Abstract
Articular cartilage is indispensable for joint function but has limited capacity for self-repair. Engineering of neocartilage in vitro is therefore a major target for autologous cartilage repair in arthritis. Previous analysis of neocartilage has targeted cellular organization and specific molecular components. However, the complexity of extracellular matrix (ECM) development in neocartilage has not been investigated by proteomics. To redress this, we developed a mouse neocartilage culture system that produces a cartilaginous ECM. Differential analysis of the tissue proteome of 3-week neocartilage and 3-day postnatal mouse cartilage using solubility-based protein fractionation targeted components involved in neocartilage development, including ECM maturation. Initially, SDS-PAGE analysis of sequential extracts revealed the transition in protein solubility from a high proportion of readily soluble (NaCl-extracted) proteins in juvenile cartilage to a high proportion of poorly soluble (guanidine hydrochloride-extracted) proteins in neocartilage. Label-free quantitative mass spectrometry (LTQ-Orbitrap) and statistical analysis were then used to filter three significant protein groups: proteins enriched according to extraction condition, proteins differentially abundant between juvenile cartilage and neocartilage, and proteins with differential solubility properties between the two tissue types. Classification of proteins differentially abundant between NaCl and guanidine hydrochloride extracts (n = 403) using bioinformatics revealed effective partitioning of readily soluble components from subunits of larger protein complexes. Proteins significantly enriched in neocartilage (n = 78) included proteins previously not reported or with unknown function in cartilage (integrin-binding protein DEL1; coiled-coil domain-containing protein 80; emilin-1 and pigment epithelium derived factor). Proteins with differential extractability between juvenile cartilage and neocartilage included ECM components (nidogen-2, perlecan, collagen VI, matrilin-3, tenascin and thrombospondin-1), and the relationship between protein extractability and ECM ultrastructural organization was supported by electron microscopy. Additionally, one guanidine extract-specific neocartilage protein, protease nexin-1, was confirmed by immunohistochemistry as a novel component of developing articular cartilage in vivo. The extraction profile and matrix-associated immunostaining implicates protease nexin-1 in cartilage development in vitro and in vivo.
The cartilage of the mammalian skeletal system has two distinct roles. The epiphyseal cartilage of the growth plate drives endochondral bone growth, and the hyaline cartilage at the weight-bearing surfaces of bones facilitates joint articulation. In both environments, chondrocyte-regulated production, assembly, and turnover of the extracellular matrix (ECM)1 are essential for the tissue to withstand compressive forces and respond to mechanical loading. The major structural constituents of cartilage ECM are the heterotypic collagen II/IX/XI fibrils and proteoglycan-glycosaminoglycan networks of aggrecan and hyaluronan. Loss of joint function in osteoarthritis (OA) is strongly associated with net loss of aggrecan and collagen breakdown caused by an imbalance of ECM homeostasis (1). In addition, many inherited human chondrodysplasias involve disruption of cartilage matrix assembly or cell-matrix interactions, resulting in abnormal skeletal development and in some cases early onset cartilage degeneration (2, 3).
The alterations in chondrocyte metabolism that occur during OA are complex and remain poorly understood (4). An early response to loss or fragmentation of ECM components is attempted tissue repair through secretion of anabolic factors, cell proliferation, and matrix remodeling (5). However, the resulting product is a fibrocartilage that does not recapitulate the composition or precise architecture of the original hyaline articular cartilage. This limited capacity of cartilage for regeneration has driven research into cartilage tissue engineering (6). Production of authentic hyaline cartilage in vitro remains challenging due to the dedifferentiation of primary chondrocytes upon removal from their three-dimensional matrix environment (7). However, improved “neocartilage” culture systems have been developed through evaluation of suitable chondroprogenitor or chondrocyte subpopulations and optimization of exogenous support matrices and growth factors (8, 9). The therapeutic target of neocartilage culture is autologous tissue repair. However, there is fundamental value in using neocartilage systems to elucidate mechanisms of protein integration into the ECM and the role of specific protein interactions during cartilage maturation.
Cartilage profiling by 2-DE and mass spectrometry-based proteomics is generating important new insight into mechanisms of cartilage degeneration in vitro and in vivo (10). For example, anabolic factors with potential roles in cartilage repair, including connective tissue growth factor and inhibin βA (activin), were identified in the secretome of human OA cartilage explants (11). Comparison of cartilage protein extracts from normal donors and OA patients revealed significantly increased levels of the serine protease Htra1 in patient cartilage (12) and that Htra1-mediated proteolysis of aggrecan may significantly contribute to OA pathology (13). Targeted analysis of the chondrocyte mitochondrial proteome highlighted OA-related changes in energy production and protection against reactive oxygen species (14). Obtaining sufficient chondrocytes from human donors for proteomics unfortunately requires expansion of the cell population with potential loss of the chondrocyte phenotype during prolonged culture. Other drawbacks encountered with human samples include the clinical heterogeneity of OA, lack of matched controls, and inherent genetic variation of human subjects (15). Alternatively, animal models that recapitulate hallmarks of progressive cartilage degeneration, such as aggrecan loss and articular surface fibrillation, are emerging as a powerful resource, particularly in mice lacking specific proteases or protease target sites (16, 17). The development of techniques for analysis of murine cartilage using proteomics has paved the way for differential analysis of normal and pathological or genetically targeted cartilage (18, 19).
Label-free methods for relative peptide quantitation, such as ion intensity measurement and spectral counting, are emerging as reliable and cost-effective alternatives to chemical modification or isotopic peptide labeling (20). Combining orthogonal protein and/or peptide fractionation with high resolution HPLC-MS can achieve proteome-wide coverage (21). Because extensive sample fractionation can introduce redundancy and variation, improved sequence/proteome coverage must be balanced against the cost of additional sample handling and lengthy LC-MS runs (22).
Here we describe a novel platform for analysis of mouse cartilage using solubility-based protein fractionation (19) combined with label-free quantitative tandem MS (LTQ-Orbitrap). Sequential extraction of 3-day postnatal (P3) mouse epiphyseal cartilage and 3-week neocartilage cultures revealed a marked transition from a high proportion of readily soluble components in P3 extracts to a greater proportion of poorly soluble proteins in neocartilage. Principal component analysis and hierarchical clustering were used to globally assess the inter-relationships between P3 cartilage and neocartilage NaCl and guanidine hydrochloride (GdnHCl) extracts. At a p value cutoff of 0.05, 403 proteins were classified as extract-specific, whereas 125 proteins were classified as tissue sample-specific. Many of the proteins significantly enriched in neocartilage were annotated by the terms cell adhesion, extracellular matrix, and cytoskeletal remodeling. Further statistical analysis identified a third important protein category in which protein solubility was altered between the P3 and neocartilage. Identification of proteins involved in neocartilage maturation has generated novel insight into the fundamental process of cartilage matrix development with potential for further analysis of engineered cartilaginous tissues with biomedical applications.
EXPERIMENTAL PROCEDURES
Cartilage Preparation and High Density Chondrocyte Culture
Tibial and femoral condyles and primary chondrocytes were obtained under sterile conditions from 3-day-old C57/Bl6 mice. Dissected knee joints were incubated for 2 h at 37 °C in 2 mg/ml bacterial collagenase (Worthington) with occasional vortex mixing to completely remove the meniscus, perichondrium, and ligaments (23). The condyles were either rinsed in PBS, frozen on dry ice, and stored at −80 °C for processing or transferred into a 10-cm2 culture dish and incubated at 37 °C overnight in fresh collagenase. Chondrocytes were filtered through a 40-μm cell strainer, collected by centrifugation (1,500 × g for 3 min), and resuspended in Dulbecco's modified Eagle's medium (DMEM) containing 10% (v/v) FCS, 20 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). Cells were counted and assessed for viability using an automated cell counter (Invitrogen) and plated into a 24-well culture dish at 2.5 × 106 cells/well (n = 4). High density multilayer cultures were maintained for up to 6 weeks in DMEM, 10% FCS supplemented with 0.25 mm sodium ascorbate with the medium replaced daily.
Protein Extraction and Sample Preparation
Serum proteins were depleted from neocartilage cultures prior to harvesting the cultures by extensive washing in serum-free DMEM (four changes of medium in 6 h). Cultures were rinsed twice in PBS, weighed, and snap frozen on dry ice. Replicate protein extracts were prepared from neocartilage and 3-day-old mouse cartilage (n = 4) using ∼10 mg of frozen tissue. Samples were pulverized into a fine powder using a liquid nitrogen-cooled tissue grinder, transferred into Eppendorf tubes containing 100 μl of Tris acetate, pH 8.0, 10 mm EDTA, and 0.1 unit of chondroitinase ABC (MP Biomedicals) and deglycosylated for 6 h at 37 °C. In this study, we used the sample preparation workflow developed for 2-DE analysis of sequential extracts of mouse femoral head cartilage (19) involving sequential extraction in 1 m NaCl followed by extraction in 4 m GdnHCl and then molecular weight cutoff centrifugal filtration of guanidine extracts. Using the same procedure allowed us to compare the neocartilage, juvenile cartilage, and femoral head cartilage extracts with the additional benefit of allowing further analysis by 2-DE if required. Sequential extracts were prepared using a non-denaturing buffer (1 m NaCl in 100 mm Tris acetate, pH 8.0) followed by extraction of the salt-insoluble fraction in a chaotropic buffer (4 m guanidine HCl, 65 mm DTT, 10 mm EDTA in 50 mm sodium acetate, pH 5.8). Each extraction was performed for 18–24 h at 4 °C with end-over-end mixing. The final insoluble fraction was incubated in 100 mm acetic acid containing 100 μg/ml pepsin (Sigma) to release pepsin-resistant collagenous polypeptides (P1 fraction). Guanidine-extracted proteins were then fractionated by molecular mass cutoff centrifugal filtration through a 100-kDa-cutoff column (Amicon) with the filtrate designated the E1 fraction and the retentate designated the E2 fraction. The E0, E1, and E2 protein extracts were precipitated with 9 volumes of ethanol, and protein pellets were washed twice in 70% (v/v) ethanol and resuspended in 200 μl of solubilization buffer containing 7 m urea, 2 m thiourea, 4% CHAPS, and 30 mm Tris, pH 8.0. Protein concentrations were estimated by Bradford assay (Pierce) against BSA diluted in solubilization buffer. The P1 fraction was divided into 10 aliquots and stored freeze-dried at −20 °C.
SDS-PAGE and Immunoblotting
Cartilage extracts were analyzed by SDS-PAGE to assess intersample consistency and quantitative/qualitative differences between juvenile epiphyseal cartilage and neocartilage cultures harvested at 7 and 21 days. Samples were loaded according to either a consistent protein amount (3 μg of protein/lane) or as a percentage of the protein yield as indicated. Aliquots of E0, E1, and E2 extracts were heated in Laemmli buffer containing 50 mm dithiothreitol for 15 min at 65 °C and resolved through 4–12% acrylamide Bis-Tris NuPAGE gels (Invitrogen), and proteins were visualized by silver staining as described (23). Aliquots of the pepsin-resistant material were resolved using 3–8% acrylamide Tris acetate NuPAGE gels and stained with Coomassie Blue to detect collagen I, II, and XI α-chains and the low molecular weight collagenous domains of collagen IX. To correlate spectral counting data with an independent method of protein measurement, proteins were also detected by fluorescence immunoblotting and laser scanning (19) and quantified using ImageQuant software (GE Healthcare).
