Abstract
Ecotropic viral integration site 1 (EVI1) is an important transcription factor for leukemogenesis. EVI1 is a member of a group of transcription factors with C-terminal binding protein (CtBP)-binding motifs that act as transcriptional co-repressors; however, we recently found that EVI1 directly activates GATA2 transcription, which is an important gene for the maintenance of hematopoietic stem cells. We show here that EVI1-activated GATA2 transcripts derive from exon 1S of GATA2, which is specifically activated in neural and hematopoietic cells. EVI1 was acetylated by the histone acetyltransferase p300/CBP association factor (P/CAF) in myeloid leukemia cells and hematopoietic progenitor cells. Acetylation at Lys564, which is adjacent to the CtBP-binding consensus sequence of EVI1, was found to be important for transcriptional activation of GATA2. Mutation of Lys564 to alanine (K564A) markedly reduced the ability of EVI1 to bind DNA and activate transcription of GATA2. Furthermore, we confirmed that Lys564 in EVI1 was specifically acetylated in leukemia and primary hematopoietic cells by using an antibody directed against an acetylated Lys564 EVI1 peptide. Moreover, co-transfection of P/CAF with EVI1 overcame the suppressive effect of the CtBP co-repressor and resulted in GATA2 transcriptional activation; nonetheless, CtBP2 was still included in the protein complex with EVI1 and P/CAF on the EVI1-binding site in the GATA2 promoter region. Thus, acetylation of EVI1 at Lys564 by P/CAF enhances the DNA binding capacity of EVI1 and thereby contributes to the activation of GATA2.
Keywords: Co-repressor Transcription, Leukemia, Transcription Co-activators, Transcription Factors, Zinc Finger, Acetylation, EVI1, P/CAF
Introduction
Ecotropic viral integration site 1 (Evi1) was first identified as a common retroviral integration site in AKXD murine myeloid tumors (1). The human homologue EVI1 is transcriptionally activated by specific chromosomal abnormalities at 3q26 such as t(3;3)(q21;q26), inv(3)(q21q26), t(3,21), or others (2, 3). These abnormalities lead to the aberrant expression of EVI1 and are associated with human acute myelogenous leukemia, myelodysplastic syndrome, and chronic myelogenous leukemia. High expression of EVI1 is detectable in around 8% of myeloid leukemia cases and is a poor prognostic indicator in myeloid leukemia (4, 5).
In EVI1, two DNA binding domains with seven and three zinc finger repeats bind DNA through specific conserved GATA-like or ETS-like sequence motifs, and they have the potential to interact with both co-repressors and co-activators as a dual transcriptional factor (6–8). EVI1 has been shown to interact directly with the known transcriptional repressor C-terminal binding protein (CtBP)2 via two CtBP-binding consensus motifs at amino acids 544–607 (9, 10). Although CtBP binding to EVI1 has been suggested to recruit histone deacetylase complexes (HDACs) and lead to transcriptional repression via chromatin remodeling, specific target genes repressed by EVI1 have not yet been found. On the other hand, the interaction of EVI1 with cAMP-responsive element-binding protein-binding protein (CBP) and p300/CBP-associated factor (P/CAF) was reported to result in the reversible acetylation of EVI1 and in their co-localization in nuclear speckles (11). However, there is no information about how the acetylation of EVI1 or the binding of EVI1 to CBP or P/CAF influences the transcriptional activity of EVI1.
We recently reported that in Evi1-deficient mouse embryos, hematopoietic stem cells are fewer and less able to proliferate just before death at embryonic day 10.5. Moreover, we found that defective hematopoietic stem cells in Evi1-deficient mice were due to down-regulation of Gata2 transcription, which is crucial for hematopoietic stem cell maintenance and development, and that Gata2 was one of the target genes transcriptionally regulated by Evi1 (12). Recently, Evi1 was shown to be necessary for maintaining hematopoiesis in the adult murine hematopoietic system and in transformed leukemic cells (13). Therefore, Evi1 is one of the transcriptional regulators essential for maintaining embryonic and adult hematopoietic stem cells, and the regulation of Gata2 by Evi1 is also an important event in hematopoietic stem cell maintenance.
In this study, we investigated whether EVI1 regulates GATA2 expression in leukemia cells with high EVI1 expression (EVI1high leukemia) and how EVI1 regulates GATA2 transcription in those leukemia cells. Initially, we found that EVI1high leukemia cells strongly expressed GATA2 mRNA and that transcriptional activation of GATA2 by EVI1 is dependent on the 1S promoter, one of the alternatives to promoter 1 in GATA2 (14, 15). After the introduction of an Evi1 expression vector into murine leukemia EML-C1 cells, GATA2 transcription was induced by EVI1, whereas knocking down EVI1 transcripts by the introduction of small hairpin RNA (shRNA) down-regulated GATA2 transcription in EVI1high leukemia cell lines. Next, we found that the histone acetyltransferase p300/CBP-associated factor (P/CAF) could bind to EVI1 and acetylated EVI1 in leukemia cells. Lys564 in EVI1 is one of the important residues for the activation of GATA2 transcription by P/CAF acetylation. Finally, we developed specific antibodies that recognize the acetylated Lys564 of EVI1, and we found acetylated EVI1 in UCSD/AML1 cells with EVI1 high expression and in CD34+ hematopoietic progenitor cells from cord blood, but EVI1 was not acetylated in NT2 neuronal precursor cells. Using chromatin immunoprecipitation PCR and gel mobility shift assays, we found that the binding of EVI1 to the GATA2 promoter region was clearly enhanced by EVI1 acetylation. Therefore, the acetylation of EVI1 is an important modification that regulates the transcriptional activity and the DNA binding activity of EVI1.
EXPERIMENTAL PROCEDURES
Cell Lines and Cell Culture
UCSD/AML1 cells derived from human acute myeloid leukemia (16) were cultured in RPMI 1640 medium (Wako) supplemented with 10% fetal calf serum and 1 ng/ml granulocyte-macrophage colony-stimulating factor. K562, THP1, HL60, and U937 were purchased from RIKEN Cell Bank. MOLM1 (17) was purchased from Hayashibara Institute. Kasumi-3 (18) was provided by Dr. Asoh (Hiroshima University); K051 was kindly provided by Dr. Nomura (Nippon Medical School); NH was kindly provided by Dr. Suzukawa (University of Tsukuba); and OIH-1 was kindly provided by Dr. Hamaguchi (Musashino Red Cross Hospital). HL60, U937, K562, MOLM1, K051, NH, OIH-1, and Kasumi-3 were cultured in RPMI 1640 medium (Wako) supplemented with 10% fetal calf serum. The UCSD/AML1, MOLM-1, and Kasumi-3 cells lines each have chromosome 3q26 abnormalities involving EVI1 expression, although HL60, U937, K562, K051, NH, and OIH-1 do not have 3q26 abnormalities and show low expression of EVI1. Murine stem cell factor-dependent EML-C1 cell lines were provided by Dr. Suda (19). The 93T, COS7, and neural NT2 cell lines were cultured in Dulbecco's modified Eagle's medium (Wako) supplemented with 10% fetal calf serum.
