Abstract
Exosomes are nanometer-sized vesicles, secreted by various cell types, present in biological fluids that are particularly rich in membrane proteins. Ex vivo analysis of exosomes may provide biomarker discovery platforms and form non-invasive tools for disease diagnosis and monitoring. These vesicles have never before been studied in the context of bladder cancer, a major malignancy of the urological tract. We present the first proteomics analysis of bladder cancer cell exosomes. Using ultracentrifugation on a sucrose cushion, exosomes were highly purified from cultured HT1376 bladder cancer cells and verified as low in contaminants by Western blotting and flow cytometry of exosome-coated beads. Solubilization in a buffer containing SDS and DTT was essential for achieving proteomics analysis using an LC-MALDI-TOF/TOF MS approach. We report 353 high quality identifications with 72 proteins not previously identified by other human exosome proteomics studies. Overrepresentation analysis to compare this data set with previous exosome proteomics studies (using the ExoCarta database) revealed that the proteome was consistent with that of various exosomes with particular overlap with exosomes of carcinoma origin. Interrogating the Gene Ontology database highlighted a strong association of this proteome with carcinoma of bladder and other sites. The data also highlighted how homology among human leukocyte antigen haplotypes may confound MASCOT designation of major histocompatability complex Class I nomenclature, requiring data from PCR-based human leukocyte antigen haplotyping to clarify anomalous identifications. Validation of 18 MS protein identifications (including basigin, galectin-3, trophoblast glycoprotein (5T4), and others) was performed by a combination of Western blotting, flotation on linear sucrose gradients, and flow cytometry, confirming their exosomal expression. Some were confirmed positive on urinary exosomes from a bladder cancer patient. In summary, the exosome proteomics data set presented is of unrivaled quality. The data will aid in the development of urine exosome-based clinical tools for monitoring disease and will inform follow-up studies into varied aspects of exosome manufacture and function.
Bladder cancer is one of the eight most frequent cancers in the Western world, and the frequency of transitional cell carcinoma (TCC),1 which accounts for 90% of bladder cancers, is second only to prostate cancer as a malignancy of the genitourinary tract. Urine cytology and cystoscopy remain the predominant clinical tools for diagnosing and monitoring the disease, but cytology is poorly sensitive, particularly for low grade tumors, and does not serve as a prognostic tool. Cystoscopy is an invasive procedure, and there is pressing need to identify informative molecular markers that can be used to replace it.
Recently, small cell-derived vesicles termed exosomes that are present in body fluids (1–5) have been proposed as a potential source of diagnostic markers (2, 6–8). These nanometer-sized vesicles, which are secreted by most cell types, originate from multivesicular bodies of the endocytic tract and reflect a subproteome of the cell. Exosomes are enriched in membrane and cytosolic proteins, and this molecular repertoire appears to be of particular functional importance to the immune system (9). Exosomes also comprise an array of lipids, mRNA, and microRNA, which are likely involved in conveying intercellular communication processes (10). Importantly, many exosomal components are simply not present as free soluble molecules in body fluids, such as certain microRNA species, which are encapsulated within the exosome lumen (6, 10). Therefore, the ability to isolate exosomes from urine (2), plasma (1), saliva (11), or other physiological sources (3) holds significant potential for obtaining novel and complex sets of biomarkers in a non-invasive manner. Exosome analysis may therefore be of value in disease diagnosis and monitoring in a variety of settings (6, 7, 12–14).
Exosomes as indicators of pathology were first documented in the context of renal injury where a differential proteomics approach revealed changes in urinary exosome phenotype following renal injury (7). The researchers identified exosomally expressed Fetuin-A as a marker that became elevated 50-fold within hours following nephrotoxin exposure in rodents. Exosomal Fetuin-A elevation was also apparent in patients with acute renal injury before changes in urinary creatinine were observed (7). Clinical exosome analysis may also prove useful for solid cancers, such as ovarian or lung cancer, where the quantity of epithelial cell adhesion molecule-positive serum exosomes may correlate with tumor stage/grade. Such disease-associated exosomes express microRNA species not detected in healthy subjects (6, 12), although in this respect, there is little correlation between microRNA and disease bulk (6, 12). Other recent examples include studies of urinary exosomes in prostate cancer with exosomes expressing protein markers 5T4 (15), prostate cancer gene 3 (PCA-3) (8), or mRNA (TMPRSS2-ERG) (8, 16) associated with prostate cancer. To our knowledge, exosomes have not yet been studied in the context of other urological malignancies such as renal cancer, and to date, only one report describes the urine-derived microparticles from bladder cancer patients (17). In that report, they examined the proteome of a highly complex mixture of microvesicles, exosomes, and other urinary constituents that can be pelleted by high speed ultracentrifugation, identifying eight proteins that may be elevated in cancer. However, given the nature of the sample analyzed, it is unknown whether these proteins are exosomally expressed.
Identification of the principal and most relevant molecular markers in these and other clinical scenarios remains a major challenge. In part, this is because exosomes present within complex body fluids originate from heterogeneous cell types. For example, plasma exosomes may be derived from platelets, lymphocytes, or endothelial cells (1), and a proportion may arise from well perfused organs such as the liver (18) and likely other organs as well (16). Similarly, exosomes present in urine arise from urothelial cells of the kidney and downstream of the renal tract (2, 8, 15).
Importantly, all proteomics studies of exosomes isolated from body fluids are unavoidably complicated by the presence of high abundance non-exosomal proteins contaminating the preparations. Examples include albumin, immunoglobulin, and complement components present in exosomes prepared from malignant effusions (5) and Tamm-Horsfall protein present in exosomes purified from urine (2). As such, great care must be taken in the interpretation of the large data sets produced by proteomics studies, requiring careful validation of the proteins of interest. The protein composition of exosomes using a single homogenous cell type is one approach that may be used to uncover the protein components of exosomes produced by various cell types.
There remain two major issues in the realm of exosome proteomics that complicate our interpretation of lists of identified proteins. Foremost are the diverse methods chosen for exosome purification that in some studies have involved attempts to remove contaminants through a key biophysical property of the vesicles, i.e. their capacity to float on sucrose (19, 20) or other dense media (21). Not all published studies, however, have taken such steps, preferring a far simpler pellet (or pellet and wash) approach. These latter preparations may be significantly contaminated by components of the cellular secretome, cell fragments, and other components. All of these factors could lead to false positive identifications of exosome proteins. The second key issue centers on the MS approaches utilized in various exosome proteomics studies. Many early examples relied only on a peptide mass fingerprinting approach, lacking robust peptide sequence data (22, 23), and more recently, search criteria that are generally recommended for MS-derived sequence data have not been specified in all studies. In this study, we have listed only those proteins identified by good quality MS/MS data for two or more peptides. Variability in the robustness and bias in bioinformatics analysis of data sets and in the steps taken to validate identified proteins is an additional factor that impacts the confidence in the identification lists produced.
In this study, we aimed to perform the first proteomics analysis of human bladder cancer exosomes. We took extensive steps to produce high purity and quality-assured exosome preparations prior to beginning proteomics workflows. Solubilizing the sample with SDS and a reducing agent (DTT) was a critical step that allowed for global protein identification using nanoscale liquid chromatography followed by MALDI-TOF/TOF mass spectrometry. In this study, we present the identification of a significant number of exosomally expressed proteins (353 in total) of unrivaled quality. Critical manual examination of these identifications revealed issues with multiple (physiologically impossible) MHC Class I identifications that were attributed to a misdesignation of nomenclature by MASCOT due to peptide (and target protein) homology. The data were subjected to unbiased overrepresentation analysis (examining ExoCarta and Gene Ontology databases) to reveal a proteome consistent with exosomes, particularly of carcinoma origin. Validation of several identified proteins, by combining ultracentrifugation on a linear sucrose gradient with Western blotting and/or analysis of exosome-coated latex beads, demonstrated correct surface orientation of several MS-identified membrane proteins at densities consistent with exosomes.
The robust approaches taken emphasize our confidence in the validity of the identifications generated and highlight that 72 (of 353) proteins have not been previously shown to be exosomally expressed by other human proteomics studies. The data will be useful for future studies in this underinvestigated disease and will form a platform not only for future clinical validation of some of these putative markers but also to aid further investigations into novel aspects of exosome function and manufacture.
EXPERIMENTAL PROCEDURES
Cell Culture
HT1376 is a cell line originating from a primary TCC of the bladder (Stage T2, Grade G4) (24). In vitro cultured HT1376 cells were used as the exosome source for this study because they have been extensively characterized previously and are representative of the behavior and phenotype of TCC (24, 25). The cells were maintained in Dulbecco's modified Eagle's medium (Lonza) supplemented with penicillin/streptomycin and 5% FBS (which had been depleted of exosomes by overnight ultracentrifugation at 100,000 × g followed by filtration through 0.2-μm and then 0.1-μm vacuum filters (Millipore)). The cells were seeded into bioreactor flasks (from Integra) and maintained at high density culture for exosome production as described (26). Cells were confirmed negative for mycoplasma contamination by monthly screening (Mycoalert, Lonza). Additional well characterized bladder cancer cell lines (HT1197, RT4, RT112, and T24) (25, 27) were obtained from ATCC or from Cancer Research UK cell bank and cultured similarly.
