Abstract
Approximately 75% of vertebrate proteins belong to protein families encoded by multiple evolutionarily related genes, a pattern that emerged as a result of gene and genome duplications over the course of vertebrate evolution. In families of genes with similar or related functions, adaptation to a strong selective agent should involve multiple adaptive changes across the entire gene family. However, we know of no evolutionary studies that have explicitly addressed this point. Here, we show how 4 taxonomically diverse species of pufferfishes (Tetraodontidae) each evolved resistance to the guanidinium toxins tetrodotoxin (TTX) and saxitoxin (STX) via parallel amino acid replacements across all 8 sodium channels present in teleost fish genomes. This resulted in diverse suites of coexisting sodium channel types that all confer varying degrees of toxin resistance, yet show remarkable convergence among genes and phylogenetically diverse species. Using site-directed mutagenesis and expression of a vertebrate sodium channel, we also demonstrate that resistance to TTX/STX is enhanced up to 15-fold by single, frequently observed replacements at 2 sites that have not previously been implicated in toxin binding but show similar or identical replacements in pufferfishes and in distantly related vertebrate and nonvertebrate animals. This study presents an example of natural selection acting upon a complete gene family, repeatedly arriving at a diverse but limited number of adaptive changes within the same genome. To be maximally informative, we suggest that future studies of molecular adaptation should consider all functionally similar paralogs of the affected gene family.
Keywords: adaptation, parallel evolution, gene families, sodium channels, tetrodotoxin, saxitoxin
Introduction
A key focus in evolutionary biology is the question of how organisms adapt to changes in their environments. Adaptation must involve evolutionary change at the molecular level but often the selective agent is complex or ill defined, and it can be difficult to pinpoint the relevant genes or the specific changes in protein sequence that affect protein function. Some of the most lucid examples of molecular adaptation come from studies of the physical interactions between channel-binding neurotoxins and the single voltage–gated sodium channel present in the genomes of most nonvertebrate animals. For example, in wild insect populations, rapid evolution of pyrethroid and DDT resistance occurred within a few decades of insecticide exposure due to natural selection for point mutations in the sodium channels of flies, cockroaches, and other species (ffrench-Constant et al. 1998; Liu et al. 2002). More recently, some populations of the soft-shell clam Mya arenaria were found to be resistant to the naturally occurring paralytic shellfish poisoning guanidinium toxin saxitoxin (STX), due to a specific amino acid replacement in the single sodium channel in their genome (Bricelj et al. 2005).
Similar studies in vertebrate animals have identified specific mutations that provide some sodium channels with resistance to the deadly, naturally occurring guanidinium toxin tetrodotoxin (TTX), an extremely potent agent that, like STX, arrests neural and muscular action potentials by binding to and blocking the sodium channel pore that allows passage of the Na+ ion. TTX has been found in a remarkably diverse variety of animals, including pufferfishes such as the genomic model species Takifugu rubripes and Tetraodon nigroviridis; related tetraodontiform and other fishes (Halstead 1967); some newts (e.g., Taricha spp. and Cynops pyrrhogaster; Hanifin et al. 1999) and frogs (Tanu et al. 2001; Pires et al. 2002); and a number of nonvertebrate species from at least 6 different phyla (Scheumack et al. 1978; Theusen 1991; Blair and Anderson 1993; Asakawa et al. 2000; Lin and Hwang 2001; Nagashima et al. 2002). Although STX and TTX are structurally different molecules, both contain a positively charged guanidinium moiety that is believed to bind to the sodium channel pore in a similar way. Thus, organisms that are resistant to one toxin (TTX or STX) are usually resistant to the other, and many of the same amino acid sites are implemented in both TTX and STX binding and resistance.
Recently, one TTX-resistant sodium channel has been described in some populations of the garter snake Thamnophis sirtalis that preys on the TTX-sequestering newt Taricha granulosa (Geffeney et al. 2005) and in pufferfishes that also sequester exceedingly high concentrations of TTX in their tissues (Yotsu-Yamashita et al. 2000; Venkatesh et al. 2005). However, such single-gene studies are incomplete because in vertebrate animals, neurotoxin resistance (i.e., survival) cannot be achieved by evolution of a single channel alone; adaptation likely requires parallel evolution of all the paralogs in the encoding gene family. As a result of 3 proposed rounds of whole-genome duplication—2 in early vertebrate ancestors and 1 in early teleosts—teleost fish (including pufferfishes) possess a family of 8 voltage-gated sodium channels (Novak et al. 2006), each of which has its own profile of expression in skeletal muscle, the heart, or the nervous system. However, to date only 2 of these channels (teleost Nav1.4a and Nav1.4b, both expressed in skeletal muscle) have been shown to be TTX/STX resistant in pufferfishes (Yotsu-Yamashita et al. 2000; Venkatesh et al. 2005).
The voltage-gated sodium channel is comprised of 4 transmembrane subunits (Domains I, II, III, and IV) that fold together to form an ion-permeable pore (fig. 1A). Protein modeling and site-directed mutagenesis experiments have demonstrated that TTX and STX bind directly to highly conserved residues in the pore region and that pore-region mutations are particularly likely to affect sensitivity to these toxins (Terlau et al. 1991). Here, we address how pufferfishes evolved genetic resistance to TTX and/or STX (Nakashima et al. 2004; Tanu et al. 2004; Jang and Yotsu-Yamashita 2006) via pore-region amino acid replacements in the whole complement of sodium channel paralogs present in their genomes. Of all the tetraodontiform fishes, the tetraodontidae are the most toxic and the most TTX resistant (Kidokoro et al. 1974); thus, we wished to test the prediction that all or most of the sodium channels in pufferfish genomes should differ from their orthologs in nontoxic teleosts. For each pore-region change we examined, the functional effects on sodium channel physiology were studied empirically using site-directed mutagenesis and toxin-binding experiments (dose–response voltage clamp). By analyzing sodium channel genes of multiple species of pufferfishes, we also ask whether amino acid replacements conferring resistance to guanidinium toxins occurred early (preceding the radiation of Tetraodontidae) or evolved in parallel.