Plasmids and Cloning
All expression vectors were constructed according to standard methods and verified by sequencing. A pFLAG-CMV26-based expression vector (pFLAG-EVI1) contained full-length human EVI1 (20) as well as a series of EVI1 deletion mutants with an N-terminal FLAG tag put it under control of the cytomegalovirus promoter (Sigma), and the mutants were kindly provided by Dr. Kurokawa (University of Tokyo) (21). After PCR amplification, two cDNA fragments (amino acids 1–232 and 534–1052) were ligated to produce the proline-rich deletion (ΔPR; amino acids 233–533) of EVI1. Deletion of the first zinc finger repeat in DNA binding domain 1, Δ1(1) (deletion of amino acids 1–43), of EVI1 was achieved by PCR amplification from pCMV26-FLAG-EVI1. Deletion of the CtBP binding domain with the repression domain (ΔCtBP + RD, deletion of amino acids 529–732) of EVI1 was inversely PCR-amplified from the plasmid pME18SΔRD (deletion of amino acids 608–732). Site-directed mutagenesis at Lys559, Lys561, or Lys564 to Ala in EVI1 (K559A, K561A, or K564A) was achieved with the PfuTurbo DNA polymerase system (Stratagene). Murine Ctbp2 cDNA provided by Dr. Crossley (9) was ligated into a pFLAG-CMV26 expression vector (Sigma) by PCR. CBP (pLNCX-FLAG-CBP), p300 (CMV-HA-p300), and P/CAF (RSV-P/CAF) expression vectors were kindly provided by Dr. Kitabayashi (National Cancer Center Research Institute) and Dr. Kurokawa (Saitama Medical College). pKD-PCAF-v2 (Millipore) and mammalian P/CAF small interfering RNA expression plasmids were purchased from Millipore.
Antibodies and Reagents
Anti-GATA2 (H-116), anti-P/CAF (H-369), anti-p300 (N-15), anti-CBP (A-22), anti-CtBP (C-1), and anti-EVI1 (N-20) antibodies were purchased from Santa Cruz Biotechnology, Inc. A rabbit polyclonal anti-HDAC1 antibody was purchased from Millipore. Mouse monoclonal anti-acetylated lysine antibody was purchased from Cell Signaling Technology. A mouse monoclonal anti-FLAG (M2) antibody was purchased from Sigma. Trichostatin A was purchased from Wako Pure Chemical Industries. An acetylated EVI1 peptide spanning Lys564 (TKRKDE(Ac)KPLTPVP) was synthesized to immunize rabbits. After immunization, a fraction of the specific immunoglobulin recognized by acetylated EVI1 was affinity-purified by binding to a column conjugated with the acetylated EVI1 peptide.
Reverse Transcriptase-PCR (RT-PCR)
Total RNA was extracted from each cell using TRIzol reagent (Invitrogen). Total RNA was converted into cDNA using avian myeloblastosis virus reverse transcriptase (Takara-Bio Inc.), and the cDNA was used as a template for PCR. The PCR was carried out by EX Taq polymerase (Takara-Bio Inc.), and the primers used were as follows: for human EVI1, forward 5′-CGAAAGCGAGAAT-3′ and reverse 5′-GGAAGACGTAGTG-3′; for human β-actin, forward 5′-AAGAGATGGCCACG-3′ and reverse 5′-TCCTTCTGCATCCTG-3′; for human GATA2, forward 5′-TGCGTCTCCAGCCTCATCTTCCGCG-3′ and reverse 5′-CGAGCCTTGCTGCGCTGCTT-3′; for human GATA2/1G, forward 5′-CCGGACCGCGTGTTGATGTACCTGG-3′ and reverse 5′-AACTCCTTCACTCTCAGAGGCC-3′; for human GATA2/1S, forward 5′-AAGCGGTCCGCTGAACACC-3′ and reverse 5′-CCACCTGCAGAAGAAGTAGT-3′; for human P/CAF, forward 5′-GCAAGGCCGAGGAGTCTTGTAAA-3′ and reverse 5′-GTTATTCACACCCTGTTCAATGCT-3′.
Quantitative Real Time RT-PCR
After extraction of total RNA from each cell line using TRIzol reagent (Invitrogen), the total RNA was converted into cDNA by avian myeloblastosis virus reverse transcriptase (Takara-Bio Inc.). The resulting first-strand cDNA was used for real time RT-PCR using a SYBR Green PCR master mix kit (Applied Biosystems). The primers used were as follows: for human GATA2, forward 5′-CTGCACAATGTTAACAGGCCACTGACCATG-3′ and reverse 5′-CATGGTCAGTGGCCTGTTAACATTGTGCAG-3′; for human EVI1, forward 5′-TTGCCAAGTAACAGCTTTGCTG-3′ and reverse 5′-GGTAGACTTTAGAGAAAGGCC-3′; and for human β-actin, forward 5′-CGGGAAATCGGTGCGTGACAT-3′ and reverse 5′-CTAGAAGCATTTGCGGTGGA-3′. The mRNA levels were detected using an ABI Prism 7000, and the data were analyzed by sequence detection systems software (Applied Biosystems) and normalized to the amount of β-actin mRNA.
Establishment of Stable UCSD/AML1 Cell Lines Expressing shEVI1
A DNA-based small hairpin (sh) RNA expression vector (pSIREN-retroQ-ZsGreen plasmid, Takara-Bio Inc.) was used in the EVI1 knockdown experiment. The following sequence was cloned into the BamHI-EcoRI site of the plasmid to create an shRNA against human EVI1 (22): 5′-GATCGCTCTAAGGCTGAACTAGCAGTTCAAGAGACTGCTAGTTCAGCCTTAGATTTTTTG-3′. A pSIREN-retroQ-ZsGreen-shLuc containing shRNA against luciferase (Takara-Bio Inc.) was used as a control. Retroviral particles were generated using the p10A1 packaging vector (Takara-Bio Inc.) and transient transfection of the 293T cell line, which was carried out by Hilymax liposome transfection reagent (Dojindo Molecular Technologies, Inc.). For retroviral infection, 1 × 106 UCSD/AML1 cells were placed in 6-cm dishes containing 5 ml of retroviral supernatant with 100 ng/ml Polybrene for 24 h. ZsGreen-positive UCSD/AML1 cells were sorted by a JSAN cell sorter (Bay Bioscience) 2 weeks after viral infection. Repression of EVI1 expression was confirmed by RT-PCR as above.