Exosome Purification
The culture medium of HT1376 cells (typically 15–30 ml) was subjected to serial centrifugation to remove cells (400 × g for 10 min) and cellular debris (2000 × g for 15 min). The supernatant was then centrifuged at 10,000 × g for 30 min, and the supernatant was further purified by underlaying with a 30% sucrose, D2O cushion and subjected to ultracentrifugation at 100,000 × g for 2 h. The cushion was collected, and exosomes were washed in PBS as described previously (20, 28, 29). Exosome pellets were resuspended in 100–150 μl of PBS and frozen at −80 °C. The quantity of exosomes was determined by the micro-BCA protein assay (Pierce/Thermo Scientific), and this gave an average of 12 μg (±2.2 S.E., n = 5) exosomes/ml of culture medium for the HT1376 cell line. Transmission electron microscopy of exosomal preparations was performed as described (29).
Determinination of Exosome Density
To quantify the density of exosomes produced by HT1376, we used a protocol similar to that described previously based on ultracentrifugation on a linear sucrose gradient (19, 30). Briefly, cell culture supernatant was subjected to differential centrifugation, and the pellet at 70,000 × g was overlaid on a linear sucrose gradient (0.2 m up to 2.5 m sucrose). Specimens were centrifuged at 4 °C overnight at 210,000 × g using an MLS-50 rotor in an Optima-Max ultracentrifuge (Beckman Coulter). The refractive index of collected fractions was measured at 20 °C using an automatic refractometer (J57WR-SV, Rudolph Scientific), and from this, the density was calculated as described previously (19). Fractions were washed in buffer (PBS or MES buffer; discussed below) by ultracentrifugation at 150,000 × g (in a TLA-110 rotor in an Optima-Max ultracentrifuge), and pellets were resuspended in MES buffer for coupling to microbeads or in SDS sample buffer for analysis by Western blot.
Flow Cytometric Analyses of Exosome-coated Beads
One microgram of purified exosomes was incubated with 1 μl of latex beads (surfactant-free, aldehyde sulfate 3.9-μm beads, Interfacial Dynamics) that had been washed twice in MES buffer (0.025 m MES, 0.154 m NaCl, pH 6). For analysis of sucrose gradient fractions, 30% of each fraction was coupled to 0.5 μl of stock beads. Exosome beads were incubated in a final volume of 100 μl of MES buffer at room temperature (RT) for 1 h on a shaking platform followed by rolling overnight at 4 °C. Beads were blocked by incubating with 1% BSA, MES buffer for 2 h at RT. Blocking buffer was washed away, and beads were resuspended in 0.1% BSA, MES buffer. Primary monoclonal antibodies were used (at 2–10 μg/ml) for 1 h at 4 °C. After one wash, goat anti-mouse Alexa Fluor 488-conjugated antibody (Invitrogen) diluted 1:200 in 0.1% BSA, MES buffer was added for 1 h. After washing, beads were analyzed by flow cytometry using a FACSCanto instrument configured with a high throughput sampling module running FACSDiva Version 6.1.2 software (BD Biosciences). The conditions used for exosome coupling to beads and subsequent antibody staining were determined experimentally as described previously (30).
One-dimensional Electrophoresis and Immunoblotting
Cell lysates were compared with exosome lysates by immunoblotting as described (31) where protein (up to 20 μg/well) was solubilized by the addition of a 30% volume of 6 m urea, 50 mm Tris-HCl, 2% SDS, 20 mm DTT, and 0.002% (w/v) bromphenol blue. Samples were electrophoresed through 4–12% Bis-Tris gels (Invitrogen) and transferred to PVDF membranes that were blocked and probed with antibodies using the Qdot® system (Invitrogen). Bands were visualized using the MiniBIS Pro imaging system (DNR Bio-Imaging Systems). The following primary monoclonal antibodies were used: TSG101, lysosome-associated membrane protein 1 (LAMP-1), hsp90, calnexin, HLA-G, galectin-3, basigin, hnRNPK, gp96, cytokeratins 18 and 17, and CD44 (Santa Cruz Biotechnology), glyceraldehyde-3-phosphate dehydrogenase (BioChain Institute, Inc.), CD9 (R&D Systems), and CD63 and CD81 (Serotec). Anti-5T4 was a gift from Dr. R. Harrop (Oxford BioMedica UK Ltd.).
Two-dimensional Electrophoresis and MS
A gel-based approach was used to examine the exosome protein profile using a standard 2DE protocol. Briefly, exosomes (750 μg) were solubilized for 1 h at RT in 150 μl of lysis buffer (7 m urea, 2 m thiourea, 20 mm DTT, 4% (w/v) CHAPS, 0.005% (w/v) bromphenol blue, and 0.5% (v/v) IPG buffer pH 3–10 non-linear (GE Healthcare)). Extracted proteins were then solvent-precipitated using the 2D Clean-Up kit (GE Healthcare) before the pellet was resuspended in lysis buffer. From this, 500 μg of protein was recovered, and this was subjected to isoelectric focusing using 18-cm pH 3–10 non-linear IPG rehydrated strips, an Ettan IPGphor III IEF system (GE Healthcare), and recommended voltages. Subsequently, the IPG strip was equilibrated for 15 min in equilibration buffer (50 mm Tris-HCl, pH 8.8, 6 m urea, 2% (w/v) SDS, 30% (v/v) glycerol, and 0.002% (w/v) bromphenol blue) containing 1% (w/v) DTT followed by 15 min in equilibration buffer containing 2.5% (w/v) iodoacetamide. Equilibrated IPG strips were subjected to second dimension separation using the EttanTM DALTsix system (GE Healthcare). Silver staining was performed, and randomly selected gel spots were excised, subjected to trypsin digestion, and MALDI-TOF/TOF mass spectrometry analysis as described previously (32). The database search settings used were the same as described later for LC-MALDI protein identification except that a precursor mass tolerance of 50 ppm was used.
Preparation of Exosome-derived Peptides for Nano-LC
HT1376-derived exosome preparations were repelleted at 118,000 × g for 45 min at 4 °C in a TLA-110 rotor in an Optima-Max ultracentrifuge (Beckman Coulter). The pellets were solubilized in 100 μl of triethylammonium bicarbonate (TEAB) lysis buffer (20 mm TEAB) containing 20 mm DTT and 1% (w/v) SDS at RT for 10 min, then heated to 95 °C for 10 min, and then left for a further 10 min at RT. The samples were subjected to an additional ultracentrifugation step (118,000 × g for 45 min at RT), and supernatants (now free of insoluble material) were subjected to solvent precipitation to remove salts, lipids, and detergent (using the 2D Clean-Up kit, GE Healthcare). The pellets were resuspended in 20 mm TEAB and left overnight at 4 °C. The protein content was then determined using a BCA protein assay kit (Sigma). Samples were then reduced, denatured, and alkylated using an Applied Biosystems iTRAQ (isobaric tags for relative and absolute quantitation) labeling kit and standard protocol. The proteins were subjected to digestion with trypsin (0.8 μg/sample) and incubated at 37 °C for 12–16 h. The samples were then dried and resuspended in water with 0.1% (v/v) TFA.
LC-MALDI and Protein Identification
Digested peptides were separated on a nano-LC system (UltiMate 3000, Dionex, Sunnyvale, CA) using a two-dimensional salt plug method as described previously (32). Mass spectrometry was performed using an Applied Biosystems 4800 MALDI-TOF/TOF mass spectrometer as described (32). The MS/MS data were used to search the Swiss-Prot database (Version 57.7; release date, September 1, 2009; 497,293 sequences; human taxonomy) using MASCOT database search engine Version 2.1.04 (Matrix Science Ltd., London, UK) embedded into GPS Explorer software Version 3.6 Build 327 (Applied Biosystems) (default GPS Explorer parameters; one missed cleavage allowed; fixed modification of methyl methanethiosulfonate (Cys); variable modifications of oxidation (Met), pyro-Glu (N-terminal Glu), and pyro-Glu (N-terminal Gln); 150-ppm mass tolerance in MS and 0.3-Da mass tolerance for MS/MS, which are recommended published tolerances for LC-MALDI (32)). For a protein to be identified, there needed to be a minimum of two peptides with MASCOT E-values less than 0.05. There was a false discovery rate (FDR) of 0%, which was determined using the same Swiss-Prot database with the entire sequence randomized. Where more than one protein was identified, the protein with the highest MOWSE (molecular weight search) score in MASCOT is reported. The analysis was performed with two biological replicates, each including a technical replicate.
MS Data Analysis
The resultant protein list was analyzed for any biological enrichment against previously defined lists using MetaCore GeneGO (Version 5.4) and selected ExoCarta submissions (33) (MS-based data containing 10 or more matching gene identifiers). For analysis using 44 studies from ExoCarta gene sets, our protein list was converted from Swiss-Prot accession numbers to EntrezGene IDs using BioMart before overrepresentation analysis (ORA) using the hypergeometric distribution in R against a background of all human genes with EntrezGene IDs. For ORA in MetaCore, data were first converted into Swiss-Prot IDs (using BioMart) before analysis, again using hypergeometric tests.
