are available, it is often difficult to use these data for identification of new proteins, particularly if EST clones lack obvious homologues in other species. Furthermore, it is often difficult to distinguish open reading frames in EST clones lacking obvious homologues from 3′-untranslated regions, intermediate splice products, or cloning artifacts. In addition, several other problems aggravate identification of peptides in partially characterized genomes, which spurred the development of a number of alternative approaches (reviewed in Refs. 1 and 2). Traditional database searches usually allow only identification of peptides that are conserved in newly detected proteins and putative homologous proteins from closely related species (3, 4). Unfortunately, such an approach is not efficient to recognize proteins that are phylogenetically distant from available reference organisms, or belong to poorly conserved protein families. The reliable identification of unknown proteins in “isolated” model organisms currently remains unsolved, although new software tools based on MS BLAST sequence-similarity searches that use multiple redundant and partially accurate candidate peptide sequences have been developed to cope with this difficulty. One potential solution for this problem is de novo protein sequencing, which, however, remains a challenging problem (reviewed in Refs. 5 and 6).
The urodele amphibian Notophthalmus viridescens, vulgo newts, which is one of the best-characterized organisms for the regeneration of complex tissues, is an example of an “isolated” model organism. Newts are able to completely regenerate limbs (7) and tail (8) after amputation, lens and retina (9) as well as inner organs such as the heart (10) and parts of the central nervous system (11) but very little sequence information is available that can be used to decipher the molecular circuits that underlie these regenerative processes. The NCBI database host only 131 protein and 114 nucleotide entries (as of March 2009), which, because of redundant information, represent less than 100 unique protein sequences. This paucity is even more dramatic in the light of the size of the newt genome, which is ∼10 times larger than the human genome. Several recent attempts have been made to understand the molecular mechanism underlying appendage regeneration in urodele amphibians (12, 13) as well as studies to characterize the transcriptome of nerve-dependent limb regeneration in axolotl (14). The current understanding of protein expression programs during the process of regeneration is far from being complete. Because proper appendage regeneration after amputation requires a number of complex steps, such as rapid closure of the limb stump by a wound epithelium, generation of a blastema (15), and differentiation of cells from a pool of progenitors (16, 17), it seems likely that a plethora of different specialized proteins are involved in this process. Furthermore these proteins are likely to have specific needs in this unique process. To identify such proteins, which might lack counterparts in other organisms as well as to increase the confidence level for the detection of potential homologues in other species we have developed a new approach. This approach is based on labeling of proteins in vivo that permits reliable peptide verification by mass spectrometry (MS) for organisms with little or no available sequence information.
So far, relatively few studies have attempted to identify proteins in organisms with an unknown genome (3). The MS BLAST technique (18), for example, allowed the identification of approximately 50 unknown proteins of the unicellular green alga (Dunaliella salina). Other more traditional approaches to characterize proteins in organisms with unknown genomes are based on the use of degenerated primers derived from homologous sequences and antibody staining. Although these strategies were undoubtedly useful for the identification of new proteins involved in regenerative processes (19), several inherent limitations of such methods exist.
Our approach is derived from the stable isotope labeling with amino acids in cell culture (SILAC1) method (20) and uses biologically produced [13C6]lysine containing proteins (21) to label newly synthesized proteins in regenerating newt appendages in vivo. We took advantage of the so called “pulsed or dynamic” SILAC approach (22), which has already been used to determine translation rates (23) or protein turnover in human cancer cell lines (24) in vitro. Mass spectrometric analysis of mixed samples from labeled and unlabeled tissue enabled us to detect a large number of proteins that were incorporated into regenerating newt tails. The recognition of SILAC-peptide pairs significantly improved the rate of protein identification.
MATERIALS AND METHODS
Animal Treatment
Adult newts 3–4 years of age were purchased from Charles Sullivan Inc. Newt Farm (Nashville, TN). Animals, kept at 20 °C in aerated single aquaria, were habituated to mouse liver diet by manual feeding in 3-day intervals for 4 weeks. During this period, animals consuming mouse liver tissue showed no discernible health effects compared with animals on a regular tubifex worm diet. We also detected no changes in weight during the feeding period with liver tissue. To incorporate [13C6]lysine into newt tissues, livers from mice were used that had been labeled with a mouse diet (SILANTES, München, Germany) containing [13C6]lysine (21). After 20 days of feeding, one group of newts was anesthetized with 0.1% ethyl 3-aminobenzoate, methanesulfonic acid solution (Sigma). After tail-tip amputation (1 cm), newts were incubated for 2 h in 0.5% sulfamerazine solution (Sigma) to prevent infections. After an additional 40 days of feeding with proteins containing [13C6]lysine, newts were deeply anesthetized, decapitated, and tail regenerates were immediately collected for further analysis.
Sample Preparation
For protein isolation we homogenized tail tissue with an Ultra-Turrax (IKA-Werke GmbH & Co. KG, Staufen, Germany) in a buffer containing 1% Nonidet P-40, 0.1% sodium deoxycholate, 150 mm NaCl, 1 mm EDTA, and 50 mm Tris, pH 7.5, supplemented with a protease inhibitor mixture (Complete tablets; Roche Applied Science). Protein concentrations were estimated by a Bradford assay. To reduce complexity, samples were resolved by SDS-PAGE, which was cut into 15 slices per lane after Coomassie Blue staining. In-gel digests (components from Sigma) were performed with the protease LysC (Wako Chemicals GmbH, Neuss, Germany), and peptides were loaded onto STAGE-tips (25, 26) for subsequent MS analysis after extraction.