Protein Reduction/Alkylation and In-solution Trypsin Digestion
Protein samples for LC-MS/MS analysis were sequentially reduced and alkylated under nitrogen by incubation in 10 mm dithiothreitol (overnight at 4 °C) and then 50 mm iodoacetamide (2 h at 25 °C in the dark). Proteins were co-precipitated with 1 μg of trypsin (Promega) overnight at −20 °C in 1 ml of methanol. The trypsin-protein precipitates were washed once with chilled methanol, dried, and reconstituted in 100 mm ammonium bicarbonate followed by trypsinization at 37 °C for 5 h with addition of 1 μg of trypsin after 2 h. Digests were terminated by freezing on dry ice.
Orbitrap Mass Spectrometry
LC-MS/MS analysis was performed using an UltiMate 3000 HPLC system (Dionex) or a Tempo nano-LC system (Eksigent) in line with an LTQ-Orbitrap XL (Thermo Fischer Scientific). Aliquots of tryptic peptides equivalent to 25% of the in-solution digests were loaded onto a 0.3 × 5-mm C18 trap column (Dionex) at 20 μl/min in 98% solvent A (0.1% (v/v) formic acid) and 2% solvent B (80% (v/v) acetonitrile, 0.1% (v/v) formic acid) for 5 min and subsequently back-flushed onto a pre-equilibrated analytical column (Vydac Everest C18 300 Å, 150 μm × 150 mm, Alltech) using a flow rate of 1 μl/min. Peptides were separated at 40 °C using three linear gradient segments (2–12% solvent B for 6 min, 12–50% solvent B for 65 min, and then 50–100% solvent B for 15 min), holding at 100% solvent B for a further 15 min before returning to 2% solvent B for 5 min.
The LTQ-Orbitrap was fitted with a dynamic nanoelectrospray ion source (Proxeon) containing a 30-μm-inner diameter uncoated silica emitter (New Objective). The LTQ-Orbitrap XL was controlled using Xcalibur 2.0 software (Thermo Electron) and operated in data-dependent acquisition mode whereby the survey scan was acquired in the Orbitrap with a resolving power set to 60,000 (at 400 m/z). MS/MS spectra were concurrently acquired in the LTQ mass analyzer on the seven most intense ions from the FT survey scan. Charge state filtering, where unassigned precursor ions were not selected for fragmentation, and dynamic exclusion (repeat count, 1; repeat duration, 30 s; exclusion list size, 500) were used. Fragmentation conditions in the LTQ were as follows: normalized collision energy, 35%; activation q, 0.25; activation time, 50 ms; and minimum ion selection intensity, 500 counts.
Database Searching
Peptide information was extracted from Xcalibur raw files using Thermo Proteome Discoverer version 1.0 (Thermo Fischer Scientific) (parent ions in the mass range of 300–5000 m/z, signal:noise ratio of 2). MS/MS data were analyzed using an in-house MASCOT server (version 2.2, Matrix Science) to search the Swiss-Prot database (version 57.6; taxonomy, mouse; 16,169 entries) assuming trypsin digestion. A parent ion tolerance of 10 ppm and a fragment ion mass tolerance of 0.8 Da were used. S-Carboxamidomethylation of cysteine residues was specified as a fixed modification, whereas cyclization of N-terminal glutamine to pyroglutamic acid, deamidation of asparagine, hydroxylation of proline, oxidation of methionine, and phosphorylation of serine and tyrosine were specified as variable modifications.
Criteria for Protein Identification
Scaffold (version Scaffold_2_05_01, Proteome Software Inc., Portland, OR) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the PeptideProphet algorithm (24). Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least two identified peptides. Protein probabilities were assigned by the ProteinProphet algorithm (25). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
Statistical and Bioinformatics Analysis of Mass Spectrometry Data
Scaffold was used to convert unweighted spectral count (SC) values into normalized SC values prior to statistical evaluation of the data set by ANOVA and Student's t test. -Fold changes between sample categories and significance (p) values were calculated based on mean normalized SC values where non-detected proteins were assigned a value of 1. The first comparison was between sequential extracts of femoral head cartilage (n = 3 in each category). The second comparison was between sequential extracts of the P3 and T21 samples (NaCl versus GdnHCl extracts, n = 8 in each category), and the third comparison was between the two sample types (P3 versus T21, n = 8 in each category). Additional analysis was carried out using R and Bioconductor software (26). Principal component analysis (PCA) was carried out on the complete data set using Made4 (27) with the resulting components plotted as a grid to illustrate the PCA transformation. This set of proteins was subsequently filtered to include only proteins that were reliably detected (≥2 peptides) in a minimum of four samples. Unsupervised hierarchical clustering of this filtered list was used to generate a heat map of relative protein abundances (log2-normalized SC values) and visualize the inter-relatedness within and between sample categories.
Ontological classification and functional enrichment analysis were performed using the database for annotation, visualization, and integrated discovery (DAVID) (28). To identify enrichment terms associated with protein subsets of interest, protein lists were uploaded to the DAVID web site using the complete mouse genome as background. Significantly enriched functional groups were ranked using the Functional Annotation Clustering tool set to the highest classification stringency, based on p values adjusted for multiple testing (29). The levels of proteins detected in different extracts, based on normalized SC values or Western blot band intensity, were represented using GraphPad software. Significant differences were measured by one-way ANOVA, and adjusted p values were calculated in the GraphPad Prism 5 statistics module using Bonferroni multiple comparison.
Immunohistochemical Analysis
Femoral heads isolated from 21-day-old mice and knee joints isolated from 3-day-old C57/Bl6 mice were fixed with 4% paraformaldehyde in PBS for 6 h followed by three washes in PBS. All tissues were embedded in TissueTek Optimal Cutting Temperature medium, frozen in liquid nitrogen, and stored at −80 °C. 6-μm sagittal sections of neocartilage, 3-day knee joints, and 21-day femoral heads were cut using a Leica CM3050 cryostat. Immunostaining of tissue sections with each antibody was undertaken simultaneously. Sections were briefly rinsed in PBS, fixed with 4% paraformaldehyde (v/v) in PBS, and then treated with 6% H2O2 (v/v) in methanol to inactivate endogenous peroxidase activity. Tissue sections were then treated with 0.8% bovine hyaluronidase (Type IV-S) to unmask antigens followed by three further PBS washes. Prior to incubation with mouse anti-collagen II (monoclonal 2B1.5 used at 1:500 dilution, Neomarkers) and mouse protease nexin-1 (monoclonal 4B3 used at 1:1,000, Santa Cruz Biotechnology), nonspecific binding was blocked by incubating sections with a mouse IgG blocking reagent for 1 h. Subsequent steps were performed using the Vector mouse-on-mouse immunodetection kit (Vector Laboratories) according to the manufacturer's instructions. Tissue sections probed with anti-collagen I (LF41 rabbit polyclonal used at 1:5,000, generously supplied by Larry Fisher) were first blocked with goat serum (Vectastain Elite ABC rabbit IgG kit) in 1% BSA in PBS for 30 min prior to incubation with primary antibody for 1 h. Control sections were probed only with the appropriate anti-mouse or anti-rabbit IgG secondary antibody at 1:1,000. Immunohistochemical staining was detected using ImmPact 3,3′-diaminobenzidine substrate (Vector Laboratories) until color was detected. Reactions were terminated by rinsing in tap water followed by dehydration of tissue sections using a graded ethanol series (30, 50, 70, 90, and 100%). Sections were cleared in xylene and mounted using Pertex mounting medium. Immunostaining was visualized with a Nikon Eclipse 80i microscope, and images were captured at 20× magnification.
Ultrastructural Analysis by Transmission Electron Microscopy
Neocartilage cultures were fixed in sodium cacodylate-buffered glutaraldehyde (Karnovsky's fixative) or with Karnovsky's fixative supplemented with 0.7% (v/v) safranin O, a modification of the standard procedure used to enhance structural preservation and stabilization of cartilage proteoglycans (30). Samples were postfixed in 1% osmium tetroxide, 1.5% potassium ferrocyanide and embedded in Spurr's resin. Semithin sections (0.5 μm) were stained with 1% methylene blue to visualize tissue structure, including metachromatic staining of glycosaminoglycans. Ultrathin sections were contrasted with uranyl acetate and Reynold's lead citrate and examined with a Philips 300 transmission electron microscope at 60 kV.
RESULTS
Extracellular Matrix Formation in Mouse Neocartilage Cultures
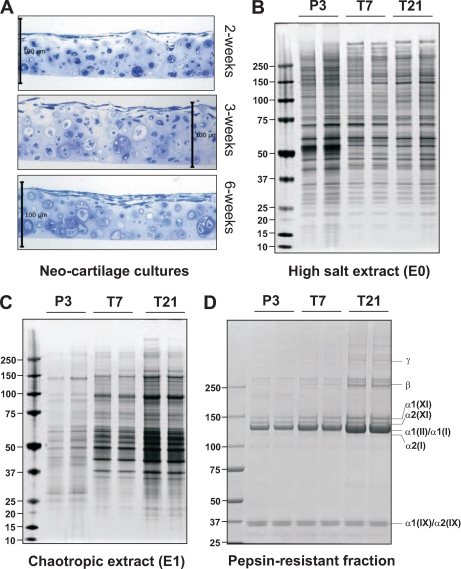
To investigate the process of cartilage matrix assembly using proteomics, we first established a mouse neocartilage culture system that maintains the chondrocyte differentiation state and produces a cartilaginous ECM. Chondrocytes were dissociated from the epiphyseal cartilage of 3-day-old (P3) mouse knees and maintained in culture for 2, 3, and 6 weeks. Semithin sections through the cultures revealed that most chondrocytes maintained a round morphology and synthesized a proteoglycan-rich ECM, essential characteristics of the cartilage phenotype (Fig. 1A).
Fig. 1.
Histology of mouse neocartilage cultures and SDS-PAGE analysis of protein extracts from neocartilage and juvenile mouse epiphyseal cartilage. A, semithin sections of mouse neocartilage cultures, harvested at the time points indicated, stained with methylene blue. Scale bar, 100 μm. B and C, sequential extracts of juvenile cartilage (P3), 1-week neocartilage (T7), and 3-week neocartilage cultures (T21) resolved by Bis-Tris 4–12% NuPAGE. Each lane contains the same proportion of the total protein yield (10 μl of the total 200 μl) to indicate changes in the relative amounts of proteins extracted in 1 m NaCl (High salt extract) or 4 m GdnHCl (Chaotropic extract). D, aliquots of the guanidine-insoluble, pepsin-resistant material (20 μl of the total 500 μl) were resolved by Tris acetate 3–8% NuPAGE. Polypeptides corresponding to the pepsin-resistant domains of collagen I (α1(I) and α2(I)), collagen II (α1(II)), collagen IX (α1(IX) and α2(IX)), and collagen XI (α1(XI) and α2(XI)) and the cross-linked collagen α-chain dimers (β) and trimers (γ) are indicated.
SDS-PAGE Analysis of Neocartilage Sequential Protein Extracts
We anticipated that the maturation of neocartilage involves changes in both protein abundance and extractability. Therefore, sequential extracts of neocartilage cultures were prepared using non-denaturing buffer (1 m NaCl) followed by chaotropic buffer (4 m GdnHCl) and finally pepsin to analyze cross-linked collagenous polypeptides (31, 32). For comparison, extracts were prepared directly from non-cultured juvenile (P3) epiphyseal cartilage. The levels of NaCl-extracted proteins changed little between 7 and 21 days of culture (Fig. 1B), whereas the yield of GdnHCl-extracted proteins increased substantially (Fig. 1C). The same trend was observed for levels of pepsin-resistant polypeptides with a marked increase in collagen II and XI α-chain monomers and cross-linked β and γ components from 7- to 21-day neocartilage culture (Fig. 1D). Levels of collagen IX, the third component of cartilage collagen fibrils, were unchanged, indicated by consistent levels of pepsin-resistant collagenous α1(IX) and α2(IX) fragments in each sample. The concomitant increase in GdnHCl-extracted material and collagen II/IX/XI deposition is strong evidence for assembly of tightly interacting ECM networks in 3-week neocartilage.
