Abstract
Mycoplasma pneumoniae belongs to the Mollicutes, the group of organisms with the smallest genomes that are capable of host-independent life. These bacteria show little regulation in gene expression, suggesting an important role for the control of protein activities. We have studied protein phosphorylation in M. pneumoniae to identify phosphorylated proteins. Two-dimensional gel electrophoresis and mass spectrometry allowed the detection of 63 phosphorylated proteins, many of them enzymes of central carbon metabolism and proteins related to host cell adhesion. We identified 16 phosphorylation sites, among them 8 serine and 8 threonine residues, respectively. A phosphoproteome analysis with mutants affected in the two annotated protein kinase genes or in the single known protein phosphatase gene suggested that only one protein (HPr) is phosphorylated by the HPr kinase, HPrK, whereas four adhesion-related or surface proteins were targets of the protein kinase C, PrkC. A comparison with the phosphoproteomes of other bacteria revealed that protein phosphorylation is evolutionarily only poorly conserved. Only one single protein with an identified phosphorylation site, a phosphosugar mutase (ManB in M. pneumoniae), is phosphorylated on a conserved serine residue in all studied organisms from archaea and bacteria to man. We demonstrate that this protein undergoes autophosphorylation. This explains the strong conservation of this phosphorylation event. For most other proteins, even if they are phosphorylated in different species, the actual phosphorylation sites are different. This suggests that protein phosphorylation is a form of adaptation of the bacteria to the specific needs of their particular ecological niche.
Bacteria of the group called Mollicutes are unique among all living organisms because of their small genome that nevertheless allows them to grow independently from any host cell. The two most intensively studied representatives of the Mollicutes are Mycoplasma genitalium and Mycoplasma pneumoniae with genome sizes of 580 and 816 kb, respectively. The approximately 475 protein-coding and 43 RNA-coding genes of M. genitalium define the lower limit of the genetic equipment that permits independent life. The small genomes of the Mollicutes reflect their adaptation to rarely changing ecosystems, such as lung epithelia for M. pneumoniae (1). Furthermore, M. pneumoniae has only limited metabolic capabilities: this bacterium can utilize only a few carbon sources (glucose, fructose, and glycerol) (2), and its only way to produce ATP is by substrate level phosphorylation in glycolysis. The citric acid cycle, respiration, and most anabolic reactions are not carried out by M. pneumoniae (3).
M. pneumoniae is a human pathogen that causes usually mild infections such as atypical pneumonia; however, the infections may be severe in children and elderly people. In addition, M. pneumoniae is involved in extrapulmonary complications, such as erythema multiforme and pediatric encephalitis (4–6).
The close adaptation of M. pneumoniae to human mucosal surfaces did not only result in a reductive evolution of metabolic capabilities but also affected the regulation of protein biosynthesis and activity: in contrast to versatile environmental bacteria such as Pseudomonas aeruginosa that reserve about 10% of their coding capacity for regulators of gene expression, M. pneumoniae encodes only three potential transcription regulators (less than 0.5% of all genes) (1). This absence of regulation at the level of gene expression suggests that the control of protein activities might play an important role in M. pneumoniae and other Mollicutes.
An important way to control the activity of a protein is post-translational modification with protein phosphorylation being the most prominent regulatory modification both in bacteria and in eukaryotes (7, 8). Studies on the phosphoproteome have been performed for several bacteria, including the model organisms Escherichia coli and Bacillus subtilis. In both bacteria, about 5% of all proteins can be phosphorylated on Ser, Thr, or Tyr residues (9–12). Interestingly, there is a high level of species specificity with respect to the phosphoproteome: a comparison of the phosphoproteomes of B. subtilis, E. coli, and Lactococcus lactis revealed only eight proteins that are phosphorylated in all three species. Six of these proteins are involved in sugar metabolism, and two are translation factors. Of these eight proteins, only one, phosphoglucosamine mutase, is phosphorylated at a conserved residue (13).
In M. pneumoniae, only the phosphorylation of the HPr protein of the phosphoenolpyruvate:sugar phosphotransferase system by the HPr kinase has been studied to some detail (2, 14, 15). Once phosphorylated, HPr can be dephosphorylated by a protein phosphatase, PrpC (16). The corresponding prpC gene is in many Gram-positive bacteria clustered with the gene for a protein kinase, prkC, suggesting that PrkC and PrpC form a functional couple with opposing activities. Evidence for this idea was provided by the observation that the two proteins from B. subtilis have identical targets in vitro (17). The prpC/prkC gene cluster also exists in M. pneumoniae; however, the targets of the corresponding proteins are so far unknown. Previous studies with M. pneumoniae demonstrated that several proteins implicated in adhesion to the host cell are subject to protein phosphorylation in vivo (18–20). These proteins, HMW1 and HMW2, are important for the virulence of M. pneumoniae because adhesion is the first step in the interaction with the host. A recent proteomics study resulted in the identification of 18 phosphorylated proteins in M. pneumoniae, among them adhesion and surface proteins and metabolic enzymes (21). Unfortunately, none of the previously known phosphoproteins were detected in this study. Moreover, Ser-46 in HPr is still the only known phosphorylation site in M. pneumoniae (with the exception of phosphorylated His and Cys residues in phosphotransferase system proteins), and the (de)phosphorylation of HPr is the only event for which the kinase and the phosphatase are known. Finally, nothing is known about protein tyrosine phosphorylation in M. pneumoniae and other Mollicutes.
We are interested in the elucidation of the molecular networks that allow life of M. pneumoniae as a minimal organism. The analysis of these bacteria is hampered by their slow growth and by the lack of efficient genetic systems that allow the elucidation of gene functions. We have established a technique, haystack mutagenesis, that can be used to isolate mutants of M. pneumoniae (16). In this work, we have studied the phosphoproteome of the M. pneumoniae wild type strain and of three isogenic mutants that are affected in the two protein kinases, HPr kinase (HPrK) and protein kinase C (PrkC), and in the protein phosphatase PrpC. We identified 63 phosphorylated proteins. However, most of these proteins are phosphorylated by so far unknown kinases. An in-depth analysis of the only universally conserved phosphoprotein, a phosphosugar mutase, revealed that this enzyme autophosphorylates.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
The M. pneumoniae strains used in this study were M. pneumoniae M129 (ATCC 29342) in the 32nd broth passage and its isogenic mutant derivatives GPM11 (prkC::mini-Tn, GmR) (20), GPM51 (hprK::mini-Tn, GmR) (16), and GPM68 (prpC::mini-Tn, GmR) (16). M. pneumoniae was grown at 37 °C in 150-cm2 tissue culture flasks containing 100 ml of modified Hayflick medium with glucose (1%, w/v) as the carbon source as described previously (2). Strains harboring transposon insertions were cultivated in the presence of 80 μg/ml gentamicin. B. subtilis 168 (trpC2; laboratory collection) was grown in LB and in minimal medium containing succinate and glutamate/ammonium as basic sources of carbon and nitrogen, respectively (22). The media were supplemented with tryptophan (at 50 mg/liter) and glucose (0.5%, w/v). B. subtilis was transformed with plasmid DNA or PCR products according to the two-step protocol (23). Transformants were selected on sporulation medium plates (22) containing erythromycin plus lincomycin (2 and 25 μg/ml, respectively) and/or spectinomycin (100 μg/ml).
Preparation of Cell Extracts
After 4 days of incubation at 37 °C, the culture medium was removed from the M. pneumoniae cell layer, and the cells were washed twice with 20 ml of cold PBS. After washing, cells were collected by scraping into 1.5 ml of PBS and subsequent centrifugation (5 min, 15,000 × g, 4 °C). The cell pellet was resuspended in 500 μl of 10 mm Tris/HCl, pH 7.5, and the cells were disrupted by sonication (3 × 10 s, 50 watts, 4 °C). Cell debris were sedimented by centrifugation (10 min, 15,000 × g, 4 °C), and the resulting supernatant was centrifuged again (30 min, 24,000 × g, 4 °C) to remove disturbing particles. The protein concentration of the supernatant was determined using the Bio-Rad dye binding assay with bovine serum albumin as the standard.
Two-dimensional SDS-Polyacrylamide Gel Electrophoresis
Two-dimensional separation of proteins was performed as described previously (24). For separation in the first dimension, IPG strips with a linear pH range of 4–7, 4.5–5.5, or 6–11 were used (ImmobilineTM DryStrip, Amersham Biosciences). The gels were subsequently stained with Pro-Q Diamond (Invitrogen) and FlamingoTM fluorescent dye (Bio-Rad) to visualize the phosphoproteins and all proteins, respectively. Image analysis, spot quantification, and determination of putative phosphorylated protein spots were performed as described previously (9). The determination of phosphorylated proteins was based on the Pro-Q/Flamingo log ratio. Phosphoprotein spots were cut from the gel and identified by mass spectroscopy.