High-performance Liquid Chromatography and Mass Spectrometry
Reversed-phase nano-LC-MS/MS was performed by using an Agilent 1200 nanoflow LC system (Agilent Technologies, Santa Clara, CA). The LC system was coupled to a LTQ Orbitrap XL instrument (Thermo Fisher Scientific, Waltham, MA) equipped with a nanoelectrospray source (Proxeon, Odense, Denmark). Chromatographic separation of peptides was performed in columns filled with reversed-phase ReproSil-Pur C18-AQ 3 μm resin (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany). The LysC-digested peptide mixtures were autosampled at a flow rate of 0.5 μl/min and then eluted with a linear gradient at a flow rate of 0.2 μl/min. The mass spectrometer was operated in the data-dependent mode to automatically measure MS and MS/MS. LTQ-FT full scan MS spectra (from m/z 350 to 1750) were acquired with a resolution of r = 60,000 at m/z 400. The five most intense ions were sequentially isolated and fragmented in the linear ion trap by using collision-induced dissociation.
Analysis of LC-MS/MS Data
Raw data files were converted to MASCOT generic format files with MaxQuant (27) and the MASCOT search engine (version 2.2.02) was used for data base searches and protein identification. The following search parameters were used in all MASCOT searches: LysC digestion, two missed cleavages, and carbamidomethylation of cysteine were set as fixed modification and oxidation of methionines was selected as variable modification.
The maximum allowed mass deviation for MS and MS/MS scans was 10 ppm and 0.5 Da, respectively. For peptide identification, we searched in cross-species data bases, including IPI 3.37 zebrafish, IPI 3.37 mouse, IPI 3.37 human, and the data bases NCB Inr protein, NCBI Xenopus laevis (18016), NCBI Ambystoma (697), and NCBI N. viridescens (110 as of October 2008). In addition, we used an in-house–generated database from regenerating newt hearts (28) for peptide assignment. This database includes 11520 N. viridescens ESTs, translated in three reading frames. Foreign organism databases were generated as DECOY target data bases (29). A minimum peptide length of six amino acids and two peptides per protein group, including one unique peptide, were used for positive output (supplemental Table 1). False discovery rates were based on reverse sequence matches in the combined DECOY target data bases. Our maximum false discovery rate was set below 1% for peptide and protein identifications. All RAW files are available as specified in Table I (30).
Table I. The following supporting data are saved at Tranche (https://proteomecommons.org/tranche). They can be accessed using the hash codes below.
| Part | Hash |
|---|---|
| 1 | 0PDUurcq0P68AonRjxMf8VADUNGp5ScmffwoVk+Ux3P1r2QyMerz3YQZdHdx6XU8rmvP2Ov0YXRovpFt9uE4rcMcSt0AAAAAAAAB0Q== |
| 2 | h9TSWy7khxraUAUIIBTluH1M97iBKB1beUacuI0Ta+vVAT3oMGirWXlLHSF/XVGSQ6GrxSPonvjSPhzabA7XMg7psbEAAAAAAAAB0w== |
| 3 | qufLZxuRiinh9U/InIoj1lfF9ZSqQVMOs1EWS0M3379qGGpF5uvsc8aUq7G35863IQT35jtDWp+PwojnEC6SIv+QUaMAAAAAAAABzg== |
| 4 | rIwZUjcs0S5V9Qk4osR6GCIwW/o05WXU2xqoSwXpkzOQ5s7UPrcwlFRhjQDO3YN8jarIqWsz9gqqvRNrxzdQfCOxXWwAAAAAAAAB0w== |
| 5 | v/Zi2cDtkc0nJ3L6cTM8O52fKVTpZmKEW3Ja6YSMzoJIY7wpCbCPkLN2t0ocXqtFSN9WOyRZwHyw98dD4H1UY0azN+IAAAAAAAAB0w== |
| 6 | iNk226+8dCZRUd678e8EFhLMNXFmmOROjhXFJ/Xw7F3hDcT16HbqludQPMSNGSqM/2z6+LcPkqiZkjGMG0H5Rp7PZeIAAAAAAAAB0w== |
| 7 | X4ySagQE9Lt2ug8zR6niHkGeT6cSzb9wj/BgWfUAATFXIR8KSFBLbxKG8Ge+joXOS261bdXU57erov9D+i6+elDK3wwAAAAAAAAB0g== |
| 8 | xUZzHOCqmELUaWtIHeWPk5Eu4xgnoT/8npnznvom3PTVyjYgTwNRnP5JlD7ICuW2X9icrfRYZ/W1r1PGlsq5nPZwu1IAAAAAAAAB0g== |
| 9 | d4GdKTrowLQW7uLxyfzEhATSBzCZDd0O1wiD0ISCfaS+WYoKHlMCWIXn5gujsOAa2TrTT6nu/Yyeq+ye/Z2bHzUeGDMAAAAAAAAB0g== |
| 10 | nSY3VlviZWHMHePz54IAlH5OGOIRQeQtmZw252n3t2hsu7LQ2bBVE2No2w51srnIHl2cg+IhYudw0kiAwqKG41B2xCUAAAAAAAAB1A== |
| 11 | gWT+hmkM0ALCrlkUD72E5Ss32ubOCHKU0zsXnWDxVAUtbITPdwDcZ6uygAbInyW17ltU2qshIDKwnNPodWQtQ2nIrd4AAAAAAAAB0w== |
| 12 | EfXsK8WKQKnCiGubVrXxCmmygO9Wbb8TYBBFZgdSBwla6M3AjwhqeHpWgNp8DijuGCbKH49+jBc625dSmgLPaNuv0JgAAAAAAAAB0w== |
| 13 | 5KHL1nDzuEIDho3vqq+hahJNVl8yTQEH/7Mks93EMX99Fv2WSfdXqhPbY/FqWNT+EI7S0DvCPNz7+zB+wUo8RJjEKMAAAAAAAAAB0w== |
| 14 | OBedeiqjUY5ccf/vF0PZ7CB5g3WaNK8QxIAGr0g+XdEUBP4fMsBsGThOPlukA3n5nRCQQysKL5n2eUnrjffzL0wuELwAAAAAAAAB0w== |
| 15 | 8KByxRzcX22OLR/0qTajKZGJ8WhyYbwRAp1o1NB4LlAO/gLvpSJh9sSWUt5aSiwfPpPBAubkfCKPdf9GDtqZHjlgw0IAAAAAAAAB0w== |
| 16 | djhmDJhXGSBV+PnImEwbuG9fYrkSeCe+hzVjIQMmJXildmpghQCTFmUoeNtbQUZD1MHZTur73PtW7Jq0+4dpz2LMBwcAAAAAAAAB0w== |
| 17 | Wo9bp89SF6LACC7VediUDTxNI6xb14qEcbuR7cUBTRD7x/FO3GBvLbnwqYL/cs1eagTpqg7KcFzKxoeckuWgAbuMAREAAAAAAAAB1A== |
| 18 | XWCE4ApnRjyTdNDuE14ptTctBXdlnItVjEQ7+noOYjHaEAgTwkIGrKjb/KVTkBUlqcKqumI1ztKM3wjOgjL8ZMeydJUAAAAAAAAB0w== |
| Experimental setting | Dt5ygdKJBalfxk9IvYjPbSCAVrZ2W5SLbVQUybkbpYT1Kg/fJcMFckvGZ1Nzab47dOjcnYCnwS9riNdTtULyHmgG5eAAAAAAAAABvA== |
Analysis of Protein Ratios
Differential incorporation of the [13C6]lysine into identical proteins from different time points of regeneration was calculated by the ratio of heavy/light peptide peaks, using the MaxQuant software tool (27). Labeled proteins were placed into different bins according to the percentage of heavy/light labeling and displayed as a function of frequency. This calculation was done for each database and both time points. (supplemental Table 2).
