Abstract
The bacterial RecN protein is involved in the recombinational repair of DNA double-stranded breaks, and recN mutants are sensitive to DNA-damaging agents. Little is known about the biochemical function of RecN. Protein sequence analysis suggests that RecN is related to the SMC (structural maintenance of chromosomes) family of proteins, predicting globular N- and C-terminal domains connected by an extensive coil-coiled domain. The N- and C-domains contain the nucleotide-binding sequences Walker A and Walker B, respectively. We have purified the RecN protein from Deinococcus radiodurans and characterized its DNA-dependent and DNA-independent ATPase activity. The RecN protein hydrolyzes ATP with a kcat of 24 min−1, and this rate is stimulated 4-fold by duplex DNA but not by single-stranded DNA. This DNA-dependent ATP turnover rate exhibits a dependence on the concentration of RecN protein, suggesting that RecN-RecN interactions are required for efficient ATP hydrolysis, and those interactions are stabilized only by duplex DNA. Finally, we show that RecN stimulates the intermolecular ligation of linear DNA molecules in the presence of DNA ligase. This DNA bridging activity is strikingly similar to that of the cohesin complex, an SMC family member, to which RecN is related.
Keywords: ATPases, DNA Enzymes, DNA Recombination, DNA Repair, Enzymes, Protein-DNA Interaction
Introduction
The cellular genome maintenance processes of DNA replication, recombination, and repair are highly interconnected, sharing multiple pathways and common enzymes. A physical understanding of the function of key enzymes involved in genome maintenance processes is critical for elucidating the basic DNA metabolic strategies of cells. The primary function of homologous DNA recombination in mitotic cells as well as bacterial cells is the nonmutagenic repair of replication forks that have stalled or collapsed at noncoding lesions or breaks (1–3). When a replication fork encounters a break in the phosphodiester backbone of one template strand, a double-stranded break (DSB)2 results (4). Recombinational DSB repair pathways promote strand invasion to regenerate a fork structure. In Escherichia coli, these pathways include the helicase and nuclease functions of the RecBCD complex, and the RecA protein. RecN, among various other proteins, is required for DSB repair in bacteria. Existing DSB repair models do not encompass RecN, because no clear function has been ascribed to the protein as yet.
The recN gene of E. coli (also known as radB (5)) was identified by two groups over 20 years ago. Lloyd et al. (6) isolated a mutation (rec-259) that caused increased sensitivity to UV light in a recBC sbcB background. Independently, Sargentini and Smith (7) isolated the radB101 mutation, which caused sensitivity to γ-irradiation and mitomycin C. Expression of the recN gene is regulated by the LexA repressor (8, 9), and the RecN protein is one of the most abundantly produced proteins induced as part of the SOS response to DNA damage (10). Once produced in response to damage, RecN protein has a very short half-life (∼10 min), because it apparently is rapidly proteolyzed by the ClpXP protease system (11).
The recN gene product was originally placed in the RecF-dependent gap repair pathway, but substantial evidence suggests that RecN is also involved in the RecBCD DSB repair pathway. Unlike other members of the RecF pathway, recN mutants are quite sensitive to ionizing radiation or mitomycin C, and the repair of multiple, site-specific DSBs does not occur in the absence of RecN (8, 12, 13). The recN gene is also required for the suppression of chromosomal rearrangements and deletions (12) during the repair of a single DSB. Given this wealth of genetic data, the RecN protein is clearly involved in the recombinational repair of DNA damage.
The recN gene is widespread in bacterial species. The only known absences are in phyla Mollicutes and Chlamydiae (14). Mutations in the recN genes of Helicobacter pylori (15), Neisseria gonorrhoeae (16), Bacillus subtilis (17), and Deinococcus radiodurans (18) have been shown to confer DNA damage repair defects. The RecN protein from the Gram-positive B. subtilis binds to single-stranded DNA (ssDNA) or duplex DNA with long ssDNA extensions in vitro (19), although no such activity has been observed with the recently purified RecN proteins from Haemophilus influenzae, Aquifex aeolicus, and Bacteriodes fragilis (20). Alonso and Graumann and co-workers (17, 21) have demonstrated that RecN localizes to sites of DSBs in live B. subtilis cells.
RecN is a member of the SMC (structural maintenance of chromosomes) family of proteins, which are fundamental to several facets of chromosome dynamics and organization, including assembly, segregation, and condensation (22). SMC proteins also have a critical role in DNA damage responses and DSB repair (23). The SMC family of proteins are found in all three branches of life and are characterized by globular domains at the N and C termini connected by a long coiled-coil domain (24, 25). The head domains each contain part of a functional site for the binding and hydrolysis of ATP, and SMC proteins become competent for ATP hydrolysis when these domains are brought together. The SMC family includes three major types of SMC-containing complexes (SMC1/3, SMC2/4, and SMC5/6) found in eukaryotic organisms. Additionally, “SMC-like” proteins (such as Rad50) share sequence homology with SMC proteins and appear to have the architecture described above. The Rad50 protein functions in the heterotrimeric complex made up of Mre11, Rad50, and Xrs2 (MRX, in yeast) or Mre11, Rad50, and Nbs1 (MRN, in humans) (26). Homologous recombinational DSB repair pathways as well as nonhomologous end joining pathways require this complex. The bacterial protein most similar to Rad50 is SbcC, which is part of the SbcCD complex involved in the cleavage of DNA hairpin ends and in removing proteins at DNA ends (27, 28).