Protein Identification by Mass Spectrometry
Gel pieces were washed twice with 200 μl of 20 mm NH4HCO3, 30% (v/v) ACN for 30 min at 37 °C and dried in a vacuum centrifuge (Concentrator 5301, Eppendorf). Trypsin solution (10 ng/μl trypsin in 20 mm ammonium bicarbonate) was added until gel pieces stopped swelling, and digestion was allowed to proceed for 16–18 h at 37 °C. Peptides were extracted from gel pieces by incubation in an ultrasonic bath for 15 min in 20 μl of HPLC grade water and transferred into microvials for mass spectrometric analysis. Peptides were separated by liquid chromatography and measured on line by ESI-mass spectrometry using a nanoACQUITY UPLCTM system (Waters) coupled to an LTQ OrbitrapTM mass spectrometer (Thermo Fisher Scientific, Waltham, MA). Peptides were desalted onto a trap column (Symmetry® C18, Waters). Elution was performed onto an analytical column (BEH130 C18, Waters) by a binary gradient of buffer A (0.1% (v/v) acetic acid) and B (100% (v/v) acetonitrile, 0.1% (v/v) acetic acid) over a period of 50 min with a flow rate of 400 nl/min. The LTQ Orbitrap was operated in data-dependent MS/MS mode using multistage activation for phosphorelevant masses. Proteins were identified by searching all MS/MS spectra in .dta format against all 689 M. pneumoniae proteins (extracted from the NCBI database) using SorcererTM-SEQUEST® (SEQUEST version 2.7 revision 11, Thermo Electron) including Scaffold_2_05_02 (Proteome Software Inc., Portland, OR). SEQUEST was searched with a fragment ion mass tolerance of 1.00 Da and a parent ion tolerance of 10 ppm. Up to two missed tryptic cleavages were allowed. Methionine oxidation (+15.99492 Da), cysteine carbamidomethylation (+57.021465 Da), and phosphorylation (+79.966331 Da) were set as variable modifications. For protein identification, a stringent SEQUEST filter for peptides was used (Xcorr versus charge state: 1.90 for singly, 2.2 for doubly, and 3.3 for triply charged ions and ΔCn value greater than 0.10), and at least two peptides per proteins were required. With this method, all identified proteins had a protein probability score greater than 99.9%. Protein probabilities were assigned by the Protein Prophet algorithm (25). Phosphorylated peptides that had a peptide probability score greater than 95.0% as specified by the Peptide Prophet algorithm (26) were examined manually and accepted only when b- or y-ions confirmed the identification. All spectra with Scaffold-annotated b- and y-ions series are provided as screen shots in the supplemental material (see supplemental Fig. S3).
Analysis of Protein Conservation and Structure Prediction
To address the potential conservation of phosphorylation sites, multiple sequence alignments were performed using the EXPRESSO alignment server that takes available structures into account (27). Structural models of the phosphoproteins were created using SWISS-MODEL in the automated mode and Swiss-PdbViewer v4.0.1 (28, 29). Ordered and disordered regions of proteins were predicted using the DisEMBL v1.5 web server (30).
Cloning Procedures
E. coli DH5α (31) was used for cloning experiments and protein expression. E. coli was grown in LB medium. LB plates were prepared by addition of 17 g of Bacto agar/liter (Difco) to LB (31). Transformation of E. coli and plasmid DNA extraction were performed using standard procedures (31). Restriction enzymes, T4 DNA ligase, and DNA polymerases were used as recommended by the manufacturers. DNA fragments were purified from agarose gels using the QIAquick Gel Extraction kit (Qiagen). Pfu DNA polymerase was used for the polymerase chain reaction as recommended by the manufacturer. DNA sequences were determined using the dideoxy chain termination method (31). Standard procedures were used to transform E. coli (31), and transformants were selected on LB plates containing ampicillin (100 μg/ml).
Plasmid Constructions
Plasmids for the overexpression and purification of the phosphosugar mutases GlmM and ManB from B. subtilis and M. pneumoniae, respectively, were constructed as follows. The coding sequence of the genes was amplified by PCR with gene-specific primers (listed in supplemental Table S1) using chromosomal DNA of B. subtilis 168 and M. pneumoniae M129 as the template. The PCR products were digested as shown in supplemental Table S2 and cloned into the appropriately linearized expression vector pWH844 (32). These plasmids allowed the expression of the phosphosugar mutases carrying an N-terminal strep tag. The resulting plasmids pGP1401 and pGP656 are listed in supplemental Table S2. In M. pneumoniae, the UGA specifies a tryptophan; however, in E. coli, it is a stop codon. The manB gene contains three UGA codons that were replaced by the multiple mutation reaction (33) using the phosphorylated mutagenic oligonucleotides SS109, SS110, and SS111 (see supplemental Table S1) and plasmid pGP656 as the template. The resulting plasmid was pGP657 (see supplemental Table S2). The phosphorylation sites of GlmM and ManB were replaced by alanine residues. For this purpose, the combined chain reaction (34) was applied with the mutagenic primers NP116 and SS112 (see supplemental Table S1) and plasmids pGP1401 and pGP657. The resulting plasmids are pGP1405 (glmM) and pGP658 (manB) (see supplemental Table S2). All plasmid inserts were verified by DNA sequencing.
The plasmids pGP400 and pGP1403 were used to express the wild type and S100A mutant forms of GlmM, respectively, in B. subtilis. For the construction of pGP400, the glmM gene was amplified using the oligonucleotides NP108/NP109. The PCR product was digested with BamHI and SalI and cloned into pBQ200 (35) digested with the same enzymes. For the construction of the plasmid encoding the S100A GlmM variant, mutagenesis was performed using the combined chain reaction with NP116 as the mutagenic primer and NP108/NP109. The resulting PCR product was cloned into pBQ200 as described for the wild type allele. The resulting plasmid was pGP1403.
Overexpression and Purification of Recombinant Proteins
E. coli DH5α transformed with the appropriate expression vector was used as host for the overexpression of recombinant proteins. Expression was induced by the addition of isopropyl 1-thio-β-d-galactopyranoside (final concentration, 1 mm) to exponentially growing cultures (A600 of 0.6). Cells were lysed using a French press (20,000 p.s.i., 138,000 kilopascals; Spectronic Instruments). After lysis, the crude extracts were centrifuged at 15,000 × g for 1 h. For purification of His-tagged proteins, the resulting supernatants were passed over a Ni2+ HiTrap chelating column (4-ml bed volume; GE Healthcare) followed by elution with an imidazole gradient (from 0 to 500 mm imidazole in a buffer containing 10 mm Tris/HCl, pH 7.5, 600 mm NaCl, 10 mm β-mercaptoethanol) over 30 ml at a flow rate of 0.5 ml/min. After elution, the fractions were tested for the desired protein using 12.5% SDS-PAGE. The relevant fractions were combined and dialyzed overnight. Protein concentration was determined using the Bio-Rad dye binding assay where bovine serum albumin served as the standard.
Assays for Protein Phosphorylation
Protein phosphorylation assays were carried out with purified proteins in assay buffer (25 mm Tris/HCl, pH 7.6, 1 mm dithiothreitol, 0.4 mm [γ-32P]ATP (480 Ci/mmol), divalent cations as indicated) using purified target proteins. The assays were carried out at 37 °C for 30 min followed by thermal inactivation of the protein (10 min at 95 °C). The assay mixtures were analyzed on 16% SDS-polyacrylamide gels. Proteins were visualized by Coomassie staining. Radioactive protein spots were detected using a phosphorimaging system (Storm 860, GE Healthcare).
Allelic Replacement of B. subtilis glmM Gene
To delete the chromosomal copy of the glmM gene, the long flanking homology PCR technique was used (36). Briefly, a cassette carrying the spc resistance gene was amplified from plasmid pDG1726 (37). DNA fragments of about 1,000 bp flanking the glmM gene at its 5′- and 3′-ends were amplified. The 3′-end of the upstream fragment as well as the 5′-end of the downstream fragment extended into glmM in a way that all expression signals of genes up- and downstream of the glmM remained intact. The joining of the two fragments to the resistance cassette was performed in a second PCR as described previously (38). In this reaction, we used the primer pair spec-fwd/spec-rev (38) for the amplification and joining of the spc cassette. The PCR product was directly used to transform B. subtilis carrying plasmids that express different alleles of glmM. The integrity of the regions flanking the integrated resistance cassette was verified by sequencing PCR products of about 1,000 bp amplified from chromosomal DNA of the resulting mutants.
RESULTS
Phosphoproteome of Growing M. pneumoniae Cells
In a previous study on protein phosphorylation in M. pneumoniae, no phosphorylation sites were determined, and no mutants affected in the players of protein phosphorylation were studied (21). To get a more comprehensive insight into protein phosphorylation in M. pneumoniae, we decided to analyze the phosphoproteome of the wild type strain M129 and of three isogenic mutants defective in the two annotated protein kinases and the single annotated protein phosphatase of M. pneumoniae.
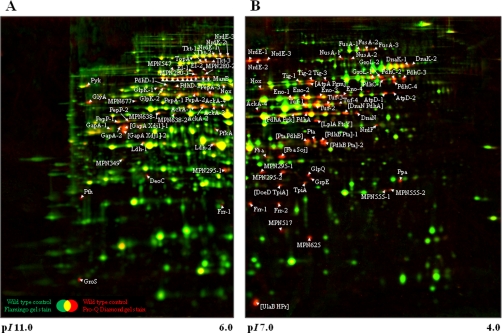
The wild type strain M129 was grown in modified Hayflick medium with glucose as the principal carbon source, and cells were harvested in the late logarithmic phase. Protein extracts were prepared as described under “Experimental Procedures,” and the proteins were separated by two-dimensional gel electrophoresis (see Fig. 1). Traditional two-dimensional gels cover only the neutral and acidic proteins (pH range, 4–7). However, the theoretical proteome of M. pneumoniae shows a high number of more basic proteins that might escape detection (see supplemental Fig. S1). To include these basic proteins, we performed the isoelectric focusing in two pH ranges, 4–7 and 6–11. The gels were stained with Pro-Q Diamond and Flamingo fluorescent dye to detect the phosphorylated proteins and the all proteins, respectively. The presence of more than 700 protein spots on the gels revealed that the proteome of M. pneumoniae was completely covered by our system (39, 40). The identification of phosphorylated proteins revealed 58 phosphoproteins (see Table I). We observed 36 phosphorylated proteins in the pH range from 4 to 7 and 27 phosphoproteins in the pH range from 6 to 11 with only five proteins (i.e. AckA, Frr, MPN295, Nox, and NrdE) detected in both gel systems. Of the proteins previously known to be phosphorylated, we detected HPr and, with the exception of the ribosomal protein S2 and the adhesin-related protein P65, all 17 phosphoproteins reported by Su et al. (21).
