Abstract
The Arp2/3 complex is essential for actin filament nucleation in a variety of cellular processes. The activation of the Arp2/3 complex is mediated by nucleation-promoting factors, such as the Wiskott-Aldrich syndrome family proteins, which share a WCA (WH2 domain, central region, acidic region) catalytic module at the C-terminal region, required for Arp2/3 activation, but diverge at the N-terminal region, required for binding to specific activators. Here, we report the characterization of WASH, a new member of the WAS family that has nucleation-promoting factor activity and recently has been demonstrated to play a role in endosomal sorting. We found that overexpression of the WASH-WCA domain induced disruption of the actin cytoskeleton, whereas overexpression of full-length WASH in mammalian cells did not affect stress fiber organization. Furthermore, our analysis has revealed that nerve growth factor treatment of PC12 cells overexpressing full-length WASH leads to disruption of the actin cytoskeleton. We have also found that WASH interacts through its N-terminal region with BLOS2, a centrosomal protein belonging to the BLOC-1 complex that functions as a scaffolding factor in the biogenesis of lysosome-related organelles. In addition to BLOS2, WASH also interacts with centrosomal γ-tubulin and with pallidin, an additional component of the BLOC-1 complex. Collectively, our data propose that WASH is a bimodular protein in which the C terminus is involved in Arp2/3-mediated actin nucleation, whereas the N-terminal portion is required for its regulation and localization in the cells. Moreover, our data suggest that WASH is also a component of the BLOC-1 complex that is associated with the centrosomes.
Keywords: Cell, Cytoskeleton, Cytoskeleton/Actin, Cytoskeleton/Microtubules, Cytoskeleton/Tubulin, Subcellular Organelles/Centrosome, Subcellular Organelles/Cytoskeleton
Introduction
The actin cytoskeleton has an important role in many cellular processes such as cell migration, endocytosis, vesicle trafficking, and cytokinesis. The core constituent of the actin cytoskeleton is monomeric globular (G)-actin that can self-assemble into filamentous (F)-actin. The dynamic assembly and disassembly of the filaments are crucial aspects of the function of actin and are therefore under the control of actin-binding proteins (1, 2). An important set of actin regulators initiates the formation of new actin filaments by a process that is called nucleation (3, 4). The first identified actin nucleation factor was the Arp2/3 complex that binds to the side of pre-existing actin filament, and through its nucleation activity produces a new branched filament (5). In mammals, the Arp2/3 complex itself requires activation by one of several nucleation promoting factors (NPFs).2 The most thoroughly characterized NPFs are the members of the Wiskott-Aldrich syndrome (WAS) protein family. These are bi-modular proteins in which the C-terminal portion contains the WCA module, whereas the N-terminal region differs between WAS family members and places each of them in a specific Arp2/3-dependent process in the cell (6). The WCA module splits into three domains as follows: the WASP-homology-2 domain (W), involved in the binding of monomeric G-actin, the central domain (C), and the acidic domain (A), which are all required for the binding and the activation of Arp2/3 complex. In contrast, although WAS members, such as WASP and N-WASP, possess an N-terminal WASP homology domain 1 (WH1) and a central GTPase binding domain (7), WAVE proteins lack these domains but possess a highly specific WAVE homology domain. Through these N-terminal domains, WASPs and WAVEs act as effectors of the specific small GTPase. Moreover, the regulation of the WASP/N-WASP and WAVE protein activity is also the result of structural variation. In fact, under resting conditions, WASPs have an autoinhibited conformation in which the C-terminal portion interacts with the N terminus, preventing the binding and the activation of the Arp2/3 complex (1, 8, 9).
WASH (Wiskott-Aldrich syndrome protein and SCAR homolog) is a recently discovered member of the WAS family (10). Like the other members of this family, WASH contains a WCA domain at the C-terminal region, although its N-terminal region does not display any significant homology to other members of the WAS family, although it contains two evolutionarily conserved regions that were recently re-named WADH1 (WASH homology domain 1) and tubulin-binding region, both required for its function (11). Two recent papers indicate that WASH is an endosomal protein that plays a role in endosome sorting by facilitating tubule fission via Arp2/3 activation and demonstrate that WASH is a part of large multiprotein complexes (11, 12).