Each of the three main types of SMC complexes consists of a specific pair of SMC proteins that form heterodimers and several other non-SMC proteins (24). First, condensin complexes (containing SMC2/SMC4 heterodimer) play a critical role in chromosome condensation. In vitro, condensin promotes the knotting of relaxed circular double-stranded DNA (dsDNA) in the presence of DNA topoisomerase (29). Second, cohesin complexes (containing the SMC1/SMC3 heterodimer) link together the sister chromatids following replication, playing a major role in chromosome segregation (30). In vitro, cohesin promotes the catenation of relaxed circular dsDNA in the presence DNA topoisomerase II and also facilitates the ligation of linear duplex DNAs (31). The third, less characterized, SMC complex (containing the SMC5/SMC6 heterodimer) has been implicated in DNA repair and DNA damage checkpoint responses (22, 32). Both cohesins and SMC5/SMC6 complexes have been implicated in the recombinational repair of DSBs. Both complexes localize to DSBs, and several mutations have been described in the yeast components of these complexes that are sensitive to DNA damage, and these phenotypes are epistatic with recombination mutants (30). Based on its association with sister chromatid cohesion, cohesin complexes are thought to contribute to homologous recombination by providing proximity between the broken chromosome and the sister chromatid (33). The role of the SMC5/SMC6 complex is not known, although smc5/smc6 mutants exhibit elevated levels of gross chromosomal rearrangements (34).
Bacterial SMCs are not ubiquitous, and no cohesin-type SMC protein has been identified in E. coli. However, the E. coli MukB protein, in complex with the MukE and MukF proteins, is a functional homolog of the eukaryotic condensin (35). MukB is involved in chromosomal partitioning, and mutants produce anucleated cells. The mukB null mutant is not sensitive to either UV light or γ-irradiation (36), indicating that MukB has no DNA repair role. An SMC protein has been found in B. subtilis (BsSMC), functioning as a homodimer in a complex with the non-SMC proteins ScpA and ScpB (37, 38). BsSMC apparently exhibits both condensin and cohesin characteristics (39, 40), but there is no evidence that BsSMC is involved in DNA damage responses or repair. There is no clear differentiation in bacterial chromosomal cycles for cohesion and condensation, and it appears that a single smc gene provides these functions.
Historically, platforms of detailed biochemical characterization of bacterial functional orthologs have been the launching pad for mechanistic studies in eukaryotic systems. Indeed most of the mechanistic details related to eukaryotic condensin function have been provided by studies using the bacterial BsSMC ortholog. However, scant biochemical data exist to help explain the specific role of SMC or SMC-like complexes in DNA repair. RecN is a bacterial SMC-like protein that localizes to DSBs (17, 21) and is demonstrably important to DSB repair. The specific biochemical function of RecN has remained enigmatic because of problems purifying the E. coli protein. We hypothesize that RecN is a bacterial cohesin-type SMC protein. Characterization of the RecN protein will likely facilitate the study of SMC proteins and their role in DSB repair.
In the current study, we have initiated the biochemical description of the RecN protein from D. radiodurans, one of the most DNA damage-resistant organisms known (41). This organism can withstand levels of γ-irradiation more than 200-fold higher than E. coli and has become a model for studying the basis of the differential levels of cellular resistance to DNA damage. A robust DSB repair pathway, including a functional recN gene, is essential to the recovery of D. radiodurans from exposure to high doses of ionizing radiation (18, 41, 42).
EXPERIMENTAL PROCEDURES
Enzymes and Biochemicals
Unless otherwise noted, all of the reagents were purchased from Fisher. Lysozyme, phosphoenolpyruvate, pyruvate kinase, lactate dehydrogenase, NAD+, NADH, ATP, polyethyleneimine, and bromphenol blue were purchased from Sigma. ATPγS was from Roche Applied Science. Proteinase K was from Amresco. All of the restriction endonucleases and E. coli DNA ligase were purchased from New England Biolabs. The T4 DNA ligase was from Fermentas. Dithiothreitol (DTT) was from Soltec Ventures. 4-(2-Aminoethyl)benzenesulfonyl fluoride-HCl (AEBSF-HCl) and pepstatin A was from AppliChem. Isopropyl β-d-1-thiogalactopyranoside was from Gold Biotechnology. DEAE-Sepharose and Source Q resin was from GE Healthcare. Hydroxyapatite resin was from Bio-Rad.
DNA Substrates
Plasmid DNA substrates pEAW324 and pEAW3 were prepared using CsCl-ethidium bromide gradients as described (43). The pEAW3 plasmid (2431 bp) has been described elsewhere (44). The pEAW324 plasmid (8716 bp) was constructed by inserting the KanR gene into the single PmeI restriction site of pEAW311 (45). Unless otherwise noted, full-length linear duplex DNA substrate was generated by the digestion of the plasmid DNA at a unique site with the PstI restriction endonuclease, using conditions suggested by the enzyme supplier. The digested DNA was extracted with phenol/chloroform/isoamyl alcohol (25:24:1), followed by ethanol precipitation. Circular plasmid DNA containing a single nick (nicked circular dsDNA) was generated using a previously described method (46). Bacteriophage ϕX174 circular ssDNA (virion) was purchased from New England Biolabs. The concentrations of ssDNA and dsDNA were determined by absorbance at 260 nm, using 36 and 50 μg ml−1 A260−1, respectively, as conversion factors. Linear ssDNA poly(dT) was purchased from GE Healthcare. The concentration of poly(dT) was determined by absorbance at 260 nm using an extinction coefficient of 8.73 mm−1 cm−1. All of the DNA concentrations are given in μm nucleotides or base pairs.