Fig. 1.
Dual channel image of total protein amount (Flamingo fluorescent dye-stained; green) and phosphorylated proteins (Pro-Q Diamond-stained; red) of stationary phase M. pneumoniae cells. About 125 μg of a total protein extract of M. pneumoniae grown in modified Hayflick medium supplemented with glucose were subjected to two-dimensional gel electrophoresis using either an 18-cm IPG strip with a linear pH gradient of pI 6–11 (A) or pI 4–7 (B) in the first dimension. Dual channel images were obtained using the software DECODON Delta2D 3.6. Spots with high Pro-Q Diamond/Flamingo log ratios representing phosphorylated proteins (9) are indicated on the gel. Multiple protein spots corresponding to the same original protein are distinguished by appended digits (e.g. Tuf-1 and Tuf-2). Protein spots were cut from the gel and identified by MS/MS (see Table I and supplemental Table S3).
Table I. Phosphoproteins of M. pneumoniae.
For detailed MS/MS data, see supplemental Tables S3 and S4 and Fig. S3. pS, phosphoserine; pT, phosphothreonine.
| Locus name | Protein name | Protein function | COGa | Phospho spots in two-dimensional gel | Phosphosite | Phosphopeptide sequence |
|---|---|---|---|---|---|---|
| MPN001 | DnaN | DNA polymerase III subunit β | L | 2 | ||
| MPN002 | Xdj1 | DnaJ-like protein | O | 2 | ||
| MPN024b | RpoE | RNA polymerase subunit δ | K | 1 | [Ser-87 or Thr-93] | Ip[SQAMFVT]KEIFEEGYEDLSNK |
| MPN025 | Fba | Fructose-bisphosphate aldolase | G | 2 | ||
| MPN050 | GlpK | Glycerol kinase | C | 2 | ||
| MPN053c | HPr | Phosphocarrier protein | G | 1 | ||
| MPN062 | DeoD | Purine-nucleoside phosphorylase | F | 1 | ||
| MPN063 | DeoC | Deoxyribose-phosphate aldolase | F | 1 | ||
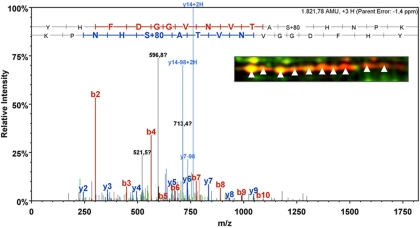
| MPN066 | ManB | Phosphomannomutase/phosphoglucomutase | G | 10 | Ser-149 | (K)YHFDGGVNVTApSHNPK |
| MPN082 | Tkt | Transketolase | G | 3 | ||
| MPN120 | GrpE | Heat-shock protein | O | 1 | ||
| MPN154 | NusA | Transcription elongation factor | K | 2 | Ser-503 | PVVKPKpSVFSITVEADDSK |
| MPN221 | Pth | Peptidyl-tRNA hydrolase | J | 1 | ||
| MPN227 | FusA | Elongation factor G | J | 3 | ||
| MPN256b | MPN256 | Uncharacterized protein | S | 2 | Thr-200 | FIDELDQIpTK |
| MPN261 | TopA | DNA topoisomerase I | L | 1 | Ser-426 | TVApSLMADCKK |
| MPN280 | MPN280 | Ribonuclease J | A | 2 | ||
| MPN295 | MPN295 | Uncharacterized protein | S | 2 | Ser-63 | YGPECEKSFLSLQpSK |
| MPN302 | PfkA | Phosphofructokinase | G | 1 | ||
| MPN303 | Pyk | Pyruvate kinase | G | 1 | ||
| MPN311b | P41 | Adhesin-related protein | M | 1 | Thr-223 | TNNSIQQLEAEIQIPpTTHIK |
| MPN322 | NrdF | Ribonucleoside-diphosphate reductase subunit β | F | |||
| MPN324 | NrdE | Ribonucleoside-diphosphate reductase subunit α | F | 3 | Thr-159, Ser-412 | FQPApTPTFLNAGR, VGNDIpSCNLGSLNIAK |
| MPN331 | Tig | Trigger factor | O | 3 | ||
| MPN349 | MPN349 | Uncharacterized protein | S | 1 | ||
| MPN389 | LplA | Lipoate-protein ligase A | H | 1 | ||
| MPN390 | PdhD | Pyruvate dehydrogenase E3 component | C | 2 | ||
| MPN391 | PdhC | Pyruvate dehydrogenase E2 component | C | 4 | ||
| MPN392 | PdhB | Pyruvate dehydrogenase E1 component subunit β | C | 3 | ||
| MPN393 | PdhA | Pyruvate dehydrogenase E1 component subunit α | C | 3 | Ser-205 | TKLEpSAVSDLSTK |
| MPN394 | Nox | NADH oxidase | H | 1 | ||
| MPN420 | GlpQ | Glycerophosphoryldiester phosphodiesterase | C | 1 | ||
| MPN425 | FtsY | Cell division protein homolog | U | 1 | ||
| MPN428 | Pta | Phosphotransacetylase | C | 4 | ||
| MPN429 | Pgk | Phosphoglycerate kinase | G | 1 | ||
| MPN430 | GapA | Glyceraldehyde-3-phosphate dehydrogenase | G | 4 | Ser-245 | VPVLTGpSIVELCVALEK |
| MPN434 | DnaK | Chaperone protein | O | 2 | ||
| MPN452b | HMW3 | Cytadherence high molecular weight protein 3 | M | 1 | ||
| MPN470 | PepP | Xaa-Pro aminopeptidase | E | 2 | Thr-317 | LLCENAVIpTIEPGIYIPSVGGIR |
| MPN474b | MPN474 | Coiled coil surface protein | M | 1 | Thr-773 | QNNEELpTDKCSNIQNELHDLNR |
| MPN495 | UlaB | Ascorbate-specific phosphotransferase enzyme IIB component | G | 1 | ||
| MPN517 | MPN517 | Reductase homolog | C | 1 | ||
| MPN528 | Ppa | Inorganic pyrophosphatase | P | 1 | ||
| MPN533 | AckA | Acetate kinase | C | 4 | ||
| MPN547 | MPN547 | Dihydroxyacetone kinase | C | 1 | ||
| MPN555 | MPN555 | Uncharacterized protein | S | 2 | ||
| MPN572 | PepA | Cytosol aminopeptidase | E | 3 | Ser-249, Thr-420 | YDMSGAAIVCpSTVLALAK, |
| EGVPLIHCDIASpTASIQDLGQGVLVR | ||||||
| MPN573 | GroL | 60-kDa chaperonin | O | 2 | ||
| MPN574 | GroS | 10-kDa chaperonin | O | 1 | ||
| MPN576 | GlyA | Serine hydroxymethyltransferase | E | 1 | ||
| MPN598 | AtpD | ATP synthase subunit β | C | 2 | ||
| MPN600 | AtpA | ATP synthase subunit α | C | 1 | ||
| MPN606 | Eno | Enolase | G | 4 | ||
| MPN625 | MPN625 | Osmotically inducible protein C-like protein | O | 1 | ||
| MPN627 | EI | Phosphotransferase system enzyme I | G | 2 | ||
| MPN628 | Pgm | Phosphoglycerate mutase | G | 1 | ||
| MPN629 | TpiA | Triose-phosphate isomerase | G | 2 | ||
| MPN636 | Frr | Ribosome-recycling factor | J | 2 | ||
| MPN638 | MPN638 | Putative type I restriction enzyme specificity protein | V | 2 | ||
| MPN665 | Tuf | Elongation factor TU | J | 4 | Thr-383 | EGGRpTVGAGSVTEVLE |
| MPN674 | Ldh | l-Lactate dehydrogenase | C | 2 | ||
| MPN677 | MPN677 | Uncharacterized protein | S | 1 | ||
| MPN688 | Soj | ParA family protein | D | 1 |
a COG, cluster of orthologous groups of proteins; A, RNA processing and modification; C, energy production and conversion; D, cell cycle control, cell division, and chromosome partitioning; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; G, carbohydrate transport and metabolism; H, coenzyme transport and metabolism; J, translation, ribosomal structure, and biogenesis; K, transcription; L, replication, recombination, and repair; M, cell wall, membrane, and envelope biogenesis; O, post-translational modification, protein turnover, and chaperones; P, inorganic ion transport and metabolism; S, function unknown; U, intracellular trafficking, secretion, and vesicular transport; V, defense mechanisms.
b Phosphoproteins only detectable in a prpC::Tn mutant (GPM68).
c HPr (MPN053) not detectable in a hprK::Tn mutant (GPM51).