Immunoprecipitation
COS7 cells were transfected with FLAG-tagged EVI1, FLAG-tagged mutant EVI1, and a P/CAF expression plasmid using HilyMax according to the manufacturer's instructions. After culturing for 48 h, the transfected cells were solubilized with lysis buffer (50 mm Tris-HCl, pH 7.8, 150 mm NaCl, 1% Nonidet P-40, 0.25% sodium deoxycholate, 1 mm EDTA) supplemented with protease inhibitor (Complete, Roche Applied Science). The lysates were then immunoprecipitated by incubation with an anti-FLAG (M2), anti-P/CAF, anti-EVI1 (N-20), anti-CtBP (C-1), or anti-HDAC1 antibody followed by incubation with protein G beads (Amersham Biosciences). After washing, the bound proteins were subjected to SDS-PAGE, blotted to a polyvinylidene difluoride membrane (Millipore), and incubated with horseradish peroxidase-conjugated anti-mouse IgG antibody or anti-rabbit IgG antibody (Invitrogen). The transfer membrane was developed by ECL PlusTM reagent (GE Healthcare).
Immunohistochemistry
Cells were fixed with 4% paraformaldehyde in Tris-buffered saline (TBS) for 15 min at room temperature, washed with 0.1 m glycine in TBS for 5 min, permeabilized by 0.1% Triton X-100 in TBS for 10 min, and washed with 0.1 m glycine in TBS for 5 min. Cells were then blocked with 1% bovine serum albumin in TBS at 4 °C for 1 h prior to incubation with a first antibody (goat anti-EVI1 antibody, rabbit anti-P/CAF antibody, mouse anti-acetyl-lysine antibody, or rabbit anti-acetylated Lys564 EVI1 antibody). Samples were incubated with each first antibody at 4 °C overnight, washed with phosphate-buffered saline, and incubated with either Alexa Fluor 488-conjugated anti-goat IgG antibody, Alexa Fluor 546-conjugated anti-mouse IgG antibody, or Alexa Fluor 555 anti-rabbit IgG antibody (Invitrogen) at 4 °C for 1 h. Nuclei were counterstained with 4′,6-diamino-2-phenylindole (Dojindo Molecular Technologies, Inc.) or TOTO-3 from Molecular Probes (Invitrogen). Samples were enclosed with Prolong Gold antifade (Invitrogen) and observed under a laser-scanning microscope (TCS SP2 AOBS, Leica Microsystems).
GATA2 Promoter Luciferase Assay
A GATA2 promoter luciferase assay was performed based on a previous report (12). In brief, the GATA2 promoter construct (pGL3-B7) with luciferase under the herpes simplex virus thymidine kinase promoter (pRL-TK) as an internal control was co-transfected with each expression plasmid, such as FLAG-EVI1 or a series of FLAG-EVI1 mutants into 5 × 103 COS7 cells/well (24 well plate) by Hilymax Lipofection reagent. Transfected cells were incubated for 48 h in a CO2 incubator and collected by centrifugation. The luciferase assay was performed using a Dual-Luciferase assay system (Promega) according to the manufacturer's protocol. Luciferase activity was measured using a MiniLumat LB9506 (Berthold).
Chromatin Immunoprecipitation (ChIP) Assay
A ChIP assay was performed with a ChIP assay kit (Upstate) according to the manufacturer's protocol. After transfection of FLAG-EVI1, FLAG-EVI1K561A, or FLAG-EVI1K564A expression plasmids with GATA2 promoter constructs (pGL2-B7) into COS7 cells with Hilymax transfection reagent, the transfected COS7 cells were cross-linked with 1% formaldehyde and sonicated with a Sonifier 450 (Branson). After cross-linked chromatin fragments were immunoprecipitated with anti-FLAG M2 antibody, immunoprecipitated chromatin was reverse cross-linked at 65 °C overnight, digested with ribonuclease A and proteinase K, and then purified. Purified DNA fragments were amplified using a pair of primers for the specific EVI1-binding site at the GATA2 promoter, forward 5′-CCTCTGCCTCTCTGCCTCTGCCTC-3′ and reverse 5′-GCATGTTTTCTAATATCCAGGCCT-3′ (12). The same ChIP experiment was done by co-transfection of the EVI1 expression vector with CtBP2 or P/CAF.
Gel Mobility Shift Assay
Gel mobility shift assay was performed using a second generation digoxigenin-labeled gel shift assay kit (Roche Applied Science) according to the manufacturer's protocol. A FLAG-EVI or a FLAG-EVI1K564A expression plasmid was introduced into COS7 cells with Hilymax Lipofection reagent. After incubation for 48 h, the transfected cells were collected by centrifuge and lysed with lysis buffer. The cell supernatants were used for immunoprecipitation with mouse monoclonal anti-FLAG M2 antibody-conjugated agarose beads. Immunoprecipitated EVI1 protein was washed with phosphate-buffered saline and eluted with 100 μg/ml FLAG peptide (Sigma). Purified FLAG-EVI1 or FLAG-EVI1K564A was mixed with 0.4 ng/μl DIG-labeled double-stranded DNA nucleotides containing the EVI1-binding sequence at the GATA2 promoter region (5′-ACAGTGGTGATGTCCATGTCCCTTTTATTT-3′) (12) or with 0.4 ng/μl DIG-labeled nucleotides containing cold excess unlabeled nucleotides (0.1 μg/μl) as a competitor. Samples were separated by electrophoresis in 0.2× TBE buffer and blotted to a positively charged nylon membrane (Hybond N+, GE Healthcare) and then cross-linked by a 120 mJ/cm2 cross-linker (UVP). The membrane was incubated with an alkaline phosphatase-conjugated anti-DIG Fab fragment (Roche Applied Science) and then developed with disodium 3-(4-methoxyspiro{1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.13,7]decan}-4-yl) phenyl phosphate reagent (Roche Applied Science).
RESULTS
EVI1 Transcriptionally Regulated GATA2 mRNA Derived from Exon 1S
Recently, we reported that Evi1 maintained murine hematopoietic stem cells through direct transcriptional activation of the Gata2 gene (12). To determine whether EVI1 regulates GATA2 transcripts in human myeloid leukemia cells, various leukemia cell lines with or without EVI1 expression were evaluated for GATA2 expression levels by semi-quantitative RT-PCR. As shown in Fig. 1A, human myeloid leukemia cell lines with chromosomal translocations or inversions of chromosome 3q26 (UCSD/AML1, Kasumi-3, and MOLM-1) strongly expressed EVI1 (EVI1high leukemia cells), whereas cell lines without a chromosome 3q26 abnormality (NH, K051, HL-60, THP-1, U937, and OIH-1) expressed very low levels of EVI1 (EVI1low leukemia cells). A GATA2/1S transcript was derived from the distal first exon (1S), which is specifically used in hematopoietic and neuronal cells, and a GATA2/1G transcript was derived from the proximal first exon (1G), which is utilized in various tissues (14, 15). Using specific primers for detecting 1S exon-derived transcripts (GATA2/1S), 1G exon-derived transcripts (GATA2/1G), or both GATA2 transcripts (GATA2/T), we performed semi-quantitative RT-PCR. GATA2/T was detected in all of the leukemia cell lines and showed relatively high expression in the EVI1high leukemia cells (Fig. 1A). Interestingly, GATA2/1G was expressed in most of the cell lines; however, GATA2/1S was only detected in three EVI1high leukemia cell lines. These results indicated that EVI1 might specifically regulate the expression of GATA2/1S transcripts in leukemia cell lines.
