Abstract
Oxidative modifications of protein tyrosines have been implicated in multiple human diseases. Among these modifications, elevations in levels of 3,4-dihydroxyphenylalanine (DOPA), a major product of hydroxyl radical addition to tyrosine, has been observed in a number of pathologies. Here we report the first proteome survey of endogenous site-specific modifications, i.e. DOPA and its further oxidation product dopaquinone in mouse brain and heart tissues. Results from LC-MS/MS analyses included 50 and 14 DOPA-modified tyrosine sites identified from brain and heart, respectively, whereas only a few nitrotyrosine-containing peptides, a more commonly studied marker of oxidative stress, were detectable, suggesting the much higher abundance for DOPA modification as compared with tyrosine nitration. Moreover, 20 and 12 dopaquinone-modified peptides were observed from brain and heart, respectively; nearly one-fourth of these peptides were also observed with DOPA modification on the same sites. For both tissues, these modifications are preferentially found in mitochondrial proteins with metal binding properties, consistent with metal-catalyzed hydroxyl radical formation from mitochondrial superoxide and hydrogen peroxide. These modifications also link to a number of mitochondrially associated and other signaling pathways. Furthermore, many of the modification sites were common sites of previously reported tyrosine phosphorylation, suggesting potential disruption of signaling pathways. Collectively, the results suggest that these modifications are linked with mitochondrially derived oxidative stress and may serve as sensitive markers for disease pathologies.
Generation of reactive oxygen species (ROS)1 and reactive nitrogen species is a normal consequence of aerobic metabolism that, in excess, results in oxidative stress that further leads to oxidative modification of proteins, lipids, and DNA, events that may lead to altered cellular function and even cell death (1, 2). Chronic oxidative stress is well recognized as having a central role in disease and is responsible for both direct alteration of biomolecular structure-function and compensatory changes in cellular processes (1–4). It is increasingly recognized that oxidative modifications of proteins can serve as potential biomarkers indicative of the physiological states and changes that occur during disease progression. Thus, the ability to quantitatively measure specific protein oxidation products has the potential to provide the means to monitor the physiological state of a tissue or organism, in particular any progression toward pathology. Given Parkinson disease (PD) as an example, a number of oxidative modifications on proteins pertinent to PD have been identified, further supporting the potential importance of oxidative modifications to disease pathogenesis (5).
Many oxidative modifications on specific amino acid residues, such as protein carbonylation (6), cysteine S-nitrosylation (7–9), cysteine oxidation to sulfinic or sulfonic acid (10–12), methionine oxidation (13, 14), and tyrosine nitration (15–21) within complex protein mixtures, have been detected by MS-based proteomics; however, their low abundance levels within complex proteomes often hinder confident identification of these potentially significant modifications (22). For example, tyrosine nitration is a well studied post-translational modification mediated by peroxynitrite (ONOO−) or nitrogen dioxide (·NO2), which commonly occur in cells during oxidative stress and inflammation; however, only a small number of nitrotyrosine proteins have been identified from a given proteome sample because of insufficient analytical sensitivity and the chance of incorrect peptide assignments (19, 23). With recent advances in high resolution MS that provide high mass measurement accuracy, the ability to confidently identify modified peptides has been significantly enhanced (24).
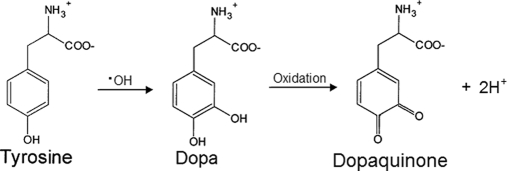
Hydroxyl radical (HO·) is one of the most reactive and major species generated under aerobic conditions in biological systems (1, 25, 26). Among several HO·-mediated oxidative modifications, the protein tyrosine modification 3,4-dihydroxyphenylalanine (DOPA) has been reported as a major product and index of HO· attack on tyrosine residues in proteins (Fig. 1) (27, 28). DOPA is also formed on protein tyrosine residues via controlled enzymatic pathways through enzymes such as tyrosinase or tyrosine hydroxylase (28). Once formed, protein-bound DOPA has the potential to initiate further oxidative reactions through binding and reducing transition metals or through redox cycling between catechol and quinone (dopaquinone) forms (29, 30). Recent studies have suggested that protein-bound DOPA is involved in triggering antioxidant defenses (30) and mediating oxidative damage to DNA (31). Moreover, elevated levels of protein-bound DOPA have been reported in several diseases, including atherosclerosis, cataracts, and myocardial disease, and in PD patients undergoing levodopa therapy (26, 32–36). However, the specific DOPA-modified proteins, which could provide mechanistic knowledge of the progression of these diseases, have not been identified (27, 28). The ability to identify site-specific protein modifications should lead to a better understanding of the role of DOPA modification in disease pathologies as well as new molecular signatures or therapeutic targets for diseases.
Fig. 1.
DOPA and dopaquinone formation from tyrosine.
Therefore, in this study, we demonstrate the ability to identify site-specific DOPA and dopaquinone (DQ) modifications on protein tyrosine residues in normal mouse brain and heart tissues and their relative stoichiometries that are present in vivo under non-stressed conditions. Such endogenous protein modifications were detected using LC-MS/MS. The results from this global proteomics survey suggests that HO· in tissues under normal conditions is generated largely from the mitochondria and metal-binding proteins where the resulting DOPA/DQ modifications have the potential to disrupt mitochondrial respiration as well as alter tyrosine phosphorylation signaling pathways such as 14-3-3-mediated signaling in brain tissue.
EXPERIMENTAL PROCEDURES
Mouse Brain and Heart Tissue Sample Processing
Brain tissue was dissected from C57BL/6J male mice (8 weeks of age, 21–27 g) obtained from The Jackson Laboratory (Bar Harbor, ME) and frozen immediately in liquid nitrogen until use. Brain tissues from four mice were homogenized in 50 mm NH4HCO3 (pH 7.8), and protein concentrations were determined by the BCA assay (Pierce). All animal procedures were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Heart tissue from 4-month-old female BALB/cByJNiaHsd mice was obtained from NIA, National Institutes of Health where tissues were frozen in liquid nitrogen after sacrifice and removal. All procedures were done on ice if not stated otherwise. Five frozen mouse hearts (1.2 g total) from five individual mice were pulverized with a mortar and pestle on dry ice followed by homogenization in 6–8 ml of 50 mm NH4HCO3 (pH 7.8), 150 mm NaCl with a Silverson L4R homogenizer (East Longmeadow, MA). Homogenates were centrifuged for 10 min at 500 × gmax, and supernatants were filtered through one layer of cheesecloth into a clean precooled centrifuge tube. The resulting supernatants (soluble fractions) were saved.