Cloning and Overexpression of RecN Protein
The D. radiodurans recN gene (locus tag DR_1477) was amplified by PCR using strain R1 genomic DNA (accession number NC_001263.1). PCR primer design for amplification was guided by the recN gene sequence (accession number NP_295200.1) annotated in the NCBI gene bank. The gene was cloned into the NdeI/BamHI sites of pET21A (Novagen) resulting in pEAW309 (AmpR). A 10-liter culture of STL2669 cells (a gift from Susan Lovett, Brandeis University) cotransformed with pEAW309 and pT7POL26 (KanR) was grown in LB broth (10 g/liter of tryptone, 5 g/liter of yeast extract, and 10 g/liter of NaCl, with pH adjusted to 7.0) to an A600 of 0.5. The pT7POL26 plasmid codes for T7 RNA polymerase under the control of a lac promoter (47). RecN protein expression was induced by the addition of isopropyl β-d-1-thiogalactopyranoside to a final concentration of 0.4 mm. Following a 6-h incubation at 37 °C, the cells were harvested by centrifugation, flash-frozen in liquid N2, and stored at −80 °C.
Purification of the RecN Protein
All of the steps of this purification were carried out at 4 °C. Cell paste was thawed and fully resuspended to a 20% (w/v) cell ratio in Tris-sucrose solution (25% (w/v) sucrose and 250 mm Tris-HCl, 80% cation, pH 7.5) supplemented with the protease inhibitors AEBSF-HCl and pepstatin A to final concentrations of 0.5 mg/ml and 0.7 μg/ml, respectively. The cells were lysed by a 60-min incubation with lysozyme in 250 mm Tris-HCl (80% cation, pH 7.5) to 2.5 mg/ml, followed by the addition of 0.4 ml of 25 mm EDTA/ml of lysed cell suspension, sonication, and centrifugation. The lysate was precipitated with polyethyleneimine, pH 7.5 (final concentration, 0.5%) and centrifuged. The pellet was washed with R buffer (20 mm Tris-HCl (80% cation, pH 7.5), 1 mm DTT, 0.1 mm EDTA, and 10% (w/v) glycerol) + 150 mm ammonium sulfate and extracted two times with R buffer + 300 mm ammonium sulfate. The protein solution was precipitated by the addition of 0.23 g of solid ammonium sulfate/ml of solution (40% saturation). The precipitant was washed two times with R buffer + 2.3 m ammonium sulfate, resuspended in R buffer + 300 mm KCl and dialyzed versus R buffer + 50 mm KCl. The following chromatography steps were carried out using the AKTA fast protein liquid chromatography system (GE Healthcare). The protein was loaded onto a DEAE-Sepharose column, washed with one column volume of R buffer + 50 mm KCl, and eluted with a linear gradient of KCl from 50 mm to 1 m KCl over 10 column volumes. The RecN protein eluted at ∼340 mm KCl. Peak fractions were analyzed and identified by SDS-PAGE, and fractions containing RecN were pooled and dialyzed versus P buffer (20 mm potassium phosphate, pH 6.8, 1 mm DTT, 0.1 mm EDTA, and 10% (w/v) glycerol). Protein was then loaded onto a hydroxyapatite column, washed with one column volume of P buffer, and eluted with a linear gradient from 20 mm to 1 m potassium-phosphate buffer, pH 6.8, over 10 column volumes. The RecN protein eluted at ∼380 mm phosphate. The fractions containing RecN were pooled and concentrated using a Source Q column. The pure RecN protein was determined to be free of nuclease contamination and was dialyzed extensively versus storage buffer (R buffer + 50 mm KCl), flash-frozen in liquid N2, and stored at −80 °C. The concentration of the RecN protein (molecular mass, 59,798 Da) was determined from the absorbance at 280 nm using the calculated extinction coefficient 29,160 m−1 cm−1.
ATPase Assay
A coupled spectrophotometric enzyme assay was used to measure the ATPase activity of the RecN protein as described (48). The regeneration of ATP from phosphoenolpyruvate and ADP (produced by hydrolysis) was coupled to the oxidation of NADH through the addition of pyruvate kinase and lactate dehydrogenase. The assay was carried out on a Varian Cary 100 Bio dual beam spectrophotometer equipped with a temperature controller and a 12-position cell changer. The cell path length was 1 cm. The NADH extinction coefficient at 380 nm of 1.21 mm−1 cm−1 was used to calculate the rate of ATP hydrolysis. The reactions were carried out at 37 °C in Buffer N (25 mm Tris-OAc (80% cation), 1 mm DTT, 3 mm potassium glutamate, 17.5 mm Mg(OAc)2, 40 mm KOAc, 5% (w/v) glycerol, 2.5% buffered polyethylene glycol 8000), an ATP regeneration system (10 units/ml pyruvate kinase, 3 mm phosphoenolpyruvate), and a coupling system (1.5 mm NADH and 10 units/ml lactate dehydrogenase). These components were incubated for 20 min with the concentration of RecN indicated in the figure in the presence or absence of DNA as indicated. The assay was initiated by the addition of ATP to 2.5 mm.