FIGURE 1.
Activation of GATA2 transcription from exon 1S by EVI1. A, levels of EVI1 and GATA2 transcripts in various types of human myeloid leukemia cell lines as determined by semiquantitative RT-PCR. GATA2/1S transcripts are derived from exon 1S of GATA2; GATA2/1G transcripts are derived from exon 1G, and GATA2/T transcripts are included in both types of transcript. B, induction of GATA2 transcripts in murine stem cell factor-dependent EML-C1 myeloid cells by EVI1 transfection. After EVI1 or Mock expression vectors were transfected into EML-C1 cells, the transfected cells were collected at the indicated time points and used for RT-PCR. C, induction of two types of GATA2 transcripts by EVI1 transfection in EML-C1 cells detected by quantitative real time RT-PCR. A statistical analysis was done using the t test (*, p < 0.05). D, down-regulation of EVI1 by introduction of small hairpin RNA against EVI1 (shEVI1) suppressed GATA2 transcription. shLuc indicates transfection of an shRNA expression vector for the firefly luciferase gene and shNega indicates transfection of an shRNA mock expression vector. Three cell lines with shEVI1 expression were established as shEVI1–1, -2, and -3. Asterisks indicate nonspecific bands.
Next, to determine whether the ectopic expression of EVI1 activates GATA2/1S and/or GATA2/1G transcription in leukemia cells, a green fluorescence protein-EVI1 expression plasmid was transfected into stem cell factor-dependent murine EML-C1 cells (19), and three kinds of GATA2 transcripts were monitored in the EML-C1 cells (Fig. 1B). EVI1, GATA2/1S, GATA2/1G, and both of the GATA2 transcripts (GATA2/T) were faintly expressed in parental EML-C1 cells. After transfection of EVI1, the levels of EVI1 and GATA2/1S transcripts increased at the same time 48 h after the transfection, but the level of the GATA2/1G transcript was unchanged before and after EVI1 transfection. An increase in the GATA2/T and GATA2/1S transcripts of ∼4-fold was detected by real time RT-PCR along with the same up-regulated level of EVI1 expression (Fig. 1C). Thus, the GATA2/1S transcript is specifically up-regulated in EML-C1 by EVI1 expression.
To determine whether the GATA2 expression in EVI1high leukemia cells is dependent on EVI1 expression, EVI1 transcripts were knocked down in UCSD/AML1 cells by the transfection of retroviral vectors with small EVI1 hairpin RNA (shEVI1). Green fluorescence protein expression was used as a transfection marker. After sorting green fluorescence protein-positive UCSD/AML1 cells, three independent cell lines (shEVI1–1, shEVI1–2, and shEVI1–3) were established. The expression of EVI1 was significantly suppressed in the shEVI1–1 to shEVI1–3 cell lines as compared with the parental UCSD/AML1 cells and with UCSD/AML1 cells transfected with shRNA for firefly luciferase (shLuc/AML1) or shRNA mock vector (shNega/AML1) as controls (Fig. 1D). Along with down-regulated EVI1 expression, the expression levels of GATA2/T, GATA2/1S, and GATA2/1G mRNA were also significantly down-regulated. Thus, EVI1 enhances GATA2 transcription in EVI1high leukemia cells.
Interaction of P/CAF and Acetylation of EVI1 Protein in EVI1high Leukemia Cells
After showing that EVI1 activates GATA2 transcription in EVI1high leukemia cells, we next examined the molecular mechanism of transcriptional activation of GATA2. Initially, we looked for an interaction between EVI1 and histone acetyltransferase/transcriptional co-activators such as p300, CBP, and P/CAF. After the EVI1 expression plasmid was transiently co-transfected with p300, CBP, or P/CAF expression plasmids into COS7 cells, cell lysates were immunoprecipitated with either anti-P/CAF, anti-CBP, anti-p300, or EVI1 antibodies. The precipitated proteins were then immunoblotted with each antibody, as above. An EVI1-P/CAF complex was precipitated by either anti-EVI1 or P/CAF antibodies (Fig. 2A), but EVI1 did not interact with p300 or CBP (supplemental Fig. S1, A–C). Next, we asked whether P/CAF could enhance the transcriptional activation of GATA2 by EVI1. Various amounts of a P/CAF expression plasmid were co-transfected with an EVI1 expression plasmid along with a GATA2-luciferase promoter/reporter plasmid into COS7 cells. GATA2 transcription was dose-dependently enhanced by P/CAF co-transfection with EVI1 (Fig. 2B), but p300 and CBP did not enhance GATA2 transcription (Fig. 2C). Because EVI1 was acetylated by P/CAF or CBP (11), we next examined the acetylation of EVI1 after P/CAF co-transfection (Fig. 2D). After transfection of FLAG-tagged EVI1 with or without P/CAF, the precipitated proteins were immunoblotted with either anti-EVI1 or anti-acetylated lysine antibody. Acetylated EVI1 protein was increased after co-transfection with P/CAF into COS7 cells, and the level of acetylated EVI1 protein was significantly enhanced after both EVI1 alone and EVI1 with P/CAF transfection upon treatment with trichostatin A, which is an inhibitor of histone deacetylases (Fig. 2D). To confirm whether endogenous EVI1 is acetylated and whether the endogenous EVI1 and P/CAF form a protein complex in EVI1high leukemia cells, cell extracts of UCSD/AML1 cells were immunoprecipitated with anti-acetylated or anti-P/CAF antibodies, and the precipitated proteins were immunoblotted with anti-EVI1 or anti-P/CAF antibodies. As shown in Fig. 2F, EVI1 was significantly acetylated and interacted with P/CAF in EVI1high leukemia cells. To confirm the co-localization of EVI1 and P/CAF and the existence of acetylated EVI1 in the UCSD/AML1 cells, endogenous EVI1 and P/CAF were stained with anti-EVI1 and anti-P/CAF antibodies (upper column), or with anti-EVI1 and anti-acetylated lysine antibodies (lower column) and examined under confocal microscopy. As shown in Fig. 2F, we confirmed that EVI1 co-localized with P/CAF in the nuclei of UCSD/AML1 cells and was significantly acetylated.
FIGURE 2.