In a next step, we tried to identify the phosphorylation sites in all phosphorylated peptides that had a peptide probability score greater than 95.0% as specified by the Peptide Prophet algorithm (see “Experimental Procedures”). This analysis identified 12 phosphorylation sites in 10 phosphoproteins (see Table I). These phosphorylations occurred on serine (eight sites) and threonine (four sites). Two proteins possessing two phosphorylation sites, NrdE and PepA, were phosphorylated on both a serine and a threonine residue. It is interesting to note that the individual NrdE molecules were phosphorylated only on one site: the two phosphopeptides for this protein were identified in different protein spots. In contrast, both PepA phosphopeptides were present in the protein from one spot, suggesting that this molecule was indeed doubly phosphorylated.
Phosphoproteome in prpC Mutant
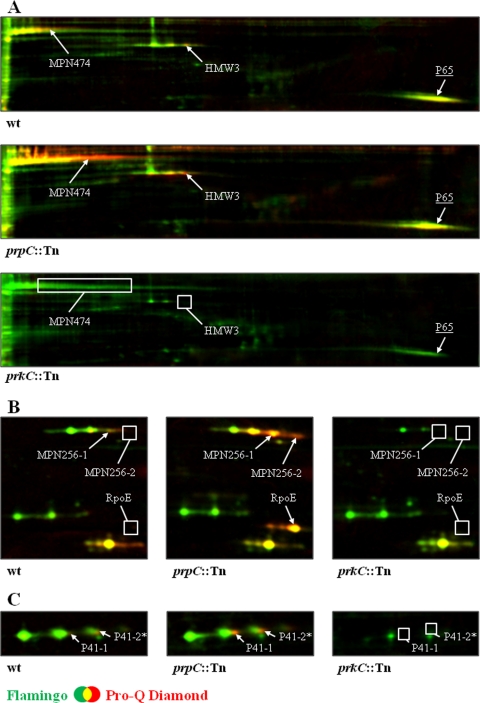
The M. pneumoniae genome encodes one protein phosphatase, PrpC. Previously, we have shown that PrpC dephosphorylates HPr(Ser-P) (16). We considered that the analysis of the phosphoproteome in a prpC mutant might allow the identification of additional phosphorylated proteins that may have escaped our attention in the experiments with the wild type strain. Therefore, we cultivated the prpC mutant strain GPM68 as described above for the wild type and analyzed the protein phosphorylation pattern by two-dimensional gel electrophoresis. A comparison of the phosphoproteomes of the two strains revealed that all phosphoproteins detected in the wild type were also present in the prpC mutant. In addition, a few additional phosphoprotein spots were detected in the prpC mutant. These proteins had isoelectric points between 4 and 5 (data not shown). Therefore, we increased the resolution for these proteins by performing the isoelectric focusing in the pH range from 4.5 to 5.5. In the prpC mutant, we observed five phosphoprotein spots that were not present in the wild type strain. The corresponding proteins were RpoE, the δ subunit of RNA polymerase, the adhesin-related protein P41, the high molecular weight cytadherence protein HMW3, the uncharacterized protein MPN256, and the cell surface protein MPN474 (see Table I and Fig. 2). With the exception of RpoE, phosphorylation of these proteins seemed likely in the wild type strain but was at the detection limit.
Fig. 2.
PrpC/PrkC-dependent modification of cytadherence proteins in M. pneumoniae Shown are sections of dual channel images of Flamingo fluorescent dye- (green) and Pro-Q Diamond-stained (red) two-dimensional gels representing selected proteins in the M. pneumoniae wild type, prpC::Tn mutant, and prkC::Tn mutant. Proteins were separated on an 18-cm IPG strip with a linear pH gradient of pI 4.5–5.5 in the first dimension. The proteins HMW3, MPN474, P65 (A), MPN256, RpoE (B), and P41 (C) are shown. Putative phosphorylated proteins in the M. pneumoniae wild type as well as proteins with increased or new phosphorylation spots in the prpC::Tn mutant are indicated. Missing phosphorylation spots in the M. pneumoniae wild type and the prkC::Tn mutant are highlighted by a box. Protein amounts of HMW3, MPN256, and P41 (all represented by a triple protein spot) seem to be reduced as well as for the P65 protein (underlined) in the prkC::Tn mutant. Note that the P41 spot “2*” is only putative because of a low Pro-Q Diamond/Flamingo log ratio in the analyzed strains. Protein spots were cut from the gel and identified by MS/MS (see Table I and supplemental Table S3).
The analysis of the phosphorylated peptides that occur exclusively in the prpC mutant led to the identification of four additional phosphorylation sites. For P41, MPN256, and MPN474, phosphorylation occurred on a threonine residue. In addition, two different phosphorylation sites (Ser-87 and Thr-93) could be validated independently on the same phosphopeptide in the case of RpoE. However, the exact site could not be determined unambiguously, so it could only be assumed that phosphorylation occurred most likely on the threonine residue.
Effect of Inactivation of Ser/Thr Kinases on Phosphoproteome
M. pneumoniae encodes two known serine/threonine kinases, i.e. the HPr kinase HPrK and the protein kinase C PrkC. To analyze the impact of these kinases on protein phosphorylation on a global level, we analyzed the phosphoproteome in the corresponding mutant strains. As described for the wild type strain M129, the isogenic mutants GPM51 (hprK) and GPM11 (prkC) were grown in modified Hayflick medium, and cell extracts separated by two-dimensional gel electrophoresis.
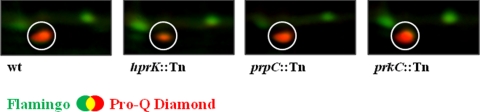
In the hprK mutant, only one phosphoprotein spot appeared to be less intense as compared with the wild type (see Fig. 3). In the wild type strain, this spot contains two phosphorylated proteins, HPr and UlaB, a putative EIIB component of the phosphotransferase system. In the hprK mutant GPM51, this spot contained only the phosphorylated UlaB protein, whereas HPr was absent. This observation is in good agreement with the previous report that HprK is the only kinase for HPr (16). On the other hand, HPr is the only substrate that is phosphorylated by HprK.
Fig. 3.
Comparison of HPr phosphorylation pattern between different M. pneumoniae strains. Section of dual channel images of Flamingo fluorescent dye- (green) and Pro-Q Diamond-stained (red) two-dimensional gels displaying the phosphorylated spot of HPr in the M. pneumoniae wild type and different mutant strains. Proteins were separated on an 18-cm IPG strip with a linear pH gradient of pI 4–7 in the first dimension. Note that the HPr spot represents a multiprotein spot containing HPr and UlaB, whereas only UlaB could be detected in the hprK::Tn mutant (much lesser intensity of the phosphorylation spot).
For the prkC mutant, we observed that the majority of phosphoproteins were not affected by the inactivation of this kinase. However, the phosphorylation or the accumulation of a few proteins was different in the prkC mutant as compared with the isogenic wild type. The large surface protein MPN474 was weakly phosphorylated in the wild type, and this protein was not at all phosphorylated in the prkC mutant, suggesting that MPN474 is a substrate for PrkC. This hypothesis is reinforced by the finding that MPN474 phosphorylation is strongly enhanced in the prpC phosphatase mutant (see Fig. 2A). Similarly, the so far uncharacterized protein MPN256 and the cytadherence proteins HMW3 and P41 were phosphorylated in the wild type but not in the prkC mutant strain GPM11. As observed with MPN474, the intensity of the phosphoprotein spot of these proteins was strongly increased in the prpC mutant (see Fig. 2, A and B). Thus, MPN256, HMW3, and P41 are novel targets of PrkC-dependent phosphorylation in M. pneumoniae.
The prkC mutation did not only affect protein phosphorylation but also the accumulation of some proteins. MPN256, a substrate of PrkC, was much less abundant in the prkC mutant as compared with the wild type. Similarly, the adhesion proteins P41 and HMW3 were present in reduced amounts in the prkC mutant. In addition, the adhesin-related protein P65 was less abundant in the prkC mutant. However, the phosphorylation signals for this protein in the wild type and the prpC mutant strains were below the threshold to conclude phosphorylation.
Classes of Phosphorylated Proteins
To get an impression of which cellular processes are affected by protein phosphorylation, we grouped the proteins according to their functional categories (cluster of orthologous groups of proteins) (see Table I). This analysis revealed that most phosphoproteins (36 proteins) are metabolic enzymes or transporters. 13 phosphorylated proteins are involved in cellular processes such as protein folding or envelope biogenesis. Nine of the phosphorylated proteins are required for the storage and processing of the genetic information, and the functions of five proteins are unknown.
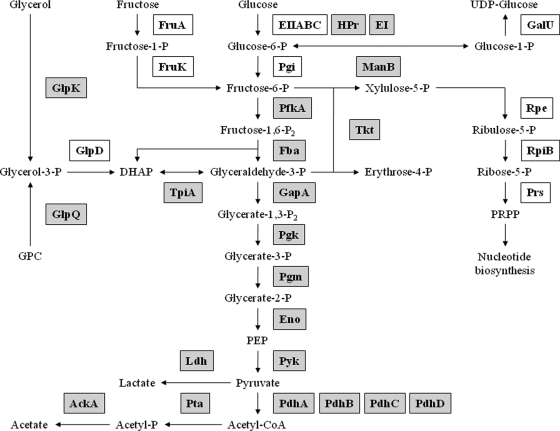
Among the phosphorylated enzymes and transporters, a large number are involved in the central carbon metabolism (see Fig. 4). Of the glycolytic enzymes, all but the phosphoglucose isomerase are phosphorylated in M. pneumoniae. Moreover, all subunits of the pyruvate dehydrogenase and all three enzymes of overflow metabolism (Ldh, AckA, and Pta) are phosphoproteins. Of the enzymes of the pentose phosphate pathway, only the transketolase (Tkt) is phosphorylated in M. pneumoniae. In addition to glucose, M. pneumoniae can utilize glycerol and fructose. Although the enzymes that introduce fructose in the central metabolism are not phosphorylated, two glycerol 3-phosphate-forming enzymes are phosphoproteins.