DNA Ligation Assay
The reactions were carried out in Buffer N, plus an ATP regeneration system (2.5 mm phosphoenolpyruvate, 10 units/ml pyruvate kinase). The RecN protein was incubated with 1.2 μm bp (80 nanograms) of linearized plasmid DNA (pEAW3, 2431 bp) in the presence of 2.5 mm ATP in a total volume of 100 μl for 30 min at 37 °C. The concentration of RecN protein used in each experiment is noted in the figure legend. After the incubation, each reaction was supplemented with fresh 10× E. coli DNA ligase reaction buffer (300 mm Tris-HCl, pH 8, 40 mm MgCl2, 10 mm DTT, 260 μm NAD+, 500 μg/ml bovine serum albumin) and 2.5 units of E. coli DNA ligase and for 30 min at 16 °C. These same conditions were used when the D. radiodurans LigA DNA ligase (a gift from Michael Cox) was used except that the ligase was added to a concentration of 1 mm, and the incubation was carried out at 30 °C as previously described (49). For experiments utilizing the T4 DNA ligase, fresh 10× supplemental buffer contained 400 mm Tris-HCl, pH 7.8, 100 mm MgCl2, 10 mm DTT, and 5 mm ATP. Ligation reactions were stopped by adding 80 μl of a termination buffer containing 20 mm Tris-HCl (80% cation), 20 mm EDTA, and 0.5% SDS and deproteinized with 100 μg of predigested proteinase K. The DNA from each reaction was extracted after 20 min of incubation at 37 °C with a phenol/chloroform/iso-amyl-alcohol solution (25:24:1) and precipitated with ethanol. The precipitated DNA was resuspended in 10 μl of TE (10 mm Tris-HCl, 1 mm EDTA) plus 5 μl of a solution containing 60 mm EDTA, 6% SDS, 25% (w/v) glycerol, and 0.2% bromphenol blue. The samples were electrophoresed in 0.7% agarose gels in 0.5× TBE (45 mm Tris-borate, 1 mm EDTA, pH 8.0) buffer and stained with ethidium bromide. The gel images were obtained using a digital CCD camera with Foto/Analyst PC Image software v10.21 (Fotodyne). Where indicated, the intensity of DNA bands was quantitated with TotalLab TL100 software v2006b (Nonlinear Dynamics).
RESULTS
Purification of the RecN Protein
The aim of this study was to characterize the biochemical activities of the bacterial RecN protein. Such characterization has been hampered by the relative insolubility and susceptibility to proteolytic breakdown of the E. coli protein (11, 50).3 Furthermore, biochemical activities have yet to be detailed for the RecN orthologs purified to date from B. subtilis, H. influenzae, A. aeolicus, and B. fragilis (see Introduction). We have identified a RecN homolog from D. radiodurans (Dr RecN) that is soluble in E. coli cells and have purified the full-length protein (564 amino acid residues) to near homogeneity (Fig. 1). The protein identification was confirmed by subjecting the final preparation sample to Edman N-terminal sequence analysis (University of Nevada, Reno INBRE Proteomics Facility). The protein was free of detectable nuclease activities. The experiments reported here were carried out using RecN protein from four different preparations, each yielding similar results. We observed a marked decrease in activity after prolonged enzyme storage at −80 °C; therefore, all of the results were obtained using protein within 3 months of the date of purification.
FIGURE 1.

D. radiodurans RecN protein expressed in E. coli. The samples were electrophoresed in a 13% SDS-polyacrylamide gel. Protein expression and purification protocols are detailed under “Experimental Procedures.” Lane M, molecular mass (kDa) markers. Lane 1, E. coli STL2669 cells transformed with pT7pol26 and pEAW309 Dr RecN expression plasmid. Lane 2, E. coli STL2669 cells transformed with pT7pol26 and pEAW309, induced with isopropyl β-d-1-thiogalactopyranoside and incubated for 6 h at 37 °C. Lane 3, purified Dr RecN protein.
The ATPase Activity of RecN Is Stimulated by Duplex DNA
Sequence analysis of bacterial RecN proteins demonstrates that RecN includes two well known motifs (Walker A and Walker B) involved in ATP binding and hydrolysis as well as significant homology to the SMC family of proteins (20, 51). Therefore, we have begun the biochemical characterization of RecN by exploring its ATP hydrolysis activity. We used an enzyme-coupled, real time assay to assess the ability of the Dr RecN protein to hydrolyze ATP. We measured an apparent turnover rate (rate of ATP hydrolyzed per RecN monomer) of 24 min−1 in the absence of DNA (Fig. 2A) for RecN concentrations above 1 μm. The ATP turnover rate increases ∼4-fold in the presence of 25 μm base pairs of linear duplex DNA, indicating that RecN interacts with the DNA. This DNA-dependent ATP hydrolysis was further dependent on enzyme concentration (Fig. 2A). The turnover rate increased linearly to a maximal rate of 75 min−1 as a function of protein concentration from 0.25 to 3 μm, suggesting that RecN-RecN interactions are required for efficient ATP hydrolysis and that DNA may stabilize a RecN complex. Multimeric RecN complexes have been observed with other RecN orthologs (19, 20). DNA is a cofactor in the RecN catalyzed ATPase reaction because the rate of hydrolysis depends on DNA concentration (Fig. 2B). The DNA-dependent rate, however, appears to be specific to duplex DNA. We observed no stimulation in RecN ATPase activity by linear or circular ssDNA (Fig. 2C) even when the ssDNA was titrated into the reaction up to a concentration of 100 μm nucleotides (data not shown). This does not rule out RecN binding to these ssDNA substrates. It may be that higher order RecN complexes do not form on these substrates. RecN also appears to interact with duplex DNA and does not require an “end” because supercoiled and relaxed plasmid DNA also stimulate the ATP hydrolysis rate (Fig. 2C).
FIGURE 2.