Pooled brain and heart tissue homogenates with 300 μg of protein each were treated with 50% (v/v) trifluoroethanol (Sigma) for 2 h at 60 °C and then with 2 mm DTT (Sigma) for 1 h at 37 °C with gentle shaking at 300 rpm. Both samples were then diluted 5-fold with 50 mm NH4HCO3 and digested with sequencing grade trypsin (Promega, Madison, WI) for 3 h at 37 °C with a 1:50 trypsin to protein ratio (w/w). 1 mm CaCl2 was applied during the digestion. Peptides were recovered using solid phase extraction C18 column (Supelco, Bellefonte, PA) cleanup, dried down with a speed vacuum concentrator dissolved in 25 mm NH4HCO3, and stored at −80 °C until time for further analysis.
In Vitro Oxidation of Heart Peptide Sample
Mouse heart peptides were resuspended in 25 mm NH4HCO3 (pH 7.2) and incubated with the iron/EDTA/ascorbate system, which consisted of 50 mm sodium phosphate buffer (pH 7.2) containing 240 μm EDTA and 1 mm ascorbic acid (Sigma). The reaction was initiated by the addition of Fe2+ (FeSO4·7H2O) to a final concentration of 200 μm in a final volume of 100 μl. The sample was then incubated at room temperature for 1 h with constant shaking at 600 rpm. The incubation tube was opened and covered with Breathe-EASIER tube membrane (Diversified Biotech, Boston, Massachusetts) during the incubation to improve oxygenation. Control samples were treated without iron, EDTA, and ascorbate. Following the reaction, peptides were recovered by solid phase extraction C18 column (Supelco) cleanup, dried down in a speed vacuum concentrator dissolved in 25 mm NH4HCO3, and stored at −80 °C for further MS analysis.
Strong Cation Exchange (SCX) Fractionation
The dried peptides from mouse brain and heart were dissolved in 25 mm NH4HCO3 and subjected to HPLC (Agilent, Palo Alto, CA)-based SCX fractionation on a 200 × 2.1-mm (5-μm particles, 300-Å pore size) Polysulfoethyl A column (PolyLC, Columbia, MD) at a flow rate of 0.2 ml/min. Mobile phase solvents consisted of 10 mm ammonium formate, 25% acetonitrile (pH 3.0) (A) and 500 mm ammonium formate, 25% acetonitrile (pH 6.8) (B). Once the column was loaded, isocratic conditions at 100% A were maintained for 10 min. Peptides were separated by using a gradient from 0 to 50% B over 40 min followed by a gradient of 50–100% B over 10 min. The gradient was then held at 100% solvent B for another 10 min. 30 fractions were collected for the brain sample, whereas 15 fractions were collected for the heart sample. Following lyophilization, the fractions were dissolved in 25 mm NH4HCO3 and stored at −80 °C until further use.
Nano-LC-MS/MS Analysis
All SCX fractions of mouse brain and heart samples as well as the in vitro oxidized heart sample were further analyzed using a fully automated custom-built four-column nano-HPLC system (37) coupled on line with an LTQ Orbitrap mass spectrometer (ThermoFinnigan, San Jose, CA) modified with an in-house manufactured electrospray ionization interface. The reversed phase capillary column was slurry-packed using 3-μm Jupiter C18 particles (Phenomenex, Torrance, CA) into a 75-μm (inside diameter) × 65-cm fused silica capillary (Polymicro Technologies, Phoenix, AZ). The mobile phases consisted of A (0.2% acetic acid and 0.05% TFA in water) and B (0.1% TFA in 90% acetonitrile). The mobile phases A and B were stably delivered using Isco LC pumps (Teledyne Isco, Lincoln, NE) at a constant pressure of 10,000 p.s.i. and were mixed in a stainless steel mixer (∼2.5 ml) before entering the separation capillary. The setup generates an exponential gradient for the separation that started with 100% A and gradually increased to 60% B over the course of 100 min with a flow rate of ∼500 nl/min for the separation column. The instrument was operated in data-dependent mode with an m/z range of 400–2000. The 10 most abundant ions from MS analysis were selected for further MS/MS analysis using a normalized collision energy setting of 35%. MS/MS fragments were measured in the linear ion trap. The ion trap full and MSn automatic gain control targets were set at 30,000 and 10,000, respectively. The automatic gain control setting for the orbitrap was 1,000,000 with a resolution of 60,000. A dynamic exclusion of 1 min was applied to reduce repetitive analysis of the same abundant precursor ion.
MS/MS Data Analysis
The LC-MS/MS raw data were converted into a .dta file using an in-house software, DeconMSn (version v2.1.4.1), which accurately calculates the parent monoisotopic mass for each spectrum from the parent isotopic distribution using a modified THRASH approach. The MS/MS data were then searched against the mouse International Protein Index (IPI) database with a total of 51,252 total protein entries (version 3.19, released July 25, 2006) with the reversed sequence decoy database searching option (for assessing false positive rate) using X!Tandem 2 software (38). The search parameters used were as follows: 60-ppm tolerance for precursor ion masses and 0.4-Da tolerance for fragment ion masses with no enzyme restraint and a maximum of three missed tryptic cleavages. Modifications for the dynamic oxidation (15.9949 Da) of methionine, proline, and tryptophan residues were applied during the database search. Three tyrosine modifications, nitrotyrosine, DOPA, and DQ, were investigated by applying dynamic modifications of 44.9851, 15.9949, and 13.9793 Da on tyrosine residues, respectively, in separate database searches due to the one-modification limitation on one residue in X!Tandem search. A 5-ppm mass tolerance cutoff was used as an initial filter for the raw X!Tandem results. Additionally, peptide expectation values (log(e)) of <−1 and <−2 were required for fully or partially tryptic peptides, respectively. Such filtering criteria were established based on decoy database search results to provide a <1% false positive rate (FPR) for overall peptide identifications (39, 40). The decoy protein database includes forward and reversed sequences of the amino acid sequences for each protein, and the FPR for peptide identifications was estimated by doubling the number of unique peptides identified from the reversed sequence (NR) and then dividing by the total number of unique peptides identified from both forward (NF) and the reversed database search (NF + NR), i.e. FPR = 2 × NR/(NF + NR). Protein Prospector and Sequest searches were also performed for DOPA-modified peptides to further confirm some of the site identifications. The in-house written software SpectrumLook (version 1.5.37) was used to manually inspect the MS/MS spectra by matching observed fragments in MS/MS spectra to masses of predicted fragments within a tolerance of ±0.5 for m/z values. Manual inspection was performed at the MS/MS spectrum level for all the DOPA-, DQ-, and nitrotyrosine-modified peptides to ensure that the major peak assignments were matched with theoretical predictions of b and y fragments as illustrated by MS/MS spectra for both modified and unmodified versions of the peptides and that the modification sites were unambiguously identified by specific fragments indicative of the modification sites. All annotated spectra are supplied as supplemental materials.