Fig. 4.
Schematic illustration of phosphorylation events in central metabolic pathways of M. pneumoniae. Of the central metabolic pathways, only glycolysis is operative in M. pneumoniae, and a few carbon sources such as glucose, fructose, and glycerol can be utilized. Pyruvate can be oxidized to acetate or reduced to lactate. An incomplete pentose phosphate pathway serves to generate phosphoribosyl pyrophosphate for nucleotide biosynthesis. Phosphorylated enzymes are highlighted in gray. DHAP, dihydroxyacetone phosphate; GPC, glycerophosphorylcholine; PEP, phosphoenolpyruvate; PRPP, phosphoribosyl pyrophosphate; P, phosphate.
Glycolytic enzymes are phosphorylated in many bacteria (see Table II). A comparison of the phosphorylation pattern of these enzymes in different bacteria, however, revealed specific sets of phosphorylated glycolytic enzymes in each species. None of the glycolytic enzymes are phosphorylated in all species, and the phosphofructokinase is only phosphorylated in M. pneumoniae but not in any other bacterium studied so far. For some of the glycolytic phosphoproteins, the phosphorylation sites have been determined in multiple species. It is interesting to note that even if the same enzyme is phosphorylated in different species, the phosphorylation sites are not conserved. An exception is the pyruvate kinase that is phosphorylated on Ser-36 in a Gram-positive and in a Gram-negative bacterium (B. subtilis and E. coli, respectively) (11, 12). However, phosphorylation of this enzyme was not detected in L. lactis (13), a close relative of both B. subtilis and M. pneumoniae (see “Discussion”).
Table II. Phosphorylation events in glycolytic pathway of bacteria.
Bracketed letters indicate putative phosphorylation sites due to identification of the phosphopeptide.
| Protein | Organism |
||||
|---|---|---|---|---|---|
| M. pneumoniae | B. subtilis | L. lactis | E. coli | C. jejuni | |
| Glucose-6-phosphate isomerase | No | Yes (Thr-39) | Yes (Ser-143) | Yes ([Ser-105], Thr-107) | No |
| Phosphofructokinase | Yes | No | No | No | Noa |
| Fructose-bisphosphate aldolase | Yes | Yes (Thr-212, Thr-234) | Yes ([Thr-49, Ser-50], Ser-216) | No | Yes |
| Triose-phosphate isomerase | Yes | Yes (Ser-213) | No | No | Yes |
| Glyceraldehyde-3-phosphate dehydrogenase | Yes (Ser-245) | Yes ([Ser-148, Ser-151, Thr-153, Thr-154]) | Yes (Ser-126, Ser-211, [Thr-212], Thr-321) | No | No |
| Phosphoglycerate kinase | Yes | Yes (Ser-183, Thr-299) | Yes (Ser-217) | Yes (Ser-192, Thr-196, Thr-199) | No |
| Phosphoglycerate mutase | Yes | Yes (Ser-62) | Yes (Thr-94, Ser-144) | Yes (Ser-146) | No |
| Enolase | Yes | Yes (Thr-141, Ser-259, Tyr-281, Ser-325) | No | Yes ([Ser-372, Thr-375, Thr-379]) | Yes ([Ser-180, Tyr-185]) |
| Pyruvate kinase | Yes | Yes (Ser-36) | No | Yes (Ser-36) | Yes |
a C. jejuni does not encode for the glycolytic enzyme 6-phosphofructokinase.
Another interesting group of phosphoproteins is involved in cytadherence. These three proteins (P41, HMW3, and MPN474) were all only weakly phosphorylated in the wild type strain, but the intensity of the phosphorylation signals was strongly increased in the prpC phosphatase mutant, whereas these phosphorylation events were completely abolished in the prkC mutant. As outlined above, these observations suggest that PrkC phosphorylates these proteins. In addition to HPr, the well known target of the HprK, these adhesion proteins and the unknown protein MPN256 are the only phosphoproteins for which the kinase could be identified.
Autophosphorylation of Single Universally Conserved Phosphoprotein
As mentioned above, there is little conservation of the phosphorylation sites even if the same proteins are phosphorylated in fairly closely related organisms. There is only one exception to this notion, the phosphosugar mutase ManB (according to the nomenclature for M. pneumoniae). Phosphosugar mutases are phosphorylated in all domains of life from the archaeon Halobacterium salinarum and bacteria such as M. pneumoniae and E. coli to eukaryotes such as yeast, Drosophila melanogaster, and humans (11, 41–44).
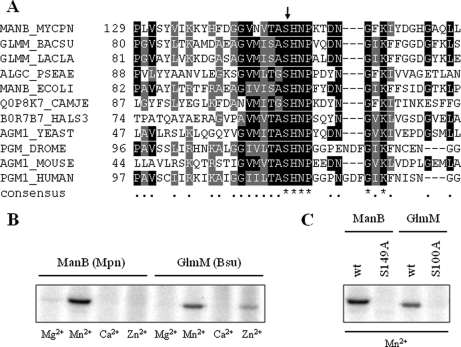
The phosphoproteome analysis showed the presence of 10 phosphospots that do all correspond to ManB. The analysis of the phosphorylation site revealed that Ser-149 was phosphorylated in ManB of M. pneumoniae (see Fig. 5). This site was detected in all 10 spots, suggesting that ManB is subject to different post-translational modifications that affect the isoelectric point of the protein. An alignment of the regions surrounding the phosphorylation site of these proteins revealed a very strong conservation of the region around the phosphorylated serine residue (see Fig. 6A). The motif (T/S)ASHN(P/R) is present in all proteins. This strong similarity suggests phylogenetic conservation of the phosphorylation site and also implies conservation of the phosphorylation mechanism (the kinase) and of the biological function of the phosphorylation event.
Fig. 5.
MS/MS spectrum of serine-phosphorylated peptide from phosphomannomutase/phosphoglucomutase ManB (MPN066). The peptide was measured on line by ESI-mass spectrometry using a nanoACQUITY UPLC system coupled to an LTQ Orbitrap mass spectrometer (see “Experimental Procedures”). The phosphorylation site (Ser-149) is located in the conserved phosphoserine signature of phosphosugar mutases. Detection of the same phosphorylated peptide in all 10 spots of ManB (enhanced image section) suggests further post-translational modifications that affect the isoelectric point of the ManB protein.
Fig. 6.
Autophosphorylation on universally conserved serine residue in active site of phosphosugar mutases. A, multiple alignment of the conserved phosphoserine signature of phosphosugar mutases from all domains of life. Amino acids with similarities in at least two of the sequences are highlighted in gray, whereas amino acids that are identical in at least two of the sequences are depicted in a black background. The UniProtKB entry names of the aligned sequences are MANB_MYCPN (ManB, M. pneumoniae), GLMM_BACSU (GlmM; B. subtilis), GLMM_LACLA (GlmM; L. lactis), ALGC_PSEAE (AlgC; P. aeruginosa), MANB_ECOLI (ManB; E. coli), Q0P8K7_CAMJE (ManB; C. jejuni), B0R7B7_HALS3 (PMM1; H. salinarum), AGM1_YEAST (AGM1; Saccharomyces cerevisiae), PGM_DROME (PGM; D. melanogaster), AGM1_MOUSE (AGM1; Mus musculus), and PGM1_HUMAN (PGM1; Homo sapiens). The conserved active site phosphoserine is indicated by an arrow. B, M. pneumoniae ManB and B. subtilis GlmM autophosphorylation assay in the presence of various divalent cations. About 5 μg of purified His-tagged ManB (lanes 1–4) or GlmM (lanes 4–8) were incubated in the presence of [γ32P]ATP in an autophosphorylation assay (see “Experimental Procedures”). Each reaction mixture was analyzed by SDS-PAGE and phosphorimaging analysis. Divalent cations (5 mm) used in the assays are indicated below the lanes. Bsu, B. subtilis; Mpn, M. pneumoniae. C, autophosphorylation assay of M. pneumoniae ManB (wild type (wt) and S149A) and B. subtilis GlmM (wild type and S100A) recombinant proteins. The four recombinant proteins were checked for autophosphorylation as described in B. Autophosphorylation reactions were conducted in the presence of Mn2+ (5 mm) as divalent cation as it showed the strongest signal in B.
Because there are no conserved protein kinases in the three domains of life, we considered the possibility that ManB might autophosphorylate as it had been shown for the phosphoglucosamine mutase from E. coli (45). To test this idea, we purified ManB carrying an N-terminal His tag and performed phosphorylation assays. As shown in Fig. 6B, the protein did indeed autophosphorylate in the presence of manganese. Only faint phosphorylation signals were observed in the presence of other divalent cations. A protein in which the phosphorylated serine residue was replaced by a non-phosphorylatable alanine did not autophosphorylate (Fig. 6C). This result demonstrates that ManB is phosphorylated on the same site in vivo and in vitro and supports the hypothesis of autophosphorylation. To demonstrate that autophosphorylation is not limited to the enzyme of M. pneumoniae, we also purified the phosphosugar mutase GlmM from B. subtilis and observed autophosphorylation for this protein as well. Again, no phosphorylation was detectable if the phosphorylated serine residue (Ser-100 (9, 12)) was replaced by an alanine. Thus, autophosphorylation is the common mechanism of phosphorylation of these enzymes. This explains the unique conservation of a phosphorylation site.