In this study, we focus on the biochemical and functional characterization of mouse WASH (mWASH). We have found that overexpression of mWASH-WCA causes stress fiber disruption, whereas overexpression of full-length mWASH does not. In contrast, mWASH overexpression causes stress fiber disruption in NGF-stimulated PC12 cells, suggesting that mWASH is inactive in resting cells and could be activated in an NGF-dependent manner. Using a yeast two-hybrid approach, we have found a new WASH-interacting protein, BLOS2, a component of the Biogenesis of Lysosome-related Organelle Complex-1 (BLOC-1 complex) that has also been found to be associated with the centrosomes (13). We show that mWASH might be activated by the interaction with BLOS2, and like BLOS2, it also interacts with γ-tubulin. Finally, we have found that mWASH interacts and partially co-localizes with pallidin, another component of the BLOC-1 complex (14), suggesting that mWASH is a new subunit of this complex.
EXPERIMENTAL PROCEDURES
Plasmids
WASH was amplified by PCR from a mouse fetal brain cDNA library (Clontech) and cloned into pcDNA3.1-HA and -FLAG (Invitrogen), pGFP-C1 (Clontech), and pCMV-Myc-tag-3C (Clontech) for expression in mammalian cells. mWASH deletion mutants were prepared by conventional PCR and cloned into pGFP-C1. The sequences of primers used in the preparation of all expression vectors are listed in supplemental Table 1.
Cell Culture and Biological Reagents
HEK293T and U2OS cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum, 100 units/ml penicillin, 100 mg/ml streptomycin, and 1% glutamine. PC12 cells were grown in RPMI 1640 medium (Invitrogen) containing 10% horse serum, 5% fetal bovine serum, 1% glutamine. and antibiotics (100 units/ml penicillin and 100 mg/ml streptomycin). The CHO cells were maintained in F-12 medium (Invitrogen) supplemented with 10% fetal bovine serum, 100 units/ml penicillin, 100 mg/ml streptomycin, and 1% glutamine.
Immunofluorescence
CHO, PC12, and U2OS cells were grown on coverslips, transiently transfected with the constructs listed in Fig. 1, and analyzed by indirect immunofluorescence 24 h after transfection. All procedures were performed as described previously (15). The anti-WASH and anti-pallidin antibodies were used at 1:50 and 1:100 dilutions, respectively. The F-actin staining was performed using rhodamine phalloidin from Molecular Probes. The images of fixed cells were acquired by a Leica confocal microscope.
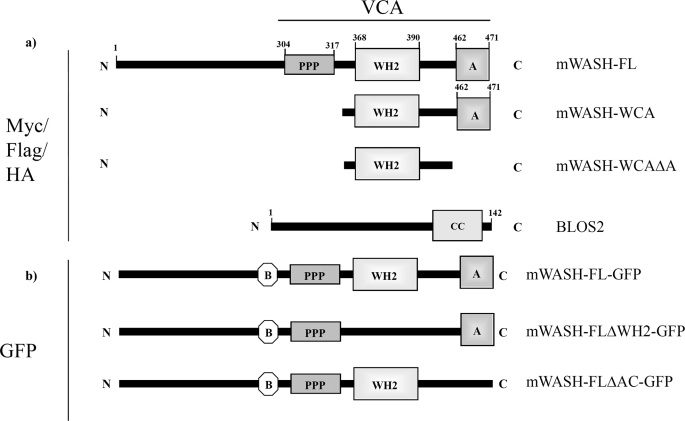
FIGURE 1.
Plasmid description. a, mWASH full-length (mWASH-FL), the mWASH WCA module (318–471 amino acids, mWASH-WCA), and the mWASH WC module, lacking the acidic region (318–462 amino acids, mWASH-WCΔA), were cloned in pCMV-Myc-tag3C, pCDNA3-FLAG, and pCDNA3-HA vectors. The full-length of BLOS2 was cloned in pCDNA3-FLAG and pCDNA3-HA vectors. b, two mWASH-derived constructs lacking the WH2 domain (mWASH-FLΔWH2) or the acidic region (mWASH-FLΔA), respectively, were fused to green fluorescent protein in pGFP-C1 plasmid. B, basic region; PPP, polyproline stretch; WH2, Wiskott-Aldrich homologous domain 2; A, acidic region; CC, coiled-coil region.
Yeast Two-hybrid Screening
The cDNA encoding the N-terminal region of mouse WASH (amino acids 1–300) was cloned in-frame into the GAL-4 DNA binding domain vector pGBKT7 (Clontech). The resulting plasmid pGBKT7-WASH was used as bait in a yeast two-hybrid screen of a mouse fetal brain cDNA library (Clontech) in Saccharomyces cerevisiae strain AH109.