EVI1 was acetylated by the histone acetyltransferase P/CAF during transcriptional activation of GATA2. A, EVI1 co-precipitated with P/CAF, but not with p300 or CBP. After transfection of EVI1 and P/CAF into COS7 cells, EVI1 and P/CAF protein complexes were immunoprecipitated (IP) with anti-EVI1 or anti-P/CAF antibody, respectively, and the precipitated proteins were immunoblotted with anti-P/CAF or anti-EVI1 antibody. B, transcriptional activation of the GATA2 promoter is dependent on P/CAF. EVI1 and/or various amounts of P/CAF expression vector were transfected into COS7 cells, and GATA2 transcriptional activity was measured by a Dual-Luciferase assay system. C, activation of GATA2 transcription by transfection of EVI1 with P/CAF, p300, or CBP was determined by using a luciferase assay system with the GATA2 promoter. D, acetylation of EVI1 was detected in COS7 cells transfected with EVI1 expression vectors using an anti-acetylated antibody. Co-transfection of P/CAF with an EVI1 expression vector and treatment with trichostatin A (TSA) as an HDAC inhibitor enhanced the acetylation of EVI1 in COS7 cells. E, endogenous EVI1 was acetylated and co-precipitated with endogenous P/CAF in UCSD/AML1 cells. After immunoprecipitation with an anti-acetylated antibody, the precipitated proteins were immunoblotted with an anti-EVI1 antibody. After immunoprecipitation with an anti-P/CAF antibody, the precipitated proteins were immunoblotted with an anti-EVI1 or anti-P/CAF antibody. F, EVI1 co-localized with P/CAF in the nuclei of UCSD/AML1 cells, and most of the EVI1 in the nucleus was acetylated, as revealed by immunohistochemical staining under confocal microscopy. Nuclear DNA was stained with TOTO-3. Anti-human EVI1 antibody was stained with Alexa Fluor 488-labeled secondary antibody (green), anti-acetylated lysine was stained with Alexa Fluor 546-labeled secondary antibody (red) and anti-P/CAF antibody was stained with Alexa Fluor 555-labeled antibody (red).
First DNA Binding Domains, CtBP Binding Domains, and C-terminal Acidic Domains of EVI1 Are Important for Activation of GATA2 Transcription
To determine which domain of EVI1 is necessary for the activation of GATA2 transcription, various kinds of EVI1 deletion mutants were transfected with a GATA2-luciferase promoter/reporter plasmid into COS7 cells (Fig. 3A). The expression of each EVI1 deletion mutant construct was confirmed by immunoblot analysis with anti-EVI1 antibody (Fig. 3B and supplemental Fig. S2). Transcription of GATA2 was highly activated by transfection of wild-type EVI1 and the EVI1 deletion mutant constructs with deletion of the 2nd to 7th zinc finger repeats in the first DNA binding domain (ΔD1(2–7)), deletion of the proline-rich domain (ΔPR), deletion of the repressor domain (ΔRD), and deletion of the 8th to 10th zinc finger repeats in the 2nd DNA binding domain (ΔD2(8–10)) (Fig. 3A). However, GATA2 transcription was not activated by transfection of EVI1 deletion mutant constructs with deletion of the 1st to 7th zinc finger repeats in the first DNA binding domain (ΔD1(1–7)), the CtBP binding domain with the repression domain (ΔCtBP + RD), or the acidic amino acid cluster domain (ΔAD). Among the three domains important for GATA2 transcriptional activation, the ΔD1(1–7) mutant of EVI1 might lose its transcriptional enhancer activity because of loss of the capacity to bind GATA2 promoter region DNA. As the ΔD1(2–7) mutant of EVI1 kept its transcriptional enhancer activity, a ΔD1(1) mutant of EVI1 with deletion of only the first zinc finger repeat was constructed and tested for transcriptional enhancer activity. The ΔD1(1) mutant lost its transcriptional enhancer activity (Fig. 3A and supplemental Fig. S2), suggesting that the first zinc finger repeat is important for enhancing GATA2 transcription.
FIGURE 3.
Acetylation of Lys564 just after the CtBP-binding consensus sequences is important for transcriptional activation of GATA2. A, various FLAG-tagged EVI1 deletion mutants (illustrated in Fig. 3A) were co-transfected with the GATA2-luciferase promoter/reporter construct into COS7 cells, and the luciferase activity was assayed. Three EVI1 mutants with lysine to alanine replacements were also used as follows: EVI1 K559A, K561A, and K564A. B, protein expression of the various kinds of transfected EVI1 deletion mutants was confirmed by immunoblotting using an anti-FLAG antibody. WT, wild type. C, characterization of anti-acetylated Lys564 EVI1 antibody made by immunization with anti-acetylated EVI1 peptides. EVI1 with K561A or K564A mutations was transfected into COS7 cells, and the cell lysates were immunoblotted with anti-FLAG antibody (α-FLAG) or anti-acetylated Lys564 EVI1 antibody (α-AcK564 EVI1). D, histochemical staining of EVI1 with anti-EVI1 or anti-AcK564 EVI1 antibody in NT2 neuronal cells, UCSD/AML1 leukemia cells, and CD34+ cord blood cells (CD34+ CBC). Each cell was stained with 4′,6-diamino-2-phenylindole or TOTO-3 for the nucleus (blue), anti-EVI1 antibody (Alexa Fluor 488, green), or anti-AcK564 EVI1 antibody (Alexa Fluor 555, red).
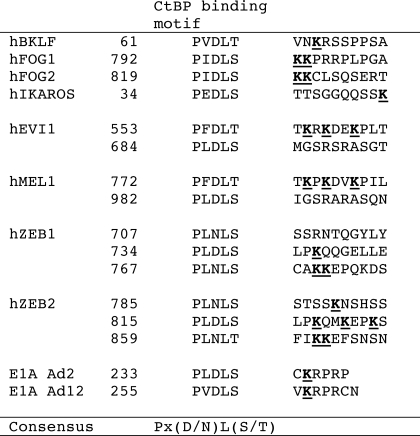
Although the ΔCtBP + RD mutant of EVI1 lost its transcriptional enhancer activity, the RD mutant of EVI1 kept its enhancer activity. Therefore, these results suggest that the CtBP binding domain is an important domain for transcriptional activation of GATA2. In the E1A protein, the acetylation of the lysine residue adjacent to the CtBP-binding consensus sequence by histone acetyltransferases might disrupt repressor complexes (including CtBP) and mediate gene activation (23). In the EVI1 protein, two CtBP-binding consensus sequences (553PFDLT557 and 584PLDLS588) were found within the CtBP binding domain. Three lysine residues (Lys559, Lys561, and Lys564) were found adjacent to the first CtBP-binding sequence, 553PFDLT557 (Table 1), and we replaced each of them with an alanine residue to construct three mutated EVI1 expression vectors (K559A, K561A, and K564A). In the transcriptional activation assay using the GATA2-luciferase promoter/reporter, EVI1-K564A completely lost its transcriptional enhancer activity (Fig. 3A), although EVI-K559A and EVI-K561A retained their activities, suggesting that Lys564 is an important residue for the maintenance of transcriptional enhancer activity.
TABLE 1.
Alignment of the lysine residues just after the CtBP-binding motif
CtBP-binding consensus sequences were aligned with the flanking lysine residues just after the CtBP-binding motifs (underlined). Genes in the list were selected from tables in the cited literature (9, 23, 36). The GenBankTM accession numbers are as follows: hBKLF, NP_057615; hFOG1, AAN45858; hFOG2, NP_036214; hIKAROS, NP_006051; hEVI1, NP_001098548; hMEL1, Q9HAZ2; hZEB1, P37275; hZEB2, O60315; E1A (human adenovirus 5), CAB40664; E1A (human adenovirus 12), CAA51877.
