Abstract
Despite much interest in regulatory evolution, how promoters have evolved remains poorly studied, mainly owing to paucity of data on promoter regions. Using a new set of high-quality experimentally determined core promoters of the human genome, we conducted a comparative analysis of 2,492 human and rhesus macaque promoters and their neighboring nearly neutral regions. We found that the core promoters have an average rate of nucleotide substitution substantially higher than that at 4-fold degenerate sites and only slightly lower than that for the assumed neutral controls of neighboring noncoding regions, suggesting that core promoters are subject to very weak selective constraints. Interestingly, we identified 24 core promoters (at false discovery rate = 50%) that have evolved at an accelerated rate compared with the neutral controls, suggesting that they may have undergone positive selection. The inferred positively selected genes show strong bias in molecular function. We also used population genetic approaches to examine the evolution of core promoters in human populations and found evidence of positive selection at some loci. Taken together, our results suggest that positive selection has played a substantial role in the evolution of transcriptional regulation in primates.
Keywords: regulatory evolution, substitution rate, positive selection, primate evolution, hitchhiking
Introduction
Transcription of a eukaryotic protein-coding gene begins with the recruitment of the transcriptional machinery onto the promoter. Thus, the promoter region, which includes various cis-regulatory elements, plays a very important role in determining the expression pattern of a gene. Since King and Wilson (1975), the evolution of gene regulation in higher primates has attracted much interest in the hope that it may provide important clues for the phenotypic divergence between humans and apes. However, the issue of what drives the evolution of primate promoters has been controversial. Based on a relatively small data set, Jareborg et al. (1999) found that the promoter regions were subject to selective constraints. In contrast, Keightley, Lercher, et al. (2005) reported that the evolutionary rate in the upstream region of a gene was similar to that in intronic sequences, and they concluded that the evolution of hominid promoters is dominated by random drift, owing to the small effective population size of hominids. More recently, comparing with neutral sites in ancient repetitive sequences, Taylor et al. (2006) detected a reduced evolutionary rate in the immediate vicinity of transcription start sites (TSSs) but an increased rate in regions further upstream that may be related to the unusual chromatin structure for transcriptional regulation.
A major challenge in studying promoter evolution is that eukaryotic promoters are often too diverse to be inferred based on gene annotation alone (Sandelin et al. 2007). For example, a cis-regulatory element can affect gene expression as far as 1 Mb away from the coding region (Lettice et al. 2002). In fact, a message from the recent ENCODE project (intensive analysis on 1% of the human genome) is that transcription activities in the human genome are far more complex than previously thought (Birney et al. 2007); moreover, many novel promoters are associated with known genes in an unexpected manner (Trinklein et al. 2007). Thus, the conventional localization of a promoter may either include too much background noise or miss the entire functional element. Therefore, instead of studying vaguely defined promoter regions or intergenic regions immediately upstream of TSS of a gene, we focus on “core” promoters that can be relatively accurately defined by experimental methods (Sandelin et al. 2007). The core promoter is the major functional region for assembling the RNA polymerase II preinitiation complex that includes the RNA polymerase II itself and several general transcription factors, such as the transcription factor IID (TFIID) (Smale and Kadonaga 2003). The core promoter is usually very short (<150 bp) and includes various functional elements that interact directly with components of preinitiation complex (Sandelin et al. 2007). Thus, genetic variation in this region may make a difference on transcriptional regulation and may be subject to selection. Taking advantage of the available high-resolution human core promoter map (Kim et al. 2005) and using both comparative genomic and population genetic approaches, we examined the evolution of core promoters in the human and macaque genomes.
Materials and Methods
Promoter Data and Substitution Rate Analysis
We obtained the coordinates of TFIID-binding sites (50 bp each) from Kim et al. (2005) and converted their hg16 version coordinates into those of the hg18 version, using the LiftOver tool from the University of California at Santa Cruz Genome Browser (Karolchik et al. 2003). To reduce the noise in our analysis, we only included the TFIID-binding sites within 1 kb of TSS of a RefSeq transcript with an unambiguous genomic location. We downloaded the RefSeq coordinates from UCSC and excluded those that are not in the main human genome assembly or correspond to multiple genomic locations.