Transfection and Immunoprecipitation Assay
Lipofectamine or Lipofectamine 2000-mediated transfections were performed according to the manufacturer's instructions (Invitrogen). All transfections included the supplemental empty vector to ensure that the total amount of transfected DNA was kept constant in each dish culture. For the immunoprecipitation of transfected proteins, HEK293T and CHO cells (4 × 106) were transiently transfected, and 24 h after transfection the cells were lysed in Triton X-100 lysis buffer (20 mm Hepes, pH 7.4, 150 mm NaCl, 10% glycerol, 1% Triton X-100, and Complete Protease Inhibitor mixture). After an additional 10 min on ice, the cell extracts were centrifuged for 10 min at 14,000 × g at 4 °C, and supernatants were incubated for 4 h at 4 °C with anti-FLAG antibodies bound to agarose beads (M2, Sigma) or poly(A) agarose-beads (Sigma) preincubated with the mWASH antibody. The immunoprecipitates were washed five times with Triton X-100 lysis buffer and subjected to SDS-PAGE.
GST Pulldown Assays
The recombinant proteins were expressed in Escherichia coli and purified as described previously (16), using a pST39 vector modified as reported (17), and codifying for recombinant BLOC-1 (GST-dysbidin and His-pallidin complex) (18). The GST and GST fusion proteins were immobilized onto 30 μl of glutathione-Sepharose 4 Fast Flow beads (Amersham Biosciences) and then incubated with cleared extracts prepared from CHO cells in 500 μl of lysis buffer (20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm EDTA, 0.5% Nonidet P-40, 2 mm dithiothreitol, complete protease inhibitor mixture) for 4 h. The amounts of GST or GST fusion protein used per sample were 50 μg. GST beads were washed three times with lysis buffer, and the bound proteins were eluted by boiling in SDS-PAGE loading buffer and detected by Western blotting.
Protein Expression and Antibody Production and Characterization
The mWASH-WCA was cloned into the pHAT2 vector (19), and this construct was expressed as His tag fusion proteins, enriched with nickel-nitrilotriacetic acid Superflow beads (Qiagen), and purified with the Q-Sepharose high performance anion-exchange column (GE Healthcare). Polyclonal antibodies against mWASH were raised by immunizing rabbits with purified His-mWASH(360–472), and then affinity-purified using the corresponding column prepared using the mWASH(360–471) fragment fixed with the AminoLink immobilization kit (Thermo Scientific). The anti-γ-tubulin monoclonal antibody was purchased from Sigma. The horseradish peroxidase-conjugated secondary antibodies were from Amersham Biosciences.
RESULTS
Overexpression of the mWASH-WCA Disrupts the Actin Cytoskeleton
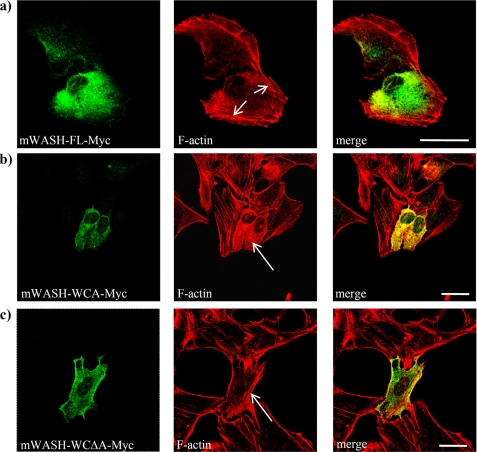
It has been demonstrated that the overexpression of the WCA module of WAVEs and WASPs causes actin cytoskeleton disruption, due to an enhanced Arp2/3-mediated actin polymerization (20). Because mWASH contains a WCA module, which is able to promote actin nucleation through the Arp2/3 complex in vitro (10), we examined whether overexpression of the WCA module of mWASH could affect the actin cytoskeleton organization in cells. We obtained several constructs encoding for either mWASH full-length (mWASH-FL), the WCA region, containing the WH2 domain and the acidic region (mWASH-WCA), or the WCAΔA, containing the WH2 domain but lacking the acidic region (mWASH-WCAΔA, see Fig. 1a). Each truncated form was fused to the Myc tag, transfected in the U2OS cells, which were subsequently stained with rhodamine-phalloidin. Confocal microscopy analysis revealed that cells overexpressing the mWASH-FL showed normal lamellipodia and stress fiber organization (Fig. 2a), although cells overexpressing mWASH-WCA displayed the disruption of the stress fibers (Fig. 2b). This phenotype was related to the ability of the WCA region of mWASH to activate actin nucleation by the Arp2/3 complex because in cells expressing WASH-WCΔA, which contains the WH2 domain but not the acidic region necessary for Arp2/3 complex activation, the stress fibers were correctly organized (Fig. 2c). The effect of WASH-WCA expression was not cell type-specific, because we observed comparable results in the CHO cells (supplemental Fig. S1a).
FIGURE 2.