Development of Label-free Quantitative LC-MS/MS Analysis of Mouse Cartilage Sequential Extracts
Our previous 2-DE-based analysis of sequential extracts prepared from 3-week-old mouse femoral head cartilage (referred to herein as P21 cartilage) generated distinct and reproducible “extraction profiles” (19). To test the feasibility of LC-MS/MS for global measurement of relative protein abundance in sequential cartilage extracts, triplicate P21 sequential extracts were fragmented into tryptic peptides, resolved by capillary HPLC, and analyzed on line by LTQ-Orbitrap tandem MS. In total, nine samples were analyzed as we isolated NaCl-soluble material (E0 extract) followed by further separation of the GdnHCl-soluble material by 100-kDa-molecular mass-cutoff filtration into filtrate (E1) and retentate (E2) fractions (19). A total of 585 non-redundant proteins were identified in the E0, E1, and E2 extracts at high confidence (≥2 peptides) of which 27% were E0-specific (152 proteins), 27% were E1-specific (156 proteins), 7% were E2-specific (43 proteins), and the remaining 39% were detected in more than one extract (234 proteins) (supplemental Fig. 1). The effective partitioning of cartilage proteins between NaCl- and GdnHCl-soluble fractions revealed by LC-MS/MS analysis is consistent with the corresponding 2-DE (19) and SDS-PAGE (supplemental Fig. 1A) analyses.
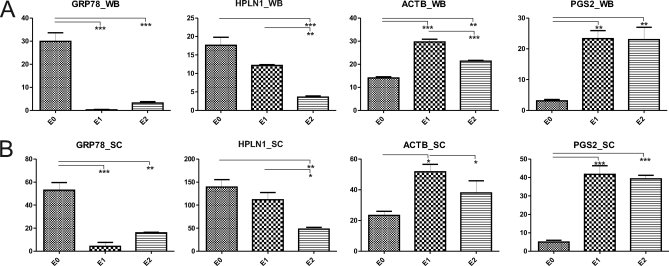
Validation of our sequential extraction approach using immunoblotting previously identified protein subsets based on their differential extractability (19). Therefore, we compared immunoblot and SC data for four proteins with different extraction profiles. The immunoblot and normalized SC data for BiP (GRP78), link protein (HPLN1), β-actin (ACTB), and decorin (PGS2) are shown in supplemental Fig. 2. The quantified relative abundance of the four proteins among the E0, E1, and E2 fractions determined by immunoblotting closely matched the relative protein abundance between extracts according to normalized SC values (Fig. 2).
Fig. 2.
Comparison of immunoblotting and spectral counting for quantitative analysis of relative protein abundance in cartilage extracts. Sequential extracts of 21-day femoral head cartilage (where E0 denotes the 1 m NaCl extract and E1 and E2 denote the filtrate and retentate, respectively after 100-kDa-molecular mass-cutoff centrifugal filtration of the 4 m GdnHCl extract) were analyzed by two independent semiquantitative methods. A, equal amounts of protein extract (3 μg/lane) were resolved by SDS-PAGE and probed with antibodies to BiP (GRP78_WB), link protein (HPLN1_WB), β-actin (ACTB_WB), and decorin (PGS2_WB). Background-subtracted volumes for each protein were calculated by densitometry and plotted as mean values (n = 3) on the y axis where error bars are S.E. p values adjusted for multiple correction are shown: *, p < 0.05; **, p < 0.01; and ***, p < 0.005. B, SC values corresponding to GRP78, HPLN1, ACTB, and PGS2 were normalized, mean values were plotted on the y axis, and significance was determined as described in A. The raw Western blot data and normalized SC values for each of the sample replicates are represented in supplemental Fig. 2.
Independent LC-MS/MS analysis of the 100-kDa-molecular mass-cutoff filtrate (E1) and retentate (E2) fractions of the GdnHCl extracts increases the number of proteins identified but does not reveal any additional information regarding protein solubility. Therefore, SC data corresponding to E1 and E2 fractions were combined using the Scaffold MudPIT (multidimensional protein identification technology) function, normalized, and reanalyzed to identify proteins differentially abundant in NaCl (E0) extracts and GdnHCl extracts (designated E3). The 253 proteins that were significantly differentially abundant (t test, p < 0.05) were almost equally partitioned between E0 and E3 extracts (46 and 54%, respectively). The complete list of proteins identified in 3-week mouse femoral head cartilage is included as supplemental Table 1A, and the list of significant differential NaCl- and GdnHCl-extracted proteins are presented separately in supplemental Table 1B.
Quantitative Label-free LC-MS/MS Analysis of Mouse Neocartilage Cultures
Neocartilage cultures accumulate a proteoglycan- and collagen-rich ECM with a concomitant increase in the amount of GdnHCl-extracted proteins relative to the more soluble NaCl-extracted proteins (Fig. 1). The small proportion of GdnHCl-extracted proteins in juvenile cartilage suggests either a lack of matrix protein expression or the absence of a tightly integrated ECM. Therefore, to investigate the process of ECM maturation and changes in chondrocyte protein expression in neocartilage cultures, we conducted large scale comparison of juvenile cartilage (P3) and 3-week neocartilage (T21) by quantitative LC-MS/MS.
Sequential E0, E1, and E2 extracts were prepared from P3 tissue (n = 4) and T21 cultures (n = 4) using the procedures described in the previous section, generating a total of 24 samples. The SDS-PAGE profiles of triplicate sequential extracts are shown in supplemental Fig. 3A. LC-MS/MS analysis of sequential extracts identified a total of 819 proteins of which 620 proteins and 706 proteins were identified in the P3 and T21 samples, respectively. Protein identifications were accepted on the basis of two or more peptides matching the Swiss-Prot database. The degree of overlap among sequential E0, E1, and E2 fractions from the two sample categories (P3 and T21) is illustrated as Venn diagrams (supplemental Fig. 3B). The main objective of this study was to profile the difference in protein abundance and, importantly, the extractability of proteins between juvenile cartilage and neocartilage cultures. Therefore, further analysis was based on four main sample categories: juvenile cartilage NaCl and GdnHCl extracts (P3E0 and P3E3) and neocartilage NaCl and GdnHCl extracts (T21E0 and T21E3) where E3 denotes combined spectral count data for E1 and E2 samples. Supplemental Table 2 provides full details of the peptides identified in all replicates (n = 4) of these sample categories.
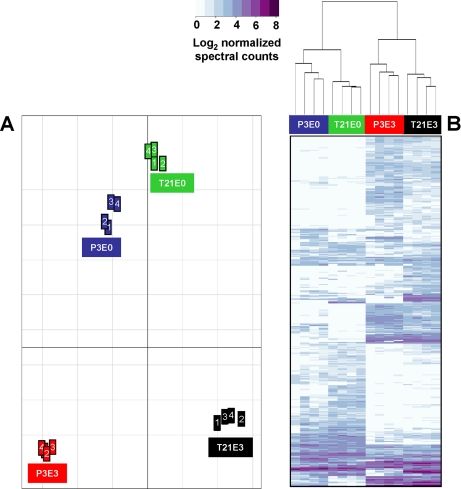
The next level of analysis was global assessment of the inter-relationships between biological replicates and the four sample categories. Principal component analysis of all the proteins identified in P3 and T21 samples revealed greatest similarity between the P3E0 and T21E0 samples and considerable difference from their respective E3 extracts, which were in turn different from each other (Fig. 3A). Protein quantitation by spectral counting is most robust for proteins above a certain level of abundance due to the semirandom sampling of low abundance peptides (20). Hierarchical clustering was therefore performed on the subset of proteins detected (≥2 peptides) in a minimum of four of the 16 samples (n = 385). Schematic representation of this filtered protein subset as a heat map (Fig. 3B) revealed relationships between the samples that were consistent with PCA of the whole data set with greatest similarity (first level of the dendrogram) between biological replicates and the next level of similarity between the E0 extracts of P3 and T21 samples and finally between E3 extracts of the P3 and T21 samples.
Fig. 3.
Statistical analysis of normalized spectral count values obtained from LC-MS of neocartilage and juvenile epiphyseal cartilage sequential extracts. A, principal component analysis of the full set of proteins identified at a high confidence level (n = 819) in the biological replicates (n = 4) of the four main sample categories: P3E0 and P3E3 denote NaCl-soluble and GdnHCl-soluble proteins extracted from juvenile cartilage, and T21E0 and T21E3 denote NaCl-soluble and GdnHCl-soluble proteins extracted from neocartilage. The placement of the groups is proportional to the divergence between the samples. B, hierarchical clustering of proteins, represented as a heat map depicting relative protein abundance (normalized SC values logged to base 2) of the protein list filtered to include only proteins detected in a minimum of four samples (n = 385).
Extraction Profiling of Juvenile Cartilage and Neocartilage
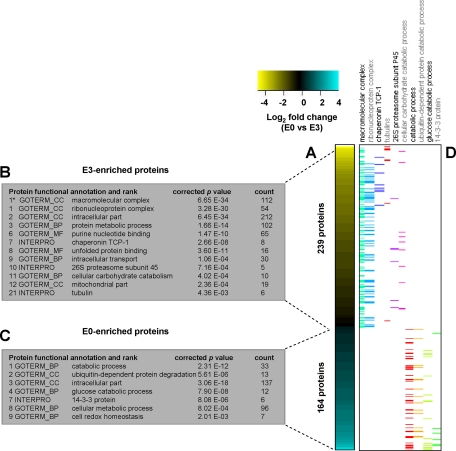
The results of PCA showed that “extract,” i.e. NaCl versus GdnHCl, accounted for the greatest distinction between the four sample categories. To further investigate the partitioning effect of our fractionation approach, extraction profiles of the juvenile cartilage and neocartilage were constructed based on proteins that showed significant differential abundance between E0 and E3 extracts independent of the sample type (n = 8 in each category). The 403 significant proteins (t test, p < 0.05) were ranked by average E0:E3 ratio and depicted as a heat map schematic (Fig. 4A).
Fig. 4.
Functional classification of significant proteins enriched in NaCl and GdnHCl extracts of juvenile epiphyseal cartilage and neocartilage. A, -fold changes were calculated for proteins detected in NaCl extracts (n = 8) and GdnHCl extracts (n = 8) of juvenile cartilage and neocartilage, generating a list of 403 differentially abundant (E0-enriched and E3-enriched, respectively) proteins (p < 0.05). The heat map depicts the -fold change values (logged to base 2) of significant proteins (listed in supplemental Table 3) from the most highly enriched proteins in the E3 extracts to the most highly enriched proteins in the E0 extracts. B and C, significant GO terms, including cell component (CC), molecular function (MF), and biological processes (BP), and protein families (INTERPRO) assigned to the E3- and E0-enriched protein subsets. Significant functional terms were ranked according to enrichment scores generated using the annotation clustering algorithm in DAVID at the highest stringency, apart from the broad term macromolecular complex (1*) identified by interrogating DAVID at high stringency. D, distribution of differentially abundant proteins (depicted as colored bars) associated with significant functional terms of interest.