Because the core promoter can extend ∼35 bp upstream and/or downstream of the transcription initiation site (Smale and Kadonaga 2003), we extended 35 bp on each side of the TFIID-binding site and defined the 120-bp region as the core promoter. For each core promoter, the RefSeq transcript containing the nearest TSS was chosen as the corresponding gene. Moreover, we restricted our analysis on a set of core promoters that reside upstream of their corresponding coding regions and do not overlap with any known coding regions. We also chose another 120-bp intergenic DNA region 1 kb upstream of each core promoter and obtained a set of “pseudopromoter” regions as the control; the regions close to any known TFIID-binding sites were excluded.
For each core promoter and its corresponding pseudopromoter and coding region (an assembly of exons), we generated the human–macaque orthologous alignments using the “reciprocal best hits” criteria based on the UCSC human/rhesus (hg18/rheMac2) and rhesus/human (rheMac2/hg18) pairwise genome alignments. Note that the frequently used MultiZ alignments (Blanchette et al. 2004) do not guarantee 1-to-1 orthologous alignment. By definition, MultiZ alignments contain the best match in an assembly (species) to a given region in the human assembly, so the same region of an assembly may align to multiple regions in the human genome. For the coding regions, we removed the alignments containing in-frame stop codons or indels that can cause a frameshift. We also excluded the alignments with more than 10% gap positions from the analysis. In addition, we confirmed the synteny of core promoters, pseudopromoters, and coding regions between the 2 species.
To rigorously compare the substitution rate in the core promoter region (Kcore), the rate in the pseudopromoter (Kpseudo), the rate at the second codon positions (K2nd) (representing the nonsynonymous substitution rate), and the rate at the 4-fold degenerate sites (K4), we further masked all the CpG sites in the alignments because the substitution rate and pattern at these sites are very different from the rest of the sequences (Eyre-Walker and Hurst 2001). Then we calculated the substitution rate of each region, using the JC69 model in the PAML package (Yang 1997). To detect positive selection, we used Fisher's exact test (1-tailed) to determine whether the proportion of substituted sites in a core promoter is significantly higher than that in its pseudopromoter, which serves as the neutral control in our analysis. The P value provides an unbiased way to quantify the difference between Kcore and Kpseudo, regardless of the number of informative nucleotide sites in the analysis. For comparison, we performed the same analyses on Kcore versus K4 and K2nd versus K4. We also compared the substitution rates of different regions within a core promoter and found no difference between the middle 50 bp and the rest of a core promoter. We also calculated the proportion of transition or AT ↔ GC substitutions in core promoters.
At false discovery rate = 50%, we chose P = 0.005 as a cutoff to identify 24 promoters that Kcore is significantly higher than Kpseudo and thus is potentially under positive selection. For these inferred positively selected core promoters, we further used BLAT (Kent 2002) to confirm their location correspondence between the human and macaque genomes. In addition, we performed the same analyses between chimpanzee (The Chimpanzee Consortium 2005) and human. However, given the short length of core promoters in our study, the orthologous sequences between these 2 species are too close to each other to have a statistical power.
GO Term Analysis
We used the Gene Ontology (GO) term analysis tools (Boyle et al. 2004) to study the biological implications of inferred positively selected core promoters. For each GO term listed in the default GOA slim file, we determined whether the positively selected genes are overrepresented using the binomial test. At false discovery rate = 40%, P = 0.05 was chosen to identify overrepresented GO terms.
Long-Range Haplotype Analysis in Human Populations
We obtained the integrated haplotype score (iHS) from the program haplotter (http://hg-wen.uchicago.edu/selection/haplotter.htm) (Voight et al. 2006). Following Voight et al. (2006), we identified the potentially positively selected single nucleotide polymorphism (SNPs) using the threshold |iHS| > 2.5, which corresponds to the most extreme 1% of iHS values in the whole genome.