Overexpression of mWASH-Myc constructs in U2OS and analysis of F-actin organization by phalloidin staining. Confocal microscopy analysis of U2OS cells transiently transfected with mWASH-FL-Myc (a); mWASH-WCA-Myc (b), and mWASH-WCΔA-Myc (c). The cells were stained with anti-Myc (green) and rhodamine phalloidin (red). The arrows show normal (a and c) and disrupted (b) stress fibers. The scale bar represents 20 μm (b) and 30 μm (b and c).
Overexpression of Full-length mWASH Causes Stress Fiber Disassembly in NGF-stimulated PC12 Cells
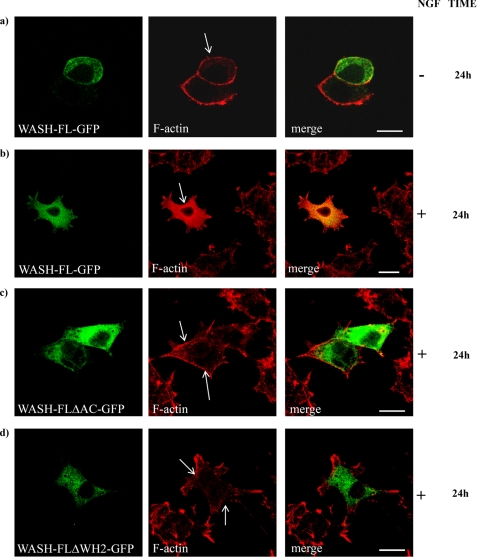
As overexpression of WASH-FL did not produce disruption of the actin cytoskeleton in unstimulated cells, we wondered whether mWASH, like WASP and WAVE, could be inactive in resting cells and require a trigger to become activated. Because the actin filament organization through the activation of the Arp2/3 complex is mediated by NGF in neuronal cells (21, 22) and Wash mRNA is mainly expressed in the central nervous system (23), we sought to verify if NGF could release WASH activity. We performed overexpression experiments of three constructs encoding for WASH full-length (mWASH-FL-ΔAC-GFP), WASH full-length lacking the acidic region (mWASH-FL-ΔAC-GFP), and WASH full-length lacking the WH2 domain (WASH-FLΔWH2-GFP, see Fig. 1b) fused to green fluorescent protein in PC12 cells. We stimulated the cells with NGF and stained them at 0-, 24-, 48- or 72-h time points with phalloidin. Confocal analysis revealed that in unstimulated PC12 cells, the WASH-FL-GFP overexpression did not cause any abnormalities in F-actin organization (Fig. 3a). However, after 24, 48, and 72 h of NGF stimulation, PC12 cells overexpressing mWASH-FL displayed an absence of stress fibers but normal neurite extension (Fig. 3b and supplemental Fig. S2a). Moreover, in NGF-stimulated cells, a large fraction of F-actin typically co-localized with mWASH at the perinuclear region (see supplemental Fig. S2b). To demonstrate that this phenotype was correlated to an enhanced actin polymerization mediated by the activation of the Arp2/3 complex, we overexpressed mWASH-FLΔA-GFP in PC12 cells stimulated with NGF for 24, 48, and 72 h. In this case, a normal organization of stress fibers was observed (Fig. 3c and supplemental Fig. S2c). Similar results were obtained in NGF-stimulated PC12 cells transfected with mWASH-FLΔWH2-GFP (Fig. 3d and supplemental Fig. S2c).
FIGURE 3.
Overexpression of mWASH constructs in PC12 cells upon NGF treatment. Confocal microscopy analysis of PC12 cells transfected with mWASH-GFP constructs followed by a NGF treatment (100 μg/ml) for the indicated time. a, untreated cells overexpressing mWASH-FL-GFP. b, cells overexpressing mWASH-FL-GFP after 24 h of NGF stimulation. c, overexpression of the mWASH-FL-ΔAC-GFP after 24 h of NGF treatment. d, overexpression of mWASH-ΔWH2-GFP after 24 h of NGF treatment (see supplemental Fig. S2 for 48- and 72-h NGF treatments). The arrows show normal (a, c, and d) and disrupted (b) stress fibers. The scale bar represents 10 μm (a, c, and d) and 20 μm (b).