FIG. 1.—
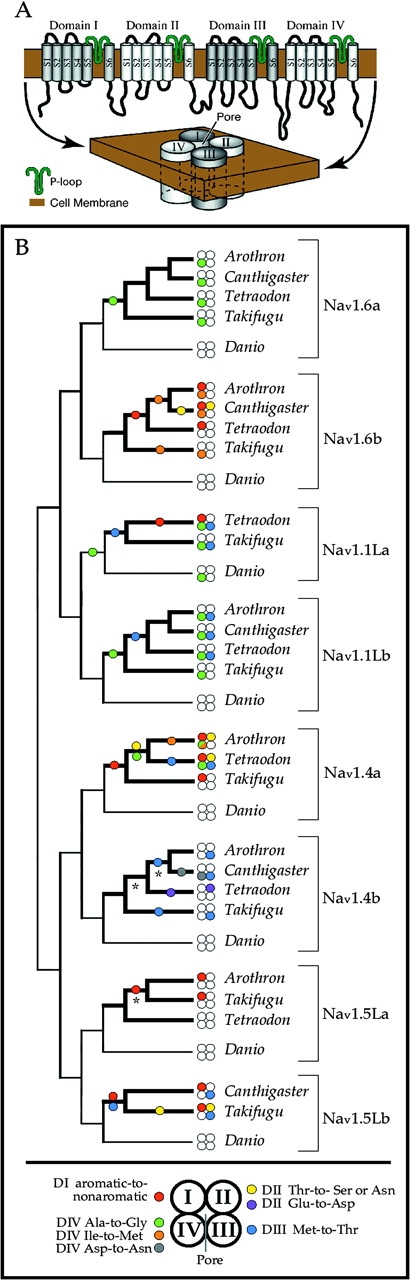
(A and B) Sodium channel protein structure and phylogeny showing parallel evolution of TTX resistance in pufferfish sodium channels. (A) Linear and folded structure of the voltage-gated sodium channel. Four short and highly conserved pore loops, 1 from each of 4 domains, come together to form the ion-permeable pore. (B) Phylogenetic reconstruction. Branches leading to genes from TTX-sequestering species are indicated by bold weight lines. The pore region of each channel is represented by 4 circles, 1 for each pore loop in each of the 4 protein domains. Circles are colored to indicate specific amino acid replacements at sites in each domain that enhance resistance to toxin blocking. Colored circles along branches indicate the most parsimonious reconstruction for the origin of the change. From these results, it is clear that pufferfishes have evolved a variety of different genotypes that result in TTX-resistant phenotypes. Asterisks indicate 3 nodes with Bayesian posterior support of less than 99%, which did not affect our conclusions (due to higher support at more basal nodes.)
Materials and Methods
Taxon Sampling
We chose to focus our study on pufferfishes (Teleostei: Tetraodontiformes: Tetraodontidae) because they typically contain high concentrations of guanidinium neurotoxins and are thus likely to have evolved under strong selection for toxin-resistant sodium channels. For taxonomic breadth, we chose exemplars from 4 different genera. Species included the existing genomic models Takifugu rubripes and Tetraodon nigroviridis and 2 additional taxa, Canthigaster solandri and Arothron nigropunctatus. The zebrafish Danio rerio (also a teleost) was selected as the outgroup and comparative species, due to the fact that it is not a member of Tetraodontiformes and because its sodium channels are not resistant to guanidinium toxins.
Cloning and Sequencing
For C. solandri and A. nigropunctatus, RNA from muscle, brain, or heart was isolated by homogenization in guanidium thiocyanate or RNA STAT-60 (Ambion, Austin, TX). Reverse transcription was performed with either the random primers or the 3′ reverse primer of the subsequent polymerase chain reaction (PCR). PCR primers and reaction parameters were optimized for each gene and species. PCR products were cloned into the TOPO TA vector (Invitrogen, Carlsbad, CA) following the addition of terminal adenines (1 unit Taq at 72 °C, 10 min). Inserts were sequenced in both directions with an ABI 3100 sequencer (Applied Biosystems, Foster City, CA), and additional rounds of PCR were performed with both specific and degenerate primers to extend products in both directions until all 4 channel domains were sequenced. Resultant contiguous sequences were up to 5,000+ bases in length. Eleven new pufferfish sequences were deposited in GenBank under accession numbers EU391401–EU391406 (for A. nigropunctatus) and EU391407–EU391411 (for C. solandri).
Orthologs of some of the 8 sodium channel genes present in the genomes of other teleost fish (Novak et al. 2006) were not isolated in Canthigaster or Arothron, despite repeated attempts using different individuals, tissues, primers, and PCR parameters. Some of these genes may have been deleted (lost) from the genomes of these species, either in the course of genome size reduction in pufferfishes (Brenner et al. 1993; Neafsey and Palumbi 2003) or possibly as a result of selection against TTX-sensitive sodium channels. Regardless of the reason some genes could not be isolated, the results and conclusions of our study are not affected by their absence from the analyses.
Phylogenetic Analyses
The alignment of nucleotide sequences included our data for C. solandri, A. nigropunctatus, and D. rerio (Novak et al. 2006), as well as assembled contiguous sequences for sodium channels from the Takifugu rubripes (version 4: http://genome.jgi-psf.org/Takru4/Takru4.home.html) and Tetraodon nigroviridis (http://www.genoscope.cns.fr/externe/tetranew/) genome databases (Aparicio et al. 2002; Jaillon et al. 2004). Takifugu rubripes exons were assembled using sequence data from version 4 scaffolds 36, 46, 47, 66, 134, 317, 502, and 559. Tetraodon nigroviridis exons were assembled using sequence data from chromosomes 2, 3, 9, 11, 15, and Un_random. Danio rerio sequences from a previous study (Novak et al. 2006) are deposited under GenBank accession numbers NM_131628, NM_001044895, NM_001039825, NM_001045183, NM_200132, NM_001045065, NM_001045123, and NM_001044922.