Results and Discussion
Detecting Selection on Core Promoters
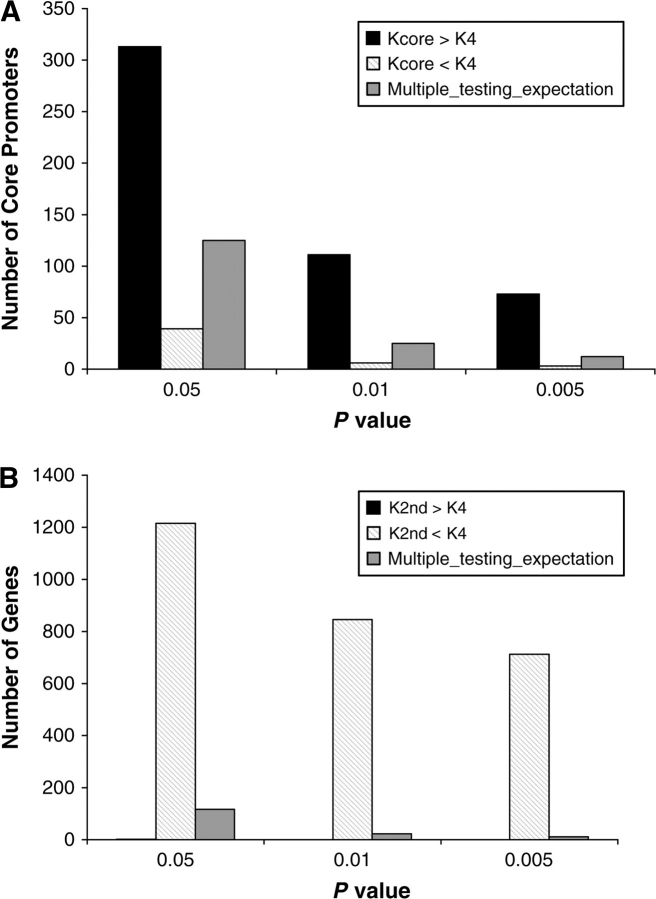
We constructed the human–macaque orthologous alignments for 2,492 high-confidence core promoters (each spanning 120 bp) (supplementary data set S1, Supplementary Material online). For each of them, we further constructed the alignments for its downstream coding region and a same size pseudopromoter, which is 1 kb upstream of the core promoter. We found that both Kpseudo and Kcore are substantially higher than K4 (table 1). Moreover, using Fisher's exact test, we found that there is a deficiency of core promoters with Kcore/K4 < 1 but a large excess of promoters with Kcore/K4 > 1 (given P = 0.05, for Kcore/K4 < 1, 86 promoters fewer than expected and for Kcore/K4 > 1, 188 more than expected), in sharp contrast to the comparison of K2nd versus K4 (given P = 0.05, for K2nd/K4 < 1, 1,098 more than expected and for K2nd/K4 > 1, 1,115 fewer than expected; fig. 1). This observation reveals that, as observed in other upstream regions (Keightley, Lercher, and Eyre-Walker 2005), mutation and random drift may have played a dominant role in nucleotide substitution even in core promoters. Note, however, that Kcore is, on average, slightly but significantly lower than Kpseudo (paired Wilcoxon rank test P = 1.8 × 10−4, table 1), suggesting the presence of purifying selection on core promoters.
Table 1.
Summary Statistics of Nucleotide Substitution Rate Analysis
| DNA Type | Number of Sites | Median K | Mean K (standard error) |
| Pseudopromoter | 274,440 | 0.0476 | 0.05597 (0.0015) |
| Core promoter | 247,071 | 0.0439 | 0.05129 (0.00078) |
| 4-fold degenerate site | 477,082 | 0.0295 | 0.03224 (0.00049) |
FIG. 1.—
Different evolutionary patterns between core promoters and coding regions. (A) Proportion of core promoters that have evolved significantly faster (black bars) or more slowly (striped bars) than the 4-fold degenerate sites in the same genes. The proportion expected from multiple testing is shown (gray bars). (B) Proportion of genes that have evolved significantly faster (black bars) or more slowly (striped bars) at the second codon positions than at the 4-fold degenerate sites in the same genes. Only 2 genes have K2nd/K4 > 1 at P value < 0.05 and no gene at P value < 0.01 or 0.005. The total number of core promoters studied is 2,492 (in 2,333 genes). The expected numbers from multiple testing are calculated using the P value × the total number of promoters (genes) in the analysis.
There are 3 possible reasons for Kcore > K4. First, 4-fold degenerate sites in coding regions appear to be under some functional constraints so that they have evolved more slowly than the neutral rate (The Chimpanzee Consortium 2005; Lu and Wu 2005). Second, due to the unusual chromatin structure related to the transcription process (Sabo et al. 2004), there may be an elevated background mutation rate in upstream noncoding regions (Taylor et al. 2006). (This potential factor, however, may not apply to genes that are not expressed in the germ line.) For these 2 reasons, it is in practice difficult to distinguish between them. Third, positive selection might have operated on some core promoters, thereby accelerating their evolution. In this case, we would expect Kcore > Kpseudo. This possibility is considered in the next section.