mWASH Interacts with BLOS2
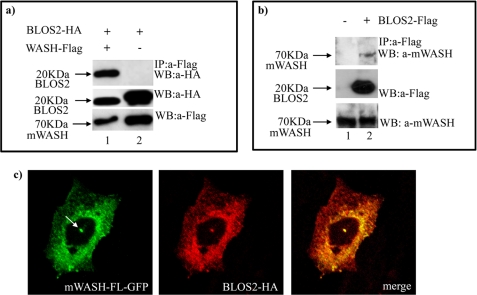
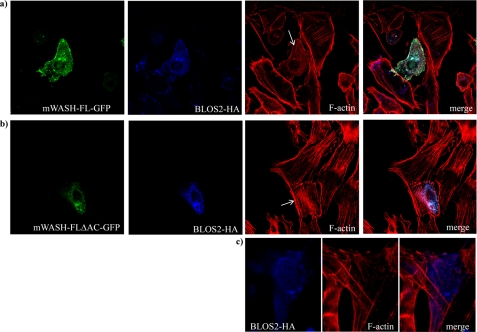
To define the cellular role of mWASH, we used the N terminus of the mWASH region (1–300 amino acids) as a bait to screen a mouse fetal brain cDNA library by the yeast two-hybrid system. Three out of 40 sequenced clones encoded for the adapter protein BLOS2 (13, 24). Co-immunoprecipitation experiments in HEK293T cells confirmed the interaction between mWASH-FL-FLAG and BLOS2-HA (Fig. 1a and Fig. 4a, lane 1).
FIGURE 4.
Endogenous mWASH interacts and co-localizes with BLOS2. Co-immunoprecipitation assay in HEK293T cells transfected with BLOS2-HA and/or mWASH-FL-FLAG constructs (a). The cell extracts were immunoprecipitated (IP) with anti-FLAG antibody, and the Western blot (WB) was stained with anti-hemagglutinin antibody. The levels of expression of transfected BLOS2-HA by Western blot were determined with anti-hemagglutinin antibody. b, HEK293T cells transfected with BLOS2-FLAG constructs. The cell extracts were immunoprecipitated with anti-FLAG antibody, and the Western blot was stained with anti-mWASH antibody. The level of expression of transfected BLOS2-FLAG was determined by Western blot with anti-FLAG. c, confocal microscopy analysis of CHO cells transfected with mWASH-FL-GFP and BLOS2-HA. The arrow shows the centrosome.
To determine whether endogenous mWASH was able to interact with BLOS2, we performed co-immunoprecipitation assays on CHO cells transfected with BLOS2-FLAG. Lysates prepared from these cells were immunoprecipitated with anti-FLAG antibodies and analyzed by Western blot with the anti-mWASH antibody. This analysis revealed that also the endogenous mWASH interacts with the BLOS2-FLAG (Fig. 4b, lane 2). To establish if mWASH co-localized with BLOS2, we performed co-immunolocalization experiments in CHO cells transfected with mWASH-FL-GFP and BLOS2-HA. We found that the two proteins co-localize in the cytoplasm in a spotted manner and that in 30% of cells both mWASH and BLOS2 co-localize at the centrosome (Fig. 4c and supplemental Fig. S3).
mWASH Interacts with γ-Tubulin
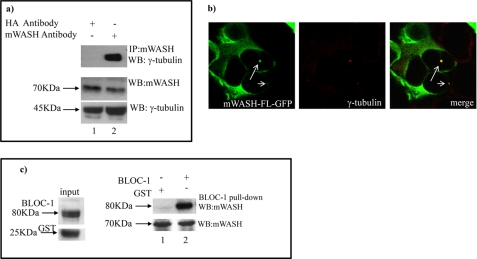
Because mWASH interacts with BLOS2, which is a well known γ-tubulin interacting protein, and both proteins localize at the centrosome in 30% of cells (Fig. 4a; supplemental Fig. S3), we also tested if mWASH interacts with γ-tubulin by using co-immunoprecipitation experiments. To this end, cell extracts prepared from CHO cells were incubated with antibodies against mWASH and analyzed after co-immunoprecipitation and Western blotting, using antibodies against γ-tubulin, thus revealing that the endogenous mWASH interacts with γ-tubulin (Fig. 5a, lane 2). Moreover, immunolocalization experiments revealed that these two proteins co-localize at the centrosome (Fig. 5b).
FIGURE 5.
mWASH interacts and co-localizes with γ-tubulin and pallidin in CHO cells. Co-immunoprecipitation assay performed in HEK293T cells. a, cell extracts were immunoprecipitated (IP) with anti-WASH antibody, and the Western blot (WB) was stained with anti-γ-tubulin. The CHO extract was also incubated with anti-hemagglutinin (HA) antibody to verify that the interaction between mWASH and γ-tubulin is specific and not due to unspecific interactions (lane 1). mWASH interacts with γ-tubulin (lane 2). The levels of expression of mWASH and γ-tubulin were determined from Western blots with anti-WASH and anti-γ-tubulin antibodies, respectively. b, confocal microscopy analysis of HEK293T cells transfected with mWASH-FL-GFP. The cells were stained with anti-γ-tubulin. c, pulldown experiments performed using BLOC-1 complex expressed as GST fusion proteins in E. coli. GST fusion protein beads and GST beads as control (input) were incubated with CHO extracts. Endogenous mWASH interacts with the recombinant BLOC-1 complex (lane 2) as revealed by a Western blot analysis using an antibody against mWASH. The level of mWASH in the CHO extracts was determined with an anti-WASH antibody (lanes 1 and 2, lower panel) and the input levels of BLOC-1-GST and GST alone produced in E. coli, were detected with an anti-GST antibody.