Protein Classification with Gene Ontology
Detected proteins in the databases IPI mouse, IPI human, and IPI zebrafish were used for GO term annotations based on Uniprot (31). The vertical position of GO terms within the acyclic graph of the GO tree was determined by indirect annotation of proteins to parental GO nodes until the root nodes biological process, cellular component, or molecular function was reached. Calculation of protein representation in GO terms was done by comparing the ratio of proteins within a GO term to the number of proteins for the root nodes biological process, cellular component, or molecular function. Over-represented GO terms were determined by comparing the protein representation ratios from newly identified newt proteins to the ratios from the entire Uniprot data base. GO term over-representation was calculated by corrected p values (Biological Network Gene Ontology tool [BiNGO]) (32).
RT-PCR Analysis
Total RNA was isolated with TRIzol (Invitrogen) according to the manual of the manufacturer. One microgram of total RNA was used for reverse transcription with SuperScript IITM (Invitrogen) according to the guidelines of the manufacturer. A list of the primers used is given in supplemental Table 3.
RESULTS
Reliable Identification of Newt Proteins by Pulsed in Vivo SILAC
Labeling of newt tissue with [13C6]lysine was achieved by maintaining adult N. viridescens for 20 days on a diet consisting of mouse liver derived from fully labeled SILAC mice (21). To accelerate labeling and to mark proteins that were newly synthesized during regeneration, we divided the animals into two groups after completion of the initial incorporation phase. One population was subjected to tail amputation and the labeling time was extended for a further 40 days. The control group was also labeled for an additional 40 days but without tail amputation. By the end of the labeling period, proteins were extracted from control and regenerating tails and analyzed by mass spectrometry (Fig. 1, a and b). After measuring more than 100 samples using an LTQ-Orbitrap, Xcalibur raw files were imported into the MaxQuant software tool and analyzed based on the MASCOT database search engine. Data were searched against different data bases (see “Cross-species database Searches Identify a Large Number of Nonredundant and Time Point-selective Pulsed SILAC Proteins in Regenerating Newt Tails”). Protein identifications on foreign organism databases were based on at least two peptides and one unique peptide. Measured SILAC ratios were calculated as % [13C6]lysine incorporation rate (% label = (SILAC-ratio × 100)/(SILAC-ratio + 1)). The presence of peptide pairs, which comprised [13C6]lysine-labeled and unlabeled peptides, greatly facilitated detection of homologous proteins in evolutionarily distant species. In total, we identified 2994 proteins in foreign organisms that had a match in at least one of the databases employed and 447 proteins in our proprietary newt EST data base. To quantify the improvement achieved by the SILAC approach, we compared our SILAC dataset with a simulated no-SILAC analysis using the IPI human, mouse, and zebrafish databases. For the simulation, we used the same measurements as for the SILAC-analysis but set [13C6]lysine as a variable modification. Inclusion of the information gained by the SILAC approach improved the detection rate in database searches by 43% (mouse and zebrafish data bases) and 32% (human database). The detection rate was even up by 56% when we used our own proprietary newt database (supplemental Table 4).
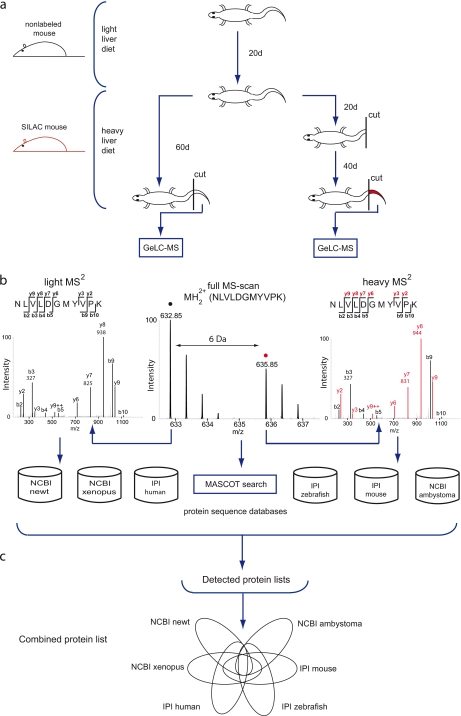
Fig. 1.