Identification of Positively Selected Core Promoters
Faster evolving core promoters are potential candidates subject to positive selection. But we need to rule out 2 possibilities before attributing positive selection to the elevated substitution rates. First, although we masked all the CpG sites in the pairwise alignments, it is possible that some “hidden” CpG sites may exist (i.e., CpG sites that have experienced substitutions in both human and macaque lineages). If the K of faster evolving core promoters is elevated mainly due to this hidden CpG effect, we would expect that faster evolving core promoters have a higher proportion of transition substitutions because deamination of methyl CpG specifically results in a transition substitution. However, this is not the case: faster evolving promoters actually have a lower proportion of transitions (supplementary fig. S1, Supplementary Material online), indicating that the hidden CpG sites do not influence our substitution rate analysis. Second, it has been recently suggested that biased gene conversion (BGC) can result in substitution hotpots (Galtier and Duret 2007). During this neutral process, AT → GC mutations have a higher probability to be transmitted to the next generation, presumably through the GC-biased repair of A:C and G:T mismatches in heteroduplexed recombination intermediates. If the K of faster evolving core promoters is increased by BGC, we would expect that faster evolving core promoters have a higher proportion of AT ↔ GC substitutions. However, this is not the case either: faster evolving promoters have a normal proportion of AT ↔ GC substitutions (supplementary fig. S1, Supplementary Material online), indicating that BGC does not distort our results.
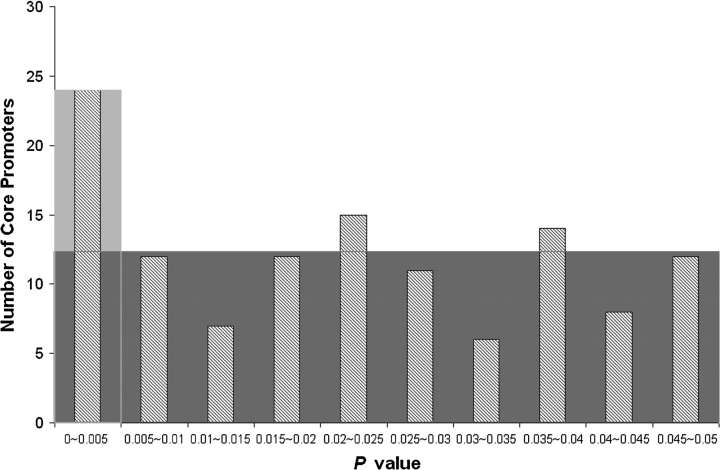
To pinpoint core promoters that have potentially been subject to positive selection, we studied the P value distribution of significantly faster evolving core promoters than the corresponding pseudopromoter regions (i.e., 121 core promoters at P < 0.05). As shown in figure 2, instead of the uniform distribution expected from chance alone, we found a significant excess of core promoters at very low P values. At false discovery rate = 50%, a P value cutoff of 0.005 was chosen to identify 24 out of 2,492 core promoters (supplementary table S1, Supplementary Material online). These core promoters also have evolved much faster than their corresponding upstream noncoding and intronic regions, indicating that the high Kcore/Kpseudo ratio (>1) in these genes was not due to purifying selection on the corresponding pseudopromoters (supplementary table S2, Supplementary Material online). Therefore, we inferred that these core promoters may have undergone positive selection, although we cannot polarize their substitutions into human- or macaque-lineage specific.
FIG. 2.—
The observed distribution of core promoters that have evolved significantly faster than neutral controls. Striped bars represent the observed numbers at different P value intervals, and the gray square represents the uniform distribution expected from chance alone. The light gray square highlights the positively selected promoters identified in this study (P < 0.005, false discovery rate = 50%).