mWASH Is a New Subunit of the BLOC-1 Complex
BLOS2 is a subunit of BLOC-1 (14). To verify if mWASH could interact with other components of this complex, we performed pulldown experiments with recombinant BLOC-1 fused to GST. CHO cell extracts were incubated with either the purified GST fusion proteins or GST alone as a control. Western blot analysis using an antibody against mWASH revealed that endogenous mWASH interacts with the BLOC-1 complex (Fig. 5c, lane 2). Moreover, we found that in CHO cells mWASH-FL-HA partially co-localized with endogenous pallidin in a spotted manner, although it did not co-localize with LAMP2, a well known lysosomal marker (supplemental Fig. S4 and data not shown).
Overexpression of mWASH and BLOS2 Induces Stress Fiber Disassembly
The interaction of WAS proteins with their partners often results in the release of the autoinhibitory conformation of these proteins. To test if the physical interaction between mWASH and BLOS2 could activate mWASH, we co-transfected mWASH-FL-GFP with BLOS2-HA in CHO cells and stained them with phalloidin. As shown in Fig. 6a and supplemental Fig. S5, co-expression of mWASH-FL-GFP with BLOS2-HA induced the disappearance of stress fibers (S5a1), a phenotype also caused by mWASH-WCA overexpression (Fig. 2b). In contrast, this phenotype was not detectable in cells expressing either mWASH-FL or BLOS2 (Figs. 6c and 2a), as well as in cells expressing both WASH-FLΔA-GFP, which lacks the acidic region, and BLOS2-HA alone (Fig. 6, b and c). These data suggest that BLOS2 overexpression activates mWASH-mediated Arp2/3 activation.
FIGURE 6.
Overexpression of BLOS2 and mWASH causes stress fiber disruption in CHO cells. Confocal microscopy analysis of CHO cells transiently transfected with mWASH-FL-GFP and/or BLOS2-HA and stained with anti-hemagglutinin (HA) antibody and phalloidin. a, overexpression of both mWASH-FL-GFP and BLOS2-HA. b, overexpression of mWASH-ΔA-GFP and BLOS2-HA. c, overexpression of BLOS2-HA.
DISCUSSION
In this study, we reveal that mouse WASH is a bi-modular protein in which the C-terminal portion is involved in Arp2/3 activation, although the N-terminal portion characterizes its localization/function. As reported previously for WASPs and WAVEs, overexpression of the C-terminal WCA module of mWASH disrupts the actin cytoskeleton in several mammalian cell lines. Importantly, overexpression of the WCA module of mWASH lacking the acidic region involved in the binding and activation of the Arp2/3 complex did not affect stress fiber organization, suggesting that mWASH WCA regulates the actin cytoskeleton organization through the Arp2/3 complex.
Interestingly, we found that overexpression of full-length mWASH did not affect cytoskeleton organization. It has been demonstrated that in PC12 cells NGF stimulation releases the autoinhibitory conformation of N-WASP, allowing its WCA module to promote Arp2/3-induced actin polymerization (20, 25, 26), and triggering the formation of protrusive structures such as the filopodia and lamellipodia necessary for neurite outgrowth (27). As for N-WASP, we found that after NGF stimulation, mWASH causes a disruption of the stress fibers accompanied by an increased diffuse perinuclear F-actin staining that co-localized with mWASH, suggesting that mWASH could be inactive in resting cells and it could become activated only upon stimulation with triggers such as NGF. However, PC12 cells expressing deleted N-WASP form were normally differentiated after 24, 48, and 72 h of NGF stimulation, suggesting that mWASH does not contribute to lamellipodia or fillopodia formation but could be important in other central nervous system-related processes. This hypothesis is supported by recent evidence showing mWASH to be involved in the sorting and recycling of the endosomes (11, 12).