Urinary Exosomes
Freshly collected urine specimens (up to 250 ml) were subjected to the same exosome purification protocol as described earlier. Fresh urine was collected from three patients with confirmed diagnoses of transitional carcinoma of the bladder. The specimens were obtained following transurethral resection of bladder tumor prior to the start of any other treatment. Purification of exosomes commenced within 30 min of sample collection. As controls, urine specimens were also collected freshly from four healthy volunteers. Ethical approval was obtained from South East Wales Ethics Committee, and institutional approval for the study was obtained from the Velindre NHS Trust Research Committee and Cardiff and Vale NHS Trust.
RESULTS
Characterization of HT1376 Exosomes
Exosomes were purified from HT1376 cells, and preparations were subjected to several forms of analysis to evaluate sample quality/purity prior to analysis using proteomics workflows.
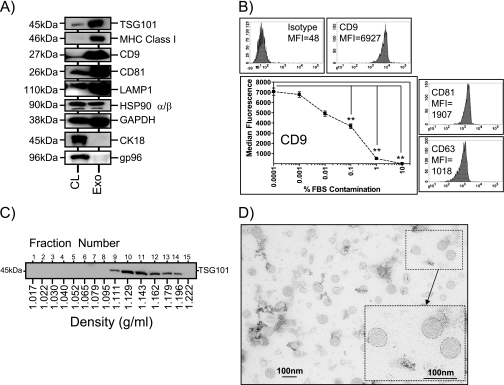
First, Western blots were performed to compare whole cell lysates with exosomes to examine the expression of expected published exosomal markers (30) and to evaluate the relative expression of these markers compared with the parent cell as a whole. As we expected, the multivesicular body marker TSG101 was strongly enriched in exosome preparations compared with cell lysates (Fig. 1A). Additionally, a number of other molecules, including MHC Class I, the tetraspanins CD9 and CD81, the lysosomal protein LAMP-1, and to some extent glyceraldehyde-3-phosphate dehydrogenase, were similarly enriched. Such features are typical of exosomes produced by varied cell types (30). The heat shock protein hsp90 was not exosomally enriched, and this is typical of cells that are not under stress conditions (26, 34, 35). Staining for cytokeratin 18 revealed a strong band in cell lysates but little or no detectable band in exosomes. Similarly, the endoplasmic reticulum-resident gp96 was readily detected in cell lysates but just detectable in exosomes, which indicated that little if any contaminating cellular debris was present in the exosome preparations.
Fig. 1.
Characterization of HT1376-derived exosomes using Western blotting, flow cytometry, and electron microscopy. Cell (CL) and exosome (Exo) lysates (5 μg/well) were compared by Western blotting using a range of antibodies as indicated. This demonstrated relative enrichment of several proteins in exosomes. Some markers, such as gp96, were absent from exosomes, indicating negligible contamination of the preparations by cellular debris (this is representative of three experiments) (A). Exosomes coupled to latex beads were analyzed by flow cytometry, and this revealed positive expression of tetraspanin molecules on the exosome surface. Median fluorescence intensity values (MFI) are shown (representative of >5 experiments) (B). Intentional contamination of purified exosomes with increasing amounts of FBS prior to coupling to latex beads reveals a decrease in signal intensity for CD9 (mean ± S.E.; n = 6; **, p < 0.001, one-way analysis of variance with Tukey's post test) (B, line graph). Material pelleted at 70,000 × g from cell-conditioned medium was overlaid on a linear sucrose gradient (0.2–2.02 m) and ultracentrifuged for 18 h at 210,000 × g. Collected fractions were analyzed by refractometry to ascertain fraction density and thereafter by Western blot using antibodies to TSG101, which is an exosome marker. TSG101 floats at typical exosome densities of between 1.1 and 1.2 g/ml (representative of four experiments) (C). Transmission electron micrograph of a typical exosome preparation reveals heterogeneous vesicles between 30 and 100 nm in diameter (D). CK, cytokeratin.
The markers present on the exosome surface were also examined, following coupling of exosomes to latex beads, by flow cytometry (Fig. 1B). This was performed to demonstrate the expression of correctly oriented proteins on the exosome surface. Tetraspanins were the choice markers for this because their expression is a well documented feature of exosomes from multiple cell types. The analyses showed very strong expression of the tetraspanin CD9 and readily detectable expression of CD81 and CD63 (Fig. 1B) for this and other bladder cancer cell lines (see Fig. 5A). Moreover, this assay can also highlight the presence of significant contaminating proteins in the preparations. When contaminants, rather than exosomes, bind to the bead surface during the coupling reaction, the assay subsequently yields low fluorescence signal for exosomal markers like CD9 (Fig. 1B, line graph). Intentional contamination of purified exosomes with FBS (the likeliest source of contaminants in a cell culture model) revealed that adding 0.01% FBS is sufficient to decrease CD9-specific staining by around 30%. Thus, we set an arbitrary threshold for the purity of exosome preparations: those with a CD9 staining below 5000 median fluorescence units were deemed low quality and not utilized further.
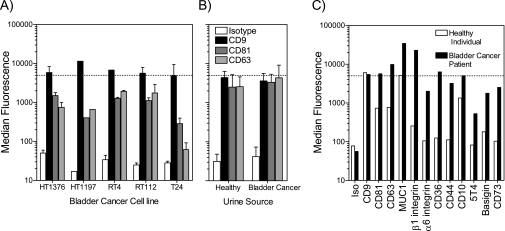
Fig. 5.
Exosomes isolated from urine specimens. The sucrose cushion method, used in this study for HT1376 exosomes, was also tested on a small panel of other bladder cancer cell lines, and the quality of preparations was assessed using the latex bead assay (A). The graph depicts median fluorescence values from the flow cytometric histogram peak (mean ± S.D. of n preparations where n = 5 for HT1376, n = 1 for HT1197, n = 2 for RT4, n = 2 for RT112, and n = 2 for T24) with beads stained for CD9, CD81, or CD63 as indicated. The dotted line indicates our arbitrary cutoff value (of 5000 units) where CD9 fluorescence above this value was indicative of a preparation of acceptable quality. Using identical methods, exosomes were prepared from urine specimens collected from healthy individuals (n = 4) or from bladder cancer patients (n = 3) (mean ± S.D.), and exosomes were analyzed as above (B). Exosome preparations from one healthy individual and one bladder cancer patient, which exceeded the quality threshold, were analyzed further for surface expression of some MS-identified proteins as indicated (C). Iso, isotype.
As well as expression of a typical exosomal molecular profile, we also investigated another key feature of exosomes, that is their density characteristics. HT1376 exosomes, pelleted at 70,000 × g, were overlaid on a linear sucrose gradient and subjected to ultracentrifugation for 18 h. Fifteen fractions were collected, and analysis by Western blot revealed the presence of TSG101 floating at a density range around 1.1–1.19 g/ml (Fig. 1C). Such analysis confirms that HT1376 cells produce exosomes of typical density similar to that described for exosomes from other cell types (19). This method, in combination with the latex microbead assay (above), was also used as a tool for validating MS protein identifications (see Results, Validation of Exosomal proteins identified in Fig. 4B). Electron microscopy of exosome preparations was also performed (Fig. 1D), revealing nanovesicular structures within a size range consistent with their definition as exosomes (30–100 nm). Taken together, the data indicate that HT1376 bladder cancer cells produce exosomes that have molecular and biophysical characteristics similar to exosomes of other cell types and that our exosome preparations are of high quality and virtually free of contaminating cellular debris.
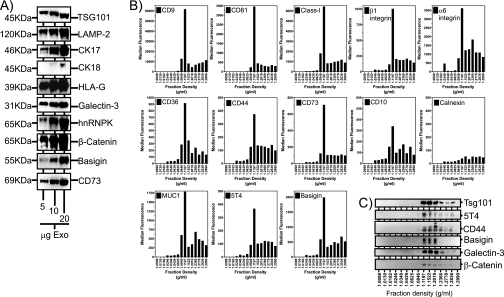
Fig. 4.
Validation of some MS-identified proteins by Western blot and flow cytometric analysis. HT1376 exosomes (5–20 μg/well), purified by the standard sucrose cushion method, were analyzed by Western blot for expression of a range of MS identified proteins as indicated (A). The 70,000 × g pellet, obtained from HT1376 cell-conditioned medium, was subjected to fractionation by centrifugation on a linear sucrose gradient (0.2–2.5 m). Fifteen total fractions were collected, and the density was measured by refractometry. Thereafter, one-third of each fraction was coupled to latex beads followed by flow cytometric analysis for exosomal surface expression as indicated (B). In parallel, the remaining two-thirds of each fraction was subjected to Western blotting for proteins as indicated (C). The data reveal proteins floating at a recognized exosomal density range (1.12–1.2 g/ml). (The data are representative of two experiments.) CK, cytokeratin.