Using the web-based integrated functional annotation algorithm DAVID (28), we identified significantly over-represented GO terms, protein families, and biological pathways in E0- and E3-enriched protein subsets (Fig. 4, B and C). The most highly ranked functional group in the 164 significant E0-enriched proteins was catabolic process, which encompassed the subcategories ubiquitin-dependent protein catabolism (13 proteins) and glucose catabolic process (12 proteins). Of the 239 significant E3-enriched proteins, 47% were classified as macromolecular complex, most of which could be grouped into the subcategories ribonucleoprotein (51 proteins), chaperonin T-complex (7 proteins), 26 S proteasomal subunits (five proteins), and tubulins (five proteins). The distribution of these proteins is indicated alongside the heat map in Fig. 4D. Several ECM proteins were significantly enriched in the E3 extracts, including proteins described by the InterPro terms von Willebrand factor, type A (MATN1, MATN3, MATN4, CO6A1, and CO6A2) and leucine-rich repeat (PGS1, EPYC, MIME, and PRELP). Details of the complete list of 403 significant proteins are provided in supplemental Table 3.
Classification of E0- and E3-enriched proteins according to the Kyoto Encyclopedia of Genes and Genomes pathway database (33) revealed some additional differences between the protein subsets. Of the ribosomal subunits identified in this study, 45 were significantly enriched in the E3 extracts (supplemental Fig. 4A) with an additional 12 ribosomal subunits detected in some of the replicate E3 samples but not reaching significance (p < 0.05). Another multisubunit complex, the 26 S proteasome, was detected as an over-represented functional group in the E0 extracts (p = 9.4e−9). These 11 proteins were exclusively the α and β subunits that comprise the 20 S core particle (supplemental Fig. 4B). In contrast, elements of the 19 S ATPase (PRS4, PRS6A, PRS7, PRS8, and PRS10) and non-ATPase (PSD7, PSD11, PSD12, PSMD2, and PSDE) regulatory particles were identified exclusively in the E3 extract. One further significant Kyoto Encyclopedia of Genes and Genomes pathway highlighted by our analysis of E0- and E3-enriched protein groups was glycolysis/gluconeogenesis (p = 1.04e−4) shown in supplemental Fig. 4C. Whereas most of the glycolytic enzymes were significantly enriched in the E0 extracts (LDHA, LDHB, ALDOA, ALDOC, PGK1, G6PI, TPIS, and PGAM1), other enzymes were significantly more abundant in the E3 extracts (ODPB, G3P, KPYM, and K6PL).
Detailed Analysis of Protein Abundance Differences in Neocartilage Relative to Juvenile Cartilage
The second component distinguishing the sample categories was sample type, i.e. 3-week neocartilage versus P3 cartilage (Fig. 3). Therefore, to generate a protein signature of mouse neocartilage formation, we identified the significant protein differences between the P3 and T21 samples, independent of extraction condition (n = 8 in each category, p < 0.05). The resulting subset of 122 differentially abundant proteins (details provided in supplemental Table 4) is smaller than the E0 versus E3 comparison as expected from PCA and hierarchical clustering. DAVID was used to identify significant protein functional groups over-represented in the P3- and T21-enriched protein cohorts.
Many proteins significantly enriched in the T21 samples compared with the P3 samples were defined by the GO term extracellular region (27 proteins, p = 3.7e−3) with subsets classed as cell adhesion proteins (15 proteins, p = 3.3e−4) and/or proteinaceous ECM (12 proteins, p = 5.9e−5). The second major functional group was cytoskeletal proteins (14 proteins, p = 3.8e−4) of which eight were actin-binding proteins (p = 3.2e−3). One protein family, the annexins, was also significantly over-represented in the T21 subgroup (four proteins, p = 3.3e−3).
Significant functional terms that applied to proteins enriched in juvenile cartilage relative to the neocartilage included transport (21 proteins, p = 3.3e−3) and endoplasmic reticulum (15 proteins, p = 1.2e−4). Of these proteins, four were related specifically to ER to Golgi vesicle-mediated protein transport, although this term was not statistically significant after adjustment for multiple testing. A further category of P3-enriched proteins was classified as extracellular matrix (nine proteins, p = 4.3e−3). Tables I, II, and III list the details for these 76 differentially abundant proteins, grouped according to their relative abundance and significant protein functional terms.
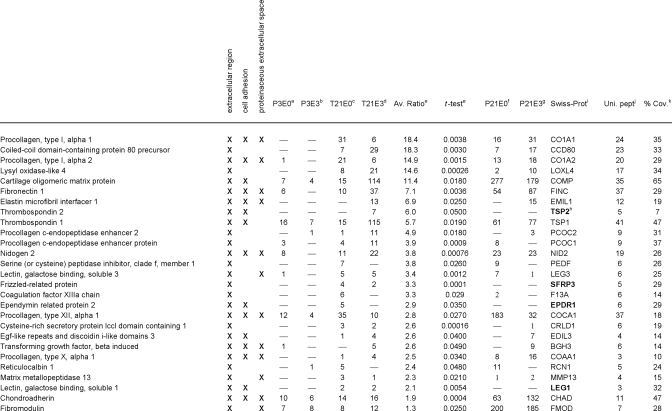
Table I. Proteins significantly increased in 3-week neo-cartilage relative to juvenile cartilage, classified into the functional groups: extracellular region, cell adhesion and proteinaceous extracellular space.
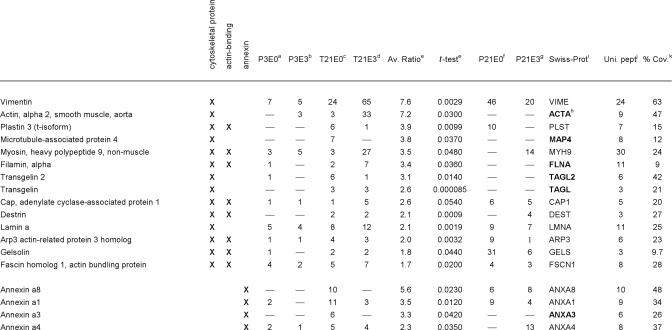
Table II. Proteins significantly increased in 3-week neo-cartilage relative to juvenile cartilage, classified into the functional groups: cytoskeletal proteins, actin-binding proteins and annexins.
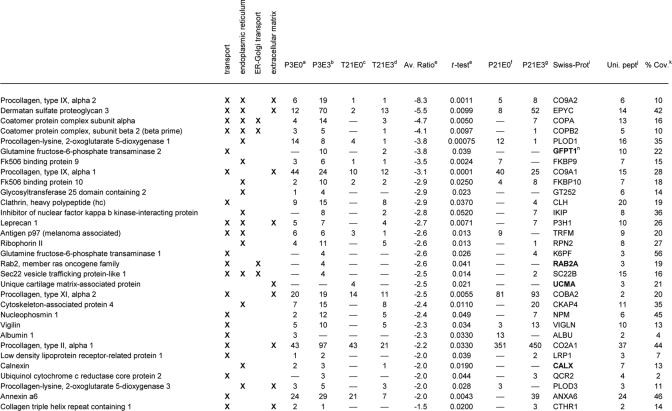
Table III. Proteins significantly decreased in 3-week neo-cartilage relative to juvenile cartilage, classified into the functional groups: transport, endoplasmic reticulum, ER-Golgi transport and extracellular matrix.
a–d Mean normalized SC values for proteins identified in juvenile cartilage NaCl extract (P3E0), juvenile cartilage GuHCl extract (P3E3), neo-cartilage NaCl extract (T21E0) and neo-cartilage GuHCl extract (T21E3), respectively.
e Fold difference (T21/P3) and significance (t-test p value) of the difference in mean normalized SC values.
f–g Mean normalized SC values, where the proteins listed were also identified in 3-week femoral head cartilage NaCl extracts (P21E0) or GuHCl extracts (P21E3).
h Proteins absent from 3-week femoral head cartilage are highlighted in bold.
i Protein identification according to the Swiss-Prot database, taxonomy mouse.
j Maximum number of unique peptides identified.
k Amino acid sequence coverage for the identified protein.
To determine how closely the protein signature of 3-week neocartilage resembles that of 3-week-old cartilage in vivo, significant proteins were cross-referenced against the proteins identified in 3-week-old femoral head cartilage extraction profiles generated in our preliminary study (P21E0 and P21E3 proteins, supplemental Table 1). Of the 27 proteins enriched in the neocartilage classed as extracellular (Table I), 22 were also detected in E0 and/or E3 extracts prepared from P21 femoral head cartilage. The five exceptions (TSP2, SFRP, CRLD1, EPDR1, and LEG1) appeared to be neocartilage-specific as they were undetected in P3 and P21 samples. Of the T21-enriched proteins classed as cytoskeletal (Table II), nine were also detected in P21 extracts, the exceptions being ACTA, FLNA, MAP4, TAGL, and TAGL2. Finally, of the annexins (Table II), all but ANXA3 were identified in P21 samples. A global statistical evaluation of the entire P3, T21, and P21 data set is beyond our core aim of identifying changes in protein abundance and extractability associated with neocartilage maturation. However, comparison of the subset of significant proteins described provides a useful snapshot of key similarities and differences between 3-week neocartilage cultures and 3-week femoral head cartilage.
Analysis of Protein Solubility Changes in Neocartilage compared with Juvenile Cartilage
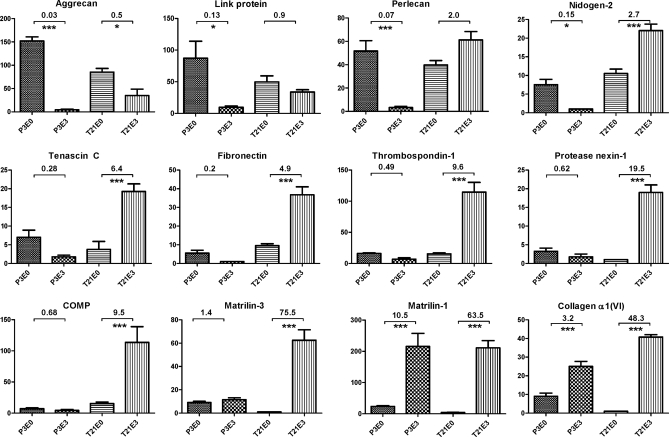
Our results have shown that cytoskeletal, matrix, and adhesion proteins are highly enriched in neocartilage relative to juvenile cartilage. These protein abundance changes are consistent with production of a cartilaginous matrix and increased cell-matrix interactions. The maturation of the cartilaginous ECM is likely to involve the formation of increasingly insoluble networks between multiple components. Therefore, to target proteins involved in this maturation process, we identified components with the greatest difference in extractability between the P3 juvenile cartilage and T21 neocartilage samples. To achieve this, the average E3:E0 ratios were calculated for all proteins detected in both P3 and T21 samples and ranked according to the greatest difference in E3:E0 ratio between the two sample types (supplemental Table 5). As anticipated, the most highly ranked proteins were all matrix components (Fig. 5). Matrilin-3 (ranked first) was detected at similar levels in the E0 and E3 extracts of P3 samples but had an average E3:E0 ratio of 75.5 in the neocartilage samples. Perlecan, protease nexin-1, fibronectin-1, thrombospondin-1, nidogen-2, and tenascin C (ranked second to seventh, respectively) had E3:E0 ratios <1 in P3 samples and >1 in T21 samples, strong evidence for differential solubility of these proteins in juvenile cartilage compared with 3-week neocartilage. Both collagen VI (ranked eighth) and matrilin-1 (ranked 22nd) were significantly enriched in the E3 extracts of both sample types, whereas aggrecan (ranked ninth) was significantly enriched in the E0 extracts of both P3 and T21 samples. However, the trend for all of these matrix components was toward decreased extractability in T21 samples, consistent with roles in maturation of the cartilage ECM.
Fig. 5.