Schematic outline of the experimental design used for the pulsed in vivo SILAC approach. a, newts were habituated to mouse liver diet for 28 days. Liver tissue from fully labeled SILAC mice was used to label newts over a period of 60 days. After 20 days, half of the newts were tail-tip amputated and allowed to regenerate; the remaining newts were left undamaged. Tissue from tail tips was isolated after 60 days and prepared for MS analysis. b, isotopic cluster pairs were identified for both damaged and undamaged tissue. The black circle in full MS spectrum defines the light peak; the red circle indicates the heavy Lys-6-labeled isotopic peak. The heavy peak is shifted by 6 Da. MS/MS spectra indicate a mass shift of 6 Da for all detected y ions, also marked in red. Masses for b3, y7, and y8 ions are displayed as examples. Peptide masses were used to perform a MASCOT search on several data bases. c, MASCOT searches for both time points were combined, and heavy light ratios were determined, resulting in several protein group lists. To identify the total number of unique protein groups, we combined all protein lists for both time points.
Pulsed in Vivo SILAC Reveals Increased Protein Turnover in Regenerating Newt Tails After Amputation
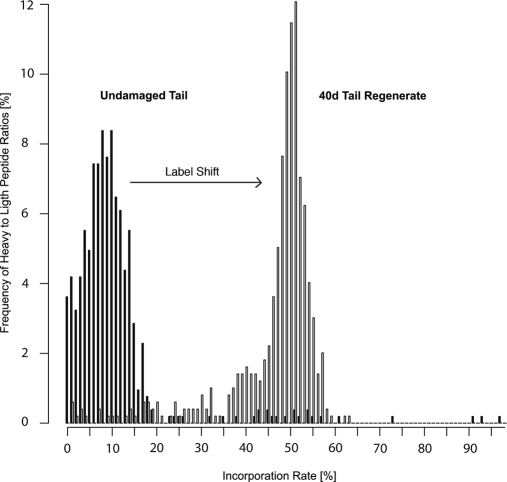
To analyze whether tail amputation resulted in an increased incorporation of [13C6]lysine, we compared the incorporation rate in the newly built tissue 40 days after amputation with undamaged controls. By the end of the feeding period, an incorporation rate of heavy amino acid isotopes of a mean of 11.5% (93.4% of all detected proteins in the mean ± S.D. interval) was measured in the undamaged tail (Fig. 2). The average labeling rate increased dramatically after induction of the regenerative process. After 40 days of regeneration, the mean incorporation rate increased to 46.7% (Fig. 2) based on MASCOT search in the IPI 3.37 mouse database. Similar incorporation rates were detected when other data bases from human, zebrafish, Xenopus, and Ambystoma species were used (data not shown). We concluded that regeneration greatly accelerated [13C6]lysine incorporation in regenerating tails, probably because of increased cell proliferation. Apparently, proteins were not put aside and recycled during regeneration but were first degraded and then resynthesized, which went along with a dilution of the pool of free amino acids by [13C6]lysine from exogenously supplied proteins. Hence, regenerating tails were not completely labeled with [13C6]lysine isotopes after 6 weeks of feeding. The probability to incorporate unlabeled free amino acids was approximately 50% after 6 weeks of feeding.2 Previous experiments in mice indicated that it takes at least three consecutive generations to obtain a relative isotope abundance of 1 (i.e. full labeling), which is difficult to achieve in newts given the relatively long generation time of these animals (21). It also seems likely that several newly synthesized proteins appeared only during regeneration, which is not completed 40 days after amputation, and were therefore not present in intact tails. A more detailed study of different time points during regeneration might also reveal additional proteins that appear only transiently and hence were already removed at the time of our analysis.
Fig. 2.
Tissue regeneration in newt results in accelerated [13C6]lysine incorporation after tail amputation. The mean incorporation rate of [13C6]lysine increased from 11.4% in undamaged tail tips to 46.7% in regenerating tails 40 days after amputation. The percentage of heavy-to-light peptide ratios is given on the y axis. The x axis displays the percentage incorporation rate of [13C6]lysine peptide pairs. A shift in the frequency distribution was observed from 93.4% of all ratios within the mean ± S.D. interval in undamaged tail tissue after 60 days of feeding to 86.1% of all ratios within the mean ± S.D. interval in 40-day tail regenerates after 60 days of feeding.
Cross-species Data Base Searches Identify a Large Number of Nonredundant and Time Point-selective Pulsed SILAC Proteins in Regenerating Newt Tails
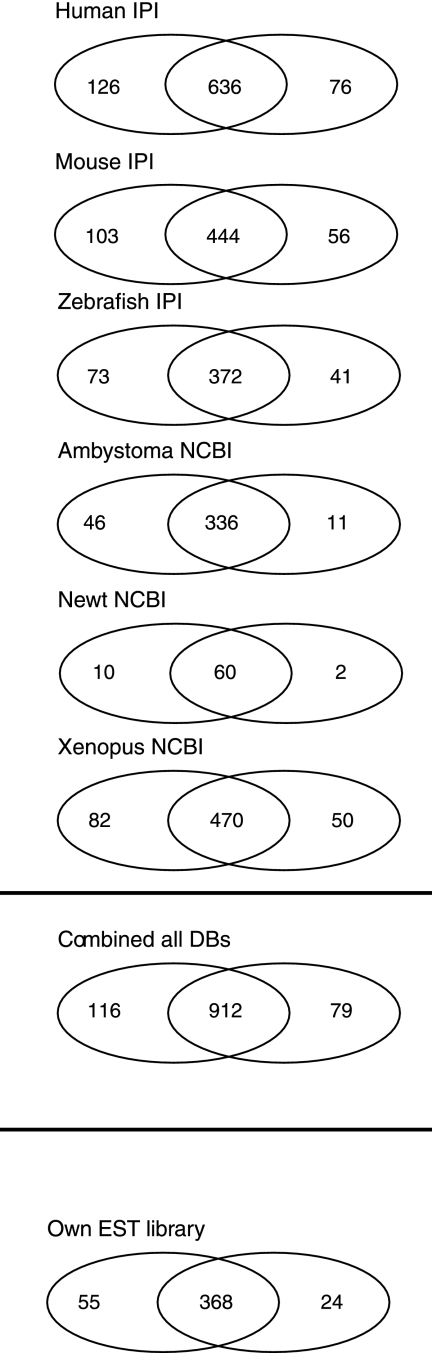
The NCBI databases contain less than 100 nonredundant protein entries for N. viridescens, which limits identification of proteins expressed in regenerating newt tails and illustrates the necessity for broader database searches. The analysis of labeled/unlabeled peptide pairs in regenerating newt tails allowed us to perform advanced screens of different databases, including mouse (IPI3.37), human (IPI3.37), zebrafish (IPI.3.37), Xenopus (NCBI), Ambystoma (NCBI), and N. viridescens (NCBI) (Fig. 3). Most individual proteins (838 proteins) were identified in the human IPI database, but numerous proteins were also identified in mouse (603 proteins) and zebrafish (486 proteins) IPI databases. Moreover, we detected 602 proteins in the Xenopus database, 393 proteins in the ambystomatoid salamander database, and 72 proteins in the newt data base.