ATP hydrolysis activity catalyzed by the Dr RecN protein. ATPase assays were carried out as described under “Experimental Procedures.” A, the turnover rate of ATP hydrolyzed as a function of RecN protein concentration in the presence (closed circles) or absence (open circles) of 25 μm bp of PstI linearized 8.7-kb plasmid DNA (pEAW324). Each titration point represents the average of nine independent trials. B, the turnover rate of ATP hydrolysis catalyzed by 3 μm RecN protein concentration as a function of Pstl linearized 8.7-kb plasmid DNA concentration. Each titration point represents the average of six independent trials. C, ATP turnover rates relative to the DNA-independent rate (No DNA) catalyzed by 3 μm RecN protein. The relative rate in the presence of ssDNA contained 30 μm nucleotide ϕX174 virion DNA (cssDNA) and represents the average of twelve independent trials or 20 μm nucleotide poly(dT) (lssDNA) and represents an average of nine independent trials. The relative rate in the presence of dsDNA contained 25 μm bp nicked circular (ncdsDNA), supercoiled (cdsDNA), or Pstl linearized (ldsDNA) 8.7-kb plasmid DNA and represents the average of nine, six, and twelve independent trials, respectively.
RecN Stimulates the Intermolecular Ligation of Linear Duplex DNA Molecules
SMC proteins have several distinctive activities, tethering DNA molecules and facilitating the formation of knots and supercoils in particular patterns depending on whether the protein is a condensin or cohesin (24). A DNA tethering activity has also been described for the RAD50/NBS1 complex (52). One activity that is characteristic of cohesins (and not displayed by condensins) is the facilitation of the ligation of linear DNA fragments (31). We tested whether Dr RecN is capable of this cohesin-like activity by incubating various amounts of RecN protein (0–4.8 μm) with 1.2 μm bp of plasmid DNA digested to produce short complementary single-stranded overhangs and adding a DNA ligase. The DNA products of this reaction are purified away from all proteins, precipitated, resuspended in a small volume, and separated by agarose gel electrophoresis (see “Experimental Procedures”). The reaction is carried out under dilute conditions such that the major product of ligation in the absence of RecN protein is intramolecular, forming circles in different topological states (Fig. 3, lane 0). RecN protein greatly stimulates the intermolecular ligation of linear duplex DNAs (2431 bp) to form dimers, trimers, etc. This indicates that RecN can hold DNA molecules in close proximity.
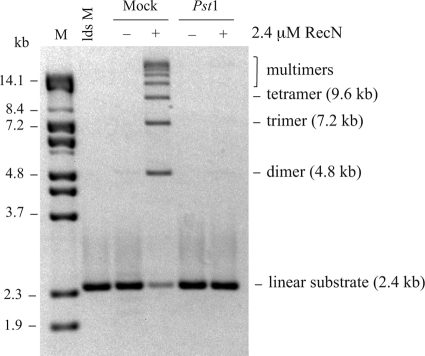
FIGURE 3.
RecN-mediated intermolecular association linear duplex DNA molecules. Ligation reactions were carried out in dilute conditions as described under “Experimental Procedures.” The indicated concentration (in μm) of Dr RecN protein was incubated with 1.2 μm bp of 2.4-kb PstI linearized plasmid DNA (pEAW3) substrate (marker, lds M) and 2.5 mm ATP for 30 min before the addition of 2.5 units of E. coli DNA ligase. The ligated DNA products were purified from the dilute reaction mixture and analyzed in a 0.7% agarose gel. Self-ligated circular plasmid represents various topoisomers of the intramolecular, covalently closed plasmid product. The migration of linear multimeric products relative to the standard (lane M) was used to identify the intermolecular ligation products as dimers (4.8 kb), trimers (7.2 kb), etc.
Because the RecN protein is titrated into the reaction to above 2.4 μm, the generation of large, higher ordered, multimers is inhibited. The reason for this is not clear; however, we have observed RecN protein precipitation when RecN protein is incubated at 37 °C at concentrations above 5 μm (data not shown). Therefore, we cannot rule out the possibility that protein aggregation is occurring (thereby decreasing the effective concentration) at the higher RecN concentration used to produce Fig. 3. Regardless, it is clear that the RecN protein concentrations from 0.6 to 2.4 μm are stimulating intermolecular DNA associations, such that the major ligation products are multimers of the 2.4-kb linear plasmid substrate. Additionally, RecN is likely stiffening the linear substrate molecule such that intramolecular ends are precluded from interacting because the DNA ligase-mediated circularization of linear DNA molecules is inhibited as the RecN concentration increases (Fig. 3).
RecN Tethers DNA Molecules Together
Under the conditions employed for the reaction in Fig. 3, RecN is stimulating the ligase-dependent intermolecular DNA ligation and inhibiting the intramolecular DNA ligations. However, RecN does not appear to increase the number of molecules that are ligated; it just changes the product of ligation from circles to multimers. This may simply be a consequence of RecN binding the length of the DNA and stiffening the molecule such that recircularization is inhibited. To test whether RecN can tether DNA molecules together, we lowered the concentration of DNA ligase 50-fold such that almost no ligation products are observed in the absence of RecN (Fig. 4, lane 0). Adding RecN greatly enhanced the intermolecular ligation of the duplex DNA and the total number of molecules ligated under these conditions (Fig. 4), suggesting that RecN is indeed tethering DNA molecules together.
FIGURE 4.
The RecN-dependent tethering of linear duplex DNA molecules. Ligation reactions were carried out in dilute conditions as described under “Experimental Procedures” and the legend to Fig. 3, except that phosphodiester bond formation was initiated with 0.05 units of E. coli DNA ligase. The ligated DNA products were purified from the dilute reaction mixture and analyzed in a 0.7% agarose gel. Self-ligated circular plasmid represents various topoisomers of the intramolecular, covalently closed plasmid product. The migration of linear multimeric products relative to the standard (M) was used to identify the intermolecular ligation products as dimers (4.8 kb), trimers (7.2 kb), etc.