A rough alignment of nucleotide sequences was initially created using default parameters in ClustalX (Thompson et al. 1997) and improved manually using known vertebrate sodium channel amino acid sequences. Amino acid sequences and sequence lengths were highly conserved for transmembrane regions; however, because the cytoplasmic loops between domains can vary extensively in length and sequence among genes, we excluded those regions from subsequent phylogenetic analyses. Phylogeny was estimated using MrBayes 3.0 (Ronquist and Huelsenbeck 2003), implementing a general time-reversible model with invariable sites and a gamma distribution for variable rate sites (GTR + I + G). Four Markov chains of 1,000,000 generations were run at the default temperature (0.2), and every 100th tree was saved to a file. The burn-in asymptote was determined by plotting tree number against log likelihood. Trees saved before the burn-in asymptote were discarded, and a majority rule consensus of the remaining trees was calculated to estimate posterior probabilities. Four replicates of these Bayesian runs were conducted to insure convergence of the posteriors. The final phylogenetic estimate was rooted to reflect the deep double-forked topology implied by 2 rounds of genome duplication in vertebrates, followed by a third round unique to teleosts (Novak et al. 2006).
We then used the phylogeny and likelihood tests in PLATO v.2.11 (Grassly and Rambaut 1999) to test for any anomalously similar regions in the alignment that could indicate nonindependent evolutionary processes such as gene conversion. For the entire alignment, 500 bp of flanking sequence immediately upstream and downstream to the pore regions of all 4 domains were analyzed using a variable sliding window from 5 to 250 bp and Monte Carlo simulation of the null distribution. No significantly anomalous regions were detected in any domain for any sodium channel gene in the analysis, supporting the conclusion that identical replacements in different genes are the product of parallel evolution and not gene conversion.
Pore region amino acid replacement events were optimized onto branches of the phylogeny using parsimony to minimize the number of independent changes (gains or losses) required to explain the phylogenetic distribution of variable sites. In 2 ambiguous cases—Domain III Met-Thr replacements for Nav1.4b and Domain IV Ile-Met replacements for Nav1.6b—alternative character state optimizations did not reduce the total number of required gains or losses. When we initially observed the phylogenetic distribution of amino acid replacements, we also noticed that parallel replacements at a particular Domain II Thr (see Results) appeared to occur chronologically after parallel replacements in Domain I. We tested the significance of this pattern using the concentrated changes test of Maddison (1990), running 1,000 evolutionary simulations with the Domain I replacement as the independent character and the Domain II replacement as the dependent character, and assuming the observed number of gains and losses.
Mutagenesis, Oocyte Expression, and Physiological Measurements
The rat skeletal muscle (Nav1.4) cDNA flanked by the Xenopus globulin 5′- and 3′-untranslated regions was provided by J. R. Moorman, University of Virginia. Site-directed mutagenesis was performed using the Quikchange Site-Directed Mutagenesis Kit (Stratagene, Cedar Creek, TX), according to the manufacturer's protocols. Mutagenic oligonucleotide primers included changes to result in the desired amino acid mutation and an additional change to introduce silent restriction sites that did not alter the coding sequence but allowed rapid identification of mutant cDNA. Mutations were confirmed by DNA sequencing. The μ1-pAlterXG vector was linearized by SalI digestion and transcribed with SP6 DNA-dependent RNA polymerase using reagents from Ambion.
Stage V and VI Xenopus oocytes were isolated from female frogs (NASCO, Ft Atkinson, WI), washed with Ca2+-free solution (90 mM NaCl, 2.5 mM KCl, 1 mM MgCl2, 1 mM NaHPO4, and 5 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid [HEPES] titrated to pH 7.6 with 1 N NaOH), and treated with 2 mg/ml collagenase (Sigma-Aldrich, St. Louis, MO) for 1.5 h; follicular cell layers were manually removed. Oocytes were injected with 5–100 ng of cRNA. Either native or mutant subunit cRNAs were injected. Oocytes were incubated at 17 °C for 1–5 days before examination. One to 7 days after injection, INa was recorded from cRNA-injected oocytes with a 2-electrode voltage clamp/bath clamp. A Dagan CA-1 (bath clamp) with a series resistance compensation circuit (TEV-208, Dagan, Minneapolis, MN) was used to make recordings. Varied concentrations of TTX were added to the bath, and dose–response curves for TTX were made using previously described methods (Penzotti et al. 1998). All recordings were made at 20–22 °C in a flowing bath solution consisting of (in mM): 96 NaCl, 2.5 KCl, 1 CaCl2, 1 MgCl2, and 5 HEPES, pH 7.5. Electrodes contained 3 M KCl and had resistances that ranged from 0.3 to 1.5 MΩ. Data were acquired using pClamp8 software (Axon CNS, Sunnyvale, CA). Data were digitized at 42 kHz and were low-pass filtered at 10 kHz (−3 dB).
Results
Phylogenetic Distribution of Pore-Region Replacements
We aligned nucleotide sequences and conducted phylogenetic analyses of all the sodium channels we could isolate from 4 species of pufferfish (Tetraodontidae)—Takifugu rubripes, Tetraodon nigroviridis, C. solandri, and A. nigropunctatus—and from an outgroup teleost, the zebrafish D. rerio. The topology of the resulting gene tree (fig. 1B) is constrained to preserve the organization of this gene family into 8 orthologs (Novak et al. 2006) and rooted to reflect the “double-forked” phylogenetic topology predicted by 2 rounds of whole-genome duplication before the divergence of teleosts and tetrapods, followed by a third whole-genome duplication in teleosts (Lopreato et al. 2001; Novak et al. 2006). All nodes except 3 had Bayesian posterior support of 99–100% (see fig. 1B). With this tree, we were also able to rule out the possibility of gene conversion between paralogs, using likelihood tests of 500 bp on either side of the pore region in all 4 domains (see Materials and Methods, above). Thus, each pore-region amino acid replacement we observe is likely to be an evolutionarily independent event. From these data alone (figs. 1B and 2), it is apparent that pufferfish sodium channels exhibit strikingly high diversity in pore-region sequence, which is normally extremely conserved across all animals (see fig. 3 for a sample comparison of Nav1.4 orthologs across vertebrates and the urochordate Halocynthia roretzi).