Next, to confirm EVI1 acetylation at Lys564, we developed a specific polyclonal antibody by immunization of rabbits with a synthetic peptide with acetylated Lys564. To confirm the existence of acetylated EVI1 at Lys564, FLAG-tagged EVI1-K561A or EVI1-K564A was transfected into COS7 cells, and each EVI1 protein was detected by immunoblot analysis using anti-FLAG antibody or anti-acetylated Lys564 EVI1 antibody (anti-AcK564-EVI1). The anti-FLAG antibody identified both the EVI1-K561A and EVI1-K564A EVI1 mutant proteins; however, the anti-AcK564-EVI1 antibody only detected EVI1-K561A and not EVI1-K564A (Fig. 3C). Next, EVI1 was immunologically stained in UCSD/AML1 cells with high EVI1 expression, CD34+ hematopoietic progenitor cells from umbilical cord blood, and an NT2 neural progenitor cell line with EVI1 expression using the anti-AcK564-EVI1 antibody. As shown in Fig. 3D, both UCSD/AML1 cells and CD34+ hematopoietic progenitor cells were stained by anti-EVI1 and anti-AcK564-EVI1 antibodies. However, EVI1 in NT2 cells was stained in the cytoplasm and nucleus by anti-EVI1 antibody but was not stained by anti-AcK564 EVI1 antibody (the lowest column in Fig. 3D), suggesting that EVI1 was not acetylated in NT2 neuronal progenitor cells. Therefore, EVI1 is specifically acetylated in hematopoietic and leukemia cells, but it can be either acetylated or not acetylated in neuron-related cells.
Acetylation of EVI1 at Lys564 Enhances Its DNA Binding Ability
To determine how the acetylation event and CtBP co-repressor binding to EVI1 is involved in the transcriptional activation of GATA2 by EVI1 in EVI1high leukemia cells, co-precipitation assays and ChIP-PCR were used. After immunoprecipitation by anti-P/CAF antibody, only endogenous P/CAF was precipitated in COS7 cells (Fig. 4A). After transfection with EVI1 and CtBP2 expression vectors, endogenous P/CAF co-precipitated with EVI1 and CtBP2. After transfection with P/CAF and EVI1, P/CAF was also precipitated with EVI1 by anti-P/CAF antibody. After transfection with EVI1, P/CAF, and CtBP2, the three proteins were equally precipitated by anti-P/CAF antibody (Fig. 4A), suggesting that these three proteins formed a ternary complex in COS7 cells and that they did not compete with each other to bind to the DNA sequence. Next, EVI1, P/CAF, CtBP2, and HDAC1 were transfected into COS7 cells to immunoprecipitate the EVI1-binding site in the GATA2 promoter. After confirmation of the expression from each protein by immunoblot (supplemental Fig. S3), ChIP-PCR revealed that EVI1, P/CAF, and CtBP2 occupied the EVI1-binding site of the GATA2 promoter, but HDAC1 was not included in the protein complex on the binding site (Fig. 4B). To determine whether the protein complex could activate GATA2 transcription, a transcriptional activation assay for GATA2 was performed after transfection of EVI1, P/CAF, and/or CtBP2 in various combinations into COS7 cells (Fig. 4C). GATA2 transcription was activated by transfection of EVI1 alone, but transfection of CtBP2 with EVI1 suppressed GATA2 transcription to its basal level. Upon co-transfection of P/CAF with EVI1 and CtBP2, GATA2 transcription was activated up to the level of transfection with EVI1 alone, suggesting that P/CAF overcame the suppressive effects of CtBP2. Moreover, EVI1-K564A lost its GATA2 transcriptional activity even if P/CAF was co-transfected with it. To determine whether the acetylation of EVI1 at Lys564 affects its DNA binding capacity, EVI1, EVI1-K561A, and EVI1-K564A were transfected, and their DNA binding capacities were determined by ChIP-PCR. As shown in Fig. 4D, EVI1-K564A displayed significantly lower DNA binding capacity at the EVI1-binding site in the GATA2 promoter, although EVI1 and EVI1-K561A retained their DNA binding capacity. Moreover, to determine whether the acetylation of EVI1 influences its DNA binding capacity, ChIP was performed after transfection of EVI1 alone, EVI1 with CtBP2, or EVI1 with P/CAF in COS7 cells. The DNA binding capacity of EVI1 was significantly enhanced by co-transfection with P/CAF, but it was not enhanced by co-transfection with CtBP2 (Fig. 4E). To confirm the results, a gel mobility shift assay using the EVI1-binding site at the GATA2 promoter region was performed after transfection with EVI1 or the EVI1-K564A mutant expression vector. A shifted band was detected with purified EVI1 protein from nuclear extracts of COS7 cells transfected with EVI1; the shifted band disappeared after adding excess cold binding oligonucleotides (D1/b), but it did not disappear after adding unrelated oligonucleotides (D2/i). However, no shifted band was detected using purified EVI1-K564A protein from COS7 cells transfected with the EVI1-K564A mutant construct (Fig. 4F), suggesting that EVI1-K564A lost its capacity to bind the EVI1-binding site in the GATA2 promoter region.
FIGURE 4.
Protein complex containing EVI1, P/CAF, and CtBP on the EVI1-binding site in the GATA2 promoter during transcriptional activation of GATA2. A, ternary protein complex containing EVI1, P/CAF, and CtBP was detected by co-immunoprecipitation (IP) Various combinations of FLAG-EVI1, P/CAF, and/or FLAG-CtBP2 expression vectors were transfected into COS7 cells, and protein complexes containing P/CAF were precipitated by anti-P/CAF antibodies. Precipitated proteins were detected by anti-EVI1, P/CAF, or CtBP antibodies. Asterisks indicate nonspecific bands. B, after transfection with four kinds of expression vectors (EVI1, P/CAF, CtBP2, or HDAC1), formalin-fixed DNA fragments were immunoprecipitated with anti-EVI1, anti-P/CAF, anti-CtBP2, or anti-HDAC1 antibody. After immunoprecipitation, purified DNA was amplified by specific pairs of primers in the GATA2 promoter region. Each species-specific IgG was used for immunoprecipitation as an isotype control. C, loss of GATA2 transcriptional activation by transfection with an EVI1-K564A mutant. EVI1 or EVI1-K564A mutant expression vectors were transfected into COS7 cells either with CtBP2 alone or CtBP2 with P/CAF expression vectors for luciferase assays of GATA2 promoter activity. D, chromatin immunoprecipitation analysis of the EVI1-binding site in the GATA2 promoter region after transfection with the lysine-mutated EVI1 expression vector. After transfection with FLAG-tagged EVI1 (WT), -K561A, or -K564A mutants with GATA2 promoter constructs, formalin-fixed DNA fragments were immunoprecipitated with anti-FLAG antibody or control mouse IgG. The precipitated DNA was then amplified using specific primers for detecting the EVI1-binding sites. IB, immunoblot. E, P/CAF enhanced the binding of EVI1 to the EVI1-binding site in the GATA2 promoter region. After co-transfection of the EVI1 expression vector with CtBP2 or P/CAF, formalin-fixed DNA fragments were precipitated with the anti-FLAG antibody or control mouse IgG. F, gel mobility shift assay for EVI1 using the EVI1-binding site (D1/b or D2/i) of the GATA2 promoter region. After transfection of EVI1 or EVI1-K564A mutant expression vectors into COS7 cells, EVI1 protein purified by anti-FLAG M2 antibody was used for gel mobility shift assay with EVI1-binding oligonucleotides specific for D1/b or D2/i. Purified EVI1 protein was detected with anti-FLAG-M2 antibody, and a 100-fold excess of cold binding oligonucleotides (D1/b or D2/i) was added to the gel mobility shift assay reaction. G, levels of EVI1, P/CAF, and GATA2 were determined in UCSD/AML1 cells during shP/CAF expression by semi-quantitative RT-PCR. UCSD/AML1 cells with an shLuc expression vector were used as a control. H, levels of P/CAF and EVI1 acetylation at Lys564, and EVI1 were determined by immunoblot using each specific antibody with β-actin as a control in both UCSD/AML1 cells with shLuc and with shP/CAF. I, chromatin immunoprecipitation analysis of the EVI1-binding site in the GATA2 promoter region after transfection of shP/CAF or shLuc expression vectors into UCSD/AML1 cells. DNA was precipitated with an anti-EVI1 antibody or control mouse IgG.