Identification of Exosomal Proteins by LC-MALDI MS
To obtain exosome-derived trypsin digest peptides for nano-LC, we used a protocol encompassing a 1% (w/v) SDS extraction that would normally be sufficient to solubilize membrane proteins (36). However, our initial attempts with this standard protocol revealed some major issues with the efficacy of exosome solubilization, resulting in very low numbers of proteins identified (three in total with multiple peptide assignments) compared with other cell types commonly processed in the laboratory (we usually identify 300–500 proteins for cultured cell lysates). We therefore modified the sample preparation protocol to achieve more efficient solubilization of exosomes by simply including DTT in the solubilization buffer.
This process resulted in the identification of 353 proteins (supplemental Table 1). Importantly, we include only proteins identified with two or more peptides and an expect value of less than 0.05, criteria that produce an FDR of 0%. By including identifications based upon a single peptide with an expect value of less than 0.0025 (an additional 261 proteins), the FDR increases to 2.6%, although inevitably some of these assignments would be valid.
Exploring these identifications revealed several proteins consistent with exosome biosynthesis. For example, members of the ubiquitin-dependent complex ESCRT (endosomal sorting complex required for transport) were present, including vacuolar protein sorting-associated protein 28 homolog (vps-28), vacuolar protein sorting-associated protein 4B (vps-4B), ubiquitin-like modifier-activating enzyme, and ubiquitin. These identifications suggest a multivesicular body origin for the sample analyzed. Proteins involved in membrane trafficking and fusion processes were also evident (clathrin heavy chain 1; Rab-11B; Rab-5A; Rab-6a; Rab-7a; Rab GDP dissociation inhibitor β; annexins A1, A2, A3, A4, A5, A6, and A7; annexin A8-like protein; and annexin A11). Markers of endosomes and lysosomes were also present (EH domain-containing proteins 1 and 2, lysosome membrane protein 2, lysosome-associated membrane protein 2, tripeptidyl-peptidase 1, cathepsin-D, and sequestosome-1), and several proteins with chaperone functions were identified (hsp70, hsc70, hsp90, stress-induced phosphoprotein 1, T-complex protein 1, and endoplasmin). Components of the cytosol are also expected to be found within the exosome lumen, a natural consequence of the membrane budding process during multivesicular body formation, and here also we found a diverse assortment of cytosolic enzymes (glyceraldehyde-3-phosphate dehydrogenase, cytosol aminopeptidase, cytosolic acetyl-CoA acetyltransferase, and nicotinate phosphoribosyltransferase) and cytoskeletal constituents (actin, α-actinin-4, cytokeratins, ezrin, tubulin, and myosin). Diverse transmembrane proteins were also abundant, including multiple integrins (β1, β4, α3, α6, and αv), MHC molecules, tetraspanins, epidermal growth factor receptor, mucin-1, CD44, syndecan-1, and various membrane transporters such as solute carrier families 2 and 3, 4F2 cell surface antigen heavy chain, choline transporter-like protein, and sodium/potassium-transporting ATPase subunit β-3. The proteome identified here is therefore broadly consistent with that expected for exosomes; it is comparable with proteomics identifications highlighted by other researchers investigating exosomes from other cellular or physiological sources (37). Of interest, when comparing our data set with MS identifications obtained from microparticles isolated from the urine of bladder cancer patients (17), proteins common to each study were only 7.5% (detailed in supplemental Fig. 1). This is perhaps not surprising given the differences in source material and sample preparation approaches, but it does indicate that some exosomal proteins are present within such microparticle preparations.
Exocarta and Gene Ontology Analysis
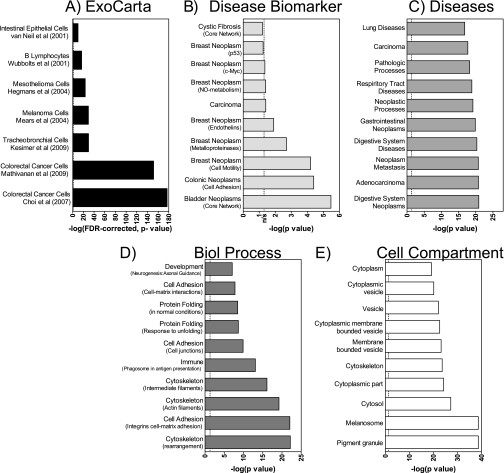
Having manually reviewed the MS/MS identifications for interesting hits related to exosome biology, we next subjected our results to a less biased assessment focused on characterizing the key biological themes within the protein list. Our 353 protein identifications were first compared with the multiple proteomics exosome studies published in ExoCarta (33) (a database collating lists extracted from exosome-related research publications), revealing that 72 proteins within this list have not previously been identified by human exosome proteomics studies (matches made using corresponding EntrezGene IDs for associated protein-encoding genes). Subsequently, ORA using the hypergeometric distribution was applied to explore whether there were more genes overlapping with ExoCarta gene sets than could be expected by chance (statistics calculated using the R environment for statistical computing). We limited the comparisons to studies utilizing MS-based proteomics approaches and to those with at least 10 matching (23, 38–43) identifications and applied an FDR correction to control for multiple testing. The results suggest that our identifications are consistent with data originating from exosomes and interestingly show a very significant overrepresentation of protein-encoding genes isolated from colorectal carcinoma cells (42, 43) (Fig. 2A).
Fig. 2.
Summary of overrepresentation analysis of nano-LC/MS-derived protein identifications against gene sets from ExoCarta and GeneGO. To facilitate comparison with ExoCarta gene sets, our protein list was first converted to an EntrezGene-identified gene list before undertaking ORA using the hypergeometric distribution. Results were filtered to include comparisons with MS-based studies only and with those reporting 10 or more matching genes, yielding seven studies (23, 38–43). This demonstrates how well our MS data compare with exosome protein profiles from specified cell types, displayed as the −log(p value) corrected for false detection rate (A). ORA analysis using MetaCore utilized the Swiss-Prot IDs for the identified protein list. For clarity, we report the top 10 overrepresented genes contained within each of the following group headings: disease biomarker (B), diseases (C), biological (Biol) process (D), and cellular compartment (E). The dotted line indicates p = 0.05; hence, columns to the left of this are not statistically significant (ns).
Similar overrepresentation analyses were performed in GeneGO MetaCore (Version 5.4), contrasting our list with gene sets derived from Gene Ontology and proprietary GeneGO data. Results in Fig. 2, B–E, show the top 10 results (gene sets ordered by ORA p value) from four gene set categories: disease biomarker, diseases in general, biological process, and cellular compartment (analyses against four other categories were uninformative). For the disease biomarker category, our data indicated the most significant association to be with bladder cancer, supporting therefore the premise that exosome analysis may well be a useful tool for disease-specific biomarker identification. Other biomarker associations included carcinomas of colon and breast (Fig. 2B). Similarly, a query examining general disease associations revealed features related to cancer of the gastrointestinal tract, metastatic cancer, respiratory tract diseases (including lung cancer), and carcinoma (Fig. 2C). Significant overrepresentation of encoding genes within our data related to genitourinary tract gene sets (including bladder neoplasm) was identified, but within the top 40. Although the accuracy of ORA can be limited by the quality and size of the gene sets queried, our analysis suggests that HT1376 exosomes express proteins strongly related to neoplastic diseases in general and to carcinomas in particular (Fig. 2, B and C).
Our proteome contained proteins whose encoding genes are located within membranous vesicles, the cytoplasm, and the cytoskeleton (Fig. 2E). Examination of the biological processes associated with this proteome revealed significant associations with the control of the cytoskeleton, intercellular adhesion, matrix adhesion processes, and protein folding-related processes (Fig. 2D). In summary, the statistically based, unbiased analyses undertaken reveal aspects of a bladder cancer exosome proteome that shows similarity to those determined from other exosome sources and emphasize a proteome particularly implicated in carcinoma.
Validation of Nano-LC Approach Using 2DE
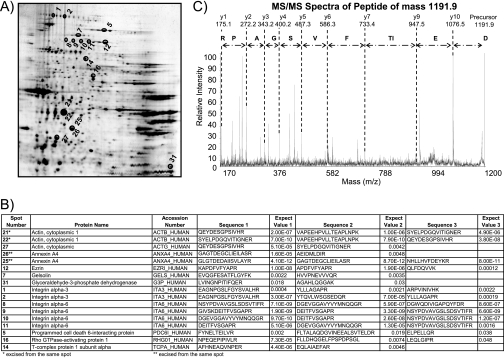
We performed 2DE with the aim of selecting random spots for MS identification and to confirm the absence/presence of these proteins in the main identification list. Running preparative gels, with 100 μg of purified exosomes per gel, was problematic because the spots picked contained too little material to yield confident protein identifications by MS. Increasing the amount of protein to 500 μg of exosomes per gel, however, resulted in an identification hit rate of >53%. Seventeen spots of intermediate staining intensity (silver-stained) were successfully identified by MS analysis (Fig. 3). These included integrins α3 and α6, gelsolin, cytosolic enzymes lactate dehydrogenase and glyceraldehyde-3-phosphate dehydrogenase, cytoskeleton proteins actin and cytokeratins, ezrin, and others. Nineteen of the 21 identifications from this gel-based approach were also identified by the nano-LC method, demonstrating excellent agreement (90%) between these different methods for resolving exosomal proteins or peptides.