FIG. 2.—
Sodium channel ion pore-loop residues from all 4 domains. Each cell shows 5 pore-loop residues for Domains I–IV (domains arranged from top to bottom), beginning with the 4 residues of the sodium selectivity filter (I-D, II-E, III-K, and IV-A). Some genes present in other teleosts were not found in pufferfishes and may be missing from their genomes—see text for possible reasons.
FIG. 3.—
Pore-loop regions of sodium channels are highly conserved, even in the most variable channel in our study. This figure illustrates the highly conserved nature of voltage-gated sodium channels by showing pore-region sequence from protein Domains I–IV for a sample of teleost and tetrapod orthologs of the mammalian channel Nav1.4, which is expressed in skeletal muscle. Pore-region sequence shows great diversity in pufferfishes but is highly conserved in both vertebrates and nonvertebrates, including the urochordate Halocynthia roretzi.
Domain I
Amino acid replacements at one well-studied site in the Domain I pore loop (Phe or Tyr to any nonaromatic residue) have been shown to provide from 190- to 2,500-fold decrease in guanidinium toxin binding (Satin et al. 1992; Kaneko et al. 1997; Yotsu-Yamashita et al. 2000; Venkatesh et al. 2005; Soong and Venkatesh 2006). This replacement is responsible for TTX resistance in one C. pyrrhogaster newt channel (Kaneko et al. 1997) and in a monophyletic cluster of 3 mammalian channels that includes Nav1.5 (cardiac) and Nav1.8 and Nav1.9 (nervous system) (Satin et al. 1992; Cummins et al. 1999). Previously, the only pufferfish sodium channels known to contain replacements at this Domain I site were the Nav1.4a (skeletal muscle) orthologs from Takifugu and Tetraodon (Yotsu-Yamashita et al. 2000; Venkatesh et al. 2005; Soong and Venkatesh 2006). In our broader survey of pufferfish sodium channels, we uncovered at this site at least 5 evolutionarily independent replacements to a nonaromatic residue (Asn, Cys, or Ala) in 5 different genes (Nav1.6b, Nav1.1La, Nav1.4a, Nav1.5La, and Nav1.5Lb) that are variously expressed in teleost neural tissue as well as cardiac and skeletal muscle. In figure 1B, these Domain I changes are indicated in red.
Domain II
For Domain II, figure 1B shows a single occurrence of a pore-loop Glu-to-Asp replacement in Nav1.4b from Tetraodon (shown in purple, also observed in the soft-shelled clam M. arenaria) that confers a 3,000-fold resistance in TTX binding (Bricelj et al. 2005; Venkatesh et al. 2005). Additionally, we note a minimum of 3 evolutionarily independent replacements of the immediately adjacent Thr to a Ser or Asn (in yellow). Choudhary et al. (2003) showed that other mutations at this Thr (to Ile, Asp, or Lys) can increase TTX resistance up to 2,000-fold. How the Domain II Thr-to-Ser or Asn replacements we observe affect toxin binding is unknown, but our phylogeny clearly shows that this particular replacement is only seen in genes that already possess earlier (evolutionarily older) replacements in Domain I (probability of observing this phylogenetic association on our tree at random, P < 0.001, using concentrated changes, Maddison 1990). Intriguingly, we found this same Domain II replacement (Thr-to-Asn) in 2 TTX-resistant channels expressed in the nervous system of mammals: Nav1.8 and Nav1.9, which also have the aforementioned nonaromatic replacement in Domain I (shared with the closely related Nav1.5). Because the different phylogenies of fish and mammalian sodium channel genes both show that this Domain II Thr changes only after previous fixation of the Domain I aromatic-to-nonaromatic mutation, it is possible that the Domain II replacement may be selected for in both pufferfishes and mammals to offset or compensate for changes in normal channel performance caused by the Domain I replacement.
Domain III
In the Domain III pore loop, we observed a minimum of 5 evolutionarily independent Met-to-Thr replacements (M1240T) in 5 different genes expressed in both skeletal muscle and neural tissues: Nav1.1La, Nav1.1Lb, Nav1.4a, Nav1.4b, and Nav1.5Lb (in blue on fig. 1B.) An alternative, equally parsimonious character optimization for Nav1.4b, as shown in figure 1B (2 independent origins, rather than 1 origin followed by a loss in Tetraodon), would imply a minimum of 6 independent Met-Thr replacements in pufferfishes. Mutation of this site to Glu or Lys increases TTX resistance by 6,000–10,000 fold but seriously impairs channel conductance (Terlau et al. 1991). We used site-directed mutagenesis to introduce the Met-to-Thr replacement into an otherwise TTX-sensitive skeletal muscle channel from rat (Nav1.4), which was then expressed in Xenopus oocytes to measure TTX binding to the mutant channel. Our results show that this replacement increases TTX resistance by approximately 15-fold (micromolar TTX to reduce current by 50% [IC50] was 0.55 for mutants, vs. 0.036 for wild type) and STX resistance by approximately 15-fold as well (micromolar STX for IC50 was 0.045 for mutants, vs. 0.003 for wild type) (see fig. 4 and table 1).
FIG. 4.—

Dose–response curves for site-directed mutants of rat sodium channel Nav1.4. The Domain III Met-to-Thr (M1240T) replacement is known only from pufferfishes and from the flatworm Bdelloura candida but has not previously been implicated in TTX/STX binding. The Domain IV Ala-to-Gly replacement (A1529G), known only from pufferfishes, the flatworm B. candida, and 1 Danio rerio channel (Nav1.1La) provides mild TTX resistance and moderate STX resistance, and produces very large currents. This may indicate a novel mechanism of guanidinium toxin resistance, such as increased channel expression or elevated channel conductance. Error bars are standard error of the mean.
Table 1.