Differential solubility properties of proteins extracted from juvenile epiphyseal cartilage and neocartilage. Differences in protein solubility between P3 and T21 samples were calculated based on the greatest difference between E3:E0 ratios of the two sample types (ranked in supplemental Table 5). The 10 highest ranking proteins (matrilin-3, perlecan, protease nexin-1, fibronectin, thrombospondin-1, nidogen-2, tenascin C, collagen α1(VI), aggrecan, and cartilage oligomeric matrix protein (COMP)) and proteins ranked 21st and 22nd (link protein and matrilin-1) are depicted. Normalized SC values for these proteins were analyzed by one-way ANOVA and plotted on the y axis of each graph as mean values (n = 4) where error bars are S.E. Corrected p values are shown for the comparisons P3E0 versus P3E3 and T21E0 versus T21E3: *, p < 0.05; **, p < 0.01; and ***, p < 0.005.
Differential Processing of Matrilin-3 and Aggrecan Isoforms in Neocartilage and Juvenile Cartilage
In addition to major differences in protein abundance and solubility between the P3 and T21 samples, examination of the peptide coverage data also revealed very specific qualitative differences that indicated potentially important processing of two significant matrix components. In the case of matrilin-3, the peptides detected in neocartilage samples spanned up to 80% of the sequence of the secreted protein, corresponding to 34–55% coverage. In contrast, the matrilin-3 peptides detected in juvenile cartilage extracts mapped exclusively to the N-terminal half of the sequence, corresponding to only 16–25% coverage (supplemental Fig. 5, A–D). Analysis of peptide data also revealed differences with respect to aggrecan isoforms detected in P3 and T21 samples. As regions of the aggrecan core protein are heavily substituted with chondroitin and keratan sulfate chains, peptide identifications are generally limited to the G1, G2, and G3 globular domains even after enzymatic removal of the glycosaminoglycan chains (34). Peptides from all three globular domains and the CS-1 region were detected in neocartilage extracts, indicating the presence of intact aggrecan. In contrast, absolutely no peptides derived from the G3 domain were detected in juvenile cartilage even though between 1.3 and 2.2% of the total spectra recorded for the P3 NaCl extracts correspond to G1- and G2-derived peptides (supplemental Fig. 5, E–H). These examples highlight striking differences in the post-translational processing of matrilin-3 and aggrecan, generating isoforms with potentially very different properties and/or functions in juvenile cartilage and neocartilage.
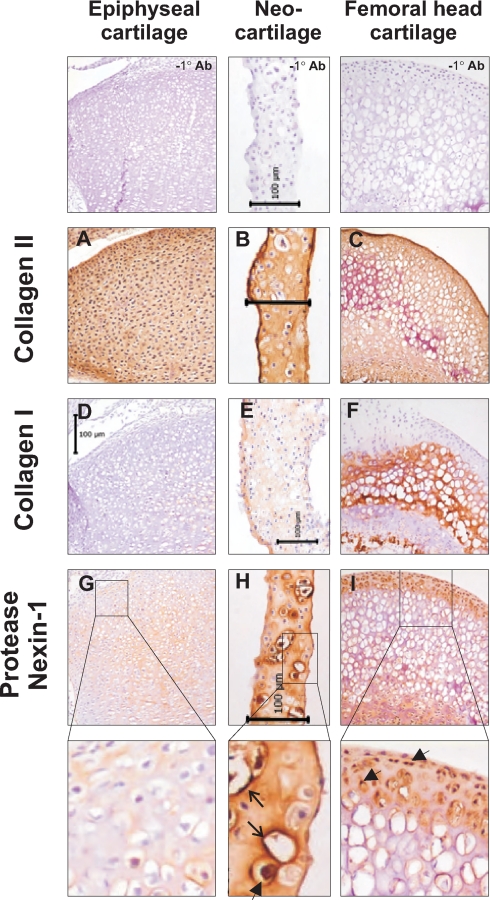
Immunohistochemical Analysis of Protease Nexin-1
The proteins that showed significant differences in solubility between neocartilage and juvenile cartilage were mostly well known cartilage matrix proteins. After matrilin-3 and perlecan, the third ranked protein was protease nexin-1 (PN-1; or glia-derived nexin), a serine protease inhibitor identified in cartilage anlagen during embryogenesis (35) but with no known role in articular or growth plate cartilage. The solubility profile observed for PN-1 suggests a possible interaction with other matrix components during maturation of the neocartilage ECM. Therefore, the localization of PN-1 in neocartilage, 3-day epiphyseal cartilage, and 21-day femoral head cartilage was investigated by immunohistochemistry.
Regional differences in PN-1 expression were assessed in relation to collagen II as a typical hyaline cartilage marker and collagen I, which was previously shown to mark the surface zone of neocartilage (9). As expected, intense collagen II staining was detected throughout the neocartilage, epiphyseal cartilage, and femoral head cartilage (Fig. 6, A–C), whereas collagen I staining was absent in epiphyseal cartilage and specific to the calcified center of the femoral head cartilage. In neocartilage, staining for collagen I was more restricted to the surface zone with some diffuse staining in the underlying tissue (Fig. 6, D–F). In epiphyseal cartilage, weak staining for PN-1 was detected in the interterritorial and pericellular matrix around some cells. In contrast, PN-1 was detected throughout the matrix of neocartilage and in femoral head cartilage with strong cell-associated and pericellular staining of some cells. The transition of PN-1 from more soluble in juvenile epiphyseal cartilage to less soluble in neocartilage suggests tight association with the neocartilage ECM. Immunohistochemical analysis suggests that PN-1 interacts strongly with both extracellular and intracellular cartilage components.
Fig. 6.
Expression and localization of collagen I, collagen II, and protease nexin-1 determined by immunohistochemistry. Cryosections of 3-day mouse knee joints, 3-week neocartilage, and 3-week-old mouse femoral heads were incubated with antibodies to collagen II (A–C), collagen I (D–F), and protease nexin-1 (G and H) using identical antibody incubations and color development for each tissue. The region of the 3-day mouse knee joint depicted is the proximal tibial epiphyseal cartilage. Positive immunolocalization of proteins is indicated by brown staining and tissue histology and nuclear localization are indicated by blue-violet hematoxylin counterstain. No immunoreactive staining was observed when primary antibodies were omitted from either the rabbit or mouse IgG detection protocol. Representative images of mouse antibody immunodetection controls are shown (−1° Ab). Each scale bar represents 100 microns. Expanded regions of protease nexin-1 images reveal intense pericellular (open arrows) and cell-associated (arrowheads) staining.
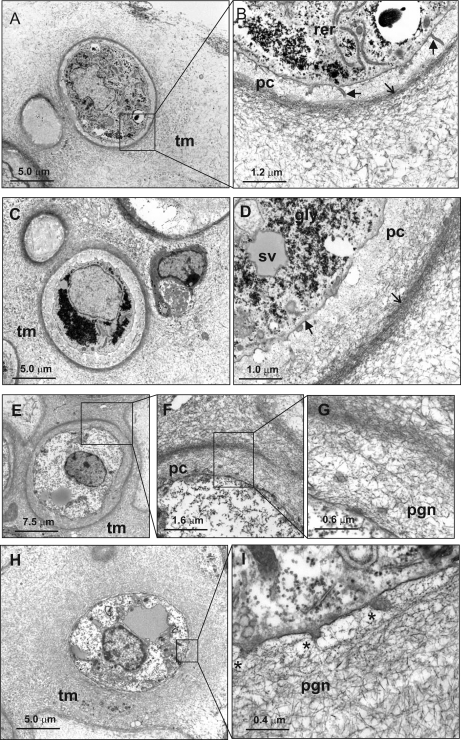
Neocartilage Ultrastructure Analysis by Transmission Electron Microscopy
Our proteomics analysis revealed complex changes in protein composition and extractability of mouse 3-week-old neocartilage relative to 3-day juvenile cartilage. Although the enrichment of GdnHCl-extracted proteins in neocartilage implies production and assembly of cartilage ECM components, it provides no information regarding the organization of these matrix components, critically important for cartilage function. Therefore, neocartilage cultures were examined at the ultrastructural level by transmission electron microscopy (Fig. 7).
Fig. 7.
Analysis of mouse neocartilage ultrastructure. A–D, ultrathin sections of 3-week mouse neocartilage cultures were examined by transmission electron microscopy after standard fixation in Karnovsky's reagent. E–I, TEM analysis of 3-week neocartilage after fixation in 0.7% safranin O. Low power micrographs display chondrocytes with a rounded or ovoid morphology (A, C, E, and H). Most chondrocytes are surrounded by a pericellular capsule (pc), defined at the outer boundary by a dense band of collagen fibrils (open arrows), which also appears to form a boundary with the territorial matrix (tm). Preservation of proteoglycans and glycosaminoglycans by fixation in safranin O reveals a dense proteoglycan network (pgn) that, at high magnification, can be observed in direct contact with the plasma membrane (I, *). Although generally smooth, finger-like protrusions are also observed projecting from the plasma membrane (B, arrowheads). Other distinctive features include abundant glycogen granules (gly), large secretory vesicles (sv), and rough endoplasmic reticulum (rer).
Many of the chondrocytes possessed a highly developed pericellular capsule with a band of densely bundled collagen fibers 1–1.5 μm from the cell surface, consistent with TEM of articular cartilage matrices in vivo (36). The region closest to the cell was occupied by only very fine filamentous material lacking the electron density of the territorial matrix with large banded collagen fibers occasionally observed adjacent to the cell surface (Fig. 7, A–D, and supplemental Fig. 6). Staining in the presence of the cationic dye safranin O revealed that this region is filled with an intricate, highly contrasted meshwork of proteoglycans/glycosaminoglycan. High magnification revealed striking organization of the proteoglycan network, very similar to that observed when authentic cartilage specimens are preserved using high pressure freezing (37) (Fig. 7, E–G). In some cases, collagen and proteoglycans formed a more homogeneous network radiating directly from the plasma membrane (Fig. 7, H and I). Although 3-week mouse neocartilage lacked the stratified arrangement of superficial, middle, and deep chondrocytes observed in other neocartilage systems (9), analysis at the ultrastructural level confirmed the presence of intricate cell-matrix contacts and clearly defined pericellular and territorial zones of cartilaginous matrix.
DISCUSSION
The dominance of poorly soluble matrix components in cartilage presents a challenge to proteomics analysis. However, the ECM is critical for cartilage function and is an important source of biomarkers of joint disease. In this study, we investigated the maturation of mouse neocartilage in vitro using a novel combination of sequential protein extraction and label-free quantitative MS. Our analysis using proteomics indicate that neocartilage formation involves production and integration of multiple ECM components into increasingly insoluble networks. Ultrastructural analysis of 3-week cultures supported our findings, revealing a neocartilaginous tissue composed of chondrocytes embedded in an ECM organized akin to that observed in cartilage. The development of a mouse neocartilage system, coupled with large scale quantitative proteomics, has generated a wealth of information on cartilage matrix biosynthesis and assembly with potential for further analysis of tissues engineered for autologous cartilage repair.
Cartilage Extraction Profiling
The cartilage extraction profiles generated in this study are, to our knowledge, the most detailed comparative analysis of a tissue proteome based on differential protein solubility to date. As expected, matrix proteins such as matrilin-1, -3, and -4 and leucine-rich repeat proteoglycans such as biglycan (PGS1) and prolargin (PRELP) were highly enriched in the guanidine-extracted (E3) fraction of neocartilage and juvenile cartilage. However, the partitioning of proteins according to extractability applied to both cellular and ECM components (supplemental Table 3). In particular, components of multisubunit complexes, such as the 40 and 60 S ribosomal subunits, were mostly E3 extract-specific, consistent with the stabilization of these complexes by strong non-covalent interactions. Unexpectedly, we observed complete separation of the 26 S proteasome regulatory particle subunits (E3-specific) from the core particle subunits (E0-specific), implying a difference in the strength of intersubunit interactions between the two complexes.