The phosphosugar mutases play a key role in metabolism as shown by the fact that they are often essential. This is true for GlmM from E. coli and B. subtilis (46, 47). To test whether ManB of M. pneumoniae is also encoded by an essential gene, we attempted to isolate a manB mutant from an ordered collection of transposon insertions. In this collection, each viable mutant is included with a probability of 99.999% (16, 48). No transposon insertion in manB was identified, suggesting that the gene is indeed essential. This finding is in good agreement with previous reports that no transposon insertion in manB could be isolated in M. pneumoniae and M. genitalium (49, 50).
The universal conservation of the phosphorylation site in phosphosugar mutases led us to conclude that this phosphorylation event might be essential. This hypothesis could not be addressed in M. pneumoniae because of the lack of genetic tools (48). Therefore, we studied the role of this phosphorylation site for GlmM B. subtilis. The glmM gene and its variant encoding the S100A protein were established on multicopy plasmids in B. subtilis. In a strain expressing the wild type glmM gene from a plasmid, the chromosomal copy could be deleted by homologous recombination using a PCR fragment obtained by long flanking homology PCR. In contrast, a resistance cassette could not replace the chromosomal copy of glmM when the glmM-S100A allele was expressed from the plasmid. Thus, the phosphorylation of GlmM on Ser-100 seems to be essential for its function and thus for the survival of the bacteria.
DISCUSSION
The analysis of the phosphoproteome of M. pneumoniae revealed substantial protein serine/threonine/tyrosine phosphorylation even in this minimal organism. We identified 63 phosphoproteins that account for nearly 10% of all proteins encoded by M. pneumoniae. This exceeds the percentage of phosphorylated proteins in other bacteria by far. For example, only 2.5% of all proteins of B. subtilis are known to be phosphorylated on a serine, threonine, or tyrosine residue (9, 10, 12). This raises the question whether protein phosphorylation is more common in M. pneumoniae than in other bacteria. It is tempting to speculate that the high rate of protein phosphorylation compensates for the obvious lack of transcriptional regulation in M. pneumoniae. However, this lack of regulation of gene expression allows the detection of the complete phosphoproteome in a single (or a few) experiment(s). Indeed, we detected about 500 CHAPS-soluble proteins that correspond to 80% of all 635 proteins that are in the pI range covered in this study (see supplemental Fig. S1). In other species with complex transcriptional regulation, only about 1,000 proteins are detectable under any specific condition, whereas the rest are required under specific growth conditions (51). For B. subtilis or E. coli, this corresponds to only 25% of all proteins. Thus, the higher coverage in our study is most probably due to the lack of transcription regulation in M. pneumoniae but does not seem to reflect a higher impact of protein phosphorylation.
A striking feature of the phosphoproteomes of different bacteria is the phosphorylation of enzymes of the central metabolic pathways. In M. pneumoniae, metabolism is strongly reduced, and the major pathways are depicted in Fig. 4. The citric acid cycle and pathways of lipid, amino acid, and nucleotide metabolisms are absent in these bacteria. As in the other Firmicutes, most glycolytic enzymes are phosphorylated in M. pneumoniae. Unfortunately, the functions of these phosphorylations as well as the responsible kinases are so far unknown. It is interesting that all four subunits of the pyruvate dehydrogenase are phosphorylated in M. pneumoniae. In B. subtilis, there are different results concerning the pyruvate dehydrogenase. One study reports phosphorylation of PdhB (12), whereas PdhC and PdhD were found to be phosphoproteins in another work (9). In contrast, none of the subunits is phosphorylated in E. coli, Campylobacter jejuni, and L. lactis (11, 13, 52).
This work is the first global analysis of the effects of mutations in the complete set of annotated protein kinases on the phosphoproteome. We have shown that the HPr kinase has only one substrate, HPr. The phosphorylation of HPr on Ser-46 is a major signal to trigger carbon catabolite repression of catabolic genes and operons in B. subtilis and other Firmicutes (53). If HPr(Ser-P) had the same function in M. pneumoniae, one would have expected major changes in the proteome of the hprK mutant as compared with the wild type strain. However, with the exception of HPr phosphorylation (see Fig. 3), no differences were detectable. This suggests that HPr phosphorylation on Ser-46 has a function different from that in the other bacteria. This conclusion is supported by three observations. First, HPr(Ser-P) acts as the cofactor for the pleiotropic transcription factor CcpA to cause catabolite repression. No protein similar to CcpA is encoded in the genome of any Mycoplasma species. Second, the phosphorylation of HPr on Ser-46 interferes with its participation in sugar transport due to the inhibition of enzyme I-dependent phosphorylation of His-15. This inhibition is relevant even in Gram-negative bacteria that also do not possess an equivalent of CcpA (54). However, in M. pneumoniae, these two phosphorylation events are not mutually exclusive (55). Finally, although the activity of the HPr kinase of B. subtilis and most other bacteria is strongly controlled by the ATP and fructose 1,6-bisphosphate concentrations, the enzyme of M. pneumoniae is constitutively active due to its high affinity to ATP (56). Thus, the phosphorylation of HPr on Ser-46 seems to serve other purposes in M. pneumoniae.
In addition to the HPr kinase, M. pneumoniae encodes only one protein kinase, PrkC. This kinase is implicated in a multitude of cellular functions in different bacteria, among them spore germination, virulence, and control of glycolysis (57–59). We found that PrkC has at least four targets in M. pneumoniae, the cell adhesion proteins HMW3 and P41, the cell surface protein MPN474, and the uncharacterized protein MPN256. These proteins are not phosphorylated in a prkC mutant, but their phosphorylation is enhanced if the protein phosphatase PrpC, the antagonist of PrkC, is not expressed (see Fig. 2). Moreover, HMW1 and P1, two very large proteins that were out of the detection window applied in this study, are likely to be phosphorylated by PrkC (20). This suggests that PrkC might be required for cell adhesion in M. pneumoniae. Indeed, a prkC mutant strain is not capable of adherent growth and has lost toxicity to human host cells, most likely due to the defect in cell adhesion (20). Thus, PrkC has a specific function in M. pneumoniae and phosphorylates a specific set of proteins.
As stated above, HPrK and PrkC are the only two annotated protein kinases in M. pneumoniae. However, they account for only five of 63 identified protein phosphorylation events. This suggests the existence of other, yet to be discovered protein kinases in M. pneumoniae. Alternatively, autophosphorylation might be more relevant than previously anticipated. Indeed, the only phosphorylation site that is universally conserved from archaea via bacteria such as M. pneumoniae to eukaryotes is the result of an autophosphorylation event in the catalytic site of phosphosugar mutases such as GlmM and ManB (Refs. 45 and 47 and this work). This autophosphorylation is essential for the enzymatic activity of these enzymes, thus providing an explanation for the universal but highly exceptional conservation of the phosphorylation event at a conserved site. Interestingly, another ubiquitous protein, the heat-shock protein DnaK (also called Hsp70) is also present as a phosphoprotein in all organisms that were studied so far. In E. coli and mycobacteria, DnaK is capable of autophosphorylating at a strongly conserved threonine residue (60–63). This site is also present in the DnaK protein of M. pneumoniae (Thr-182), but it is not conserved in the archaeal DnaK of H. salinarum (41). It is therefore possible that DnaK autophosphorylates at a conserved site in bacteria and eukaryotes, whereas another phosphorylation mechanism occurs in archaea.
In this study, we unambiguously identified 16 phosphorylation sites. Phosphorylation on serine or threonine residues is equally distributed (eight phosphorylation sites each). Serine and threonine are also the predominant phosphorylation sites in other bacteria, and they are used with similar frequency as well (11–13). For eukaryotic phosphoproteins, it was shown that phosphorylations occur often in disordered regions and that the actual phosphorylation sites are not completely fixed in these regions (64, 65). Indeed, seven of the 16 phosphorylation sites identified in this study are predicted to be localized in disordered regions (data not shown). An analysis of the regions surrounding the phosphorylation sites revealed the absence of defined amino acid sequence motifs. Rapid evolution and a corresponding lack of conservation of phosphorylation sites have recently also been observed for protein phosphorylation in yeast and other eukaryotes (66–68). Taken together, these findings suggest that either the responsible protein kinases are quite nonspecific or that a large number of so far unidentified protein kinases exist in M. pneumoniae. An alternative explanation is offered by an analysis of the positions of the phosphorylation sites: structural information is available for seven of the phosphoproteins with identified phosphorylation sites (see supplemental Fig. S2). All nine phosphorylation sites in these proteins are buried in the protein and are thus poorly accessible for protein kinases. This would suggest that these phosphorylation events are autophosphorylations as observed for ManB. Based on the structure of the P. aeruginosa phosphomannomutase/phosphoglucomutase (69) (see Fig. 6), the site of autophosphorylation of ManB and GlmM is also buried in the inside of the protein.
The lack of conservation of phosphorylation sites was also observed in similar phosphoproteins from different bacteria. Only two strongly conserved phosphorylation sites were detected, Ser-46 in HPr and Ser-149 in ManB. Although HPr is phosphorylated by the ATP-dependent HPr kinase that is conserved in many bacteria (Ref. 15 and see Fig. 3), ManB is subject to autophosphorylation. Even among the glycolytic enzymes, only one identified phosphorylation site was conserved between two species (Ser-36 in the pyruvate kinases from B. subtilis and E. coli). Because this latter site is conserved in M. pneumoniae, it is tempting to speculate that it is also phosphorylated in the Mollicutes.