We also hypothesized that the N-terminal portion of mWASH, through binding to specific adapter proteins, may specify the cellular processes in which mWASH is recruited. We found that BLOS2, a centrosome-associated protein, is an mWASH-interacting protein. BLOS2 is an adapter protein with a heterogeneous subcellular localization that has distinct roles in different cellular compartments. In addition, BLOS2 co-localizes and interacts with γ-tubulin (13). Accordingly, we provide evidence that both mWASH and BLOS2 localize in the cytoplasm in a spotted manner and also concentrate at the centrosome, where they co-localize with γ-tubulin. mWASH also physically interacts with γ-tubulin, revealing that mWASH is the first member of the WAS family that may have a role in the centrosome function.
A recent study has shown that BLOS2 is also a subunit of the BLOC-1 complex involved in the formation and/or trafficking of lysosome-related endosomal compartments (14). This complex is associated with filamentous actin, through pallidin, another subunit of this complex (28). We have found that mWASH interacts with pallidin and co-localizes with it in the nonlysosomal vesicles, suggesting that mWASH could be a new subunit of the BLOC-1 complex and the actin binding intermediate that couples the BLOC-1 complex with F-actin. Supporting this evidence, Liu et al. (29) have reported a genetic interaction between the Drosophila ortholog of WASH and the Drosophila ortholog of cappuccino, an additional component of the BLOC-1 complex.
The presence of mWASH at the centrosome, its ability to interact with γ-tubulin, its feature as a new subunit of the BLOC-1 complex, and its function as an NPF support the view that this protein might connect actin nucleation and vesicle transport processes. Taken together, those findings suggest that mWASH exert this function by participating with a multimolecular complex.
On the other hand, the expression of BLOS2 and mWASH in the central nervous system, the ability of NGF to activate mWASH and of BLOS2 to interact with Snapin (30), a protein involved in synaptosomal processes, the presence of BLOC-1 on the transferrin receptor-positive endosome where it regulates membrane protein targeting to synaptic vesicles (31) are evidence pointing to a role of this mWASH-containing complex in synaptic vesicle transport. Finally, we propose that WASH may have a role in cellular processes involving an interplay between actin and microtubules through its interaction with γ-tubulin. mWASH in fact is the first member of the WAS family showing a centrosome localization. Two recent papers further support this hypothesis. Liu et al. (29) have shown that dWASH is involved in cross-linking F-actin and microtubules, and it is essential for the actin cytoskeleton organization in the Drosophila egg chamber. Gomez and Billadeau (11) have shown that human WASH co-localizes with microtubules and appears to regulate the retrograde trafficking through this microtubule linkage.
Collectively, our results show that mWASH contains at its C terminus a WCA module, involved in Arp2/3 activation that in resting conditions could be blocked in an inactive conformation. The overexpression of both mWASH and BLOS2 causes disruption of the actin cytoskeleton and, like NGF stimulation, can activate mWASH, suggesting that this protein might be active only upon stimulation and/or interaction with specific proteins. Consistently, we found that this protein is a subunit of a new multiprotein complex, supporting the model that mWASH may link the actin and microtubule cytoskeletons and have a role in endosomal transport.
Supplementary Material
Acknowledgments
We thank Juan S. Bonifacino for giving us the pST39 plasmid codifying for the recombinant BLOC-1 and anti-pallidin antibodies; Salvatore Arbucci for help in confocal microscopy experiments; and M. G. Miano, A. Pescatore, F. Fusco, and Giorgio Scita for helpful discussion and critical reading of the manuscript.
This work was supported by Telethon Grant GGP08125 (to M. V. U.) and Istituto Banco di Napoli Fondazione.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5 and Table S1.
- NPF
- nucleation promoting factor
- WAS
- Wiskott-Aldrich syndrome
- WASP
- Wiskott-Aldrich syndrome protein
- WH1
- Wiskott-Aldrich homology domain 1
- WASH
- Wiskott-Aldrich syndrome homology
- mWASH
- mouse WASH
- NGF
- nerve growth factor
- CHO
- Chinese hamster ovary
- U2OS
- human osteosarcoma cell line
- HEK293T
- human embryonic kidney 293
- HA
- homagglutinin.