Fig. 3.
Analysis of HT1376-derived exosomes using 2DE and MS. Protein extracts from HT1376 exosomes were resolved by 2DE on a pH 3–10 non-linear gradient. Proteins were visualized by silver staining (A). Thirty-two spots were randomly chosen, gel plugs were excised, and peptides were recovered following trypsin digestion. Of these, successful identifications were obtained for 17 spots (annotated in A), and the details of the MS identifications are listed (B). A representative MS/MS analysis from the data set is shown in C; the peptide is from integrin α6 (spot 10). The peptide has a precursor mass of 1191.9 and is annotated to show the derived peptide sequence.
Validation of Proteins Identified: Anomalous MHC Class I Identifications
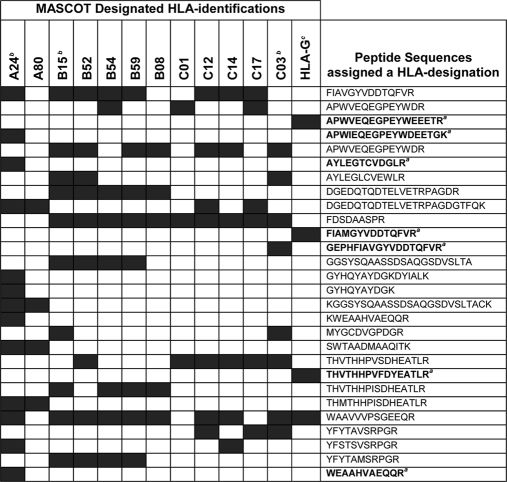
As with any such proteomics data set, it is important to evaluate the list manually for any unexpected or unexplainable MS identifications and to question the validity of any anomalies discovered in the data. In the current analysis, the LC-MALDI MS data contained multiple identifications for HLA molecules that passed our quality criteria (Expect values <0.05 and IDs based on more than one peptide). These identifications, however, were not physiologically possible as they included five HLA-B alleles and five HLA-C alleles (Table I) from a homogenous cell line. Explanations for this could include contamination of the source cell line with other cells from different donor(s), inadvertent contamination of the specimen by researchers, or issues related to how MASCOT designated HLA haplotypes nomenclature based on the peptide sequences generated from MS. To address these possibilities, a clinical diagnostic service (Welsh Blood Service, Llantrisant, Wales, UK) carried out haplotype analysis of the researcher and the HT1376 cell line. The researcher had no HLA alleles that corresponded to those in the MS list, whereas the HT1376 cells were haplotyped as HLA-A*24; -B*15(62); -Cw*03(9), confirming a homogenous cell line. This led us to examine in more detail the peptide sequences obtained and to evaluate how these were assigned by MASCOT to a given HLA nomenclature (Table I). It was apparent that several peptide sequences had been assigned to multiple HLA types. For example, sequence FDSDAASPR was designated to HLA-B15, -B52, -B54, and -B59 and to HLA-C01, -C12, -C17, and -C03. In contrast, however, there were some peptides that appeared in only a single designation. These unique sequences were assigned to HLA-A24 (APWIEQEGPEYWDEETGK, AYLEGTCVDGLR, and WEAAHVAEQQR), HLA-C03 (GEPHFIAVGYVDDTQFVR), and HLA-G (APWVEQEGPEYWEEETR, FIAMGYVDDTQFVR, and THVTHHPVFDYEATLR). There were no unique peptides for any HLA-B allele, although of the HLA-B subtypes identified, HLA-B15 was assigned the greatest number of peptides. In conclusion, manual analysis of peptides designated as MHC Class I identifications is recommended to clarify potential confusion arising from such MASCOT results.
Table I. Examination of MASCOT-designated MHC class I identifications, highlighting assignment of peptide sequences to more than one protein identification.
a Peptide sequences highlighted in bold represent those assigned to a single HLA identification.
b HLA haplotype of HT1376 cells by PCR.
c Positive expression confirmed by Western blot.
Validation of Exosomal Expression of Proteins Identified
It is also important to determine the validity of some MS-identified proteins by confirming their presence in the sample by other techniques. With a list as large as 353 proteins, it was not possible to do this wholesale so we restricted such validation to a set of proteins that may be of biological interest.
We performed a series of Western blot panels, analyzing up to 20 μg of HT1376 exosomes per well, to determine whether some MS-identified proteins were detectable in our exosome preparations. We stained for TSG101 as our choice marker for multivesicular bodies and hence exosomes. This protein was incidentally detected by MS by only a single peptide sequence and was therefore excluded from our data on this basis. LAMP-2, a molecule we expected to be present in exosomes, was detected in our sample by MS and was confirmed here to be strongly positive by Western blot (Fig. 4A). Among the MS identifications were numerous cytokeratin identifications (type I cytoskeletal keratins 1, 7, 13, 14, 16, 17, 18, and 19). We confirmed expression of cytokeratin 17 and cytokeratin 18 in the preparations, revealing abundant expression of exosomal cytokeratin 17. Cytokeratin 18, however, was only detectable with 20 μg of exosomes per well, suggesting that exosomes genuinely do express multiple cytoskeletal constituents and that the LC-MALDI MS approach is sufficiently sensitive to detect molecules such as cytokeratin 18 that are difficult to reveal by traditional Western blotting methods. Because of the anomalous issues surrounding MHC identifications, it was important to determine whether or not HLA-G was in fact expressed by HT1376 exosomes as this was not included in the PCR haplotyping of HT1376 cells. HLA-G was unequivocally confirmed to be present in exosomes by Western blot. Other membrane-associated (galectin-3, basigin, and CD73) or soluble (hnRNPK and β-catenin) molecules with documented associations in varied aspects of cancer biology were confirmed to be positively expressed by HT1376 exosomes.
Validation of Flotation Characteristics of Identified Exosomal Proteins
Although the standard exosome purification method used here is robust, it remains possible that some non-exosomal contaminating material is present in the preparations and that some of these MS identifications are not genuinely exosomally expressed proteins. To demonstrate that these proteins were exosomally expressed, we ultracentrifuged HT1376 culture medium at 70,000 × g, and the resuspended pellet was subjected to a second ultracentrifugation on a linear sucrose gradient. This was done to determine the capacity of the identified proteins to float at exosomal densities. Each of 15 fractions collected from the gradient was split: one-third was used for analysis by flow cytometry of exosome-coated beads, and two-thirds was used for Western blotting. The former method would reveal possible expression of candidate proteins at the exosome surface, whereas solubilizing exosomes for Western blot would allow surface and intraluminal constituents to be revealed. In the flow cytometry assay, exosome-containing fractions were identified by strong staining for tetraspanins CD9 and CD81 and for MHC Class I, which are known to be expressed on the surface of HT1376 exosomes, revealing a clear and principal peak at a density of 1.12 g/ml (Fig. 4B), which is within the expected exosomal density (Fig. 1C). This fraction, containing most of the exosomes, therefore also revealed positive surface staining for the MS-identified proteins β1 and α6 integrins, CD36 (lysosome membrane protein 2), CD44, CD73 (5′-nucleotidase), CD10 (neprilysin), MUC1, and basigin (CD147). 5T4, a protein not previously identified by any other exosome proteomics study, was included in this panel, demonstrating positive surface expression. The same fractions were also stained with a calnexin-specific antibody, revealing low level expression predominantly at densities greater than the exosome-containing fractions. This confirms the specificity of staining for the other markers tested and the absence of calnexin in exosome-containing fractions as expected (Fig. 4B). To reveal relevant fractions in the Western blot panel, we stained for TSG101, highlighting densities of 1.12–1.2 g/ml as exosome-containing (Fig. 4C). There was some positive staining at hyperdense fractions (>1.2g/ml), but this was relatively weak and may be due to exosome or protein aggregates. The proteins 5T4, CD44, basigin, galectin-3, and β-catenin all co-localized at the same density range, consistent with their exosomal expression. Overall, these data show that 18 of the MS protein identifications achieved in this study are confirmed to be expressed by HT1376 exosomes and that membrane-associated molecules, often difficult to solubilize and identify by MS approaches, have been successfully identified and validated as localized to the exosome membrane.
Preliminary Validation of Presence of Some MS-identified Proteins Present on Urinary Exosomes from Bladder Cancer Patients
Although we have previously examined exosomes present in the urine of prostate cancer patients (15), there are no studies to date specifically describing urinary exosomes of bladder cancer patients, although as we have mentioned, there is one report about urinary microparticles from this disease setting (17). Although a thorough examination of this question is outside the scope of this report, we have made preliminary efforts to ascertain the feasibility of doing such analyses of exosomes with bladder cancer patient-derived urine using the sucrose cushion purification method.