RecN Is Not Likely Participating Directly in the DNA Ligation Reaction
The DNA phosphodiester bond formation shown in Figs. 3 and 4 was carried out using the DNA ligase from E. coli. We next tested whether the RecN protein stimulates intermolecular DNA associations using DNA ligases from other organisms. The reaction is efficient whether E. coli, T4 or D. radiodurans (LigA) ligases are used to catalyze the phosphodiester bond (Fig. 5, lanes 1–6). We also tested whether this effect was DNA end-specific, that is, whether the 3′ overhang used in Figs. 3 and 4 was necessary for the activity by comparing the ligation of DNA ends having either a 3′ single-stranded overhang (generated by the PstI endonuclease), a 5′ one (generated by the EcoRI endonuclease), or a blunt one (generated by the SspI endonuclease). Whereas the reaction was efficient with substrate DNA ends having either 3′ or 5′ overhang (Fig. 5, lanes 7–10), there was no reaction observed when blunt-ended DNA was used (Fig. 5, lanes 11 and 12). However, this result appears to be due to the fact that the E. coli DNA ligase is deficient at blunt end ligation reactions because the RecN-mediated intermolecular stimulation of blunt end ligation is efficient when the T4 DNA ligase is utilized (Fig. 5, lanes 17 and 18).
FIGURE 5.
Stimulation of intermolecular DNA associations by RecN protein is not dependent on a particular DNA ligase or type of DNA end. Ligation reactions were carried out in dilute conditions as described under “Experimental Procedures” and the legend to Fig. 3, except that T4 DNA ligase (lanes 3–4 and 13–18) or the D. radiodurans LigA DNA ligase (lanes 5 and 6) were used to catalyze phosphodiester bond formation in addition to the E. coli DNA ligase (lanes 1, 2, and 7–12). The 2.4-kb plasmid (pEAW3) was linearized by cleavage with the restriction endonucleases PstI (3′ overhang, lanes 1–8, 13, and 14), EcoRI (5′ overhang, lanes 9, 10, 15, and 16) or SspI (blunt, lanes 11, 12, 17, and 18), generating the DNA ends indicated. 1.2 μm bp of 2.4-kb PstI linearized plasmid DNA (pEAW3) substrate (marker, lds M) was incubated with 2.5 mm ATP in the presence or absence of 2.4 μm Dr RecN protein for 30 min before the addition of 2.5 units of E. coli DNA ligase. The ligated DNA products were purified from the dilute reaction mixture and analyzed in a 0.7% agarose gel. Self-ligated circular plasmid represents various topoisomers of the intramolecular, covalently closed plasmid product. The migration of linear multimeric products relative to the standard (lane M) was used to identify the intermolecular ligation products as dimers (4.8 kb), trimers (7.2 kb), etc.
Together, the reactions of Figs. 4 and 5 indicate that the RecN protein holds different DNA molecules in close proximity, reducing the degrees of freedom of end to end association, without contributing to the phosphodiester bond chemistry. Further, to determine whether the RecN protein was covalently modifying the ends of the linear duplex DNA prior to ligation, we digested the multimeric products of the RecN-facilitated intermolecular ligation reactions with the same endonuclease used to produce the substrate molecules. If the ends were being processed in some manner, we would expect that the product multimers would be insensitive to the digestion. However, as shown in Fig. 6, all of the multimeric products can be digested back the original substrate, indicating that RecN holds DNA in close proximity without providing any DNA end modification.
FIGURE 6.
Duplex DNA ends are not modified by RecN protein. Ligation reactions were carried out in dilute conditions as described under “Experimental Procedures” except that the reaction volume was doubled to 200 μl. 1.2 μm bp of 2.4-kb PstI linearized plasmid DNA (pEAW3) substrate (marker, lds M) was incubated with 2.5 mm ATP in the presence or absence of 2.4 μm Dr RecN protein for 30 min before the addition of 2.5 units of E. coli DNA ligase. Following the ligation step, the E. coli DNA ligase was heat-inactivated (45 min at 65 °C), and each reaction was split into two samples. Both samples were supplemented with PstI restriction endonuclease reaction buffer. The samples were then incubated in the presence (PstI label) or absence (Mock label) of 25 units of PstI endonuclease enzyme for 2 h at 37 °C. The reaction was stopped, and the products were purified as described under “Experimental Procedures.”
RecN Stimulation of Intermolecular DNA Interactions Is Greatly Enhanced by ATP Nucleotide
As shown in Fig. 2, the Dr RecN protein is a modest ATPase, and the rate of ATP hydrolyzed is stimulated by duplex DNA. Each of the DNA ligation experiments above included ATP cofactor (see “Experimental Procedures”), but we wished to determine whether ATP and/or ATP hydrolysis was required for the facilitation of intermolecular ligation of linear duplex molecules. Because the E. coli DNA ligase is dependent on NAD+ cofactor, the ATP can be omitted from the reaction without affecting the DNA ligase (Fig. 7A, compare lanes 1 and 3). In the absence of nucleotide cofactor, RecN (0.9 μm) mediates the intermolecular association of the linear DNA molecules (Fig. 7A, lane 2), but fewer higher order, multimeric DNA products are produced in the absence of nucleotide than when ATP is present (Fig. 7A, lane 4). This lack of large DNA multimers is also observed when a slowly hydrolyzable ATP analog is added to the reaction instead of ATP (Fig. 7A, lane 6). In the presence of ATPγS, many intermolecular ligation products are observed; however, the distribution of ligated products is modified such that the major products are dimers instead of higher order multimers. Furthermore, intramolecular ligation products (self-ligated, circular plasmids) are inhibited by RecN to a greater extent with ATPγS than in the presence or absence of ATP. This may indicate that the RecN protein is binding the DNA with higher affinity in the presence of the ATP analog. To test this possibility, we titrated the RecN protein in the ligation experiments using either no nucleotide, ATP or ATPγS and quantitated the proportion of intermolecular, multimeric linear DNA products (dimers, trimers, etc.) formed as a function of RecN concentration (Fig. 7B). When ATP is provided as a cofactor, RecN concentrations above 1.5 μm are necessary to facilitate the formation of nearly 90% intermolecular products. However, a comparable level of multimeric products are observed with as little as 0.3 μm RecN when ATPγS is supplied to RecN. Much higher concentrations of RecN are required when no nucleotide is added to the reaction. These results show that nucleotide reduces the amount of RecN required to effectively interact with DNA, perhaps by increasing the affinity of RecN for DNA.