Fig. 3.
Pulsed in vivo SILAC enables efficient identification of peptides in the newt proteome. Venn diagram showing the number of peptide pairs identified in both undamaged newt tail and 40-day tail regenerate after 60 days of feeding with [13C6]lysine-labeled newt liver proteins (union middle) and peptide pairs identified in either undamaged (left), or regenerating tail (right). Human, mouse, and zebrafish IPI databases, as well as NCBI protein entries for Xenopus and Ambystoma, were used for cross-species database searches. N. viridescens NCBI protein entries were used to match the percentage of known newt proteins identified in labeled tail tissue.
Although 72 proteins might seem low, it represents more than 90% of all publicly available nonredundant protein sequences for this organism (supplemental Table 5). In total, we identified 2994 heavy labeled proteins. Some of the proteins were represented in several databases, which led to a certain redundancy. Subtraction of redundant sequences left 1035 new and 72 known newt proteins expressed in regenerating newt tail tissues. Total numbers of identified peptide pairs and corresponding protein numbers are given in (supplemental Table 6). To demonstrate the level of protein sequence conservation, some examples of selected peptide spectra, which represent at least two unique peptide pairs, are shown in supplemental Fig. 1. A comparison of SILAC pairs found in regenerating and control tissue revealed 116 proteins that were expressed solely in undamaged tails and 79 proteins present solely in regenerating tails (Fig. 2) from which we were able to annotate 98 and 63 proteins, respectively, to a specific GO term (supplemental Table 7). In regenerating tails, we detected an enrichment of GO-terms associated with wounding, inflammatory response, and cell migration that were completely missing in undamaged tails. A typical example was the glycoprotein fibronectin (supplemental Table 7). Peptide pairs, which were present only in undamaged newt tails, belonged mostly to proteins involved in regulation of striated muscle contraction, Ca2+ transport, and regulation of cholinergic synaptic transmission.
To prove that the increase of the labeling rate by pulsed in vivo SILAC did not only indicate an increased protein turnover but also a change in expression levels, we performed RT-PCR analysis of selected mRNAs. We found a 4.5-fold up-regulation of fibronectin in regenerating limbs, whereas ATPase Na+/K+ α1, ATP2A2, HSP27, Laminin α2, and Slc25α4, which were enriched at the protein level in nondamaged tails, were absent in regenerating tails (supplemental Fig. 2). These results suggest that an increase in the labeling index usually reflects a change in expression levels, although other parameters, such as protein stability and cellular proliferation rate, will also affect the protein labeling index.
Detection of SILAC Peptide Pairs Permits Characterization of Newt ESTs with and without Annotatable Homologies
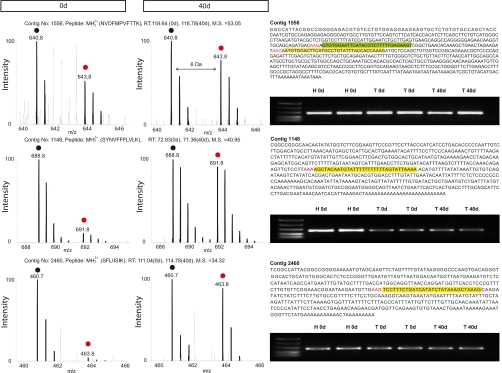
Data from EST libraries most often contain ambiguous sequences that result from single reads and from the assembly of sequences that lack 100% matches. To cope with these problems, space holders are inserted into assembled sequences, which generate undefined proteins upon translation that are difficult to use for protein database searches. The lack of a characterized genome and proteome enhances this dilemma. In principle, direct comparison of peptide sequence data generated by MS analysis to protein sequences generated by translation of EST clones in all reading frames should allow identification of new proteins even in uncharacterized genomes (Fig. 4 and supplemental Table 8). The identification of new proteins from organisms with unknown genomes has already been demonstrated for unlabeled peptides by filtering peptide spectra against a non-annotated library of background spectra and automated de novo interpretation by specific algorithms followed by MS BLAST (3, 4, 18).
Fig. 4.
Identification of previously unknown proteins with a dynamic labeling profile during tail regeneration. Peptide pairs of three selected contigs from undamaged tail and regenerating tail tissue 40 days after amputation (left panel) with no similarity to entries in public data bases with the corresponding peptide sequence in headline including charge, retention time and mascot score (M.S.). Left column displays peptide spectra from undamaged newt tail, and middle column displays spectra from 40-day regenerating tail tissue. Black circles indicate light isotopic peptide peaks, and red circles indicate heavy isotopic peptide peaks, shifting peak clusters by 6 Da in mass. Newt EST sequences with the corresponding sequence region matching to one unique peptide are highlighted in yellow, preceding codons for lysine are highlighted in red (right column, top). Additional identifying unique peptides are indicated in green. RT-PCR analysis from undamaged newt heart (H 0d), undamaged tail (T 0d), and 40-day regenerating tail (T 40d) for the corresponding EST sequences was used to verify expression on the mRNA level (right column bottom). The nucleotide sequences had been deposited in the GenBank database under GenBank Accession numbers GO934291, GO928959, and GO934397.