We first confirmed that the ultracentrifugation approach (sucrose cushion method) would be effective in isolating quality exosomes from other bladder cancer cell lines. We used the latex bead assay (as shown in Fig. 1) to evaluate the quality of the exosomes purified. Analysis of an additional four bladder cancer cell lines revealed it was possible to achieve purifications of quality comparable to those from HT1376 cells. The method should therefore be well suited for capturing bladder cancer exosomes irrespective of nuances of density that may differ slightly across different cells (Fig. 5A).
We next embarked on purifying exosomes from urine specimens using this method. Urine specimens were collected from four healthy individuals and from three patients with transitional cell carcinoma of the bladder prior to the commencement of any treatment. The latex bead assay was performed (Fig. 5B), revealing good levels of signals for the three tetraspanin molecules tested (CD9, CD81, and CD63), but only one preparation from each cohort passed our arbitrary quality threshold (over 5000 units for CD9 staining). These high quality specimens were examined further for expression of surface-oriented membrane proteins (similar to the panel in Fig. 4B), revealing that it is possible to detect positive expression (above isotype-stained controls) for most of these proteins tested. Importantly, some differences in the exosome profile between health and disease may be apparent using such a comparative test, such as elevated exosomal CD36, CD44, 5T4, basigin, and CD73 in cancer (Fig. 5C). We emphasize caution, however, in overinterpretation of these data based on the few clinical specimens that were available to us. Nevertheless, this aspect appears promising and warrants future follow-up studies.
DISCUSSION
Exosomes are highly complex nanometer-sized vesicles that are ubiquitous in biological systems. There is considerable research interest in understanding the physiological functions of exosomes in various settings, no more so than in elucidating their role in diseases like cancer. Studies of exosomes in various malignancies like prostate (8, 15, 16), breast (28), and colorectal cancers (44); melanoma (40, 45); pleural mesothelioma (5); malignancies of the central nervous system (21, 46); and others have begun. Collectively, these studies highlight expression of multiple tumor-related antigens by exosomes and perhaps general roles in immune modulation or in other aspects of cancer biology. In recent years, there has been particular interest in utilizing exosomes, isolated from patients, as tools for diagnosing disease (8, 11–14) or for discovering novel molecular markers (7). To date, however, there is only one report in the context of bladder cancer that highlights some disease-related differences in the protein constituents of urine-derived microparticles (17). Whether or not these proteins relate specifically to the exosomes present in such complex samples remains unclear.
This is an area that calls for some attention as managing and monitoring this disease are challenging, involving highly invasive and expensive procedures. Development of non-invasive tools for bladder cancer would therefore be particularly welcome. The direct contact between bladder urothelial cells and urine presents ample opportunity for secretion of exosomes directly into the urinary space (2). Collection and analysis of urinary exosomes in this disease setting hold promise as a novel diagnostic platform. However, exploitation of urinary exosomes requires a substantive high quality proteomic description of bladder cancer exosomes to be performed. Thus, this study provides essential solid ground that will greatly facilitate future developments in this understudied disease.
We have examined exosomes isolated from the HT1376 cell line, a good example of transitional cell carcinoma of bladder, and they seem typical of exosomes from other cellular sources. They are classically nanometer-sized vesicles that express high levels of tetraspanins, MHC molecules, markers of the endocytic tract, and adhesion molecules, and the vesicles exhibit the capacity to float at characteristic densities on sucrose gradients (19). This latter property is a very useful feature that can be used as a mode of separating exosomes from non-exosomal protein material that may co-pellet under high speed ultracentrifugation, an aspect that has perhaps not been fully appreciated in all exosome proteomics studies (2, 39, 40, 47, 48), resulting in possible false positive identifications. Our choice protocol is based on the method of Lamparski et al. (20) that provides a good exosome yield of very high purity. Similar approaches have also been used by other recent proteomics studies (21, 43, 49), and this vital investment in deriving maximal purity of input sample is essential for confidence in data arising from downstream proteomics analyses.
A major difficulty in the field, however, is to accurately estimate levels of sample contamination. One approach used by us and others is to perform Western blots for molecules not putatively expressed by exosomes, such as markers of the endoplasmic reticulum (e.g. calnexin or gp96), mitochondria, and nucleus. The difficulty here is that although such compartments may be relatively poorly represented in the exosome proteome it is not clear to what degree this rule is absolutely true (i.e. they may be present in exosomes at low levels). Our MS approach has indeed identified several proteins that are normally located to the endoplasmic reticulum or other compartments not well represented by exosomes, and this is also true of other exosome proteomics studies (37). These identifications may reflect the higher sensitivity of MS workflows for detecting these relatively low abundance contaminants that may not be detected efficiently by Western blotting. An alternative explanation, however, may be that these constituents are genuinely expressed by exosomes. If not directly loaded into/onto exosomes during manufacture, it may be possible that some proteins may be present at low levels at the outer surface of the cell and subsequently become taken up into the endosomal system and packaged into exosomes (50). In addition, a host of poorly understood cellular alterations occurring in cancer cells may modify trafficking of some proteins, resulting in inappropriate distributions, such as hnRNPK, which may become cytoplasmically rather than nuclearly located in certain cancers (51, 52). The absolute exclusion of endoplasmic reticulum-resident proteins (or proteins related to other cellular compartments) from exosomes may be a rule that is bent or broken in cancerous cells. Our use of “sticky” latex microbeads is a simple but effective method for estimating the degree of contamination of preparations by non-exosomal soluble proteins. Preparations replete with contaminants would result in beads poorly coated with exosomes, giving poor signal strength by flow cytometry for exosome surface markers like CD9. An arbitrary threshold value for this assay was set intentionally high as a means of assuring that the highest quality preparations only were subjected to proteomics. However, developing methods that discriminate well between exosomally expressed proteins and low abundance contaminants remains a challenge. Furthermore, establishing the precise route by which proteins are loaded into exosomes is far from straightforward. These may be aspects to be examined on a protein by protein basis during subsequent validation steps.
We used an LC-MALDI MS workflow, successfully identifying 353 proteins. This number of identifications is among the highest in the exosome proteomics field, and we are confident about the quality of such identifications because of the nature of the sample analyzed and because we report identifications based on a minimum of two peptides. A key issue for us in analyzing the data was to understand how our study compares with other exosome proteomics studies. We found ExoCarta (33), a database repository for exosome proteomics studies, to be a useful tool. By obtaining gene lists from these studies and our own, we were able to perform an overrepresentation analysis of the data, that is to discover whether there were more genes overlapping with ExoCarta gene sets than could be expected by chance. This statistical method, therefore, represents an unbiased approach for bioinformatics examination of our MS data. In essence, this revealed substantial data matching, particularly with high quality studies of colorectal cancer exosomes (42, 43), showing the HT1376 exosome proteome to be strongly consistent with exosomes of carcinoma origin. Similar results were evident using GeneGO MetaCore, emphasizing strong associations with carcinoma (of various sites) above other disease types. The GeneGO analysis, however, has also raised some issues that were not entirely expected, such as the predominant associations with control of the cytoskeleton. This statistically discovered association seems valid because multiple cytokeratins (cytokeratins 1, 5, 6B, 8, 13, 14, 16, 17, 18, and 19), actins (cytoplasmic 1, actin-like protein 6A, actin-related protein 2, and α cardiac muscle 1), myosin (myosin-1c, -10, and -14), tubulin (putative tubulin-like protein α-4B and tubulin β chain), and cytoskeletal linking proteins (filamin-A and -B, α-actinin-1 and -4, and plectin-1) were present in the identifications, and we confirmed expression of some by Western blot. The importance of these proteins in terms of exosome biology is unknown. Exploring exosomal cytokeratin profiles ex vivo may be clinically useful in bladder cancer as these can change with epithelial differentiation and with invasive properties and may help predict the outcome or assist differential diagnoses (27). An additional unexpected aspect, arising from the cell compartment GeneGO query, was the apparent strong associations with specific compartments, the melanosome and pigment granule. These tissue-specific organelles are responsible for the manufacture and storage of melanin in pigment cells of skin and eye. One would certainly not expect bladder cancer cells to strongly resemble pigment cells, and as such, this was a surprising finding. However, the melanosome is a specialized endosomal compartment that is derived from early endosomal intermediates that strongly resemble multivesicular bodies in morphology and composition. Thus, the highly evolutionally conserved molecular machinery involved in protein chaperone functions, in membrane fusion and budding events, and in transportation of proteins to the melanosome bears significant resemblance to the generic machinery giving rise to exosomes (53), which may explain this association.