FIGURE 7.
Effect of ATP nucleotide on RecN-dependent tethering of linear duplex DNA molecules. Ligation reactions were carried out in dilute conditions as described under “Experimental Procedures” except that 1.2 μm bp of 2.4-kb PstI linearized plasmid DNA (pEAW3) substrate (marker, lds M) was incubated with various amounts of Dr RecN protein (from 0 to 3.6 μm) with either no nucleotide cofactor, 2.5 mm ATP or 2.5 mm ATPγS for 30 min before the addition of 2.5 units of E. coli DNA ligase. The ligated DNA products were purified from the dilute reaction mixture as described under “Experimental Procedures” and analyzed in a 0.7% agarose gel. A, a representative gel illustrating the intermolecular ligation products stimulated by 0.9 μm RecN protein in the absence of nucleotide cofactor (none, lane 2), and the presence of ATP (lane 4) or ATPγS (lane 6). B, graph representing the proportion of multimeric intermolecular ligation products as a function of RecN concentration. Multimeric intermolecular ligation products are defined as the sum of all DNA band intensities in a particular lane that correspond in mobility to dimers, trimers, tetramers, and higher ordered multimers of the linear duplex plasmid that were detectable by the TotalLab gel quantitation software package. This sum was divided by the sum of all band intensities in the same lane (multimer products, substrate, and self-ligated circles).
DISCUSSION
The primary conclusion of this study is that the D. radiodurans RecN protein has cohesin-like function. It is a modest ATPase that bridges different DNA molecules together. Extensive genetic evidence underscores the importance of RecN to bacterial genome maintenance. Here, we provide the first characterization of the effectiveness of RecN associations with duplex DNA. It is also the first demonstration of SMC protein-like function in a bacterial recombinational DNA repair protein.
The purified RecN protein stimulates the intermolecular association of linear duplex DNA (Fig. 3). This activity was observed by utilizing a DNA ligase to catalyze phosphodiester bond formation under conditions that favor circular products in the absence of RecN protein. Intramolecular DNA associations were inhibited by RecN because ligase-dependent circularization was reduced in a RecN concentration-dependent manner. Further, RecN protein stimulates the end-to-end ligation of the linear DNA molecules, resulting in dimers, trimers, and higher order multimeric linear duplex DNA products. This activity is remarkably similar to that observed with eukaryotic cohesin proteins (31). As suggested by Losada and Hirano (31) for the human cohesin protein complex, the lack of intramolecular joining of duplex plasmid may reflect an active stiffening of the DNA molecule upon RecN binding. An alternative although not mutually exclusive explanation is that RecN protein may be increasing the number of DNA ends from different molecules that are in close proximity. This active DNA tethering activity is a likely mechanism because RecN stimulates these intermolecular associations even under ligation conditions where intramolecular recircularization is rarely observed (Fig. 4). Therefore, RecN binding does not simply alter the flexibility of the individual duplex molecule, which would explain the shift of ligation products from circles to multimeric dimers observed in Fig. 3. Rather, the RecN protein likely increases the local concentration of DNA ends because only multimeric linear duplex DNA products are observed when conditions preclude intramolecular DNA end ligation (Fig. 4). It should be emphasized, however, that our experimental design is not meant to suggest a functional interaction between RecN and a DNA ligase during recombinational DNA repair in vivo. In fact, it is unlikely that the D. radiodurans RecN protein is interacting specifically with the ligase because this reaction is efficient whether E. coli, T4 or D. radiodurans (ligA) ligases are used to catalyze the phosphodiester bond formation (Fig. 5, lanes 1–6). Further, the efficiency of the ligation chemistry as a function of the type of overhang present on the DNA substrate is dictated by the ligase and not by the RecN protein (Fig. 5, lanes 7–18). The DNA tethering activity of RecN is observed with DNA ends having either a 3′ or 5′ single-stranded overhang or with blunt ends.