We performed the following 2 analyses on the inferred positively selected core promoters. First, we conducted visual and manual examination of these core promoters to confirm that the signals we detected are not due to misalignments. Note that our method is robust to the noise in the experimentally determined data set because a nonfunctional region is unlikely to be under positive selection and thus is unlikely to exceed the local neutral rate. Second, to understand the biological functions of the genes with positively selected core promoters, we performed the GO term analysis and found that the corresponding genes are significantly biased in some biological processes, functional categories, and cellular components (table 2; supplementary table S3, Supplementary Material online). For example, positively selected genes are enriched in biosynthetic and metabolism processes. These results are generally consistent with a very recent study revealing that positive selection has targeted the transcriptional regulation of genes involved in nutrition and metabolism (Haygood et al. 2007).
Table 2.
GO Term Analysis of Inferred Positively Selected Genes
| GO Term | Category Description | Number of Positively Selected Genes | Number of Annotated Genes | P Value for Overrepresentation |
| Total | 20 | 1,856 | / | |
| Biological process | ||||
| GO:0008152 | Metabolic process | 16 | 1,113 | 0.05 |
| GO:0009058 | Biosynthetic process | 5 | 176 | 0.035 |
| Molecular function | ||||
| GO:0016491 | Oxidoreductase activity | 3 | 77 | 0.048 |
| Cellular component | ||||
| GO:0005622 | Intracellular | 18 | 1,312 | 0.041 |
| GO:0005737 | Cytoplasm | 12 | 640 | 0.015 |
NOTE.—The P value is the raw value from the binomial test. In total, 17 different biological processes, 15 molecular functions, and 8 cellular components were tested. At false discovery rate = 40%, P = 0.05 was chosen as a cutoff. The result of all GO terms in the analysis was provided in Supplementary Material online.
Population Genetic Analyses
We have also attempted to detect recent positive selection on core promoters within human populations. We employed the long-range haplotype test to identify individual positively selected SNPs. Based on the recently developed iHS (Voight et al. 2006), we identified 10 SNPs with an extreme iHS value (|iHS| > 2.5, within the top 1% genome-wide outliers), a signature of very recent positive selection (<30,000 years) (Sabeti et al. 2002) (supplementary table S4, Supplementary Material online). However, we cannot rule out the possibility that a high |iHS| value is due to the effect of a neighboring mutation. In particular, in the European population, 6 out of the 166 SNPs are associated with a high |iHS| value, which represents a significant enrichment over genome-wide expectation (P < 0.0069). We further examined the correspondence between these positively selected SNPs and the positively selected core promoters inferred in the previous section. Interestingly, we find that EEF1A1 contains a positively selected core promoter and a high-iHS SNP (rs3806980) at the same time, suggesting that this gene in which positive selection appears to be ongoing in contemporary human populations has also experienced positive selection after the human–macaque split.
Concluding Remarks
In this study, we found that core promoters in primates have evolved much faster than synonymous sites (table 1). This result and those of Keightley, Lercher, et al. (2005) and Taylor et al. (2006) clearly show that promoter sequences evolve much faster than protein-coding sequences. Of course, these results do not necessarily imply that regulatory evolution has played a more important role than protein-coding sequence evolution in the phenotypic evolution of higher primates because it remains unclear what proportion of promoter sequence changes cause phenotypic divergence.
Compared with rodents, relaxed constraints on primate noncoding regions have been detected, which probably can be attributed to a smaller effective population size in primates (Bush and Lahn 2005; Keightley, Kryukov, et al. 2005; Kryukov et al. 2005). This notwithstanding, several recent studies have reported adaptive evolution in primate noncoding regions (Pollard et al. 2006; Prabhakar et al. 2006; Birney et al. 2007; Kim and Pritchard 2007), which is consistent with the present study in a broad sense. However, it should be emphasized that there is a fundamental difference between our work and these published studies. In the published studies, the authors only searched for the signature of positive selection within most conserved noncoding sequences (CNSs). Thus, these studies are highly biased by computationally defined “conservation” and provide little information on primate-specific regulatory regions. Moreover, the inferred positively selected CNSs may contain various regulatory elements (e.g. small RNAs), most of which remain functionally unknown. In contrast, our study starts with a set of high-quality experimentally determined core promoters that have a well-defined role in assembling transcription machinery. Thus, the genes we identified with a positively selected core promoter may serve a better starting point for further investigation into their biological function and their significance in human evolution.
Supplementary Material
Supplementary data set S1, figure S1, and tables S1–S4 available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
We thank Drs Jian Lu and Kai Zeng and 3 reviewers for their valuable suggestions. This study is supported by the National Institutes of Health grants to W.H.L.