We also, albeit briefly, investigated whether exosomes could be isolated from other bladder cancer cell lines and more importantly from urine specimens using the same exosome purification method. This was highly successful, giving comparable levels of purity assessed by our latex bead assay, when using an additional four bladder cancer cell lines. When applying the techniques to freshly collected urine specimens, a source that is significantly more complex and more variable than cell culture supernatant as we described previously (15), most preparations did not reach our “exosome quality threshold.” Nevertheless, preparations from healthy donors and bladder cancer patient urine stained strongly for the tetraspanin proteins CD9, CD81, and CD63. This aspect is of particular note because it may be attractive in future clinical studies to move away from the reliance on ultracentrifugation methods, which are cumbersome and impractical for large sample sets, and replace these with an affinity approach. Such tetraspanins, therefore, would be a good choice for antibody-mediated exosome capture either directly onto microtiter plates or onto microbeads (1) followed by an analysis for additional protein markers of interest (i.e. those within our MS identifications). In fact, we were able to demonstrate that several MS-identified exosome membrane proteins were expressed at relatively higher levels in exosomes isolated from a bladder cancer patient compared with exosomes isolated from a healthy donor. Although not exhaustive, such data suggest that urinary exosome analysis in the context of bladder cancer may prove fruitful and is certainly worthy of further attention.
In summary, we have achieved the first high quality proteomic description of bladder cancer cell-derived exosomes and have learned three key lessons that have wide applicability to other proteomics studies. First, we show the usefulness of DTT as part of a solubilization buffer. This agent increased the number of identifications more than 100-fold. Second, we highlight that care must be taken with identifications of proteins showing considerable homology (with HLA proteins being an excellent case study). Our work demonstrates that MASCOT is not capable of distinguishing between the incorrect and correct HLA molecules present in exosomes. We believe this problem has probably affected many other proteomics studies. Third, we emphasize the importance of careful sample preparation both as part of a proteomics workflow and for validation. The particular example in our study was our use of latex microbeads. This allowed us to quality control our specimens before commencing proteomics and, using this approach, also allowed us to validate, quantify, and confirm the orientation of 12 proteins in a way that is impossible with just Western blotting. Follow-up investigations, informed by this report, are now planned to identify the presence of candidate markers in the urine of bladder cancer patients with the ultimate goal of replacing highly invasive procedures currently utilized in diagnosis and monitoring with a fully non-invasive urinary exosome-based technique.
Acknowledgments
We thank Dr. Richard Harrop, Oxford BioMedica UK Ltd., for providing the antibody against 5T4. We also thank Vickie Nixon, Cardiff University Central Biotechnology Services Proteomics Facility and Jacquelyn Court, Velindre NHS Trust, Cardiff, for technical assistance. We are also grateful to Dr. Zsuzsanna Tabi, Cancer Immunology Group, Section of Oncology, School of Medicine Cardiff University, for valued scientific comments and proofreading the manuscript.
* This work was supported by a Cancer Research Wales Ph.D. studentship (to J. L. W.) and by the Velindre NHS Trust.
 This article contains supplemental Fig. 1 and Table 1.
This article contains supplemental Fig. 1 and Table 1.
1 The abbreviations used are:
- TCC
- transitional cell carcinoma
- 5T4
- trophoblast glycoprotein
- BCA
- bicinchoninic acid
- FDR
- false discovery rate
- HLA
- human leukocyte antigen
- hsp90
- heat shock protein 90
- LAMP
- lysosome-associated membrane protein
- MHC
- major histocompatability complex
- TEAB
- triethylammonium bicarbonate
- TSG101
- tumor susceptibility gene 101
- NHS
- National Health Service
- FBS
- fetal bovine serum
- RT
- room temperature
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- 2DE
- two-dimensional electrophoresis
- ID
- identifier
- ORA
- overrepresentation analysis.
REFERENCES
- 1.Caby M. P., Lankar D., Vincendeau-Scherrer C., Raposo G., Bonnerot C. (2005) Exosomal-like vesicles are present in human blood plasma. Int. Immunol 17, 879–887 [DOI] [PubMed] [Google Scholar]
- 2.Pisitkun T., Shen R. F., Knepper M. A. (2004) Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. U.S.A 101, 13368–13373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Admyre C., Johansson S. M., Qazi K. R., Filén J. J., Lahesmaa R., Norman M., Neve E. P., Scheynius A., Gabrielsson S. (2007) Exosomes with immune modulatory features are present in human breast milk. J. Immunol 179, 1969–1978 [DOI] [PubMed] [Google Scholar]
- 4.Gatti J. L., Métayer S., Belghazi M., Dacheux F., Dacheux J. L. (2005) Identification, proteomic profiling, and origin of ram epididymal fluid exosome-like vesicles. Biol. Reprod 72, 1452–1465 [DOI] [PubMed] [Google Scholar]
- 5.Bard M. P., Hegmans J. P., Hemmes A., Luider T. M., Willemsen R., Severijnen L. A., van Meerbeeck J. P., Burgers S. A., Hoogsteden H. C., Lambrecht B. N. (2004) Proteomic analysis of exosomes isolated from human malignant pleural effusions. Am. J. Respir. Cell Mol. Biol 31, 114–121 [DOI] [PubMed] [Google Scholar]
- 6.Taylor D. D., Gercel-Taylor C. (2008) MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol 110, 13–21 [DOI] [PubMed] [Google Scholar]
- 7.Zhou H., Pisitkun T., Aponte A., Yuen P. S., Hoffert J. D., Yasuda H., Hu X., Chawla L., Shen R. F., Knepper M. A., Star R. A. (2006) Exosomal Fetuin-A identified by proteomics: a novel urinary biomarker for detecting acute kidney injury. Kidney Int 70, 1847–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nilsson J., Skog J., Nordstrand A., Baranov V., Mincheva-Nilsson L., Breakefield X. O., Widmark A. (2009) Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br. J. Cancer 100, 1603–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Théry C., Ostrowski M., Segura E. (2009) Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol 9, 581–593 [DOI] [PubMed] [Google Scholar]
- 10.Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J. J., Lötvall J. O. (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol 9, 654–659 [DOI] [PubMed] [Google Scholar]
- 11.Ogawa Y., Kanai-Azuma M., Akimoto Y., Kawakami H., Yanoshita R. (2008) Exosome-like vesicles with dipeptidyl peptidase IV in human saliva. Biol. Pharm. Bull 31, 1059–1062 [DOI] [PubMed] [Google Scholar]
- 12.Rabinowits G., Gerçel-Taylor C., Day J. M., Taylor D. D., Kloecker G. H. (2009) Exosomal microRNA: a diagnostic marker for lung cancer. Clin. Lung Cancer 10, 42–46 [DOI] [PubMed] [Google Scholar]
- 13.Taylor D. D., Akyol S., Gercel-Taylor C. (2006) Pregnancy-associated exosomes and their modulation of T cell signaling. J. Immunol 176, 1534–1542 [DOI] [PubMed] [Google Scholar]
- 14.Skriner K., Adolph K., Jungblut P. R., Burmester G. R. (2006) Association of citrullinated proteins with synovial exosomes. Arthritis Rheum 54, 3809–3814 [DOI] [PubMed] [Google Scholar]
- 15.Mitchell P. J., Welton J., Staffurth J., Court J., Mason M. D., Tabi Z., Clayton A. (2009) Can urinary exosomes act as treatment response markers in prostate cancer? J. Transl. Med 7, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jansen F. H., Krijgsveld J., van Rijswijk A., van den Bemd G. J., van den Berg M. S., van Weerden W. M., Willemsen R., Dekker L. J., Luider T. M., Jenster G. (2009) Exosomal secretion of cytoplasmic prostate cancer xenograft-derived proteins. Mol. Cell. Proteomics 8, 1192–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smalley D. M., Sheman N. E., Nelson K., Theodorescu D. (2008) Isolation and identification of potential urinary microparticle biomarkers of bladder cancer. J. Proteome Res 7, 2088–2096 [DOI] [PubMed] [Google Scholar]
- 18.Conde-Vancells J., Rodriguez-Suarez E., Embade N., Gil D., Matthiesen R., Valle M., Elortza F., Lu S. C., Mato J. M., Falcon-Perez J. M. (2008) Characterization and comprehensive proteome profiling of exosomes secreted by hepatocytes. J. Proteome Res 7, 5157–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raposo G., Nijman H. W., Stoorvogel W., Liejendekker R., Harding C. V., Melief C. J., Geuze H. J. (1996) B Lymphocytes secrete antigen-presenting vesicles. J. Exp. Med 183, 1161–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamparski H. G., Metha-Damani A., Yao J. Y., Patel S., Hsu D. H., Ruegg C., Le Pecq J. B. (2002) Production and characterization of clinical grade exosomes derived from dendritic cells. J. Immunol. Methods 270, 211–226 [DOI] [PubMed] [Google Scholar]