RESULTS
Endogenous DOPA- and DQ-modified Peptide Identifications from Mouse Brain and Heart Tissues
For the identification of DOPA- and DQ-modified proteins in heart and brain, we utilized a global profiling strategy, i.e. SCX fractionation coupled off line with reversed phase LC-MS/MS with high resolution parent mass measurements, which permits confident identification of several types of modified peptides and their native analogues in a complex mixture by applying a 5-ppm mass measurement accuracy constraint for the parent masses. Tryptic peptides from mouse brain or heart homogenates were fractionated by SCX, and the resulting peptide fractions were individually analyzed by high resolution nano-LC-MS/MS. Peptides were identified from MS/MS spectra by database searching using the X!Tandem algorithm (38). To identify the modified peptides, we included variable modifications on tyrosine residues with additional masses of 44.985, 15.995, and 13.979 Da, corresponding to nitrotyrosine, DOPA, and DQ modifications, respectively. Variable oxidative modifications (15.995 Da) on methionine, proline, and tryptophan were also included because these residues are also prone to oxidation (41). The inclusion of both DOPA- and nitrotyrosine-related modifications allows us to compare the relative prevalence between DOPA modification and nitration on protein tyrosine. The samples were analyzed on an LTQ Orbitrap mass spectrometer; the achievable ppm level mass accuracy on parent mass measurements allows us to achieve high confidence identifications and to differentiate the exact forms of DOPA and DQ modifications. One of the challenges in identifying low abundance modified peptides is the potential of false positive identifications. High FPR is often encountered if only a small number of modified peptides are identified among a large number of total peptide identifications. Following the application of 5-ppm mass tolerance and peptide expectation value cutoffs, we achieved <1% FPR for total peptide identifications in both tissues and a 1–5% FPR for DOPA- and DQ-modified peptide identifications based on decoy searching results; however, for nitrotyrosine-modified peptides, we observed ∼40% FPR following the application of the same criteria due to the identification of significantly fewer nitrotyrosine peptides. Upon applying a more stringent filtering criteria requiring peptide expectation values (log(e)) of <−2 and <−3 for fully or partially tryptic peptides and further manual spectrum examination, we only identified two nitrotyrosine sites from the brain tissue (Table I).
Table I. Total number of peptides and proteins from global profiling and number of unique tyrosine modified sites corresponding to three modifications (DOPA, dopaquinone, and nitrotyrosine) identified from mouse heart and brain tissue.
| Number of reversed hits/total number of peptides | FPR | Total number of proteins | Number of reversed hits/total number of modified peptides |
Number of tyrosine modified sites after manual inspection |
|||||
|---|---|---|---|---|---|---|---|---|---|
| DOPA | DQ | NO2a | DOPA | DQ | NO2 | ||||
| % | |||||||||
| Brain | 74/26,572 | 0.6 | 4,656 | 2/131 | 2/70 | 0/4 | 50 | 20 | 2 |
| Heart | 27/9,409 | 0.6 | 1,914 | 1/48 | 2/48 | 1/4 | 14 | 12 | 0 |
a Peptide expectation values (log(e)) of <−2 and <−3 were required for fully or partially tryptic NO2-modified peptides.
Table I lists the total number of peptides and proteins from global profiling of both tissues and the number of reverse hits as well as the number of unique modified peptides identified for each type of modification before and after manual inspection from mouse brain and heart tissues. The potential of oxidative modifications on multiple amino acid residues including methionine, proline, tryptophan, and tyrosine often makes it a challenge to determine the exact modification site for a given peptide even if the peptide sequence along with the modification mass is correctly identified. In the manual inspection process, only those peptides with unambiguous site identifications were retained as final identified DOPA- and DQ-modified peptides as listed in supplemental Tables 1 and 2. For the global profiling, we identified 26,572 peptides and 4,656 proteins from brain and 9,409 peptides and 1,914 proteins from heart. A total of 63 unique modified tyrosine sites was identified in the mouse brain, corresponding to 53 proteins; 50 and 20 of these sites were modified with DOPA and DQ, respectively. For mouse heart, a total of 23 unique modified tyrosine sites was identified, corresponding to 19 proteins; 14 and 12 of these sites were modified with DOPA and DQ, respectively. Thus, the percentage of DOPA/DQ-modified peptides relative to the total number of identified peptides is similar for both tissues, i.e. ∼0.24%. The relative stoichiometry of modified tyrosine sites was approximated by comparing spectral count (the number of MS/MS spectra resulting in the identification of a given peptide) information for modified and unmodified versions of each peptide; the majority of tyrosine sites were observed with a stoichiometry of less than 10%. Moreover, only one or two tyrosine modification sites were observed per protein as expected for endogenous modifications in normal tissues. When data sets for mouse brain and heart were combined, 81 unique tyrosine sites modified from 67 proteins were observed of a total 880 tyrosine sites from these proteins (supplemental Tables 4 and 5). It is interesting to note that we only identified two nitrotyrosine sites from the same brain tissue. The results again illustrate the challenge for effectively identifying tyrosine nitration using global proteome analyses without enrichment, presumably due to the low abundance of endogenous protein tyrosine nitration. This observation is in good agreement with our previous effort for nitrotyrosine profiling where the majority of nitrotyrosine-containing peptides were identified from a cysteinyl peptide-enriched sample from mouse brain (19). On the other hand, DOPA and DQ modifications on tyrosine are more readily detectable and should be more feasible for exploring the role of these modifications as potential biomarkers for oxidative stress and inflammation.
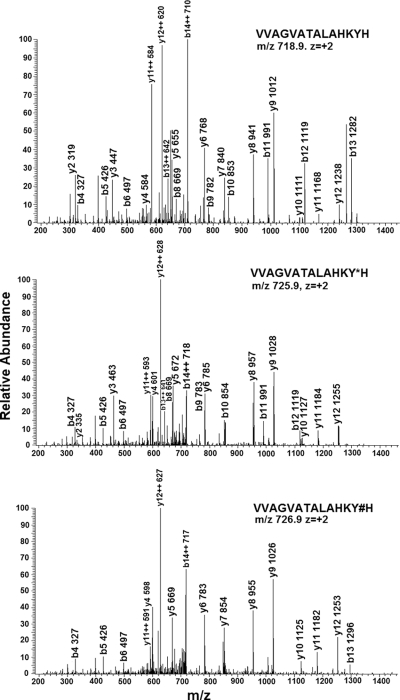
Fig. 2 shows an example MS/MS spectra of one peptide, VVAGVATALAHKYH, from the hemoglobin β subunit in both its unmodified and DOPA- and DQ-modified versions. Note that the b and y fragment ions for the three versions of this peptide match with the exception of b and y ions that contain modified tyrosine residues, i.e. b13 and y2–y12, where DOPA- or DQ-modified peptides have masses that are 15.99 or 13.98 Da, respectively, higher than those of the corresponding ions from the non-modified peptide.