To investigate whether this approach is applicable for our newt EST database, we translated 9696 high-quality ESTs from a library generated from regenerating newt tissue (28) and searched for corresponding peptide pairs in our MS data set to allow retrograde assembly of identified peptides. The nucleotide sequences had been deposited in the GenBank database under GenBank Accession Numbers GO925352 to GO935047. The use of SILAC peptide pairs allowed us to restrict the search to real peptides excluding nonpeptide peaks and other artifacts. We identified 447 protein groups, which represent 15.44% of the 2894 contigs that were assembled at the nucleotide level from the 9696 ESTs; 412 protein groups corresponded to a single contig, 22 protein groups corresponded to two contigs, and 13 protein groups corresponded to more than two contigs, which reflects an imperfect assembly on the nucleotide level for these contigs (scheme displayed in Fig. 5). Protein database searches identified proteins with different degrees of similarity for 438 of 447 peptide groups. Nine peptide groups that corresponded to open reading frames in ESTs lacked any significant match to existing protein sequences in other species. This might be due to a low degree of conservation of such proteins, which are either unique for newts or underwent a rapid evolutionary drift.
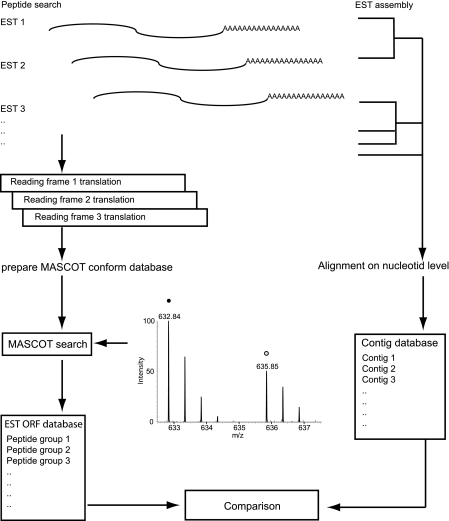
Fig. 5.
Workflow of peptide to contig assignment for newt ESTs. Individual sequence tags (EST, 5′ reads) from newt tissue were translated into three possible reading frames to generate an in silico peptide database (left) and aligned into contigs on the nucleotide level (right). In silico-translated peptides were identified via MASCOT search, and resulting peptide groups were compared with aligned contigs. More than 90% of peptide/nucleotide alignments were 1:1 assignments.
We next asked whether proteins, which appeared to be unique to newts, displayed changes in the labeling rate during tail regeneration. Three proteins (encoded by contigs 1148, 2460, and 595) showed an incorporation rate that corresponded roughly to the mean of all proteins for both time points, indicating that these proteins might not change their relative expression levels during the time course of regeneration. Furthermore, two proteins (encoded by contigs 724 and 1556) showed a higher labeling index in undamaged newt tails compared with the mean of all other proteins, suggesting a down-regulation during regeneration. The remaining four proteins were either only detected in undamaged tail tissue (contigs 2000 and 2804), or in 40 day regenerating tail (1949 and 1982). To verify that changes of the labeling rate did also indicate changes in expression levels on the mRNA level, we performed RT-PCR analysis for three selected contigs (1148, 2460, and 1556), which corroborated the results obtained by pulsed in vivo SILAC (Fig. 3). Taken together, pulsed in vivo SILAC proved to be an efficient tool to identify novel proteins in newts and to detect dynamic changes in their concentration.
DISCUSSION
MS-based proteomics has become an increasingly powerful tool (33) to identify large sets of proteins in complex samples, allowing proteome-wide quantification in cell cultures (20), differential proteome analysis in protozoans (34), and target prediction of miRNA (35). However, proteomic methods applied to complex multicellular organisms is a relatively new application (21), which so far has not been used to study organisms with an uncharacterized genome and proteome. We have developed a new approach to identify proteins that are expressed during tail regeneration of the newt N. viridescens, using [13C6]lysine-labeled mouse tissue (“pulsed in vivo SILAC”). In our case, the pulsed in vivo SILAC method served two different purposes.
First, it distinguished peptides from nonpeptide peaks and generated the number of lysines for each peptide, which significantly decreased the complexity of database searching and thereby increased the number of statistically significant peptide identifications. This feature proved to be particularly helpful in the analysis of a virtually unknown proteome for which no peptide database is available and all protein identifications have to be achieved by comparison to sequences from evolutionary distant organisms.
Second, it helped to compare protein turnover between intact and regenerating newt limbs. Because protein turnover is affected by several features, including synthesis, degradation, proliferation, and apoptosis, it is not possible to directly obtain information about the translation rate from the ratio of heavy to light peptides. In principle, a high heavy/light ratio might indicate either a high translation rate of a stable protein or a low translation rate of an unstable protein. Despite these restrictions we found a good correlation between [13C6]lysine incorporation and mRNA expression, which makes pulsed in vivo SILAC a valuable tool to estimate changes in protein concentration within regenerating newt tails.
We would like to emphasize that other approaches might be used to identify new proteins from organisms with unknown or partially characterized genomes. Most of these techniques are based on filtering of spectra from unlabeled peptides against a non-annotated library of background spectra and automated de novo interpretation by software methods followed by MS BLAST (4, 18).