The observations reported in this work have important implications for the evolution of protein phosphorylation in bacteria: protein phosphorylation events seem to be highly specific for each individual bacterial species; even among members of one bacterial phylum such as the Firmicutes, there is little conservation of protein phosphorylation. Moreover, one protein kinase may phosphorylate distinct sets of proteins in the different species. Finally, there may be a large number of protein kinases that have so far escaped discovery, and these kinases are likely to be specific for small groups of bacteria. In conclusion, protein phosphorylation may be one of the major features in the adaptation of bacteria to their individual ecological niche.
Acknowledgments
We are grateful to Julia Busse for excellent technical assistance. Robert Hertel is acknowledged for help with some experiments.
Footnotes
* This work was supported by grants from the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie (to D. B., M. H., and J. S.).
 This article contains supplemental Figs. S1–S3 and Tables S1–S4.
This article contains supplemental Figs. S1–S3 and Tables S1–S4.
REFERENCES
- 1.Stülke J., Eilers H., Schmidl S. R. (2009) Mycoplasma and spiroplasma, in Encyclopedia of Microbiology (Schaechter M. ed) pp. 208–219, Elsevier, Oxford [Google Scholar]
- 2.Halbedel S., Hames C., Stülke J. (2004) In vivo activity of enzymatic and regulatory components of the phosphoenolpyruvate:sugar phosphotransferase system in Mycoplasma pneumoniae. J. Bacteriol 186, 7936–7943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halbedel S., Hames C., Stülke J. (2007) Regulation of carbon metabolism in the mollicutes and its relation to virulence. J. Mol. Microbiol. Biotechnol 12, 147–154 [DOI] [PubMed] [Google Scholar]
- 4.Atkinson T. P., Balish M. F., Waites K. B. (2008) Epidemiology, clinical manifestations, pathogenesis and laboratory detection of Mycoplasma pneumoniae infections. FEMS Microbiol. Rev 32, 956–973 [DOI] [PubMed] [Google Scholar]
- 5.Jacobs E. (1997) Mycoplasma infections of the human respiratory tract. Wien. Klin. Wochenschr 109, 574–577 [PubMed] [Google Scholar]
- 6.Waites K. B., Talkington D. F. (2004) Mycoplasma pneumoniae and its role as a human pathogen. Clin. Microbiol. Rev 17, 697–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boekhorst J., van Breukelen B., Heck A. J., Snel B. (2008) Comparative phosphoproteomics reveals evolutionary and functional conservation of phosphorylation across eukaryotes. Genome Biol 9, R144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jers C., Soufi B., Grangeasse C., Deutscher J., Mijakovic I. (2008) Phosphoproteomics in bacteria: towards a systemic understanding of bacterial phosphorylation networks. Expert Rev. Proteomics 5, 619–627 [DOI] [PubMed] [Google Scholar]
- 9.Eymann C., Becher D., Bernhardt J., Gronau K., Klutzny A., Hecker M. (2007) Dynamics of protein phosphorylation on Ser/Thr/Tyr in Bacillus subtilis. Proteomics 7, 3509–3526 [DOI] [PubMed] [Google Scholar]
- 10.Lévine A., Vannier F., Absalon C., Kuhn L., Jackson P., Scrivener E., Labas V., Vinh J., Courtney P., Garin J., Séror S. J. (2006) Analysis of the dynamic Bacillus subtilis Ser/Thr/Tyr phosphoproteome implicated in a wide variety of cellular processes. Proteomics 6, 2157–2173 [DOI] [PubMed] [Google Scholar]
- 11.Macek B., Gnad F., Soufi B., Kumar C., Olsen J. V., Mijakovic I., Mann M. (2008) Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol. Cell. Proteomics 7, 299–307 [DOI] [PubMed] [Google Scholar]
- 12.Macek B., Mijakovic I., Olsen J. V., Gnad F., Kumar C., Jensen P. R., Mann M. (2007) The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol. Cell. Proteomics 6, 697–707 [DOI] [PubMed] [Google Scholar]
- 13.Soufi B., Gnad F., Jensen P. R., Petranovic D., Mann M., Mijakovic I., Macek B. (2008) The Ser/Thr/Tyr phosphoproteome of Lactococcus lactis IL1403 reveals multiply phosphorylated proteins. Proteomics 8, 3486–3493 [DOI] [PubMed] [Google Scholar]
- 14.Allen G. S., Steinhauer K., Hillen W., Stülke J., Brennan R. G. (2003) Crystal structure of HPr kinase/phosphatase from Mycoplasma pneumoniae. J. Mol. Biol 326, 1203–1217 [DOI] [PubMed] [Google Scholar]
- 15.Steinhauer K., Jepp T., Hillen W., Stülke J. (2002) A novel mode of control of Mycoplasma pneumoniae HPr kinase/phosphatase activity reflects its parasitic life style. Microbiology 148, 3277–3284 [DOI] [PubMed] [Google Scholar]
- 16.Halbedel S., Busse J., Schmidl S. R., Stülke J. (2006) Regulatory protein phosphorylation in Mycoplasma pneumoniae: a PP2C-type phosphatase serves to dephosphorylate HPr(Ser-P). J. Biol. Chem 281, 26253–26259 [DOI] [PubMed] [Google Scholar]
- 17.Absalon C., Obuchowski M., Madec E., Delattre D., Holland I. B., Séror S. J. (2009) CpgA, EF-Tu and the stressosome protein YezB are substrates of the Ser/Thr kinase/phosphatase couple, PrkC/PrpC, in Bacillus subtilis. Microbiology 155, 932–943 [DOI] [PubMed] [Google Scholar]
- 18.Dirksen L. B., Krebes K. A., Krause D. C. (1994) Phosphorylation of cytadherence-accessory proteins in Mycoplasma pneumonia. J. Bacteriol 176, 7499–7505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krebes K. A., Dirksen L. B., Krause D. C. (1995) Phosphorylation of Mycoplasma pneumoniae cytadherence-accessory proteins in cell extracts. J. Bacteriol 177, 4571–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidl S. R., Gronau K., Hames C., Busse J., Becher D., Hecker M., Stülke J. (2010) The stability of cytadherence proteins in Mycoplasma pneumoniae requires activity of the protein kinase PrkC. Infect. Immun 78, 184–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su H. C., Hutchison C. A., 3rd, Giddings M. C. (2007) Mapping phosphoproteins in Mycoplasma genitalium and Mycoplasma pneumoniae. BMC Microbiol 7, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faires N., Tobisch S., Bachem S., Martin-Verstraete I., Hecker M., Stülke J. (1999) The catabolite control protein CcpA controls ammonium assimilation in Bacillus subtilis. J. Mol. Microbiol. Biotechnol 1, 141–148 [PubMed] [Google Scholar]
- 23.Kunst F., Rapoport G. (1995) Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J. Bacteriol 177, 2403–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halbedel S., Eilers H., Jonas B., Busse J., Hecker M., Engelmann S., Stülke J. (2007) Transcription in Mycoplasma pneumoniae: Analysis of the promoters of the ackA and ldh genes. J. Mol. Biol 371, 596–607 [DOI] [PubMed] [Google Scholar]
- 25.Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 26.Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 27.Armougom F., Moretti S., Poirot O., Audic S., Dumas P., Schaeli B., Keduas V., Notredame C. (2006) Expresso: automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee. Nucleic Acids Res 34, W604–W608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guex N., Peitsch M. C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 29.Kiefer F., Arnold K., Künzli M., Bordoli L., Schwede T. (2009) The SWISS-MODEL Repository and associated resources. Nucleic Acids Res 37, D387–D392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Linding R., Jensen L. J., Diella F., Bork P., Gibson T. J., Russell R. B. (2003) Protein disorder prediction: implications for structural proteomics. Structure 11, 1453–1459 [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 32.Schirmer F., Ehrt S., Hillen W. (1997) Expression, inducer spectrum, domain structure, and function of MopR, the regulator of phenol degradation in Acinetobacter calcoaceticus NCIB8250. J. Bacteriol 179, 1329–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hames C., Halbedel S., Schilling O., Stülke J. (2005) Multiple-mutation reaction: a method for simultaneous introduction of multiple mutations into the glpK gene of Mycoplasma pneumoniae. Appl. Environ. Microbiol 71, 4097–4100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bi W., Stambrook P. J. (1998) Site-directed mutagenesis by combined chain reaction. Anal. Biochem 256, 137–140 [DOI] [PubMed] [Google Scholar]
- 35.Martin-Verstraete I., Débarbouillé M., Klier A., Rapoport G. (1994) Interaction of wild-type truncated LevR of Bacillus subtilis with the upstream activating sequence of the levanase operon. J. Mol. Biol 241, 178–192 [DOI] [PubMed] [Google Scholar]
- 36.Wach A. (1996) PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12, 259–265 [DOI] [PubMed] [Google Scholar]
- 37.Guérout-Fleury A. M., Shazand K., Frandsen N., Stragier P. (1995) Antibiotic resistance cassettes for Bacillus subtilis. Gene 167, 335–336 [DOI] [PubMed] [Google Scholar]