Fig. 2.
MS/MS spectra of non-modified (top), DOPA- (middle), and DQ-modified (bottom) peptide VVAGVATALAHKYH from hemoglobin subunit β (Hbb-b2) in mouse heart. Y*, DOPA-modified Tyr; Y#, DQ-modified Tyr.
Enhanced DOPA Modification following in Vitro HO· Exposure
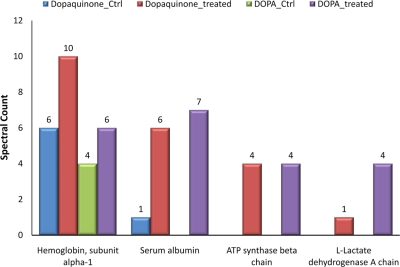
Because DOPA has been suggested to be the major product of HO· attack on tyrosine residues (27, 42), we applied an in vitro exposure to HO· using iron/EDTA/ascorbate to generate HO· (28). About twice as many unique modified peptides (seven DOPA- and six DQ-modified peptides) were identified from single dimensional LC-MS/MS analyses of the iron/EDTA/ascorbate-treated heart sample as compared with the number of unique peptides (two DOPA- and four DQ-modified peptides) identified in the control. In addition, the spectral count of many modified peptides also increased following the chemical treatment, again reflecting enhanced tyrosine hydroxylation. Fig. 3 shows the total spectral count comparison of selected proteins before and after in vitro hydroxylation treatment where the results clearly illustrate the increased extent of modifications following the chemical treatment. A detailed report of this result is available as supplemental Table 3. Thus, these in vitro results suggest that endogenous DOPA and DQ modifications reflect the in vivo formation of HO·.
Fig. 3.
Spectral counts of DOPA- and DQ-modified proteins identified from mouse heart tissue before and after chemical treatment by iron/EDTA/ascorbate system. Dopaquinone_Ctrl, DQ-modified proteins from control sample; Dopaquinone_treated, DQ-modified proteins from treated sample; DOPA_Ctrl, DOPA-modified proteins from control sample; DOPA_treated, DOPA-modified proteins from treated sample.
Preferential DOPA/DQ Modification of Mitochondrial Proteins
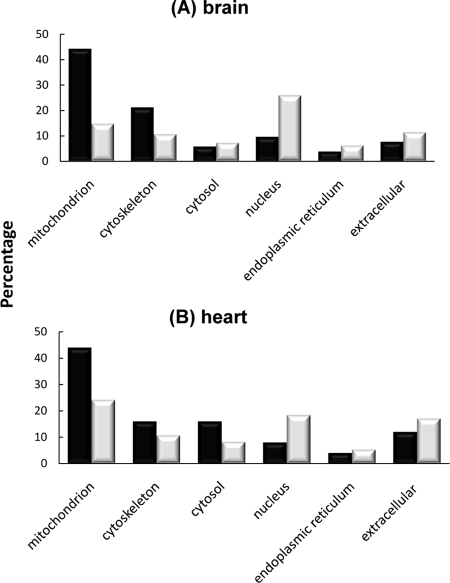
To examine what types of proteins are more sensitive to HO· attack, we categorized the cellular localization of modified proteins from mouse brain and heart based on their gene ontology annotations. Fig. 4 shows that ∼44% of modified proteins were localized in the mitochondria for both tissues. These values are nearly twice the percentage of the unmodified mitochondrial proteins identified in these global profiling data, showing a clear preference for DOPA/DQ modification of mitochondrial proteins, in good agreement with the pivotal role of mitochondria in generating ROS (43).
Fig. 4.
Preferential modifications on mitochondrial and cytoskeletal proteins from brain (A) and heart (B). Gene ontology analysis of the cellular locale of DOPA/DQ-modified proteins (black) compared with that of the total identified proteins (gray) is shown. The number of proteins in each category is expressed as a percentage of the total.
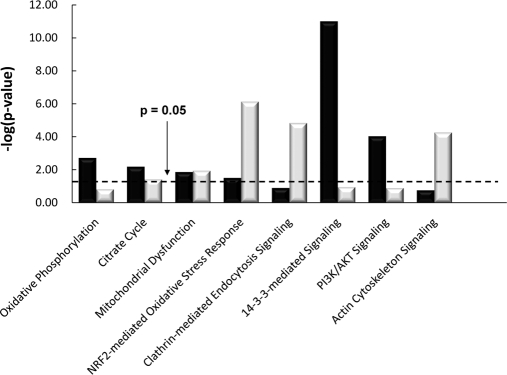
Fig. 5 shows the top pathways represented by DOPA- and DQ-modified proteins from mouse brain and heart tissues based on analysis using the Ingenuity Pathways Analysis tool (44). Consistent with the large percentage of modified proteins in mitochondria (Fig. 4), several mitochondrially associated pathways, including oxidative phosphorylation, citrate cycle, and mitochondrial dysfunction, were observed to be significant. For example, malate dehydrogenase 2 (Mdh2), a key enzyme in the citrate cycle, was modified in brain tissue. Multiple ATP synthases (Atp5b, Atp5o, Atp5h, Atp6v0d1, and Atp6v0a1) and ubiquinol-cytochrome c (Uqcrb) involved in the oxidative phosphorylation pathway were also modified.
Fig. 5.
Top canonical pathways from mouse brain (black) and heart (gray) represented by DOPA/DQ-modified proteins. The p value is a measure of the likelihood that the association between the set of modified proteins with the given pathway is due to random chance. PI3K, phosphatidylinositol 3-kinase.
In addition to mitochondrion-related pathways, several other signaling pathways were observed with significance, for example the nuclear factor-like 2 (Nrf2)-mediated oxidative stress response pathway in heart tissue. For this pathway, modifications occur on both cytoskeletal proteins (e.g. actin β, α, and γ1) and cytosolic enzymes (e.g. 90-kDa heat shock proteins (Hsp90aa1 and Hsp90ab1)). Interestingly, 14-3-3-mediated signaling and phosphatidylinositol 3-kinase/AKT signaling pathways were only observed with significance in mouse brain tissue. A significant contribution to these pathways is the modifications observed on several members of the 14-3-3 protein complex (Table II). The 14-3-3 proteins are known to be abundantly expressed in the brain and play important function in the nervous system (45). The observed modifications on multiple isoforms of α- and β-tubulin associated with the 14-3-3-mediated signaling are also interesting because the role of 14-3-3 proteins in cytoskeletal regulation has been reported (46).
Table II. DOPA/DQ modification sites identified from 14-3-3 proteins in mouse brain.