To elucidate whether the acetylation of EVI1 at Lys564 is dependent on the protein binding to P/CAF, we introduced an expression vector with a small hairpin RNA against P/CAF (shP/CAF) into UCSD/AML1 cells to determine the acetylation status of EVI1 and its DNA binding ability at the GATA2 promoter. The expression of P/CAF was significantly reduced in the UCSD/AML1 cells after shP/CAF transfection along with a reduction in GATA2 expression (Fig. 4G). The reduction in P/CAF down-regulated the level of acetylation at Lys564 of EVI1 (Fig. 4H) and reduced the ability of EVI1 to bind the GATA2 promoter region (Fig. 4I). Therefore, the acetylation of EVI1 at Lys564 is dependent on P/CAF expression and has an effect on the DNA binding ability of EVI1.
DISCUSSION
In this study, we found that transcriptional activation of GATA2 by EVI1 is dependent on the acetylation of the EVI1 protein at Lys564. EVI1 acetylation is due to its interaction with P/CAF, and the acetylation of Lys564 adjacent to the CtBP-binding consensus sequence is an important modification for the transcriptional activation of GATA2. The acetylation of Lys564 significantly enhances the ability of EVI1 to bind the GATA2 promoter region. In the transcriptional activation complex on the EVI1 DNA-binding site, the co-repressor CtBP2 was still included with P/CAF and EVI1, but HDAC was not included, suggesting that this complex of EVI1, P/CAF, and CtBP2 may exist to aid in the transcriptional activation of GATA2.
The interaction of E1A with CtBP1 is reduced by the p300-mediated acetylation of Lys239 of E1A (23), and the acetylation of Lys239 is also important for nuclear localization and importin-α binding (24). Although Lys239 in E1A is located at the second amino acid away from the CtBP-binding consensus sequence of PX(D/N)L(S/T) (CtBP-binding motif) and the lysine residue in the same position is conserved in many other transcriptional repressors with CtBP binding (23), the locations of the lysine residues in transcription factors with a CtBP motif are at a variety of positions (from first to tenth) after the CtBP-binding motif (Table 1). ZEB1 belongs to a family of CtBP-binding transcription factors with a homeobox and zinc finger DNA binding domains, and P/CAF recruitment and acetylation of ZEB1 has been reported to lead to the displacement of CtBP1 from ZEB1 (25). Although the responsible lysine residue was not identified in ZEB1, ZEB1 has three CtBP-binding motifs with many lysine residues (at Lys741, Lys774, and Lys775) just after the CtBP-binding motif (Table 1), and these three Lys residues are conserved at the same places in ZEB2 (26). The lysine residues of Lys559, Lys561, and Lys564 in EVI1 are also conserved in MEL1/PRDM16 (a member of the EVI1 gene family) (27), suggesting that the conserved lysine residues in these transcription factors possibly have an important function in the modulation of transcriptional regulation. Moreover, although Lys239 in E1A is the second amino acid next to the CtBP-binding motif and the lysine residues were frequently found at the second, third, and fourth positions after the CtBP-binding motif, such as PXDLS X(k/r)(k/r)(k/r) (28), Lys564 in EVI1 is not the first lysine but the third lysine, and it is the seventh amino acid after the CtBP-binding motif (Table 1). Therefore, the exact position of the acetylated lysine residue relevant to the transcriptional regulation is not important, but the important lysine residue is probably localized within a few amino acids just after the CtBP-binding motif. On the other hand, it has been reported that the acetylation at Lys239 adjacent to the CtBP-binding motif in E1A causes a limited reduction in its CtBP binding ability as revealed by in vitro binding assays using synthetic peptides (29). In EVI1, CtBP2 was retained in the protein complex with EVI1 and P/CAF on the DNA at the GATA2 promoter region; however, HDAC1 was not included in this protein complex. Therefore, the acetylation of EVI1 does not influence the EVI1-CtBP interaction, but it may influence the EVI1-HDAC or CtBP-HDAC interactions. By contrast, the Drosophila homologue dCtBP directly activates the transcription of certain Wnt target genes after stimulation with Wnt, but it represses others in the absence of Wnt (30). Therefore, post-translational modifications after the activation of intracellular signaling, including Wnt stimulation, may be important regulators for switching the system of transcriptional regulation by transcription factors.
Because transcription factors themselves can be acetylated and/or deacetylated by histone acetyltransferases and/or HDACs (31), respectively, this modification may influence their regulation of transcription or their DNA binding activity. The acetylation of NF-E2, TAL1, GATA2, or c-Myb may increase their transcriptional activity by increasing their DNA binding activity (32–36). On the other hand, the acetylation of GATA1 (37) and EKLF (38) enhances their transcriptional activity without increasing their DNA binding activity. The reasons behind the difference between these two groups of transcription factors are not known; however, the acetylation of a transcription factor could change its protein conformation as well as its DNA-binding affinity. Because the acetylation of GATA2 and TAL1 results in erythroid differentiation, the acetylation of EVI1 is thought to have an important biological function in hematopoietic stem cells and/or leukemia cells. Therefore, it is important to determine how the acetylation of EVI1 is regulated in hematopoietic stem cells and leukemia cells.
Supplementary Material
This work was supported in part by grants-in-aid for scientific research of priority areas from the Ministry of Education, Culture, Sports, Science and Technology in Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- CtBP
- C-terminal binding protein
- P/CAF
- p300/CBP association factor
- HDAC
- histone deacetylase complex
- shRNA
- small hairpin RNA
- CBP
- cAMP-response element-binding protein-binding protein
- TBS
- Tris-buffered saline
- RT
- reverse transcriptase
- ChIP
- chromatin immunoprecipitation.