Extract-specific protein groups also reflected cellular localization. For example, a significant cluster of E3-enriched proteins was classified as mitochondrial (n = 19, p = 2.4e−4). These included components of the mitochondrial inner and outer membranes (ADT2, IDHP, THIL, VDAC1, VDAC2, and VDAC3), ATP synthase F0F1 complex (ATPA, ATPB, ATP5H, ATPG, and ATPO), and mitochondrial matrix (ECHA, ECHB, ETFA, and ETFB). Other extract-specific protein subsets were defined by their molecular function, e.g. enrichment of the 14-3-3 signal transduction proteins in the NaCl-extracted (E0) fraction (n = 6, p = 8.1e−6). In the E3 fraction, the significant enrichment term “unfolded protein binding” encompassed the cytosolic HSP90A and -B subunits, the ER-resident chaperone GRP94 (ENPL), and the mitochondrial stress-70 protein (GRP75). Peptides corresponding to T-complex chaperonin subunits (n = 8, p = 2.7e−8) were identified exclusively in the E3 extract, a strong indication of tight intersubunit interactions. In contrast, ERP29 and BiP (GRP78) were enriched in the E0 extract, whereas the 71-kDa heat shock cognate protein (HSP7C) was divided almost equally between NaCl and GdnHCl extracts. One possible explanation is that the relative solubility of different chaperones is influenced by their target binding properties. In the case of HSC7C, these properties may differ according to the activity of distinct nuclear and cytoplasmic protein pools.
The identification of many glycolytic enzymes in juvenile cartilage and neocartilage reflects the primary role of glycolysis in chondrocyte energy metabolism (38). Somewhat surprising was the distribution of glycolytic enzymes between E0 and E3 fractions (supplemental Fig. 4C). However, the sequestration and tight association of glycolytic enzymes to actin filaments and microtubules are reported to alter cytoskeletal organization and may compartmentalize energy production (39–41). In muscle cells, direct or indirect binding to actin is observed for most if not all glycolytic enzymes but appears to be enzyme-specific and temporally regulated in other cell types. In our study, the enrichment of G3P, but not PGAM1 or LDH, in the guanidine extract of both P3 and T21 samples is the same pattern of differential solubility observed during morphogenesis of epithelial cells in vitro (42).
Differential Protein Expression in Mouse Neocartilage Cultures
Some of the most prominent differences in protein abundance between juvenile cartilage and neocartilage were related to collagen biosynthesis, processing, and maturation. For example, the procollagen C-endopeptidase enhancer proteins PCOLCE and PCOLCE 2, which facilitate propeptide processing and collagen fibrillogenesis, were both increased in neocartilage (Table I, PCOC1 and PCOC2). Accumulation of pepsin-resistant fibrillar collagens II and XI (Fig. 1D) with concomitant reduction in nascent collagen II and XI chains by 1.5–2-fold (Table II) was another indication of collagen deposition and maturation. Although collagen I was a minor fraction of the pepsin-resistant fibrillar collagen in 3-week neocartilage, nascent collagen I chains were detected in neocartilage E0 and E3 extracts (Table I). As shown in bovine neocartilage (9), collagen I staining was most prominent at the neocartilage surface (Fig. 6). Further analysis of long term cultures will determine whether these collagen I-expressing cells represent dedifferentiated chondrocytes or more closely resemble superficial zone chondrocytes.
In addition to changes in fibrillar collagens, we detected differential abundance of FACIT (fibril-associated collagens with interrupted triple helices) collagens IX and XII. Essential for cartilage development (43), collagen IX contributes to matrix stability by cross-linking collagen II fibrils (44) and via interactions with COMP and matrilin-3 (45). Although collagen IX (CO9A1 and CO9A2) was reduced in E0 and E3 extracts of neocartilage relative to P3 cartilage, detection of pepsin-resistant collagen IX polypeptides (Fig. 1D) signified ongoing collagen IX synthesis and incorporation into the matrix. The function of collagen XII in cartilage is less well known, although it shows distinctive co-localization with collagen II toward the articular surface as opposed to underlying epiphyseal cartilage and within the prehypertrophic and hypertrophic zones of the growth plate (46). Collagen XII is also known to associate with collagen I (47), and in mouse neocartilage, collagen XII distribution was similar to that of collagen I (data not shown).
Collagen X was one of several markers of chondrocyte hypertrophy identified in mouse neocartilage cultures, albeit in low amounts compared with collagen II. Other putative markers of chondrocyte hypertrophy included coiled-coil domain-containing protein 80 (CCD80) (48), lysyl oxidase C (LOXL4) (49), tissue transglutaminase-2 (F13A) (50), galectin-3 (LEG3) (51), and annexin 8 (ANXA8) (52). Further analysis of these proteins in mouse neocartilage may shed insight into their roles in hypertrophic chondrocytes in vivo as these proteins were also detected in 3-week mouse femoral head cartilage (supplemental Table 1A).
The most highly differentially abundant non-collagenous matrix protein in neocartilage was COMP, a protein intensively studied as a biomarker for cartilage degeneration and for the dominant negative effect of COMP mutations on cartilage development (45, 53). Two other members of the thrombospondin family, TSP1 and TSP2, were also induced in neocartilage, although their roles in cartilage matrix development and stability are less well understood (54).
The pericellular environment, or the chondron, is an active site of tissue repair in OA (55, 56). Pericellular matrix components such as nidogen-2, perlecan, and collagen VI were prominent in mouse neocartilage. Although collagen VI and biglycan, a known collagen VI binding partner, were poorly soluble, perlecan and nidogen-2 were detected as NaCl- and GdnHCl-soluble populations. This may reflect physically or biochemically distinct populations in the cartilage matrix, a question that could be addressed for example by immunogold TEM. Similarly, the small leucine-rich repeat proteoglycans chondroadherin and fibromodulin were divided between E0 and E3 extracts. In the case of chondroadherin, this may relate to distinct roles and tissue localization, i.e. integrin-bound at the cell surface but associated with collagen network in the territorial matrix (57). Other chondrocyte-associated proteins with potential roles in cell adhesion or modulating cell-matrix interactions in neocartilage cultures included galectin-1 (LEG1), a component of both articular and growth plate cartilage (58), and the integrin-binding protein Del1 (EDIL3), previously identified by 2-DE analysis of human articular cartilage (59) but with no ascribed function.
In addition to the pericellular matrix, intracellular structural networks are vital to sensing mechanical forces and mediating the response of cartilage to altered loading (60). Proteins related to organization of the actin, tubulin, and intermediate filament networks were also highly represented in 3-week neocartilage cultures. The most highly enriched protein in this group was the smooth muscle isoform of actin, more commonly associated with auricular cartilage but recently detected in the superficial zone of normal human articular cartilage and increased at sites of tissue repair (61, 62). The actin-bundling proteins fascin-1 and plastin-3 and actin-depolymerizing/capping proteins destrin and gelsolin were detected in both neocartilage and femoral head cartilage extracts, indicating potential roles in actin organization and chondrocyte morphology in vivo. Other actin-associated proteins were detected in neocartilage but not in femoral head cartilage. Transgelin-1 and -2 have been reported previously in human chondrocyte cultures and may be related to morphological changes associated with monolayer culture (63). The complex relationship between the cytoskeleton, chondrocyte morphology, and cartilage-specific gene expression remains to be fully elucidated (64). However, vimentin is emerging as a key player in both regulating SOX genes and the cartilage markers COL2A1 and AGC during chondrogenesis in vitro (65) and in cytoskeletal reorganization in OA human chondrocytes (66). The induction of cytoskeletal elements such as vimentin, together with cell adhesion and pericellular components, indicates that development of the ECM and cell-matrix interface in neocartilage is highly coordinated.
Differential Extractability and Post-translational Processing of Matrix Components
The major strength of our experimental approach was the ability to compare the solubility properties of proteins as well as protein abundance. Most of the proteins that followed the transition from “more soluble” in juvenile cartilage to “less soluble” in neocartilage were well known cartilage matrix components (Fig. 5). One protein not previously associated with the cartilage ECM was PN-1 (glia-derived nexin), detected in the NaCl- and GdnHCl-soluble extracts of P3 cartilage but only in the guanidine extract of neocartilage and femoral head cartilage (supplemental Tables 3 and 1B). Further analysis by immunohistochemistry revealed both intracellular and matrix-associated staining for PN-1, consistent with its role in binding and inactivation of extracellular proteases followed by internalization (67). The activity of PN-1 is regulated by several matrix components, including fibronectin and cell surface heparan sulfate (68, 69), both potential ligands in cartilage. Given the role of serine proteases in cartilage pathology (13, 70), endogenous serine protease inhibitors such as PN-1 are potentially important therapeutic targets.
A further novel finding was the differential processing of matrilin-3 and aggrecan. In neocartilage, matrilin-3 was detected exclusively in the guanidine extract, consistent with tight interactions with matrix components such as collagen II and aggrecan (71). Matrilin-3 oligomer formation is mediated via C-terminal coiled-coil domains, whereas intermolecular interactions form via N-terminal von Willebrand factor A domains. Therefore, proteolytic processing will profoundly affect matrilin binding properties and generate matrilin isoforms with potentially different functions (72, 73). We identified a novel processed form of matrilin-3, based on the finding that peptides derived from matrilin-3 in juvenile cartilage map exclusively to the von Willebrand factor A domain (residues 71–257; Ref. 74) (supplemental Fig. 5, A–D). This putative cleavage product is clearly distinct from those generated by ADAMTS (a disintegrin and metalloprotease with thrombospondin motifs)-mediated processing of matrilin in the hinge region adjacent to the coiled-coil domain (75, 76).
Stabilization of the aggrecan network in cartilage involves strong non-covalent interactions with link protein and hyaluronan at the N-terminal G1 globular domain and G3-mediated interactions at the C terminus that may be influenced by alternative splicing (77, 78). Given the abundance of aggrecan and link protein in cartilage, it was not surprising that these two components generated the highest spectral count values in juvenile cartilage. What we did not anticipate was that aggrecan in juvenile cartilage completely lacked the G3 domain, indicated by the absence of peptides corresponding to this region of the core protein (supplemental Fig. 5, E–H). This was in stark contrast to the neocartilage extracts in which G3 domain of aggrecan was detected in all samples analyzed. The release of G3-containing aggrecan fragments in vitro and in vivo is a well documented indication of cartilage breakdown (34, 79). Whether aggrecan network formation during early development of the cartilage ECM also involves processing of the G3 domain is a subject for further investigation.
Benefits and Future Applications of Our Experimental Approach
In this study, we identified 819 cartilage proteins using solubility-based protein fractionation and LTQ-Orbitrap MS/MS. This compares favorably with recent analysis of human articular cartilage where proteins were resolved by SDS-PAGE prior to LTQ MS/MS (12). However, our analytical platform offers two advantages. First, separating proteins into fewer fractions reduces cost and sample handling associated with SDS-PAGE and in-gel digestion. Second, solubility-based fractionation retains important information related to the biophysical properties of the proteins, allowing differences in protein solubility to be interpreted in the context of the biological system being studied. This approach may be particularly valuable in other tissues dominated by poorly soluble matrix components such as meniscus and tendon.