Spectral counts for both modified and unmodified versions are indicated along with those sites previously known to be phosphorylated. Y*, modified Tyr.
| Tyr site | IPI no. | Gene symbol | Peptide sequence | Spectral count |
Known Tyr(P) site | ||
|---|---|---|---|---|---|---|---|
| Unmodified | DOPA | DQ | |||||
| Tyr-131 | IPI00118384 | Ywhae | R↓Y*LAEFATGNDR↓K | 2 | 1 | ||
| Tyr-152 | IPI00118384 | Ywhae | K↓EAAENSLVAY*K↓A | 5 | 1 | ||
| Tyr-214 | IPI00118384 | Ywhae | K↓AAFDDAIAELDTLSEESY*KDSTLIMQLLR↓D | 121 | 7 | Yes | |
| Tyr-132 | IPI00227392 | Ywhah | R↓Y*LAEVASGEKK↓N | 11 | 1 | Yes | |
| Tyr-153 | IPI00227392 | Ywhah | K↓NSVVEASEAAY*K↓E | 2 | 1 | ||
| Tyr-128 | IPI00116498 | Ywhaz | R↓Y*LAEVAAGDDKK↓G | 20 | 4 | ||
| Tyr-211 | IPI00116498 | Ywhaz | K↓TAFDEAIAELDTLSEESY*KDSTLIMQLLR↓D | 382 | 5 | Yes | |
Of particular interest was the identification of a number of known tyrosine phosphorylation sites that were modified by DOPA and DQ, suggesting that 3-hydroxylation of tyrosine represents a new mechanism for disrupting cell signaling regulated by tyrosine phosphorylation. By mapping the identified DOPA-modified tyrosine sites to the Phosphosite database, we observed 21 and three unique tyrosine sites from brain and heart known as phosphorylated, corresponding to 19 and three proteins, respectively (see supplemental Tables 1 and 2). Among these, three of seven modification sites observed in 14-3-3 proteins comprise known tyrosine phosphorylation sites (Table II). We also note that the numbers of mapped known phosphorylated sites are limited by the current available database for which new sites are continually being added.
DISCUSSION
As oxidative stress has been recognized as central to many pathologies, so have oxidative modifications of proteins been implicated in the pathogenesis of multiple human diseases (22). Of particular interest are oxidative modifications on protein tyrosine because of their potential role in disrupting catalysis, protein-protein interactions, and cell signaling mediated by tyrosine phosphorylation. To date, most studies have been focused on the characterization of tyrosine nitration where elevated levels of nitrotyrosine have been observed in multiple inflammatory diseases (2, 47). Similarly, increases in total protein DOPA levels have been linked with several diseases, but information regarding specific modified proteins has been lacking (32, 33). In the present work, we have performed what we believe to be the first extensive proteomics survey of endogenous DOPA and DQ modifications on protein tyrosines in mouse brain and heart, two tissues with relevance to neurological and cardiovascular diseases. We found that the modification of proteins by DOPA/DQ is much more prevalent in animal tissues than has been previously appreciated, and we have identified sensitive in vivo protein targets. In total, we have been able to identify 85 unique tyrosine sites with these modifications based on extensive LC-MS/MS profiling. This large data set along with site-specific modification information permits insights regarding the susceptibility of such modifications in terms of cellular components and pathways as well as the potential implications in neurological and cardiovascular diseases.
The characteristics of DOPA/DQ-modified proteins are consistent with their origin as resulting from HO· exposures rather than from enzymes such as tyrosinase or tyrosine hydroxylase. For example, the number and abundance of DOPA/DQ-modified peptides was enhanced after in vitro exposure of heart homogenate peptides to HO· through an iron/EDTA/ascorbate-generating system (supplemental Table 3 and Fig. 3), whereas the relative percentage of DOPA/DQ-modified peptides in brain, where tyrosinase and tyrosine hydroxylase are more highly expressed, was not greater than that in the heart (Table I). The observed preferential modification of mitochondrial proteins by DOPA/DQ further suggests their origins from short lived and/or non-diffusive ROS generated in mitochondria. These criteria are fulfilled by the mitochondrial generation of superoxide anion (·O2−), which can be readily converted to the oxidant H2O2 by superoxide dismutase followed by its further reaction to HO· by Fenton chemistry in the presence of transition metals. Neither superoxide nor HO· is highly diffusive; in particular, the high reactivity of HO· means that it diffuses only the diameter of a typical protein within its lifetime (2). Mitochondria are noted as the repositories of cellular pools of iron, copper, manganese, and zinc, all required as cofactors for mitochondrial respiration. In particular, in the case of copper, ∼80% of this metal is not bound to proteins but localized within the mitochondrial matrix as a soluble ligand complex (48, 49). Thus, the high levels of both protein-bound and exchangeable metals within mitochondria could provide a source of potential Fenton chemistry to produce HO·. Indeed, in our proteomics analysis, a substantial amount of mitochondrial metalloproteins was observed to be modified by DOPA/DQ (50). Some examples are the cytochrome c (Cyct), cytochrome bc1 complex (Uqcrb), and the ATP synthases (Atp5h, Atp5b, and Atp5o). We also note that multiple isoforms of hemoglobin, an extracellular metalloprotein, incorporated significant levels of DOPA/DQ modifications. Additionally, the mitochondrially expressed antioxidant enzyme manganese-superoxide dismutase (Sod2) was found to be modified at Tyr-34 (within the peptide HHAAY*VNN) in both brain and heart; nitration of this same site has been demonstrated to inhibit enzyme activity (2).
Besides mitochondrial proteins, we observed modifications on many cytoskeletal proteins, reflecting known metal binding properties of cytoskeletal proteins, such as actin and tubulin, as well as the proximity of the cytoskeleton to mitochondria in both brain and heart (51). In mouse brain, DOPA/DQ modifications were also observed on several members of the 14-3-3 protein complex, a class of proteins also reported to have metal ion binding properties (52); however, there is a potential role of tyrosine hydroxylase for these modifications because the 14-3-3 proteins are known to interact with proteins that display tyrosine hydroxylase activity. Given the importance of metal-binding proteins in protein structural stability, cell signaling, regulation, transport, immune response, metabolism control, and metal homeostasis, selective DOPA modification on tyrosine sites in these proteins should play a significant role in cellular functions.
As the major source of cellular ROS, mitochondria are not only sensitive to oxidative damage but also play a crucial role in disease pathogenesis because mitochondrial dysfunction and oxidative stress are recognized as important contributing factors in the development of neurodegenerative, cardiovascular, and aging-related diseases (43, 53–55). The high percentage of identified modified proteins localized in mitochondria suggests that DOPA modification may represent a significant mechanism contributing to mitochondrial dysfunction as evidenced by several mitochondrion-related pathways that appear significantly affected by these modifications, e.g. mitochondrial dysfunction and oxidative phosphorylation. Elevated levels of HO· were also observed in the striatum and ventral midbrain regions of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-treated mice (56), a commonly used model for PD, supporting that many mitochondrial proteins may have an elevated level of DOPA/DQ as induced by an increased level of HO·.