REFERENCES
- 1.Morishita K., Parker D. S., Mucenski M. L., Jenkins N. A., Copeland N. G., Ihle J. N. (1988) Cell 54, 831–840 [DOI] [PubMed] [Google Scholar]
- 2.Morishita K., Parganas E., William C. L., Whittaker M. H., Drabkin H., Oval J., Taetle R., Valentine M. B., Ihle J. N. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 3937–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitani K., Ogawa S., Tanaka T., Miyoshi H., Kurokawa M., Mano H., Yazaki Y., Ohki M., Hirai H. (1994) EMBO J. 13, 504–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lugthart S., van Drunen E., van Norden Y., van Hoven A., Erpelinck C. A., Valk P. J., Beverloo H. B., Löwenberg B., Delwel R. (2008) Blood 111, 4329–4337 [DOI] [PubMed] [Google Scholar]
- 5.Barjesteh van Waalwijk van Doorn-Khosrovani S., Erpelinck C., van Putten W. L., Valk P. J., van der Poel-van de Luytgaarde S., Hack R., Slater R., Smit E. M., Beverloo H. B., Verhoef G., Verdonck L. F., Ossenkoppele G. J., Sonneveld P., de Greef G. E., Löwenberg B., Delwel R. (2003) Blood 101, 837–845 [DOI] [PubMed] [Google Scholar]
- 6.Perkins A. S., Fishel R., Jenkins N. A., Copeland N. G. (1991) Mol. Cell. Biol. 11, 2665–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delwel R., Funabiki T., Kreider B. L., Morishita K., Ihle J. N. (1993) Mol. Cell. Biol. 13, 4291–4300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morishita K., Suzukawa K., Taki T., Ihle J. N., Yokota J. (1995) Oncogene 10, 1961–1967 [PubMed] [Google Scholar]
- 9.Turner J., Crossley M. (1998) EMBO J. 17, 5129–5140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmer S., Brouillet J. P., Kilbey A., Fulton R., Walker M., Crossley M., Bartholomew C. (2001) J. Biol. Chem. 276, 25834–25840 [DOI] [PubMed] [Google Scholar]
- 11.Chakraborty S., Senyuk V., Sitailo S., Chi Y., Nucifora G. (2001) J. Biol. Chem. 276, 44936–44943 [DOI] [PubMed] [Google Scholar]
- 12.Yuasa H., Oike Y., Iwama A., Nishikata I., Sugiyama D., Perkins A., Mucenski M. L., Suda T., Morishita K. (2005) EMBO J. 24, 1976–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goyama S., Yamamoto G., Shimabe M., Sato T., Ichikawa M., Ogawa S., Chiba S., Kurokawa M. (2008) Cell Stem Cell 3, 207–220 [DOI] [PubMed] [Google Scholar]
- 14.Minegishi N., Ohta J., Suwabe N., Nakauchi H., Ishihara H., Hayashi N., Yamamoto M. (1998) J. Biol. Chem. 273, 3625–3634 [DOI] [PubMed] [Google Scholar]
- 15.Pan X., Minegishi N., Harigae H., Yamagiwa H., Minegishi M., Akine Y., Yamamoto M. (2000) J. Biochem. 127, 105–112 [DOI] [PubMed] [Google Scholar]
- 16.Oval J., Jones O. W., Montoya M., Taetle R. (1990) Blood 76, 1369–1374 [PubMed] [Google Scholar]
- 17.Matsuo Y., Adachi T., Tsubota T., Imanishi J., Minowada J. (1991) Hum. Cell 4, 261–264 [PubMed] [Google Scholar]
- 18.Asou H., Suzukawa K., Kita K., Nakase K., Ueda H., Morishita K., Kamada N. (1996) Jpn. J. Cancer Res. 87, 269–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsai S., Bartelmez S., Sitnicka E., Collins S. (1994) Genes Dev. 8, 2831–2841 [DOI] [PubMed] [Google Scholar]
- 20.Morishita K., Parganas E., Douglass E. C., Ihle J. N. (1990) Oncogene 5, 963–971 [PubMed] [Google Scholar]
- 21.Kurokawa M., Mitani K., Irie K., Matsuyama T., Takahashi T., Chiba S., Yazaki Y., Matsumoto K., Hirai H. (1998) Nature 394, 92–96 [DOI] [PubMed] [Google Scholar]
- 22.Liu Y., Chen L., Ko T. C., Fields A. P., Thompson E. A. (2006) Oncogene 25, 3565–3575 [DOI] [PubMed] [Google Scholar]
- 23.Zhang Q., Yao H., Vo N., Goodman R. H. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 14323–14328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madison D. L., Yaciuk P., Kwok R. P., Lundblad J. R. (2002) J. Biol. Chem. 277, 38755–38763 [DOI] [PubMed] [Google Scholar]
- 25.Postigo A. A., Depp J. L., Taylor J. J., Kroll K. L. (2003) EMBO J. 22, 2453–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Comijn J., Berx G., Vermassen P., Verschueren K., van Grunsven L., Bruyneel E., Mareel M., Huylebroeck D., van Roy F. (2001) Mol. Cell 7, 1267–1278 [DOI] [PubMed] [Google Scholar]
- 27.Mochizuki N., Shimizu S., Nagasawa T., Tanaka H., Taniwaki M., Yokota J., Morishita K. (2000) Blood 96, 3209–3214 [PubMed] [Google Scholar]
- 28.Turner J., Crossley M. (2001) BioEssays 23, 683–690 [DOI] [PubMed] [Google Scholar]
- 29.Molloy D., Mapp K. L., Webster R., Gallimore P. H., Grand R. J. (2006) Virology 355, 115–126 [DOI] [PubMed] [Google Scholar]
- 30.Fang M., Li J., Blauwkamp T., Bhambhani C., Campbell N., Cadigan K. M. (2006) EMBO J. 25, 2735–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huo X., Zhang J. (2005) J. Cell. Mol. Med. 9, 103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hung H. L., Kim A. Y., Hong W., Rakowski C., Blobel G. A. (2001) J. Biol. Chem. 276, 10715–10721 [DOI] [PubMed] [Google Scholar]
- 33.Hayakawa F., Towatari M., Ozawa Y., Tomita A., Privalsky M. L., Saito H. (2004) J. Leukocyte Biol. 75, 529–540 [DOI] [PubMed] [Google Scholar]
- 34.Huang S., Qiu Y., Stein R. W., Brandt S. J. (1999) Oncogene 18, 4958–4967 [DOI] [PubMed] [Google Scholar]
- 35.Huang S., Qiu Y., Shi Y., Xu Z., Brandt S. J. (2000) EMBO J. 19, 6792–6803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tomita A., Towatari M., Tsuzuki S., Hayakawa F., Kosugi H., Tamai K., Miyazaki T., Kinoshita T., Saito H. (2000) Oncogene 19, 444–451 [DOI] [PubMed] [Google Scholar]
- 37.Hung H. L., Lau J., Kim A. Y., Weiss M. J., Blobel G. A. (1999) Mol. Cell. Biol. 19, 3496–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang W., Bieker J. J. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 9855–9860 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.