- 21.Graner M. W., Alzate O., Dechkovskaia A. M., Keene J. D., Sampson J. H., Mitchell D. A., Bigner D. D. (2009) Proteomic and immunologic analyses of brain tumor exosomes. FASEB J 23, 1541–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Théry C., Boussac M., Véron P., Ricciardi-Castagnoli P., Raposo G., Garin J., Amigorena S. (2001) Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J. Immunol 166, 7309–7318 [DOI] [PubMed] [Google Scholar]
- 23.Wubbolts R., Leckie R. S., Veenhuizen P. T., Schwarzmann G., Möbius W., Hoernschemeyer J., Slot J. W., Geuze H. J., Stoorvogel W. (2003) Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J. Biol. Chem 278, 10963–10972 [DOI] [PubMed] [Google Scholar]
- 24.Rasheed S., Gardner M. B., Rongey R. W., Nelson-Rees W. A., Arnstein P. (1977) Human bladder carcinoma: characterization of two new tumor cell lines and search for tumor viruses. J. Natl. Cancer Inst 58, 881–890 [DOI] [PubMed] [Google Scholar]
- 25.Masters J. R., Hepburn P. J., Walker L., Highman W. J., Trejdosiewicz L. K., Povey S., Parkar M., Hill B. T., Riddle P. R., Franks L. M. (1986) Tissue culture model of transitional cell carcinoma: characterization of twenty-two human urothelial cell lines. Cancer Res 46, 3630–3636 [PubMed] [Google Scholar]
- 26.Mitchell J. P., Court J., Mason M. D., Tabi Z., Clayton A. (2008) Increased exosome production from tumour cell cultures using the Integra CELLine Culture System. J. Immunol. Methods 335, 98–105 [DOI] [PubMed] [Google Scholar]
- 27.Southgate J., Harnden P., Trejdosiewicz L. K. (1999) Cytokeratin expression patterns in normal and malignant urothelium: a review of the biological and diagnostic implications. Histol. Histopathol 14, 657–664 [DOI] [PubMed] [Google Scholar]
- 28.Andre F., Schartz N. E., Movassagh M., Flament C., Pautier P., Morice P., Pomel C., Lhomme C., Escudier B., Le Chevalier T., Tursz T., Amigorena S., Raposo G., Angevin E., Zitvogel L. (2002) Malignant effusions and immunogenic tumour-derived exosomes. Lancet 360, 295–305 [DOI] [PubMed] [Google Scholar]
- 29.Clayton A., Mitchell J. P., Court J., Mason M. D., Tabi Z. (2007) Human tumour-derived exosomes selectively impair lymphocyte responses to interleukin-2. Cancer Res 67, 7458–7466 [DOI] [PubMed] [Google Scholar]
- 30.Théry C., Amigorena S., Raposo G., Clayton A. (2006) Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol Chapter 3, Unit 3.22 [DOI] [PubMed] [Google Scholar]
- 31.Clayton A., Harris C. L., Court J., Mason M. D., Morgan B. P. (2003) Antigen presenting cell exosomes are protected from complement mediated lysis by expression of CD55 and CD59. Eur. J. Immunol 33, 522–531 [DOI] [PubMed] [Google Scholar]
- 32.Brennan P., Shore A. M., Clement M., Hewamana S., Jones M. C., Giles P., Fegan C., Pepper C., Brewis I. A. (2009) Quantitative nuclear proteomics reveals new phenotypes altered in lymphoblastoid cells. Proteomics Clin. Appl 3, 359–369 [DOI] [PubMed] [Google Scholar]
- 33.Mathivanan S., Simpson R. J. (2009) ExoCarta: a compendium of exosomal proteins and RNA. Proteomics 9, 4997–5000 [DOI] [PubMed] [Google Scholar]
- 34.Clayton A., Turkes A., Navabi H., Mason M. D., Tabi Z. (2005) Inducible heat shock protein expression in B cell exosomes. J. Cell Sci 118, 3631–3638 [DOI] [PubMed] [Google Scholar]
- 35.Dai S., Wan T., Wang B., Zhou X., Xiu F., Chen T., Wu Y., Cao X. (2005) More efficient induction of HLA-A*0201-restricted and carcinoembryonic antigen (CEA)-specific CTL response by immunization with exosomes prepared from heat-stressed CEA-positive tumor cells. Clin. Cancer Res 11, 7554–7563 [DOI] [PubMed] [Google Scholar]
- 36.Tan S., Tan H. T., Chung M. C. (2008) Membrane proteins and membrane proteomics. Proteomics 8, 3924–3932 [DOI] [PubMed] [Google Scholar]
- 37.Simpson R. J., Lim J. W., Moritz R. L., Mathivanan S. (2009) Exosomes: proteomic insights and diagnostic potential. Expert Rev. Proteomics 6, 267–283 [DOI] [PubMed] [Google Scholar]
- 38.van Niel G., Raposo G., Candalh C., Boussac M., Hershberg R., Cerf-Bensussan N., Heyman M. (2001) Intestinal epithelial cells secrete exosome-like vesicles. Gastroenterology 121, 337–349 [DOI] [PubMed] [Google Scholar]
- 39.Hegmans J. P., Bard M. P., Hemmes A., Luider T. M., Kleijmeer M. J., Prins J. B., Zitvogel L., Burgers S. A., Hoogsteden H. C., Lambrecht B. N. (2004) Proteomic analysis of exosomes secreted by human mesothelioma cells. Am. J. Pathol 164, 1807–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mears R., Craven R. A., Hanrahan S., Totty N., Upton C., Young S. L., Patel P., Selby P. J., Banks R. E. (2004) Proteomic analysis of melanoma-derived exosomes by two-dimensional polyacrylamide gel electrophoresis and mass spectrometry. Proteomics 4, 4019–4031 [DOI] [PubMed] [Google Scholar]
- 41.Kesimer M., Scull M., Brighton B., DeMaria G., Burns K., O'Neal W., Pickles R. J., Sheehan J. K. (2009) Characterization of exosome-like vesicles released from human tracheobronchial ciliated epithelium: a possible role in innate defense. FASEB J 23, 1858–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mathivanan S., Lim J. W., Tauro B. J., Ji H., Moritz R. L., Simpson R. J. (2010) Proteomic analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol. Cell. Proteomics 9, 197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi D. S., Lee J. M., Park G. W., Lim H. W., Bang J. Y., Kim Y. K., Kwon K. H., Kwon H. J., Kim K. P., Gho Y. S. (2007) Proteomic analysis of microvesicles derived from human colorectal cancer cells. J. Proteome Res 6, 4646–4655 [DOI] [PubMed] [Google Scholar]
- 44.Dai S., Wei D., Wu Z., Zhou X., Wei X., Huang H., Li G. (2008) Phase I clinical trial of autologous ascites-derived exosomes combined with GM-CSF for colorectal cancer. Mol. Ther 16, 782–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Escudier B., Dorval T., Chaput N., André F., Caby M. P., Novault S., Flament C., Leboulaire C., Borg C., Amigorena S., Boccaccio C., Bonnerot C., Dhellin O., Movassagh M., Piperno S., Robert C., Serra V., Valente N., Le Pecq J. B., Spatz A., Lantz O., Tursz T., Angevin E., Zitvogel L. (2005) Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: results of the first phase I clinical trial. J. Transl. Med 3, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Potolicchio I., Carven G. J., Xu X., Stipp C., Riese R. J., Stern L. J., Santambrogio L. (2005) Proteomic analysis of microglia-derived exosomes: metabolic role of the aminopeptidase CD13 in neuropeptide catabolism. J. Immunol 175, 2237–2243 [DOI] [PubMed] [Google Scholar]
- 47.Gonzales P. A., Pisitkun T., Hoffert J. D., Tchapyjnikov D., Star R. A., Kleta R., Wang N. S., Knepper M. A. (2009) Large-scale proteomics and phosphoproteomics of urinary exosomes. J. Am. Soc. Nephrol 20, 363–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gonzalez-Begne M., Lu B., Han X., Hagen F. K., Hand A. R., Melvin J. E., Yates J. R. (2009) Proteomic analysis of human parotid gland exosomes by multidimensional protein identification technology (MudPIT). J. Proteome Res 8, 1304–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fauré J., Lachenal G., Court M., Hirrlinger J., Chatellard-Causse C., Blot B., Grange J., Schoehn G., Goldberg Y., Boyer V., Kirchhoff F., Raposo G., Garin J., Sadoul R. (2006) Exosomes are released by cultured cortical neurones. Mol. Cell. Neurosci 31, 642–648 [DOI] [PubMed] [Google Scholar]
- 50.Papp K., Végh P., Prechl J., Kerekes K., Kovács J., Csikós G., Bajtay Z., Erdei A. (2008) B lymphocytes and macrophages release cell membrane deposited C3-fragments on exosomes with T cell response-enhancing capacity. Mol. Immunol 45, 2343–2351 [DOI] [PubMed] [Google Scholar]
- 51.Barboro P., Repaci E., Rubagotti A., Salvi S., Boccardo S., Spina B., Truini M., Introini C., Puppo P., Ferrari N., Carmignani G., Boccardo F., Balbi C. (2009) Heterogeneous nuclear ribonucleoprotein K: altered pattern of expression associated with diagnosis and prognosis of prostate cancer. Br. J. Cancer 100, 1608–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carpenter B., McKay M., Dundas S. R., Lawrie L. C., Telfer C., Murray G. I. (2006) Heterogeneous nuclear ribonucleoprotein K is over expressed, aberrantly localised and is associated with poor prognosis in colorectal cancer. Br. J. Cancer 95, 921–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raposo G., Marks M. S. (2007) Melanosomes-dark organelles enlighten endosomal membrane transport. Nat. Rev. Mol. Cell Biol 8, 786–797 [DOI] [PMC free article] [PubMed] [Google Scholar]