A number of cytosolic oxidative stress response proteins, such as 90-kDa heat shock proteins and 14-3-3 signaling proteins, appeared susceptible to modifications. 90-kDa heat shock proteins (HSP90) are essential chaperones (stress proteins) for promoting cell survival under oxidative stress conditions and targets for halting neurodegeneration (57). Moreover, all five observed tyrosine sites in HSP90 that were modified by DOPA/DQ are reported to be sites of phosphorylation. The observation of DOPA/DQ modifications on three members of the 14-3-3 protein complex (Table II) is of particularly interest because these proteins are known to have important roles in the nervous system by interacting with more than 100 binding partners and have been implicated in many neurological disorders (45). These proteins are also known as tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation proteins that bind and regulate tyrosine hydroxylase (58), which mediates the rate-limiting step of dopamine and neurotransmitter synthesis in the brain. The observed modifications on multiple isoforms of α- and β-tubulin associated with 14-3-3-mediated signaling are also of interest because of the role of 14-3-3 proteins in cytoskeletal regulation (46). Again, nearly half of the DOPA-modified tyrosine sites are reported phosphorylation sites (Table II).
The modifications observed on a number of cytoskeletal proteins may also have implications in disease because cytoskeleton integrity is essential for normal cell functions; abnormalities in cytoskeleton organization have been observed in many diseases, especially neurodegenerative diseases (59). In particular, actin is sensitive to oxidant attack and is a key regulator of apoptosis and aging (60, 61). DOPA modifications were observed in multiple isoforms of actin and tubulin. Several actin and tubulin proteins were also observed with tyrosine nitration in our previous study of mouse brain (19). Because many of the identified DOPA modification sites in these single proteins are the same sites reported to be phosphorylated or nitrated, tyrosine DOPA modification and nitration may represent two mechanisms for disrupting oxidative stress response signaling, 14-3-3 signaling, and cytoskeletal organization in disease pathogenesis by either disrupting tyrosine phosphorylation signaling or diminishing enzymatic activities. The much higher abundance of observed DOPA modifications compared with nitrotyrosine suggests that DOPA modification represents a better marker for mitochondrial dysfunction and oxidative stress than nitrotyrosine; however, nitrotyrosine should still be a complementary marker to DOPA because nitrotyrosine is an inflammation-specific marker for nitric oxide-derived oxidation (19).
In summary, the observation of prevalent DOPA and DQ modifications on protein tyrosine, their preferential localization in mitochondria, their selectivity for metal-binding proteins, the associations with important cell signaling pathways, and their potential implications in diseases suggest that these modifications are linked to mitochondrial dysfunction and oxidative stress. Moreover, the fact that the sites of DOPA modifications are frequently reported as sites of tyrosine phosphorylation and nitration further supports these modifications as significant in disrupting cell signaling. DOPA/DQ tyrosine modifications are also complementary to those of nitrotyrosine because DOPA/DQ constitute a marker of HO· chemistries in contrast to nitrotyrosine, which reflects the presence of reaction nitrogen species, thus distinguishing inflammatory from non-inflammatory physiological states.
Acknowledgments
The experimental work was performed in the Environmental Molecular Sciences Laboratory, a United States Department of Energy (DOE) Office of Biological and Environmental Research national scientific user facility on the Pacific Northwest National Laboratory (PNNL) campus. PNNL is a multiprogram national laboratory operated by Battelle for the DOE under Contract DE-AC05-76RLO 1830.
* This work was supported, in whole or in part, by National Institutes of Health Grant ES016015 from the Center for Novel Biomarkers of Response (to J. G. P.), Grant RR018522 from the Center of Proteomics Research Resource for Integrative Biology (to R. D. S.), Grant R01 NS050148 (to D. J. S.), and Grant R01 DK074795 (to R. D. S.). This work was also supported by the Pacific Northwest National Laboratory Directed Research Development program (to W.-J. Q.).
 This article contains supplemental Tables 1–5 and annotated spectra.
This article contains supplemental Tables 1–5 and annotated spectra.
1 The abbreviations used are:
- ROS
- reactive oxygen species
- DOPA
- 3,4-dihydroxyphenylalanine
- DQ
- dopaquinone
- HO·
- hydroxyl radical
- FPR
- false positive rate
- PD
- Parkinson disease
- SCX
- strong cation exchange
- IPI
- International Protein Index.
REFERENCES
- 1.Berlett B. S., Stadtman E. R. (1997) Protein oxidation in aging, disease, and oxidative stress. J. Biol. Chem 272, 20313–20316 [DOI] [PubMed] [Google Scholar]
- 2.Pacher P., Beckman J. S., Liaudet L. (2007) Nitric oxide and peroxynitrite in health and disease. Physiol. Rev 87, 315–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benz C. C., Yau C. (2008) Ageing, oxidative stress and cancer: paradigms in parallax. Nat. Rev. Cancer 8, 875–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stadtman E. R. (1992) Protein oxidation and aging. Science 257, 1220–1224 [DOI] [PubMed] [Google Scholar]
- 5.Danielson S. R., Andersen J. K. (2008) Oxidative and nitrative protein modifications in Parkinson's disease. Free Radic. Biol. Med 44, 1787–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grimsrud P. A., Xie H., Griffin T. J., Bernlohr D. A. (2008) Oxidative stress and covalent modification of protein with bioactive aldehydes. J. Biol. Chem 283, 21837–21841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derakhshan B., Wille P. C., Gross S. S. (2007) Unbiased identification of cysteine S-nitrosylation sites on proteins. Nat. Protoc 2, 1685–1691 [DOI] [PubMed] [Google Scholar]