Despite these achievements, certain limitations of existing methods are evident, which prompted us to explore the benefits of an in vivo SILAC approach. The pulsed in vivo SILAC allowed us to identify 2994 heavy labeled proteins that had a hit in at least one of the databases employed. Furthermore, we found that the SILAC approach increased the peptide identification rate significantly, although the degree of the improvement was hard to quantify because it was difficult to include the increased probability of true positive peptide identifications based on SILAC peptide pairs into the calculation. Using a mock calculation that used the same measurements as for the SILAC-analysis but employed [13C6]lysine as a variable modification, we increased the total number of identified proteins by nearly 11% (464 of 4233, supplemental Table 9). It became evident that even this calculation underestimated the true benefit of our approach when we used the information gained by the SILAC versus the no-SILAC approach for individual database searches. We calculated an approximate increase of the peptide identification rate between 32% (human) and 43% (mouse and zebrafish) for the IPI databases and an increase of approximately 56% for the newt EST database. Comparison of hits from different data bases also revealed that some sequences matched to the same protein identifier because of a high degree of conservation of a subset of newt proteins in other organisms.
Surprisingly, the largest sets of homologues were detected in mammalian databases (838 proteins in human, 603 proteins in mouse) and not in evolutionarily closer organisms, such as Xenopus (602 proteins), zebrafish (486 proteins), and salamander (393 proteins), which might be due to the more comprehensive knowledge of mammalian genomes compared with amphibians and teleost fish. The limited degree of database redundancy and the benefit of multiple cross-database searches became apparent when we calculated the intersections of the three external databases used. It is noteworthy that a large set of proteins was detected only in fish and amphibian databases, which also emphasizes the nonredundant nature of the databases and the limited degree of conservation of several proteins. As expected, most of the known newt proteins (>90%) were contained in our data set. It seems likely that missing proteins were either not expressed in regenerating or intact tails or were lost during probe preparation.
The identification of proteins without any significant similarities in other available databases suggests that newts express classes of proteins that are not present in mammalian organisms and that have not yet been detected in other amphibian species or in zebrafish because of incompleteness of sequence data. Pulse labeling with heavy isotopes not only helped to decrease the complexity of data base searching and to increase the confidence level for detection of homologous peptide sequences in other organisms but was also instrumental in the identification of completely new proteins by comparison with ESTs, which lack similarities to known proteins or ESTs.
In our current analysis, we focused mostly on highly conserved proteins using strict settings for peptide identification. We reasoned that evolutionarily distant proteins exist that will not be detected by our strict parameters despite a high degree of conservation. To reduce this problem, we employed five different databases from different organisms. Using this approach, we were able to increase the peptide identification rate significantly (supplemental Table 4). It is evident that this strategy will exclude proteins that are only weakly conserved during evolution. Such proteins might only be detectable in databases from closely related organisms such as Xenopus and Ambystoma using error-tolerant search parameters.
Yet the use of such search parameters will increase the search time considerably and result in combinatorial explosion (36). It has been pointed out by Shevchenko et al. (37) that error-tolerant searches typically produce large hit lists that require manual inspections, thereby limiting the usefulness of this method for organisms with unknown proteomes. Fixed false discovery rates (DECOY approaches) are often used to prevent manual inspections, but this approach decreases the total number of identified proteins and increases false negative rates.
In summary, we conclude that the improved peptide identification achieved by pulsed in vivo SILAC is a valuable tool to analyze proteomes of model organisms with uncharacterized genomes. MS-based proteomics enables analysis of proteins in absence of specific antibodies and allows comprehensive expression profiling of processes in a yet sparsely characterized organisms. Potential alternative approaches such as de novo protein sequencing still remain challenging and might not represent a realistic option for several organisms in the near future.
Acknowledgments
We thank J. Cox (Max-Planck-Institute for Biochemistry, Martinsried, Germany) for his skillful technical support concerning MaxQuant Software.
* This work was supported by the Max-Planck-Society, the Deutsche Forschungsgemeinschaft (Br1416), the European Union Commission (Multi-Organismic Approach to Study Normal and Aberrant Muscle Development [MYORES] Network of Excellence), and the Kerckhoff Foundation.
 This article contains supplemental Figs. 1 and 2 and supplemental Tables 1–9.
This article contains supplemental Figs. 1 and 2 and supplemental Tables 1–9.
2 M. Looso, unpublished observations.
1 The abbreviations used are:
- SILAC
- stable isotope labeling with amino acids in cell culture
- MS/MS
- tandem mass spectrometry
- GO
- gene ontology.
REFERENCES
- 1.Yates J. R., 3rd (1998) Database searching using mass spectrometry data. Electrophoresis 19, 893–900 [DOI] [PubMed] [Google Scholar]
- 2.Choudhary J. S., Blackstock W. P., Creasy D. M., Cottrell J. S. (2001) Matching peptide mass spectra to EST and genomic DNA databases. Trends Biotechnol 19, S17–S22 [DOI] [PubMed] [Google Scholar]
- 3.Habermann B., Oegema J., Sunyaev S., Shevchenko A. (2004) The power and the limitations of cross-species protein identification by mass spectrometry-driven sequence similarity searches. Mol. Cell. Proteomics 3, 238–249 [DOI] [PubMed] [Google Scholar]