Naturally Occurring Amino Acid Replacements That Enhance Sodium Channel Resistance to TTX and STX
| Mutation | Known From | Toxin Resistance | |
| Domain I aromatic-to-nonaromatic | D(F/Y)WEN | Pufferfishesa–c, Cynops newtsd, 3 mammalian channelse, f | Depending on replacement, ∼190- to 2,000-fold for TTX/STXa–e,g |
| Domain II Glu-Asp | EWIET | Tetraodon Nav1.4ba, also soft-shell clam Mya arenariah | STX: ∼3,000-foldh |
| Domain II Thr-Ser or -Asn | EWIET | Pufferfish and mammal channels with Domain I nonaromatic | Unknown |
| Domain III Met-Thr | KGWMD | Several pufferfish genes, also flatworm Bdelloura candidaa,i | TTX: 15.2-fold STX: 15-fold |
| Domain IV Ala-Gly | AGWDG | Several pufferfish genes, flatworm B. candidaj, and Nav1.1La in Danio rerio | TTX: 1.5-fold STX: 11-fold |
| Domain IV Ile | I1561k | Pufferfish Nav1.6b and Nav1.4a, also Thamnophis garter snakesk | TTX: 2- to 5-foldk |
| Domain IV Asp-Asn | AGWDG | Canthigaster Nav1.4b, also some Thamnophis garter snakesk | TTX: ∼300-foldl |
Bricelj et al. (2005).
Venkatesh et al. (2005) made note of this replacement in Tetraodon Nav1.4b but considered it unlikely to affect TTX binding based on a comparison with the (presumed) TTX-sensitive sodium channels of the flatworm Bdelloura candida (Blair and Anderson 1993). However, the IC50 of TTX for sodium currents in this species is ∼100 nM (Theusen 1991), which is 10- to 100-fold more resistant than most invertebrate sodium channels (Romey et al. 1976; Willmer and Harrison 1979; Kao and Yasumoto 1985; Daemers-Lambert and Grandfils 1988; Warmke et al. 1997). Therefore, in contrast to what has been suggested by Venkatesh et al. (2005), this replacement does contribute to TTX resistance in pufferfishes (and likely B. candida, which parasitizes horseshoe crabs—some of which sequester TTX; Ho et al. 1994). A further search of available protein sequence shows this Domain III Met-to-Thr replacement in the medicinal leech Hirudo medicinalis (which has TTX resistant Na+ currents; Blackshaw et al. 2003) and in 3 pyrethroid-resistant pest acarids (ticks and mites), Sarcoptes scabei, Varroa destructor, and Boophilus microplus. However, because the sodium channel binding site for guanidinium and pyrethroid toxins is different, the role (if any) of these replacements in chelicerate arthropods is unknown. One possibility is that this site represents a surviving plesiomorphy from early TTX-resistant chelicerates; horseshoe crabs, the most basal surviving chelicerates, are known to sequester TTX and show some degree of TTX resistance as well (Ho et al. 1994).
Domain IV
For Domain IV, the zebrafish channel Nav1.1La and its pufferfish orthologs show replacement of the inner pore Ala to a Gly (A1529G—in green on fig. 1B). In addition, we observed a minimum of 3 additional evolutionarily independent Ala-to-Gly replacements in the inner pore of pufferfish genes Nav1.1Lb, Nav1.4a, and Nav1.6a. This was a particularly surprising finding because the fundamental sodium ion selectivity of sodium channels is structurally defined by the conserved inner pore signature DI Asp, DII-Glu, DIII-Lys, and DIV-Ala (DEKA motif), even though the Na+ ion does not interact with the Domain IV Ala directly (Lipkind and Fozzard 2000; Tikhonov and Zhorov 2005). A thorough survey of other published sodium channel sequences found no other instances of this inner pore Ala-Gly replacement in any metazoan species except for the flatworm B. candida (Jeziorski et al. 1997), further suggesting that this site is likely to contribute to TTX resistance. Our site-directed mutagenesis and oocyte expression of this Ala-to-Gly replacement in mammalian Nav1.4 showed that this mutation provides only low resistance to TTX and moderate resistance to STX (micromolar TTX for IC50 was 0.056 for mutants, vs. 0.036 for wild type, about 1.5-fold; and micromolar STX for IC50 was 0.033 for mutants, vs. 0.003 for wild type, about 11-fold) (see fig. 4 and table 1). However, we consistently found that channels containing this replacement produced extraordinarily large currents, such that the mRNA had to be diluted to between 0.05 and 0.02 of the amount of wild-type RNA (or RNA for any other mutant) to give currents of comparable magnitude. This interesting result implies either an increase in single channel conductance or an increase in channel expression levels, either of which would represent a previously undescribed mechanism of rendering the channel current less sensitive to TTX and STX.
Also in Domain IV, we observed one instance of an Asp-to-Asn replacement that exists in some populations of the garter snake T. sirtalis (Geffeney et al. 2005) and is already known to provide up to 300-fold resistance to TTX (Choudhary et al. 2003, shown in gray). We also observed at least 2 evolutionarily independent replacements of a particular Ile residue near the pore that is thought to contribute to TTX resistance in T. sirtalis (I1561 from Geffeney et al. 2005). This Ile change was found in Nav1.6b for 3 of our pufferfish species (in orange on fig. 1B), as well as in Arothron Nav1.4a. In Thamnophis, the change appears as Ile-to-Val; in pufferfishes, it appears all 3 times as Ile-to-Met. Results from Geffeney et al. (2005) suggest that this replacement may increase TTX resistance by approximately 5-fold but because that study used chimaeric mutants, additional experiments on this site in isolation would be needed to confirm this effect.
Discussion
Natural selection appears to have favored several diverse solutions to the problem of ambient lethal TTX and STX in the natural environments of animals. In the case of pufferfishes, our results show that whole-organism resistance to guanidinium toxins was achieved through the emergence of diverse pore-region configurations involving sites in all 4 domains from all 8 sodium channels and that multiple sodium channel configurations coexist within the same genome. Although a reasonable hypothesis might be that TTX-/STX-resistant channels first arose in the sodium channels of an early tetraodontid ancestor before the diversification of the Tetraodontidae, our phylogenetic analyses indicate that for the most part, guanidinium toxin resistance arose after the diversification of Tetraodontidae via parallel evolution of replacements at homologous pore-region residues in different gene paralogs, and independently in different species. In addition, the fact that only a handful of sites are implicated in pufferfishes and in distantly related species as diverse as newts, mammals, snakes, flatworms, and clams, suggests that pore-region evolution may be strongly constrained by purifying selection for normally functioning sodium channels.