- 8.Hao G., Derakhshan B., Shi L., Campagne F., Gross S. S. (2006) SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc. Natl. Acad. Sci. U.S.A 103, 1012–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torta F., Usuelli V., Malgaroli A., Bachi A. (2008) Proteomic analysis of protein S-nitrosylation. Proteomics 8, 4484–4494 [DOI] [PubMed] [Google Scholar]
- 10.Paron I., D'Elia A., D'Ambrosio C., Scaloni A., D'Aurizio F., Prescott A., Damante G., Tell G. (2004) A proteomic approach to identify early molecular targets of oxidative stress in human epithelial lens cells. Biochem. J 378, 929–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rabilloud T., Heller M., Gasnier F., Luche S., Rey C., Aebersold R., Benahmed M., Louisot P., Lunardi J. (2002) Proteomics analysis of cellular response to oxidative stress. Evidence for in vivo overoxidation of peroxiredoxins at their active site. J. Biol. Chem 277, 19396–19401 [DOI] [PubMed] [Google Scholar]
- 12.Petyuk V. A., Qian W. J., Hinault C., Gritsenko M. A., Singhal M., Monroe M. E., Camp D. G., 2nd, Kulkarni R. N., Smith R. D. (2008) Characterization of the mouse pancreatic islet proteome and comparative analysis with other mouse tissues. J. Proteome Res 7, 3114–3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brock J. W., Cotham W. C., Ames J. M., Thorpe S. R., Baynes J. W. (2005) Proteomic method for the quantification of methionine sulfoxide. Ann. N.Y. Acad. Sci 1043, 284–289 [DOI] [PubMed] [Google Scholar]
- 14.Cullingford T. E., Wait R., Clerk A., Sugden P. H. (2006) Effects of oxidative stress on the cardiac myocyte proteome: modifications to peroxiredoxins and small heat shock proteins. J. Mol. Cell. Cardiol 40, 157–172 [DOI] [PubMed] [Google Scholar]
- 15.Aulak K. S., Miyagi M., Yan L., West K. A., Massillon D., Crabb J. W., Stuehr D. J. (2001) Proteomic method identifies proteins nitrated in vivo during inflammatory challenge. Proc. Natl. Acad. Sci. U.S.A 98, 12056–12061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castegna A., Thongboonkerd V., Klein J. B., Lynn B., Markesbery W. R., Butterfield D. A. (2003) Proteomic identification of nitrated proteins in Alzheimer's disease brain. J. Neurochem 85, 1394–1401 [DOI] [PubMed] [Google Scholar]
- 17.Kanski J., Hong S. J., Schöneich C. (2005) Proteomic analysis of protein nitration in aging skeletal muscle and identification of nitrotyrosine-containing sequences in vivo by nanoelectrospray ionization tandem mass spectrometry. J. Biol. Chem 280, 24261–24266 [DOI] [PubMed] [Google Scholar]
- 18.Butt Y. K., Lo S. C. (2008) Detecting nitrated proteins by proteomic technologies. Methods Enzymol 440, 17–31 [DOI] [PubMed] [Google Scholar]
- 19.Sacksteder C. A., Qian W. J., Knyushko T. V., Wang H., Chin M. H., Lacan G., Melega W. P., Camp D. G., 2nd, Smith R. D., Smith D. J., Squier T. C., Bigelow D. J. (2006) Endogenously nitrated proteins in mouse brain: links to neurodegenerative disease. Biochemistry 45, 8009–8022 [DOI] [PubMed] [Google Scholar]
- 20.Bigelow D. J., Qian W. J. (2008) Quantitative proteome mapping of nitrotyrosines. Methods Enzymol 440, 191–205 [DOI] [PubMed] [Google Scholar]
- 21.Zhang Q., Qian W. J., Knyushko T. V., Clauss T. R., Purvine S. O., Moore R. J., Sacksteder C. A., Chin M. H., Smith D. J., Camp D. G., 2nd, Bigelow D. J., Smith R. D. (2007) A method for selective enrichment and analysis of nitrotyrosine-containing peptides in complex proteome samples. J. Proteome Res 6, 2257–2268 [DOI] [PubMed] [Google Scholar]
- 22.Dalle-Donne I., Scaloni A., Giustarini D., Cavarra E., Tell G., Lungarella G., Colombo R., Rossi R., Milzani A. (2005) Proteins as biomarkers of oxidative/nitrosative stress in diseases: the contribution of redox proteomics. Mass Spectrom. Rev 24, 55–99 [DOI] [PubMed] [Google Scholar]
- 23.Stevens S. M., Jr., Prokai-Tatrai K., Prokai L. (2008) Factors that contribute to the misidentification of tyrosine nitration by shotgun proteomics. Mol. Cell. Proteomics 7, 2442–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haas W., Faherty B. K., Gerber S. A., Elias J. E., Beausoleil S. A., Bakalarski C. E., Li X., Villén J., Gygi S. P. (2006) Optimization and use of peptide mass measurement accuracy in shotgun proteomics. Mol. Cell. Proteomics 5, 1326–1337 [DOI] [PubMed] [Google Scholar]
- 25.Garrison W. M. (1987) Reaction mechanisms in the radiolysis of peptides, polypeptides, and proteins. Chem. Rev 87, 381–398 [Google Scholar]
- 26.Dean R. T., Fu S., Stocker R., Davies M. J. (1997) Biochemistry and pathology of radical-mediated protein oxidation. Biochem. J 324, 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gieseg S. P., Simpson J. A., Charlton T. S., Duncan M. W., Dean R. T. (1993) Protein-bound 3,4-dihydroxyphenylalanine is a major reductant formed during hydroxyl radical damage to proteins. Biochemistry 32, 4780–4786 [DOI] [PubMed] [Google Scholar]
- 28.Cohen G., Yakushin S., Dembiec-Cohen D. (1998) Protein L-dopa as an index of hydroxyl radical attack on protein tyrosine. Anal. Biochem 263, 232–239 [DOI] [PubMed] [Google Scholar]
- 29.Rodgers K. J., Dean R. T. (2000) Metabolism of protein-bound DOPA in mammals. Int. J. Biochem. Cell Biol 32, 945–955 [DOI] [PubMed] [Google Scholar]
- 30.Nelson M., Foxwell A. R., Tyrer P., Dean R. T. (2007) Protein-bound 3,4-dihydroxy-phenylanine (DOPA), a redox-active product of protein oxidation, as a trigger for antioxidant defences. Int. J. Biochem. Cell Biol 39, 879–889 [DOI] [PubMed] [Google Scholar]
- 31.Morin B., Davies M. J., Dean R. T. (1998) The protein oxidation product 3,4-dihydroxyphenylalanine (DOPA) mediates oxidative DNA damage. Biochem. J 330, 1059–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu S., Davies M. J., Stocker R., Dean R. T. (1998) Evidence for roles of radicals in protein oxidation in advanced human atherosclerotic plaque. Biochem. J 333, 519–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fu S., Dean R., Southan M., Truscott R. (1998) The hydroxyl radical in lens nuclear cataractogenesis. J. Biol. Chem 273, 28603–28609 [DOI] [PubMed] [Google Scholar]