- 4.Junqueira M., Spirin V., Balbuena T. S., Thomas H., Adzhubei I., Sunyaev S., Shevchenko A. (2008) Protein identification pipeline for the homology-driven proteomics. J. Proteomics 71, 346–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Standing K. G. (2003) Peptide and protein de novo sequencing by mass spectrometry. Curr. Opin. Struct. Biol 13, 595–601 [DOI] [PubMed] [Google Scholar]
- 6.Liska A. J., Popov A. V., Sunyaev S., Coughlin P., Habermann B., Shevchenko A., Bork P., Karsenti E., Shevchenko A. (2004) Homology-based functional proteomics by mass spectrometry: application to the Xenopus microtubule-associated proteome. Proteomics 4, 2707–2721 [DOI] [PubMed] [Google Scholar]
- 7.Wong C. J., Liversage R. A. (2005) Limb developmental stages of the newt Notophthalmus viridescens. Int. J. Dev. Biol 49, 375–389 [DOI] [PubMed] [Google Scholar]
- 8.Tassava R. A., Huang Y. (2005) Tail regeneration and ependymal outgrowth in the adult newt, Notophthalmus viridescens, are adversely affected by experimentally produced ischemia. J Exp. Zool. A Comp. Exp. Biol 303, 1031–1039 [DOI] [PubMed] [Google Scholar]
- 9.Kimura Y., Madhavan M., Call M. K., Santiago W., Tsonis P. A., Lambris J. D., Del Rio-Tsonis K. (2003) Expression of complement 3 and complement 5 in newt limb and lens regeneration. J. Immunol 170, 2331–2339 [DOI] [PubMed] [Google Scholar]
- 10.Borchardt T., Braun T. (2007) Cardiovascular regeneration in non-mammalian model systems: what are the differences between newts and man? Thromb. Haemost 98, 311–318 [PubMed] [Google Scholar]
- 11.Parish C. L., Beljajeva A., Arenas E., Simon A. (2007) Midbrain dopaminergic neurogenesis and behavioural recovery in a salamander lesion-induced regeneration model. Development 134, 2881–2887 [DOI] [PubMed] [Google Scholar]
- 12.Stoick-Cooper C. L., Weidinger G., Riehle K. J., Hubbert C., Major M. B., Fausto N., Moon R. T. (2007) Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development 134, 479–489 [DOI] [PubMed] [Google Scholar]
- 13.Kumar A., Godwin J. W., Gates P. B., Garza-Garcia A. A., Brockes J. P. (2007) Molecular basis for the nerve dependence of limb regeneration in an adult vertebrate. Science 318, 772–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monaghan J. R., Epp L. G., Putta S., Page R. B., Walker J. A., Beachy C. K., Zhu W., Pao G. M., Verma I. M., Hunter T., Bryant S. V., Gardiner D. M., Harkins T. T., Voss S. R. (2009) Microarray and cDNA sequence analysis of transcription during nerve-dependent limb regeneration. BMC Biol 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yokoyama H. (2008) Initiation of limb regeneration: the critical steps for regenerative capacity. Dev. Growth Differ 50, 13–22 [DOI] [PubMed] [Google Scholar]
- 16.Muneoka K., Fox W. F., Bryant S. V. (1986) Cellular contribution from dermis and cartilage to the regenerating limb blastema in axolotls. Dev. Biol 116, 256–260 [DOI] [PubMed] [Google Scholar]
- 17.Echeverri K., Clarke J. D., Tanaka E. M. (2001) In vivo imaging indicates muscle fiber dedifferentiation is a major contributor to the regenerating tail blastema. Dev. Biol 236, 151–164 [DOI] [PubMed] [Google Scholar]
- 18.Waridel P., Frank A., Thomas H., Surendranath V., Sunyaev S., Pevzner P., Shevchenko A. (2007) Sequence similarity-driven proteomics in organisms with unknown genomes by LC-MS/MS and automated de novo sequencing. Proteomics 7, 2318–2329 [DOI] [PubMed] [Google Scholar]
- 19.Kumar A., Gates P. B., Brockes J. P. (2007) Positional identity of adult stem cells in salamander limb regeneration. C. R. Biol 330, 485–490 [DOI] [PubMed] [Google Scholar]
- 20.Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 21.Krüger M., Moser M., Ussar S., Thievessen I., Luber C. A., Forner F., Schmidt S., Zanivan S., Fässler R., Mann M. (2008) SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell 134, 353–364 [DOI] [PubMed] [Google Scholar]
- 22.Doherty M. K., Hammond D. E., Clague M. J., Gaskell S. J., Beynon R. J. (2009) Turnover of the human proteome: determination of protein intracellular stability by dynamic SILAC. J. Proteome Res 8, 104–112 [DOI] [PubMed] [Google Scholar]
- 23.Schwanhäusser B., Gossen M., Dittmar G., Selbach M. (2009) Global analysis of cellular protein translation by pulsed SILAC. Proteomics 9, 205–209 [DOI] [PubMed] [Google Scholar]
- 24.Milner E., Barnea E., Beer I., Admon A. (2006) The turnover kinetics of major histocompatibility complex peptides of human cancer cells. Mol. Cell. Proteomics 5, 357–365 [DOI] [PubMed] [Google Scholar]
- 25.Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 26.Andersen J. S., Lam Y. W., Leung A. K., Ong S. E., Lyon C. E., Lamond A. I., Mann M. (2005) Nucleolar proteome dynamics. Nature 433, 77–83 [DOI] [PubMed] [Google Scholar]
- 27.Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 28.Borchardt T., Looso M., Bruckskotten M., Weis P., Kruse J., Braun T. (2010) Analysis of newly established EST databases reveals similarities between heart regeneration in newt and fish. BMC Genomics 11, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 30.Falkner J. A., Falkner J. W., Andrews P. C. (2007) ProteomeCommons. org IO Framework: reading and writing multiple proteomics data formats. Bioinformatics 23, 262–263 [DOI] [PubMed] [Google Scholar]
- 31.Barrell D., Dimmer E., Huntley R. P., Binns D., O'Donovan C., Apweiler R. (2009) The GOA database in 2009–an integrated Gene Ontology Annotation resource. Nucleic Acids Res 37, D396–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maere S., Heymans K., Kuiper M. (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449 [DOI] [PubMed] [Google Scholar]
- 33.Aebersold R., Mann M. (2003) Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- 34.Prieto J. H., Koncarevic S., Park S. K., Yates J., 3rd, Becker K. (2008) Large-scale differential proteome analysis in Plasmodium falciparum under drug treatment. PLoS One 3, e4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. (2008) Widespread changes in protein synthesis induced by microRNAs. Nature 455, 58–63 [DOI] [PubMed] [Google Scholar]
- 36.Nesvizhskii A. I., Vitek O., Aebersold R. (2007) Analysis and validation of proteomic data generated by tandem mass spectrometry. Nat. Methods 4, 787–797 [DOI] [PubMed] [Google Scholar]
- 37.Shevchenko A., Valcu C. M., Junqueira M. (2009) Tools for exploring the proteomosphere. J. Proteomics 72, 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]