- 38.Jordan S., Junker A., Helmann J. D., Mascher T. (2006) Regulation of LiaRS-dependent gene expression in Bacillus subtilis: identification of inhibitor proteins, regulator binding sites, and target genes of a conserved cell envelope stress-sensing two-component system. J. Bacteriol 188, 5153–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jaffe J. D., Berg H. C., Church G. M. (2004) Proteogenomic mapping as a complementary method to perform genome annotation. Proteomics 4, 59–77 [DOI] [PubMed] [Google Scholar]
- 40.Regula J. T., Ueberle B., Boguth G., Görg A., Schnölzer M., Herrmann R., Frank R. (2000) Towards a two-dimensional proteome map of Mycoplasma pneumoniae. Electrophoresis 21, 3765–3780 [DOI] [PubMed] [Google Scholar]
- 41.Aivaliotis M., Macek B., Gnad F., Reichelt P., Mann M., Oesterhelt D. (2009) Ser/Thr/Tyr protein phosphorylation in the archaeon Halobacterium salinarum—a representative of the third domain of life. PLoS ONE 4, e4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X., Gerber S. A., Rudner A. D., Beausoleil S. A., Haas W., Villén J., Elias J. E., Gygi S. P. (2007) Large-scale phosphorylation analysis of alpha-factor-arrested Saccharomyces cerevisiae. J. Proteome Res 6, 1190–1197 [DOI] [PubMed] [Google Scholar]
- 43.Yu L. R., Zhu Z., Chan K. C., Issaq H. J., Dimitrov D. S., Veenstra T. D. (2007) Improved titanium dioxide enrichment of phosphopeptides from HeLa cells and high confident phosphopeptide identification by cross-validation of MS/MS and MS/MS/MS spectra. J. Proteome Res 6, 4150–4162 [DOI] [PubMed] [Google Scholar]
- 44.Zhai B., Villén J., Beausoleil S. A., Mintseris J., Gygi S. P. (2008) Phosphoproteome analysis of Drosophila melanogaster embryos. J. Proteome Res 7, 1675–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jolly L., Pompeo F., van Heijenoort J., Fassy F., Mengin-Lecreulx D. (2000) Autophosphorylation of phosphoglucosamine mutase from Escherichia coli. J. Bacteriol 182, 1280–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kobayashi K., Ehrlich S. D., Albertini A., Amati G., Andersen K. K., Arnaud M., Asai K., Ashikaga S., Aymerich S., Bessieres P., Boland F., Brignell S. C., Bron S., Bunai K., Chapuis J., Christiansen L. C., Danchin A., Débarbouille M., Dervyn E., Deuerling E., Devine K., Devine S. K., Dreesen O., Errington J., Fillinger S., Foster S. J., Fujita Y., Galizzi A., Gardan R., Eschevins C., Fukushima T., Haga K., Harwood C. R., Hecker M., Hosoya D., Hullo M. F., Kakeshita H., Karamata D., Kasahara Y., Kawamura F., Koga K., Koski P., Kuwana R., Imamura D., Ishimaru M., Ishikawa S., Ishio I., Le Coq D., Masson A., Mauël C., Meima R., Mellado R. P., Moir A., Moriya S., Nagakawa E., Nanamiya H., Nakai S., Nygaard P., Ogura M., Ohanan T., O'Reilly M., O'Rourke M., Pragai Z., Pooley H. M., Rapoport G., Rawlins J. P., Rivas L. A., Rivolta C., Sadaie A., Sadaie Y., Sarvas M., Sato T., Saxild H. H., Scanlan E., Schumann W., Seegers J. F., Sekiguchi J., Sekowska A., Séror S. J., Simon M., Stragier P., Studer R., Takamatsu H., Tanaka T., Takeuchi M., Thomaides H. B., Vagner V., van Dijl J. M., Watabe K., Wipat A., Yamamoto H., Yamamoto M., Yamamoto Y., Yamane K., Yata K., Yoshida K., Yoshikawa H., Zuber U., Ogasawara N. (2003) Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. U.S.A 100, 4678–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mengin-Lecreulx D., van Heijenoort J. (1996) Characterization of the essential gene glmM encoding phosphoglucosamine mutase in Escherichia coli. J. Biol. Chem 271, 32–39 [DOI] [PubMed] [Google Scholar]
- 48.Halbedel S., Stülke J. (2007) Tools for the genetic analysis of Mycoplasma. Int. J. Med. Microbiol 297, 37–44 [DOI] [PubMed] [Google Scholar]
- 49.Glass J. I., Assad-Garcia N., Alperovich N., Yooseph S., Lewis M. R., Maruf M., Hutchison C. A., 3rd, Smith H. O., Venter J. C. (2006) Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. U.S.A 103, 425–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hutchison C. A., Peterson S. N., Gill S. R., Cline R. T., White O., Fraser C. M., Smith H. O., Venter J. C. (1999) Global transposon mutagenesis and a minimal mycoplasma genome. Science 286, 2165–2169 [DOI] [PubMed] [Google Scholar]
- 51.Eymann C., Dreisbach A., Albrecht D., Bernhardt J., Becher D., Gentner S., Tam L. T., Büttner K., Buurman G., Scharf C., Venz S., Völker U., Hecker M. (2004) A comprehensive proteome map of growing Bacillus subtilis cells. Proteomics 4, 2849–2876 [DOI] [PubMed] [Google Scholar]
- 52.Voisin S., Watson D. C., Tessier L., Ding W., Foote S., Bhatia S., Kelly J. F., Young N. M. (2007) The cytoplasmic phosphoproteome of the Gram-negative bacterium Campylobacter jejuni: evidence for modification by unidentified protein kinases. Proteomics 7, 4338–4348 [DOI] [PubMed] [Google Scholar]
- 53.Görke B., Stülke J. (2008) Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol 6, 613–624 [DOI] [PubMed] [Google Scholar]
- 54.Krausse D., Hunold K., Kusian B., Lenz O., Stülke J., Bowien B., Deutscher J. (2009) Essential role of the hprK gene in Ralstonia eutropha H16. J. Mol. Microbiol. Biotechnol 17, 146–152 [DOI] [PubMed] [Google Scholar]
- 55.Halbedel S., Stülke J. (2005) Dual phosphorylation of Mycoplasma pneumoniae HPr by Enzyme I and HPr kinase suggests an extended phosphoryl group susceptibility of HPr. FEMS Microbiol. Lett 247, 193–198 [DOI] [PubMed] [Google Scholar]
- 56.Merzbacher M., Detsch C., Hillen W., Stülke J. (2004) Mycoplasma pneumoniae HPr kinase/phosphorylase. Eur. J. Biochem 271, 367–374 [DOI] [PubMed] [Google Scholar]
- 57.Kristich C. J., Wells C. L., Dunny G. M. (2007) A eukaryotic-type Ser/Thr kinase in Enterococcus faecalis mediates antimicrobial resistance and intestinal persistence. Proc. Natl. Acad. Sci. U.S.A 104, 3508–3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lomas-Lopez R., Paracuellos P., Riberty M., Cozzone A. J., Duclos B. (2007) Several enzymes of the central metabolism are phosphorylated in Staphylococcus aureus. FEMS Microbiol. Lett 272, 35–42 [DOI] [PubMed] [Google Scholar]
- 59.Shah I. M., Laaberki M. H., Popham D. L., Dworkin J. (2008) A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell 135, 486–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCarty J. S., Walker G. C. (1991) DnaK as a thermometer: threonine-199 is site of autophosphorylation and is critical for ATPase activity. Proc. Natl. Acad. Sci. U.S.A 88, 9513–9517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peake P., Winter N., Britton W. (1998) Phosphorylation of Mycobacterium leprae heat-shock 70 protein at threonine 175 alters its substrate binding characteristics. Biochim. Biophys. Acta 1387, 387–394 [DOI] [PubMed] [Google Scholar]
- 62.Preneta R., Papavinasasundaram K. G., Cozzone A. J., Duclos B. (2004) Autophosphorylation of the 16 kDa and 70 kDa antigens (Hsp 16.3 and Hsp 70) of Mycobacterium tuberculosis. Microbiology 150, 2135–2141 [DOI] [PubMed] [Google Scholar]
- 63.Zylicz M., LeBowitz J. H., McMacken R., Georgopoulos C. (1983) The dnaK protein of Escherichia coli possesses an ATPase and autophosphorylating activity and is essential in an in vitro DNA replication system. Proc. Natl. Acad. Sci. U.S.A 80, 6431–6435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Iakoucheva L. M., Radivojac P., Brown C. J., O'Connor T. R., Sikes J. G., Obradovic Z., Dunker A. K. (2004) The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res 32, 1037–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Landry C. R., Levy E. D., Michnick S. W. (2009) Weak functional constraints on phosphoproteomes. Trends Genet 25, 193–197 [DOI] [PubMed] [Google Scholar]
- 66.Beltrao P., Trinidad J. C., Fiedler D., Roguev A., Lim W. A., Shokat K. M., Burlingame A. L., Krogan N. J. (2009) Evolution of phosphoregulation: comparison of phosphorylation patterns across yeast species. PLoS Biol 7, e1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Holt L. J., Tuch B. B., Villén J., Johnson A. D., Gygi S. P., Morgan D. O. (2009) Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325, 1682–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tan C. S., Bodenmiller B., Pasculescu A., Jovanovic M., Hengartner M. O., Jørgensen C., Bader G. D., Aebersold R., Pawson T., Linding R. (2009) Comparative analysis reveals conserved protein phosphorylation networks implicated in multiple diseases. Sci. Signal 2, ra39. [DOI] [PubMed] [Google Scholar]
- 69.Regni C., Tipton P. A., Beamer L. J. (2002) Crystal structure of PMM/PGM: an enzyme in the biosynthetic pathway of P. aeruginosa virulence factors. Structure 10, 269–279 [DOI] [PubMed] [Google Scholar]