- 34.Woods A. A., Linton S. M., Davies M. J. (2003) Detection of HOCl-mediated protein oxidation products in the extracellular matrix of human atherosclerotic plaques. Biochem. J 370, 729–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Molnár G. A., Nemes V., Biró Z., Ludány A., Wagner Z., Wittmann I. (2005) Accumulation of the hydroxyl free radical markers meta-, ortho-tyrosine and DOPA in cataractous lenses is accompanied by a lower protein and phenylalanine content of the water-soluble phase. Free Radic Res 39, 1359–1366 [DOI] [PubMed] [Google Scholar]
- 36.O'Neill C. A., Fu L. W., Halliwell B., Longhurst J. C. (1996) Hydroxyl radical production during myocardial ischemia and reperfusion in cats. Am. J. Physiol. Heart Circ. Physiol 271, H660–H667 [DOI] [PubMed] [Google Scholar]
- 37.Livesay E. A., Tang K., Taylor B. K., Buschbach M. A., Hopkins D. F., LaMarche B. L., Zhao R., Shen Y., Orton D. J., Moore R. J., Kelly R. T., Udseth H. R., Smith R. D. (2008) Fully automated four-column capillary LC-MS system for maximizing throughput in proteomic analyses. Anal. Chem 80, 294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fenyö D., Beavis R. C. (2003) A method for assessing the statistical significance of mass spectrometry-based protein identifications using general scoring schemes. Anal. Chem 75, 768–774 [DOI] [PubMed] [Google Scholar]
- 39.Qian W. J., Liu T., Monroe M. E., Strittmatter E. F., Jacobs J. M., Kangas L. J., Petritis K., Camp D. G., 2nd, Smith R. D. (2005) Probability-based evaluation of peptide and protein identifications from tandem mass spectrometry and SEQUEST analysis: the human proteome. J. Proteome Res 4, 53–62 [DOI] [PubMed] [Google Scholar]
- 40.Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 41.Davies M. J., Fu S., Wang H., Dean R. T. (1999) Stable markers of oxidant damage to proteins and their application in the study of human disease. Free Radic. Biol. Med 27, 1151–1163 [DOI] [PubMed] [Google Scholar]
- 42.Ito S., Kato T., Shinpo K., Fujita K. (1984) Oxidation of tyrosine residues in proteins by tyrosinase. Formation of protein-bonded 3,4-dihydroxyphenylalanine and 5-S-cysteinyl-3,4-dihydroxyphenylalanine. Biochem. J 222, 407–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin M. T., Beal M. F. (2006) Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443, 787–795 [DOI] [PubMed] [Google Scholar]
- 44.Calvano S. E., Xiao W., Richards D. R., Felciano R. M., Baker H. V., Cho R. J., Chen R. O., Brownstein B. H., Cobb J. P., Tschoeke S. K., Miller-Graziano C., Moldawer L. L., Mindrinos M. N., Davis R. W., Tompkins R. G., Lowry S. F. (2005) A network-based analysis of systemic inflammation in humans. Nature 437, 1032–1037 [DOI] [PubMed] [Google Scholar]
- 45.Berg D., Holzmann C., Riess O. (2003) 14-3-3 proteins in the nervous system. Nat. Rev. Neurosci 4, 752–762 [DOI] [PubMed] [Google Scholar]
- 46.Jin J., Smith F. D., Stark C., Wells C. D., Fawcett J. P., Kulkarni S., Metalnikov P., O'Donnell P., Taylor P., Taylor L., Zougman A., Woodgett J. R., Langeberg L. K., Scott J. D., Pawson T. (2004) Proteomic, functional, and domain-based analysis of in vivo 14-3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr. Biol 14, 1436–1450 [DOI] [PubMed] [Google Scholar]
- 47.Yeo W. S., Lee S. J., Lee J. R., Kim K. P. (2008) Nitrosative protein tyrosine modifications: biochemistry and functional significance. BMB Rep 41, 194–203 [DOI] [PubMed] [Google Scholar]
- 48.Leary S. C., Winge D. R., Cobine P. A. (2009) “Pulling the plug” on cellular copper: the role of mitochondria in copper export. Biochim. Biophys. Acta 1793, 146–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pierrel F., Cobine P. A., Winge D. R. (2007) Metal ion availability in mitochondria. Biometals 20, 675–682 [DOI] [PubMed] [Google Scholar]
- 50.Hosler J. P., Ferguson-Miller S., Mills D. A. (2006) Energy transduction: proton transfer through the respiratory complexes. Annu. Rev. Biochem 75, 165–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hedberg K. K., Birrell G. B., Griffith O. H. (1991) Phorbol ester-induced actin cytoskeletal reorganization requires a heavy metal ion. Cell Regul 2, 1067–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Athwal G. S., Huber J. L., Huber S. C. (1998) Biological significance of divalent metal ion binding to 14-3-3 proteins in relationship to nitrate reductase inactivation. Plant Cell Physiol 39, 1065–1072 [DOI] [PubMed] [Google Scholar]
- 53.Molavi B., Mehta J. L. (2004) Oxidative stress in cardiovascular disease: molecular basis of its deleterious effects, its detection, and therapeutic considerations. Curr. Opin. Cardiol 19, 488–493 [DOI] [PubMed] [Google Scholar]
- 54.Andersen J. K. (2004) Oxidative stress in neurodegeneration: cause or consequence? Nat Med 10, (suppl.) S18–S25 [DOI] [PubMed] [Google Scholar]
- 55.Ballinger S. W. (2005) Mitochondrial dysfunction in cardiovascular disease. Free Radic. Biol. Med 38, 1278–1295 [DOI] [PubMed] [Google Scholar]
- 56.Smith T. S., Bennett J. P., Jr. (1997) Mitochondrial toxins in models of neurodegenerative diseases. I: In vivo brain hydroxyl radical production during systemic MPTP treatment or following microdialysis infusion of methylpyridinium or azide ions. Brain Res 765, 183–188 [DOI] [PubMed] [Google Scholar]
- 57.Gallo K. A. (2006) Targeting HSP90 to halt neurodegeneration. Chem. Biol 13, 115–116 [DOI] [PubMed] [Google Scholar]
- 58.Obsilova V., Nedbalkova E., Silhan J., Boura E., Herman P., Vecer J., Sulc M., Teisinger J., Dyda F., Obsil T. (2008) The 14-3-3 protein affects the conformation of the regulatory domain of human tyrosine hydroxylase. Biochemistry 47, 1768–1777 [DOI] [PubMed] [Google Scholar]
- 59.Brandt R. (2001) Cytoskeletal mechanisms of neuronal degeneration. Cell Tissue Res 305, 255–265 [DOI] [PubMed] [Google Scholar]
- 60.Dalle-Donne I., Rossi R., Milzani A., Di Simplicio P., Colombo R. (2001) The actin cytoskeleton response to oxidants: from small heat shock protein phosphorylation to changes in the redox state of actin itself. Free Radic. Biol. Med 31, 1624–1632 [DOI] [PubMed] [Google Scholar]
- 61.Gourlay C. W., Ayscough K. R. (2005) The actin cytoskeleton in ageing and apoptosis. FEMS Yeast Res 5, 1193–1198 [DOI] [PubMed] [Google Scholar]