Voltage-gated sodium channels are highly conserved proteins essential to muscle, heart, and neural function, where ion selectivity, channel activation and inactivation, and conductance are strictly dependent on the channel structure. Mutations that enhance TTX/STX resistance are likely to affect normal channel function because they are typically found in the region of the channel (the pore) that is most conserved and most critical for proper ion selection and flow. Additionally, our data show that in most TTX-/STX-resistant channels, primary structural changes occur not only at single sites within these proteins but also at multiple functionally related sites within the same protein. These observations raise a number of questions about the additive or synergistic effects of combined mutations, when evolutionary change at one site is accompanied by other functionally related changes elsewhere in the protein that may enhance, compensate for, or be enabled by the first change. For example, our repeat observations in both pufferfishes and mammals of a phylogenetic association between a specific Domain II Thr replacement and chronologically previous Domain I aromatic-to-nonaromatic replacements point strongly to the possibility that replacement at that Domain I site alters normal channel function.
Throughout this study, we were interested in determining whether toxin resistance might arise as a result of a chronological sequence of multiple, time-ordered evolutionary changes—for example, one hypothesis was that replacements providing weak-to-moderate toxin resistance might have appeared first, followed later by replacements providing stronger resistance. However, in general we do not observe such patterns, with the exception of the time-ordered association we note between Domain I aromatic/nonaromatic replacements and subsequent replacement of the Domain II pore loop Thr. More interesting is the fact that mutations providing weak-to-moderate toxin resistance arise at all, in channels that already contain mutations providing strong resistance (such as a replacement in Domain I). Although mutations providing weak resistance can still evolve under selection by neurotoxins alone, their fixation after prior fixation of highly resistant mutations may be more indicative of the strong functional association among pore-loop residues in all 4 domains.
Amino acid replacements that influence TTX binding may have profound consequences for ion selectivity, the rates of channel opening, and movement into or recovery from the slow inactivated state. Perturbations of these processes are likely to be detrimental. For example, replacement of the Ala in Domain IV (where we observe 3 independent replacements to Gly) to Asp causes sodium channels to enter into a prolonged state of slow inactivation (Hilber et al. 2001). A mutation that similarly enhances slow inactivation is associated with Brugada syndrome, a heart disease (Amin et al. 2005). Thus, it is likely that in some channels, the evolution of TTX resistance has resulted in functional trade-offs, as in Thamnophis snakes with TTX-resistant skeletal muscle that show impaired motor function (Geffeney et al. 2002)
From an ecological standpoint, it is important to point out that the highly similar binding activity of the guanidinium toxins STX and TTX makes it impossible to know, in any particular case, which of these toxins was the selective agent during evolution in the marine environment. Na+ channels that are classified as TTX resistant may have actually evolved under selection for STX resistance in environments where high concentrations of STX may be common—such as during “red tide” events corresponding with dinoflagellate algal blooms, or in animal species where a specialized diet results in high consumption of STX. Therefore, TTX resistance in some pufferfishes and other marine organisms may in fact be a secondary property of mutations that were originally selected for by the environmental presence of STX.
The massive scale of parallel evolution we observe across many Na+ channel genes and distantly related species presents a remarkable example of natural selection repeatedly arriving at a diverse but limited number of adaptive solutions to the same selective agent. Because approximately 75% of vertebrate genes are nonunique members of multigene families (Duret et al. 1994), whole-organism adaptation in vertebrates may often (if not usually) involve parallel evolution across entire gene families. Based on our results, we predict that similar patterns of parallel evolution will be observed in other vertebrate gene families subjected to strong natural selection, and we propose that future studies in molecular adaptation will be most informative when they examine all functionally similar paralogs of the affected gene family.
Acknowledgments
We would like to thank James J. Bull, members of the Hillis and Bull laboratories, and Elizabeth Kellogg for helpful comments on the manuscript. This research was supported by National Science Foundation Grant # IBN-0236147 to H.H.Z., D.M.H., and M.C.J. and NIH Grant ROI HL-06561 to H.A.F.
References
- Amin AS, Verkerk AO, Bhuiyan ZA, Wilde AAM, Tan HL. Novel Brugada syndrome-causing mutations in ion conducting pore of cardiac Na channel does not affect ion selectivity properties. Acta Physiol Scand. 2005;185:291–301. doi: 10.1111/j.1365-201X.2005.01496.x. [DOI] [PubMed] [Google Scholar]
- Aparicio S, Chapman J, Stupka E, et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- Asakawa M, Toyoshima T, Shida Y, Noguchi T, Miyazawa K. Paralytic toxins in a ribbon worm Cephalothrix species (Nemertean) adherent to cultured oysters in Hiroshima Bay, Hiroshima Prefecture, Japan. Toxicon. 2000;38:763–773. doi: 10.1016/s0041-0101(99)00172-5. [DOI] [PubMed] [Google Scholar]