References
- Birney E, Stamatoyannopoulos JA, Dutta A, et al. (313 co-authors) Identification and analysis of functional elements in 1% of the human genome by the encode pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M, Kent WJ, Riemer C, et al. (12 co-authors) Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. GO::Termfinder—open source software for accessing gene ontology information and finding significantly enriched gene ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush EC, Lahn BT. Selective constraint on noncoding regions of hominid genomes. PLoS Comput Biol. 2005;1:e73. doi: 10.1371/journal.pcbi.0010073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A, Hurst LD. The evolution of isochores. Nat Rev Genet. 2001;2:549–555. doi: 10.1038/35080577. [DOI] [PubMed] [Google Scholar]
- Galtier N, Duret L. Adaptation or biased gene conversion? Extending the null hypothesis of molecular evolution. Trends Genet. 2007;23:273–277. doi: 10.1016/j.tig.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Haygood R, Fedrigo O, Hanson B, Yokoyama KD, Wray GA. Promoter regions of many neural- and nutrition-related genes have experienced positive selection during human evolution. Nat Genet. 2007;39:1140–1144. doi: 10.1038/ng2104. [DOI] [PubMed] [Google Scholar]
- Jareborg N, Birney E, Durbin R. Comparative analysis of noncoding regions of 77 orthologous mouse and human gene pairs. Genome Res. 1999;9:815–824. doi: 10.1101/gr.9.9.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Baertsch R, Diekhans M, et al. (13 co-authors) The UCSC genome browser database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley PD, Kryukov GV, Sunyaev S, Halligan DL, Gaffney DJ. Evolutionary constraints in conserved nongenic sequences of mammals. Genome Res. 2005;15:1373–1378. doi: 10.1101/gr.3942005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley PD, Lercher MJ, Eyre-Walker A. Evidence for widespread degradation of gene control regions in hominid genomes. PLoS Biol. 2005;3:e42. doi: 10.1371/journal.pbio.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT—the blast-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Pritchard JK. Adaptive evolution of conserved noncoding elements in mammals. PLoS Genet. 2007;3:1572–1586. doi: 10.1371/journal.pgen.0030147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Kryukov GV, Schmidt S, Sunyaev S. Small fitness effect of mutations in highly conserved non-coding regions. Hum Mol Genet. 2005;14:2221–2229. doi: 10.1093/hmg/ddi226. [DOI] [PubMed] [Google Scholar]
- Lettice LA, Horikoshi T, Heaney SJ, et al. (21 co-authors) Disruption of a long-range cis-acting regulator for shh causes preaxial polydactyly. Proc Natl Acad Sci USA. 2002;99:7548–7553. doi: 10.1073/pnas.112212199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Wu CI. Weak selection revealed by the whole-genome comparison of the X chromosome and autosomes of human and chimpanzee. Proc Natl Acad Sci USA. 2005;102:4063–4067. doi: 10.1073/pnas.0500436102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, King B, et al. (13 co-authors) Forces shaping the fastest evolving regions in the human genome. PLoS Genet. 2006;2:e168. doi: 10.1371/journal.pgen.0020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhakar S, Noonan JP, Paabo S, Rubin EM. Accelerated evolution of conserved noncoding sequences in humans. Science. 2006;314:786. doi: 10.1126/science.1130738. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Reich DE, Higgins JM, et al. (17 co-authors) Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- Sabo PJ, Humbert R, Hawrylycz M, Wallace JC, Dorschner MO, McArthur M, Stamatoyannopoulos JA. Genome-wide identification of DNase I hypersensitive sites using active chromatin sequence libraries. Proc Natl Acad Sci USA. 2004;101:4537–4542. doi: 10.1073/pnas.0400678101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat Rev Genet. 2007;8:424–436. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu Rev Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- Taylor MS, Kai C, Kawai J, Carninci P, Hayashizaki Y, Semple CA. Heterotachy in mammalian promoter evolution. PLoS Genet. 2006;2:e30. doi: 10.1371/journal.pgen.0020030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Chimpanzee Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Trinklein ND, Karaoz U, Wu J, et al. (12 co-authors) Integrated analysis of experimental data sets reveals many novel promoters in 1% of the human genome. Genome Res. 2007;17:720–731. doi: 10.1101/gr.5716607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Kudaravall S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.