This study is the first to use proteomics for comprehensive biochemical analysis of cartilage engineered in vitro, leading to identification of a cohort of proteins involved in chondrocyte metabolism, cell-matrix interactions, and maturation of the cartilage ECM. Although the neocartilage produced by mouse chondrocytes lacked the zonal organization of bovine neocartilage (9), the benefit of developing a mouse system is the potential for further analysis of models of inherited cartilage disorders. Sequential extraction and proteomics studies in mouse neocartilage lacking novel or important matrix components or incorporating proteins harboring dominant negative mutations will provide information on the role of specific cartilage components and the effect of disease-causing mutations on cartilage function in vitro and in vivo. Furthermore, our investigation has paved the way for proteomics of neocartilage grafts generated from human chondrocytes or chondroprogenitor cells. Comprehensive analysis of human neocartilage developed for autologous transplantation will generate important insight into both chondrocyte protein expression and properties of the neocartilage extracellular matrix.
Acknowledgments
We thank Kumara Kaluarachchi for expert advice in immunohistochemistry and Amanda Fosang for critical reading of the manuscript.
* This work was supported by National Health and Medical Research Council of Australia Grant 419237.
 This article contains supplemental Tables 1–5 and Figs. 1–6.
This article contains supplemental Tables 1–5 and Figs. 1–6.
1 The abbreviations used are:
- ECM
- extracellular matrix
- OA
- osteoarthritis
- PN-1
- protease nexin-1
- GdnHCl
- guanidine hydrochloride
- LTQ
- linear trap quadrupole
- SC
- spectral count
- 2-DE
- two-dimensional electrophoresis
- DAVID
- database for annotation, visualization, and integrated discovery
- PCA
- principal component analysis
- DMEM
- Dulbecco's modified Eagle's medium
- P3
- 3-day postnatal
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- ANOVA
- analysis of variance
- GO
- gene ontology
- ER
- endoplasmic reticulum
- LDH
- lactate dehydrogenase
- ACT
- actin
- TEM
- transmission electron microscopy
- PCOLCE
- procollagen C-endopeptidase enhancer protein
- COMP
- cartilage oligomeric matrix protein
- BiP
- binding immunoglobulin protein
- EPYC
- epiphycan
- MIME
- mimecan
- PRELP
- prolargin
- PSDE
- 26S proteasome non-ATPase regulatory subunit 14
- TPIS
- triosephosphate isomerase
- ODPB
- pyruvate dehydrogenase E1 component subunit beta, mitochondrial
- ALDO
- fructose-bisphosphate aldolase
- KPYM
- pyruvate kinase isozymes M1/M2
- SFRP
- secreted frizzled-related protein 3
- FLNA
- filamin-A
- TAGL
- transgelin
- IDHP
- isocitrate dehydrogenase [NADP], mitochondrial
- THIL
- acetyl-CoA acetyltransferase, mitochondrial
- ATPA
- ATP synthase subunit alpha, mitochondrial
- ECHA
- trifunctional enzyme subunit alpha.
REFERENCES
- 1.Martel-Pelletier J., Boileau C., Pelletier J. P., Roughley P. J. (2008) Cartilage in normal and osteoarthritis conditions. Best Pract. Res. Clin. Rheumatol 22, 351–384 [DOI] [PubMed] [Google Scholar]
- 2.Newman B., Wallis G. A. (2003) Skeletal dysplasias caused by a disruption of skeletal patterning and endochondral ossification. Clin. Genet 63, 241–251 [DOI] [PubMed] [Google Scholar]
- 3.Kannu P., Bateman J. F., Belluoccio D., Fosang A. J., Savarirayan R. (2009) Employing molecular genetics of chondrodysplasias to inform the study of osteoarthritis. Arthritis Rheum 60, 325–334 [DOI] [PubMed] [Google Scholar]
- 4.Aigner T., Bartnik E., Sohler F., Zimmer R. (2004) Functional genomics of osteoarthritis: on the way to evaluate disease hypotheses. Clin. Orthop. Relat. Res 427 (suppl.) S138–S143 [PubMed] [Google Scholar]
- 5.Goldring M. B. (2006) Update on the biology of the chondrocyte and new approaches to treating cartilage diseases. Best Pract. Res. Clin. Rheumatol 20, 1003–1025 [DOI] [PubMed] [Google Scholar]
- 6.van Osch G. J., Brittberg M., Dennis J. E., Bastiaansen-Jenniskens Y. M., Erben R. G., Konttinen Y. T., Luyten F. P. (2009) Cartilage repair: past and future—lessons for regenerative medicine. J. Cell. Mol. Med 13, 792–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benya P. D., Padilla S. R., Nimni M. E. (1978) Independent regulation of collagen types by chondrocytes during the loss of differentiated function in culture. Cell 15, 1313–1321 [DOI] [PubMed] [Google Scholar]
- 8.Liu G., Kawaguchi H., Ogasawara T., Asawa Y., Kishimoto J., Takahashi T., Chung U. I., Yamaoka H., Asato H., Nakamura K., Takato T., Hoshi K. (2007) Optimal combination of soluble factors for tissue engineering of permanent cartilage from cultured human chondrocytes. J. Biol. Chem 282, 20407–20415 [DOI] [PubMed] [Google Scholar]
- 9.Hayes A. J., Hall A., Brown L., Tubo R., Caterson B. (2007) Macromolecular organization and in vitro growth characteristics of scaffold-free neocartilage grafts. J. Histochem. Cytochem 55, 853–866 [DOI] [PubMed] [Google Scholar]
- 10.Wilson R., Whitelock J. M., Bateman J. F. (2009) Proteomics makes progress in cartilage and arthritis research. Matrix Biol 28, 121–128 [DOI] [PubMed] [Google Scholar]
- 11.Hermansson M., Sawaji Y., Bolton M., Alexander S., Wallace A., Begum S., Wait R., Saklatvala J. (2004) Proteomic analysis of articular cartilage shows increased type II collagen synthesis in osteoarthritis and expression of inhibin betaA (activin A), a regulatory molecule for chondrocytes. J. Biol. Chem 279, 43514–43521 [DOI] [PubMed] [Google Scholar]
- 12.Wu J., Liu W., Bemis A., Wang E., Qiu Y., Morris E. A., Flannery C. R., Yang Z. (2007) Comparative proteomic characterization of articular cartilage tissue from normal donors and patients with osteoarthritis. Arthritis Rheum 56, 3675–3684 [DOI] [PubMed] [Google Scholar]
- 13.Chamberland A., Wang E., Jones A. R., Collins-Racie L. A., LaVallie E. R., Huang Y., Liu L., Morris E. A., Flannery C. R., Yang Z. (2009) Identification of a novel HtrA1-susceptible cleavage site in human aggrecan: evidence for the involvement of HtrA1 in aggrecan proteolysis in vivo. J. Biol. Chem 284, 27352–27359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruiz-Romero C., Calamia V., Mateos J., Carreira V., Martínez-Gomariz M., Fernández M., Blanco F. J. (2009) Mitochondrial dysregulation of osteoarthritic human articular chondrocytes analyzed by proteomics: a decrease in mitochondrial superoxide dismutase points to a redox imbalance. Mol. Cell. Proteomics 8, 172–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Ceuninck F., Berenbaum F. (2009) Proteomics: addressing the challenges of osteoarthritis. Drug Discov. Today 14, 661–667 [DOI] [PubMed] [Google Scholar]
- 16.Little C. B., Meeker C. T., Golub S. B., Lawlor K. E., Farmer P. J., Smith S. M., Fosang A. J. (2007) Blocking aggrecanase cleavage in the aggrecan interglobular domain abrogates cartilage erosion and promotes cartilage repair. J. Clin. Investig 117, 1627–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glasson S. S. (2007) In vivo osteoarthritis target validation utilizing genetically-modified mice. Curr. Drug Targets 8, 367–376 [DOI] [PubMed] [Google Scholar]
- 18.Pecora F., Forlino A., Gualeni B., Lupi A., Giorgetti S., Marchese L., Stoppini M., Tenni R., Cetta G., Rossi A. (2007) A quantitative and qualitative method for direct 2-DE analysis of murine cartilage. Proteomics 7, 4003–4007 [DOI] [PubMed] [Google Scholar]
- 19.Wilson R., Bateman J. F. (2008) A robust method for proteomic characterization of mouse cartilage using solubility-based sequential fractionation and two-dimensional gel electrophoresis. Matrix Biol 27, 709–712 [DOI] [PubMed] [Google Scholar]
- 20.Old W. M., Meyer-Arendt K., Aveline-Wolf L., Pierce K. G., Mendoza A., Sevinsky J. R., Resing K. A., Ahn N. G. (2005) Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics 4, 1487–1502 [DOI] [PubMed] [Google Scholar]
- 21.de Godoy L. M., Olsen J. V., Cox J., Nielsen M. L., Hubner N. C., Fröhlich F., Walther T. C., Mann M. (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455, 1251–1254 [DOI] [PubMed] [Google Scholar]
- 22.Slebos R. J., Brock J. W., Winters N. F., Stuart S. R., Martinez M. A., Li M., Chambers M. C., Zimmerman L. J., Ham A. J., Tabb D. L., Liebler D. C. (2008) Evaluation of strong cation exchange versus isoelectric focusing of peptides for multidimensional liquid chromatography-tandem mass spectrometry. J. Proteome Res 7, 5286–5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson R., Belluoccio D., Bateman J. F. (2008) Proteomic analysis of cartilage proteins. Methods 45, 22–31 [DOI] [PubMed] [Google Scholar]
- 24.Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 25.Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 26.Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Hornik K., Hothorn T., Huber W., Iacus S., Irizarry R., Leisch F., Li C., Maechler M., Rossini A. J., Sawitzki G., Smith C., Smyth G., Tierney L., Yang J. Y., Zhang J. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Culhane A. C., Thioulouse J., Perrière G., Higgins D. G. (2005) MADE4: an R package for multivariate analysis of gene expression data. Bioinformatics 21, 2789–2790 [DOI] [PubMed] [Google Scholar]
- 28.Huang D. W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 29.Hochberg Y., Benjamini Y. (1990) More powerful procedures for multiple significance testing. Stat. Med 9, 811–818 [DOI] [PubMed] [Google Scholar]
- 30.Engfeldt B., Reinholt F. P., Hultenby K., Widholm S. M., Müller M. (1994) Ultrastructure of hypertrophic cartilage: histochemical procedures compared with high pressure freezing and freeze substitution. Calcif. Tissue Int 55, 274–280 [DOI] [PubMed] [Google Scholar]
- 31.Mendler M., Eich-Bender S. G., Vaughan L., Winterhalter K. H., Bruckner P. (1989) Cartilage contains mixed fibrils of collagen types II, IX, and XI. J. Cell Biol 108, 191–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fernandes R. J., Weis M., Scott M. A., Seegmiller R. E., Eyre D. R. (2007) Collagen XI chain misassembly in cartilage of the chondrodysplasia (cho) mouse. Matrix Biol 26, 597–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanehisa M., Goto S., Furumichi M., Tanabe M., Hirakawa M. (2010) KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res 38, D355–D360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stevens A. L., Wishnok J. S., White F. M., Grodzinsky A. J., Tannenbaum S. R. (2009) Mechanical injury and cytokines cause loss of cartilage integrity and up-regulate proteins associated with catabolism, immunity, inflammation, and repair. Mol. Cell. Proteomics 8, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mansuy I. M., van der Putten H., Schmid P., Meins M., Botteri F. M., Monard D. (1993) Variable and multiple expression of Protease Nexin-1 during mouse organogenesis and nervous system development. Development 119, 1119–1134 [DOI] [PubMed] [Google Scholar]