REFERENCES
- 1.Pollard T. D., Borisy G. G. (2003) Cell 112, 453–465 [DOI] [PubMed] [Google Scholar]
- 2.Goley E. D., Welch M. D. (2006) Nat. Rev. Mol. Cell Biol. 7, 713–726 [DOI] [PubMed] [Google Scholar]
- 3.Carlier M. F., Pantaloni D. (2007) J. Biol. Chem. 282, 23005–23009 [DOI] [PubMed] [Google Scholar]
- 4.Chhabra E. S., Higgs H. N. (2007) Nat. Cell Biol. 9, 1110–1121 [DOI] [PubMed] [Google Scholar]
- 5.Stradal T. E., Scita G. (2006) Curr. Opin. Cell Biol. 18, 4–10 [DOI] [PubMed] [Google Scholar]
- 6.Paavilainen V. O., Bertling E., Falck S., Lappalainen P. (2004) Trends Cell Biol. 14, 386–394 [DOI] [PubMed] [Google Scholar]
- 7.Callebaut I., Cossart P., Dehoux P. (1998) FEBS Lett. 441, 181–185 [DOI] [PubMed] [Google Scholar]
- 8.Higgs H. N., Pollard T. D. (2001) Annu. Rev. Biochem. 70, 649–676 [DOI] [PubMed] [Google Scholar]
- 9.Derivery E., Lombard B., Loew D., Gautreau A. (2009) Cell Motil. Cytoskeleton 66, 777–790 [DOI] [PubMed] [Google Scholar]
- 10.Linardopoulou E. V., Parghi S. S., Friedman C., Osborn G. E., Parkhurst S. M., Trask B. J. (2007) PLoS Genet. 3, e237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gomez T. S., Billadeau D. D. (2009) Dev. Cell 17, 699–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derivery E., Sousa C., Gautier J. J., Lombard B., Loew D., Gautreau A. (2009) Dev. Cell 17, 712–723 [DOI] [PubMed] [Google Scholar]
- 13.Wang Z., Wei H., Yu Y., Sun J., Yang Y., Xing G., Wu S., Zhou Y., Zhu Y., Zhang C., Zhou T., Zhao X., Sun Q., He F. (2004) J. Mol. Biol. 343, 71–82 [DOI] [PubMed] [Google Scholar]
- 14.Dell'Angelica E. C. (2004) Curr. Opin. Cell Biol. 16, 458–464 [DOI] [PubMed] [Google Scholar]
- 15.Monfregola J., Cevenini A., Terracciano A., van Vlies N., Arbucci S., Wanders R. J., D'Urso M., Vaz F. M., Ursini M. V. (2005) J. Cell Physiol. 204, 839–847 [DOI] [PubMed] [Google Scholar]
- 16.Peränen J., Auvinen P., Virta H., Wepf R., Simons K. (1996) J. Cell Biol. 135, 153–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan S. (2001) Protein Expr. Purif. 21, 224–234 [DOI] [PubMed] [Google Scholar]
- 18.Moriyama K., Bonifacino J. S. (2002) Traffic 3, 666–677 [DOI] [PubMed] [Google Scholar]
- 19.Mattila P. K., Salminen M., Yamashiro T., Lappalainen P. (2003) J. Biol. Chem. 278, 8452–8459 [DOI] [PubMed] [Google Scholar]
- 20.Machesky L. M., Insall R. H. (1998) Curr. Biol. 8, 1347–1356 [DOI] [PubMed] [Google Scholar]
- 21.Pommereit D., Wouters F. S. (2007) J. Cell Sci. 120, 2694–2705 [DOI] [PubMed] [Google Scholar]
- 22.Goldberg D. J., Foley M. S., Tang D., Grabham P. W. (2000) J. Neurosci. Res. 60, 458–467 [DOI] [PubMed] [Google Scholar]
- 23.Gianfrancesco F., Falco G., Esposito T., Rocchi M., D'Urso M. (2001) Cytogenet. Cell Genet. 94, 98–100 [DOI] [PubMed] [Google Scholar]
- 24.Felten A., Leister P., Burgdorf S., Uhlmann L., Scheidtmann K. H. (2007) Biol. Chem. 388, 569–582 [DOI] [PubMed] [Google Scholar]
- 25.Rohatgi R., Ho H. Y., Kirschner M. W. (2000) J. Cell Biol. 150, 1299–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Millard T. H., Sharp S. J., Machesky L. M. (2004) Biochem. J. 380, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banzai Y., Miki H., Yamaguchi H., Takenawa T. (2000) J. Biol. Chem. 275, 11987–11992 [DOI] [PubMed] [Google Scholar]
- 28.Starcevic M., Dell'Angelica E. C. (2004) J. Biol. Chem. 279, 28393–28401 [DOI] [PubMed] [Google Scholar]
- 29.Liu R., Abreu-Blanco M. T., Barry K. C., Linardopoulou E. V., Osborn G. E., Parkhurst S. M. (2009) Development 136, 2849–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Falcón-Pérez J. M., Starcevic M., Gautam R., Dell'Angelica E. C. (2002) J. Biol. Chem. 277, 28191–28199 [DOI] [PubMed] [Google Scholar]
- 31.Ryder P. V., Faudez V. (2009) Sci. Signal. 2, 66 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.