- Blackshaw SE, Henderson LP, Malek J, Porter DM, Gross RH, Angstadt JD, Levasseur SM, Maue RA. Single-cell analysis reveals cell-specific patterns of expression of a family of putative voltage-gated sodium channel genes in the leech. J Neurobiol. 2003;55:355–371. doi: 10.1002/neu.10214. [DOI] [PubMed] [Google Scholar]
- Blair KL, Anderson PAV. Properties of voltage-activated ionic currents in cells from the brains of the triclad flatworm Bdelloura candida. J Exp Biol. 1993;185:267–286. [Google Scholar]
- Brenner S, Elgar G, Sandford R, Macrae A, Venkatesh B, Aparicio S. Characterization of the pufferfish (Fugu) genome as a compact model vertebrate genome. Nature. 1993;366:265–268. doi: 10.1038/366265a0. [DOI] [PubMed] [Google Scholar]
- Bricelj VM, Connell L, Konoki K, Macquarrie SP, Scheuer T, Catterall WA, Trainer VL. Sodium channel mutation leading to saxitoxin resistance in clams increases risk of PSP. Nature. 2005;434:763–767. doi: 10.1038/nature03415. [DOI] [PubMed] [Google Scholar]
- Choudhary G, Yotsu-Yamashita M, Shang L, Yasumoto T, Dudley SC. Interactions of the C-11 hydroxyl of tetrodotoxin with the sodium channel outer vestibule. Biophys J. 2003;84:287–294. doi: 10.1016/S0006-3495(03)74849-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins TR, Dibb-Haj SD, Black JA, Akopian AN, Wood JN, Waxman SG. A novel persistent tetrodotoxin-resistant sodium current in SNS-null and wild-type small primary sensory neurons. J Neurosci. 1999;19(24):RC43:1–6. doi: 10.1523/JNEUROSCI.19-24-j0001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daemers-Lambert C, Grandfils C. Action potential of the median giant fibre of the earthworm, Lumbricus terrestris, as a pharmacological test of the radio-labelled TTX-derivatives activity. Comp Biochem Physiol C Pharmacal Toxicol Endocrinol. 1988;1:107–112. [Google Scholar]
- Duret L, Mouchiroud D, Gouy M. HOVERGEN, a database of homologous vertebrate genes. Nucleic Acids Res. 1994;22:2360–2365. doi: 10.1093/nar/22.12.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ffrench-Constant RH, Pittendrigh B, Vaughan A, Anthony N. Why are there so few resistance associated mutations in insecticide target genes? Philos Trans R Soc Lond B Biol Sci. 1998;353:1685–1693. doi: 10.1098/rstb.1998.0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geffeney S, Brodie ED, Ruben PC, Brodie ED. Mechanisms of adaptation in a predator-prey arms race: tTX resistant sodium channels. Science. 2002;297:1336–1339. doi: 10.1126/science.1074310. [DOI] [PubMed] [Google Scholar]
- Geffeney SL, Fujimoto E, Brodie ED, Brodie ED, Ruben PC. Evolutionary diversification of TTX-resistant sodium channels in a predator-prey interaction. Nature. 2005;434:759–763. doi: 10.1038/nature03444. [DOI] [PubMed] [Google Scholar]
- Grassly N, Rambaut A. PLATO—Partial likelihoods assessed through optimisation (v.2.11) University of Oxford; 1999. software available from authors. [Google Scholar]
- Halstead B. Poisonous and venomous marine animals of the world. Washington, DC: The US Government. 1967. [Google Scholar]
- Hanifin CT, Yotsu-Yamashita M, Yasumoto T, Brodie ED, Brodie ED. Toxicity of dangerous prey: variation of tetrodotoxin levels within and among populations of the newt Taricha granulosa. J Chem Ecol. 1999;25(9):2161–2175. [Google Scholar]
- Hilber K, Sandtner W, Kudlacek O, Glaaser IW, Weisz E, Kyle JW, French RJ, Fozzard HA, Dudley SC, Todt H. Interaction between fast and ultra-slow inactivation in the voltage-gated sodium channel. J Biol Chem. 2001;276:27831–27839. doi: 10.1074/jbc.M101933200. [DOI] [PubMed] [Google Scholar]
- Ho B, Yeo DSA, Ding JL. A tetrodotoxin neutralizing system in the haemolymph of the horseshoe crab, Carcinoscorpius rotundicauda. Toxicon. 1994;32:755–762. doi: 10.1016/0041-0101(94)90001-9. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Brunet F, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Jang J, Yotsu-Yamashita M. Distribution of tetrodotoxin, saxitoxin, and their analogs among tissues of the pufferfish Fugu pardalis. Toxicon. 2006;48:980–987. doi: 10.1016/j.toxicon.2006.07.034. [DOI] [PubMed] [Google Scholar]
- Jeziorski MC, Greenberg RM, Anderson PAV. Cloning of a putative voltage-gated sodium channel from the turbellarian flatworm Bdelloura candida. Parasitology. 1997;115:289–296. doi: 10.1017/s0031182097001388. [DOI] [PubMed] [Google Scholar]
- Kaneko Y, Matsumoto G, Hanyu Y. TTX resistivity of Na+ channel in newt retinal neuron. Biochem Biophys Res Commun. 1997;240:651–656. doi: 10.1006/bbrc.1997.7696. [DOI] [PubMed] [Google Scholar]
- Kao CY, Yasumoto T. Interactions of the C-11 hydroxyl of tetrodotoxin with the sodium channel outer vestibule. Toxicon. 1985;23:725–729. [Google Scholar]
- Kidokoro Y, Grinnell AD, Eaton DC. Tetrodotoxin sensitivity of muscle action potentials in pufferfishes and related fishes. J Comp Physiol A. 1974;89:59–72. [Google Scholar]
- Lin SJ, Hwang DF. Possible source of tetrodotoxin in the starfish Astropecten scoparius. Toxicon. 2001;39:573–579. doi: 10.1016/s0041-0101(00)00171-9. [DOI] [PubMed] [Google Scholar]
- Lipkind GM, Fozzard HA. KcsA crystal structure as framework for a molecular model of the Na1 channel pore. Biochemistry. 2000;39:8161–8170. doi: 10.1021/bi000486w. [DOI] [PubMed] [Google Scholar]