- 36.Poole C. A., Flint M. H., Beaumont B. W. (1984) Morphological and functional interrelationships of articular cartilage matrices. J. Anat 138, 113–138 [PMC free article] [PubMed] [Google Scholar]
- 37.Hunziker E. B., Schenk R. K. (1984) Cartilage ultrastructure after high pressure freezing, freeze substitution, and low temperature embedding. II. Intercellular matrix ultrastructure: preservation of proteoglycans in their native state. J. Cell Biol 98, 277–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bowie M. A., Rosenthal O., Wagoner G. (1941) Observations on respiration in articular cartilage. Ann. Rheum. Dis 2, 216–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minaschek G., Gröschel-Stewart U., Blum S., Bereiter-Hahn J. (1992) Microcompartmentation of glycolytic enzymes in cultured cells. Eur. J. Cell Biol 58, 418–428 [PubMed] [Google Scholar]
- 40.Knull H. R., Walsh J. L. (1992) Association of glycolytic enzymes with the cytoskeleton. Curr. Top. Cell. Regul 33, 15–30 [DOI] [PubMed] [Google Scholar]
- 41.Waingeh V. F., Gustafson C. D., Kozliak E. I., Lowe S. L., Knull H. R., Thomasson K. A. (2006) Glycolytic enzyme interactions with yeast and skeletal muscle F-actin. Biophys. J 90, 1371–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cao F., Yanagihara N., Burke J. M. (1999) Progressive association of a “soluble” glycolytic enzyme with the detergent-insoluble cytoskeleton during in vitro morphogenesis of MDCK epithelial cells. Cell Motil. Cytoskeleton 44, 133–142 [DOI] [PubMed] [Google Scholar]
- 43.Fässler R., Schnegelsberg P. N., Dausman J., Shinya T., Muragaki Y., McCarthy M. T., Olsen B. R., Jaenisch R. (1994) Mice lacking alpha 1 (IX) collagen develop noninflammatory degenerative joint disease. Proc. Natl. Acad. Sci. U.S.A 91, 5070–5074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eyre D. R., Pietka T., Weis M. A., Wu J. J. (2004) Covalent cross-linking of the NC1 domain of collagen type IX to collagen type II in cartilage. J. Biol. Chem 279, 2568–2574 [DOI] [PubMed] [Google Scholar]
- 45.Zaucke F., Grässel S. (2009) Genetic mouse models for the functional analysis of the perifibrillar components collagen IX, COMP and matrilin-3: implications for growth cartilage differentiation and endochondral ossification. Histol. Histopathol 24, 1067–1079 [DOI] [PubMed] [Google Scholar]
- 46.Gregory K. E., Keene D. R., Tufa S. F., Lunstrum G. P., Morris N. P. (2001) Developmental distribution of collagen type XII in cartilage: association with articular cartilage and the growth plate. J. Bone Miner. Res 16, 2005–2016 [DOI] [PubMed] [Google Scholar]
- 47.Nishiyama T., McDonough A. M., Bruns R. R., Burgeson R. E. (1994) Type XII and XIV collagens mediate interactions between banded collagen fibers in vitro and may modulate extracellular matrix deformability. J. Biol. Chem 269, 28193–28199 [PubMed] [Google Scholar]
- 48.Liu Y., Monticone M., Tonachini L., Mastrogiacomo M., Marigo V., Cancedda R., Castagnola P. (2004) URB expression in human bone marrow stromal cells and during mouse development. Biochem. Biophys. Res. Commun 322, 497–507 [DOI] [PubMed] [Google Scholar]
- 49.Ito H., Akiyama H., Iguchi H., Iyama K., Miyamoto M., Ohsawa K., Nakamura T. (2001) Molecular cloning and biological activity of a novel lysyl oxidase-related gene expressed in cartilage. J. Biol. Chem 276, 24023–24029 [DOI] [PubMed] [Google Scholar]
- 50.Nurminskaya M. V., Linsenmayer T. F. (2002) Immunohistological analysis of transglutaminase factor XIIIA expression in mouse embryonic growth plate. J. Orthop. Res 20, 575–578 [DOI] [PubMed] [Google Scholar]
- 51.Colnot C., Sidhu S. S., Poirier F., Balmain N. (1999) Cellular and subcellular distribution of galectin-3 in the epiphyseal cartilage and bone of fetal and neonatal mice. Cell. Mol. Biol 45, 1191–1202 [PubMed] [Google Scholar]
- 52.White A. H., Watson R. E., Newman B., Freemont A. J., Wallis G. A. (2002) Annexin VIII is differentially expressed by chondrocytes in the mammalian growth plate during endochondral ossification and in osteoarthritic cartilage. J. Bone Miner. Res 17, 1851–1858 [DOI] [PubMed] [Google Scholar]
- 53.Andersson M. L., Thorstensson C. A., Roos E. M., Petersson I. F., Heinegård D., Saxne T. (2006) Serum levels of cartilage oligomeric matrix protein (COMP) increase temporarily after physical exercise in patients with knee osteoarthritis. BMC Musculoskelet. Disord 7, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Posey K. L., Hankenson K., Veerisetty A. C., Bornstein P., Lawler J., Hecht J. T. (2008) Skeletal abnormalities in mice lacking extracellular matrix proteins, thrombospondin-1, thrombospondin-3, thrombospondin-5, and type IX collagen. Am. J. Pathol 172, 1664–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poole C. A. (1997) Articular cartilage chondrons: form, function and failure. J. Anat 191, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kruegel J., Sadowski B., Miosge N. (2008) Nidogen-1 and nidogen-2 in healthy human cartilage and in late-stage osteoarthritis cartilage. Arthritis Rheum 58, 1422–1432 [DOI] [PubMed] [Google Scholar]
- 57.Shen Z., Gantcheva S., Mânsson B., Heinegârd D., Sommarin Y. (1998) Chondroadherin expression changes in skeletal development. Biochem. J 330, 549–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marsich E., Mozetic P., Ortolani F., Contin M., Marchini M., Vetere A., Pacor S., Semeraro S., Vittur F., Paoletti S. (2008) Galectin-1 in cartilage: expression, influence on chondrocyte growth and interaction with ECM components. Matrix Biol 27, 513–525 [DOI] [PubMed] [Google Scholar]
- 59.Vincourt J. B., Lionneton F., Kratassiouk G., Guillemin F., Netter P., Mainard D., Magdalou J. (2006) Establishment of a reliable method for direct proteome characterization of human articular cartilage. Mol. Cell. Proteomics 5, 1984–1995 [DOI] [PubMed] [Google Scholar]
- 60.Benjamin M., Archer C. W., Ralphs J. R. (1994) Cytoskeleton of cartilage cells. Microsc. Res. Tech 28, 372–377 [DOI] [PubMed] [Google Scholar]
- 61.Kana R., Dundr P., Tvrdík D., Necas E., Povýsil C. (2006) Expression of actin isoforms in human auricular cartilage. Folia Biol 52, 167–172 [DOI] [PubMed] [Google Scholar]
- 62.Povýsil C., Kana R., Dundr P., Tvrdík D., Horák M., Vaculík J., Podskubka A., Kubes R. (2008) Distribution of chondrocytes containing alpha-smooth muscle actin in human normal, osteoarthrotic, and transplanted articular cartilage. Pathol. Res. Pract 204, 883–890 [DOI] [PubMed] [Google Scholar]
- 63.Ruiz-Romero C., López-Armada M. J., Blanco F. J. (2005) Proteomic characterization of human normal articular chondrocytes: a novel tool for the study of osteoarthritis and other rheumatic diseases. Proteomics 5, 3048–3059 [DOI] [PubMed] [Google Scholar]
- 64.Blain E. J. (2009) Involvement of the cytoskeletal elements in articular cartilage homeostasis and pathology. Int. J. Exp. Pathol 90, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bobick B. E., Tuan R. S., Chen F. H. (2010) The intermediate filament vimentin regulates chondrogenesis of adult human bone marrow-derived multipotent progenitor cells. J. Cell. Biochem 109, 265–276 [DOI] [PubMed] [Google Scholar]
- 66.Lambrecht S., Verbruggen G., Verdonk P. C., Elewaut D., Deforce D. (2008) Differential proteome analysis of normal and osteoarthritic chondrocytes reveals distortion of vimentin network in osteoarthritis. Osteoarthritis Cartilage 16, 163–173 [DOI] [PubMed] [Google Scholar]
- 67.Low D. A., Baker J. B., Koonce W. C., Cunningham D. D. (1981) Released protease-nexin regulates cellular binding, internalization, and degradation of serine proteases. Proc. Natl. Acad. Sci. U.S.A 78, 2340–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Farrell D. H., Cunningham D. D. (1987) Glycosaminoglycans on fibroblasts accelerate thrombin inhibition by protease nexin-1. Biochem. J 245, 543–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Farrell D. H., Wagner S. L., Yuan R. H., Cunningham D. D. (1988) Localization of protease nexin-1 on the fibroblast extracellular matrix. J. Cell. Physiol 134, 179–188 [DOI] [PubMed] [Google Scholar]
- 70.Milner J. M., Patel A., Rowan A. D. (2008) Emerging roles of serine proteinases in tissue turnover in arthritis. Arthritis Rheum 58, 3644–3656 [DOI] [PubMed] [Google Scholar]
- 71.Wagener R., Ehlen H. W., Ko Y. P., Kobbe B., Mann H. H., Sengle G., Paulsson M. (2005) The matrilins—adaptor proteins in the extracellular matrix. FEBS Lett 579, 3323–3329 [DOI] [PubMed] [Google Scholar]
- 72.Klatt A. R., Nitsche D. P., Kobbe B., Macht M., Paulsson M., Wagener R. (2001) Molecular structure, processing, and tissue distribution of matrilin-4. J. Biol. Chem 276, 17267–17275 [DOI] [PubMed] [Google Scholar]
- 73.Klatt A. R., Nitsche D. P., Kobbe B., Mörgelin M., Paulsson M., Wagener R. (2000) Molecular structure and tissue distribution of matrilin-3, a filament-forming extracellular matrix protein expressed during skeletal development. J. Biol. Chem 275, 3999–4006 [DOI] [PubMed] [Google Scholar]
- 74.Wagener R., Kobbe B., Paulsson M. (1997) Primary structure of matrilin-3, a new member of a family of extracellular matrix proteins related to cartilage matrix protein (matrilin-1) and von Willebrand factor. FEBS Lett 413, 129–134 [DOI] [PubMed] [Google Scholar]
- 75.Ehlen H. W., Sengle G., Klatt A. R., Talke A., Müller S., Paulsson M., Wagener R. (2009) Proteolytic processing causes extensive heterogeneity of tissue matrilin forms. J. Biol. Chem 284, 21545–21556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hills R., Mazzarella R., Fok K., Liu M., Nemirovskiy O., Leone J., Zack M. D., Arner E. C., Viswanathan M., Abujoub A., Muruganandam A., Sexton D. J., Bassill G. J., Sato A. K., Malfait A. M., Tortorella M. D. (2007) Identification of an ADAMTS-4 cleavage motif using phage display leads to the development of fluorogenic peptide substrates and reveals matrilin-3 as a novel substrate. J. Biol. Chem 282, 11101–11109 [DOI] [PubMed] [Google Scholar]
- 77.Heinegård D., Lohmander S., Thyberg J. (1978) Cartilage proteoglycan aggregates. Electron-microscopic studies of native and fragmented molecules. Biochem. J 175, 913–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Day J. M., Olin A. I., Murdoch A. D., Canfield A., Sasaki T., Timpl R., Hardingham T. E., Aspberg A. (2004) Alternative splicing in the aggrecan G3 domain influences binding interactions with tenascin-C and other extracellular matrix proteins. J. Biol. Chem 279, 12511–12518 [DOI] [PubMed] [Google Scholar]
- 79.Struglics A., Larsson S., Hansson M., Lohmander L. S. (2009) Western blot quantification of aggrecan fragments in human synovial fluid indicates differences in fragment patterns between joint diseases. Osteoarthritis Cartilage 17, 497–506 [DOI] [PubMed] [Google Scholar]