- Liu ZQ, Tan JG, Valles SM, Dong K. Synergistic interaction between two cockroach sodium channel mutations and a tobacco budworm sodium channel mutation in reducing channel sensitivity to a pyrethroid insecticide. Insect Biochem Mol Biol. 2002;32:397–404. doi: 10.1016/s0965-1748(01)00116-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopreato GF, Lu Y, Southwell A, Atkinson N, Hillis DM, Wilcox T, Zakon HH. Evolution and divergence of sodium channel genes in vertebrates. Proc Natl Acad Sci USA. 2001;98:7588–7592. doi: 10.1073/pnas.131171798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison WP. A method for testing the correlated evolution of two binary characters: are gains or losses concentrated on certain branches of a phylogenetic tree? Evolution. 1990;44(3):539–557. doi: 10.1111/j.1558-5646.1990.tb05937.x. [DOI] [PubMed] [Google Scholar]
- Nagashima Y, Yamamoto K, Shimakura K, Shiomi K. A tetrodotoxin-binding protein in the hemolymph of shore crab Hemigrapsus sanguineus: purification and properties. Toxicon. 2002;40:753–760. doi: 10.1016/s0041-0101(01)00278-1. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Arakawa O, Taniyama S, Nonaka M, Takatani T, Yamamori K, Fuchi Y, Noguchi T. Occurrence of saxitoxins as a major toxin in the ovary of a marine puffer Arothron firmamentum. Toxicon. 2004;43:207–212. doi: 10.1016/j.toxicon.2003.05.001. [DOI] [PubMed] [Google Scholar]
- Neafsey DE, Palumbi SR. Genome size evolution in pufferfish: a comparative analysis of diodontid and tetraodontid pufferfish genomes. Genome Res. 2003;13:821–830. doi: 10.1101/gr.841703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak AE, Jost MC, Lu Y, Taylor AD, Zakon HH, Ribera AB. Gene duplications and evolution of function in vertebrate voltage-gated sodium channels. J Mol Evol. 2006;63:208–221. doi: 10.1007/s00239-005-0287-9. [DOI] [PubMed] [Google Scholar]
- Penzotti JL, Fozzard HA, Lipkind GM, Dudley SC. Differences in saxitoxin and tetrodotoxin binding revealed by mutagenesis of the Na+ channel outer vestibule. Biophys J. 1998;75:2647–2657. doi: 10.1016/S0006-3495(98)77710-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires OR, Jr, Sebben A, Schwartz EF, Largura SW, Bloch C, Jr, Morales RA, Schwartz CA. Occurrence of tetrodotoxin and its analogues in the Brazilian frog Brachycephalus ephippium (Anura: Brachycephalidae) Toxicon. 2002;40:761–766. doi: 10.1016/s0041-0101(01)00279-3. [DOI] [PubMed] [Google Scholar]
- Romey G, Abita JP, Schweitz H, Wunderer G, Lazdunski M. Sea anemone toxin: a tool to study molecular mechanisms of nerve conduction and excitation-secretion coupling. Proc Nat Acad Sci USA. 1976;73(11):4055–4059. doi: 10.1073/pnas.73.11.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Satin J, Kyle JW, Chen M, Bell P, Cribbs LL, Fozzard HA, Rogart RB. A mutant of TTX-resistant cardiac sodium channels with TTX-sensitive properties. Science. 1992;256:1202–1205. doi: 10.1126/science.256.5060.1202. [DOI] [PubMed] [Google Scholar]
- Scheumack D, Howden M, Spence I, Quiin R. Maculotoxin: a neurotoxin from the venom glands of the octopus Hapalochlaena maculosa identified as tetrodotoxin. Science. 1978;199:188–189. doi: 10.1126/science.619451. [DOI] [PubMed] [Google Scholar]
- Soong TW, Venkatesh B. Adaptive evolution of tetrodotoxin resistance in animals. Trends Genet. 2006;22:621–626. doi: 10.1016/j.tig.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Tanu MB, Mahmud Y, Arakawa O, Takatani T, Kajihara H, Kawatsu K, Hamano Y, Asakawa M, Miyazawa K, Noguchi T. Immunoenzymatic visualization of tetrodotoxin (TTX) in Cephalothrix species (Nemertea: Anopla: Palaeonemertea: Cephalotrichidae) and Planocera reticulata (Platyhelminthes: Turbellaria: Polycladida: Planoceridae) Toxicon. 2004;44:515–520. doi: 10.1016/j.toxicon.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Tanu MB, Mahmud Y, Tsuruda K, Arakawa O, Noguchi T. Occurrence of tetrodotoxin in the skin of a rhacophoridid frog Polypedates sp. from Bangladesh. Toxicon. 2001;39:937–941. doi: 10.1016/s0041-0101(00)00231-2. [DOI] [PubMed] [Google Scholar]
- Terlau H, Heinemann SH, Stuhmer W, Pusch M, Conti F, Imoto K, Numa S. Mapping the site of block by tetrodotoxin and saxitoxin of sodium channel II. FEBS Lett. 1991;293:93–96. doi: 10.1016/0014-5793(91)81159-6. [DOI] [PubMed] [Google Scholar]
- Theusen EV. The tetrodotoxin venom of chaetognaths. In: Bone Q, Kapp H, Pierrot-Bults AC, editors. The Biology of Chaetognaths. London: Oxford University Press; 1991. pp. 50–60. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov DB, Zhorov BS. Modeling P-loops domain of sodium channel: homology with potassium channels and interaction with ligands. Biophys J. 2005;88:184–197. doi: 10.1529/biophysj.104.048173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh B, Lu SQ, Dandona N, See SL, Brenner S, Soong TW. Genetic basis of tetrodotoxin resistance in pufferfishes. Curr Biol. 2005;15:2069–2072. doi: 10.1016/j.cub.2005.10.068. [DOI] [PubMed] [Google Scholar]
- Warmke JW, Reenan RAG, Wang PY, et al. Functional expression of Drosophila para sodium channels. Modulation by the membrane protein TipE and toxin pharmacology. J Gen Physiol. 1997;110:119–133. doi: 10.1085/jgp.110.2.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmer PG, Harrison JB. Cation accessibility of the peripheral nervous system in Limulus polyphemus—an electrophysiological study. J Exp Biol. 1979;82:373–376. doi: 10.1242/jeb.82.1.373. [DOI] [PubMed] [Google Scholar]
- Yotsu-Yamashita M, Nishimoto K, Nitanai Y, Isemura M, Sugimoto A, Yasumoto T. Binding properties of 3H-PbTx-3 and 3H-saxitoxin to brain membranes and to skeletal muscle membranes of pufferfish Fugu pardalis and the primary structure of a voltage-gated Na+ channel a-subunit (fMNa1) from skeletal muscle of F. pardalis. Biochem Biophys Res Commun. 2000;267:403–412. doi: 10.1006/bbrc.1999.1974. [DOI] [PubMed] [Google Scholar]