The primary sequence of bacterial RecN proteins includes highly conserved nucleotide-binding motifs. The importance of the Walker A motif, and presumably ATP binding, to the function of RecN is underscored by recent data showing that a single point mutation in the E. coli recN gene (replacing the P-loop lysine to an alanine) results in a strain that is at least as sensitive to DSBs as a recN deletion strain (20). Purified RecN proteins from B. subtilis, H. influenzae, and A. aeolicus reportedly possess weak ATPase activity (19, 20). Therefore, it is not surprising that the D. radiodurans protein indeed hydrolyzes ATP (Fig. 2). Under our optimized conditions, the Dr RecN protein hydrolyzes ATP with a turnover rate of 24 min−1 in the absence of DNA. This modest rate of hydrolysis is stimulated 3–5-fold in the presence of duplex DNA. The fact that the turnover rate of ATP hydrolysis is strongly dependent on protein concentration (Fig. 2A) suggests that RecN-RecN interactions are required for efficient ATP hydrolysis. The B. subtilis RecN ortholog is reportedly an octomer (19), and various multimeric species were observed for purified H. influenzae, A. aeolicus, and B. fragilis RecN orthologs (20). Interestingly, of the various RecN proteins tested, only the ATPase rate of Dr RecN appears stimulated by duplex DNA. Either other RecN orthologs are not stimulated by DNA (20), or the rate of ATP hydrolysis is only stimulated by ssDNA (19). We observed no significant difference in the ATPase rate catalyzed by Dr RecN in the presence of either circular of linear ssDNA compared with the absence of DNA (Fig. 2C). The B. subtilis RecN ortholog binds to ssDNA substrates (19). The fact that the ATP hydrolysis activity of the Dr RecN protein is not stimulated by ssDNA substrates does not rule out the possibility that the protein interacts with ssDNA. Clearly, duplex DNA is a cofactor in the ATP hydrolysis reaction catalyzed by the Dr RecN protein because ATP hydrolysis depends on DNA concentration (Fig. 2B). Moreover, the fact that this titration yields a hyperbolic plot may allow us to use ATP hydrolysis as an indirect measure of RecN affinity for duplex DNA and approximate an apparent dissociation constant of 5–10 μm bp. Direct DNA binding measurements will be needed to confirm this estimation. It is unclear whether the DNA specificity differences observed for the various RecN orthologs reflect substantial differences in DSB repair mechanisms.
ATP nucleotide has a profound effect on the efficiency of intermolecular DNA associations mediated by RecN protein, as measured by the ligation assay (Fig. 7). The presence of ATP or an ATP analog significantly decreased the concentration of RecN required for the DNA ligase-dependent multimerization of linear duplex DNA molecules. It is likely that ATP increases the affinity of RecN for DNA. It is interesting to note that the products of ligation are significantly different in the presence of ATPγS than when ATP is supplied to RecN (Fig. 7A). In the presence of ATP very large DNA multimers (greater than 10 kb) are the major ligation products. This may indicate that enough RecN protein or RecN complexes exist in solution to hold many DNA pieces together simultaneously. This is unlikely because we never observe only low order multimeric ligated DNA products (i.e. dimers and trimers) at low concentrations of RecN in the presence of ATP (Figs. 3 and 4), although the number of complexes in our experiments is difficult to estimate currently because we do not know the plurality of the Dr RecN functional complex. Alternatively, a few DNA molecules may be held together initially; the ends are ligated followed by the disassociation of RecN from DNA and movement of the RecN unit to another DNA molecule. This “off and on” dynamic may be facilitated by the hydrolysis of ATP. This would explain why the major ligation products are dimers when ATPγS is present.
To date, the precise mechanistic role of ATP hydrolysis for the SMC family of proteins is unclear. This is especially true for the SMC proteins and SMC-like proteins with roles in recombinational DNA repair, such as the cohesin complex and Rad50. ATP binding has been linked to conformational changes that effect dimerization and/or DNA binding dynamics (24, 53, 54). The human cohesin complex stimulates the intermolecular association of duplex DNA as we have shown here for RecN protein. However, no dependence of this activity on ATP binding was observed for cohesin complex (31), whereas ATP is stimulatory to the RecN protein (see above). Most SMC proteins, either eukaryotic or prokaryotic, exist as complexes that include non-SMC subunits (22, 51). This can complicate attribution of functional characteristics to the SMC protein itself. For example, the ATPase activity of cohesin (SMC1/3) is thought to be modulated by a non-SMC component, Scc1 (55). We have no evidence that RecN protein functions as part of a heterologous complex in vivo, but we have shown here that the D. radiodurans ortholog is capable of mediating an SMC-like function, DNA tethering, without any other cognate proteins present in the reaction. Thus, the D. radiodurans RecN protein can be utilized in future studies to model the specific capabilities of SMC-type proteins in recombinational DNA repair.
The RecN-mediated intermolecular DNA interactions observed in this study may help explain the need for RecN in the repair of DNA double-stranded breaks. This type of DNA damage may occur when the replication machinery encounters a single-stranded nick in the template DNA strand. If RecN functions to hold together the duplex products of replication (sister chromatids) after the replication fork has passed, the repair of double-stranded breaks will be facilitated by the nearby presence of a homologous DNA molecule. Double-stranded breaks in DNA may also occur when multiple single-stranded nicks occur on both strands of the duplex in close proximity. This is thought to contribute to a large fraction of DSBs when cells are subjected to high doses of ionizing radiation (41). Repair of chromosomes fragmented in this manner may be facilitated by the RecN protein holding the DNA together at the site of the damage such that the DNA ends are not free to separate.
These proposals predict that the RecN protein is expressed at sufficient levels in the cell prior to DNA damage. In E. coli, expression of RecN is highly regulated and is induced as part of the SOS response (8–10). Based on this, it is thought unlikely that RecN has a role in the early steps of DSB repair (12, 20). However, in both B. subtilis and D. radiodurans, RecN expression does not rely on a response to DNA damage, suggesting housekeeping roles for RecN in genome maintenance. The B. subtilis ortholog has been directly shown to be constitutively expressed (17, 21), and D. radiodurans recN transcripts do not change in response to DNA damage (56), even though the gene is required for DNA damage tolerance (18). Although it is still unclear exactly which step in DSB repair RecN facilitates, the cohesin-like function demonstrated here for the Dr RecN protein suggests a structural role for the protein and represents a first step in understanding the precise function of bacterial RecN proteins.
Acknowledgment
We thank Elizabeth Wood for cloning the D. radiodurans recN gene, for providing plasmids, and for her expert technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants SC2 GM083697 and P20 RR016480 (to S. L. L.), R25 GM06122 (to E. D. R. and L. A. U.), and F31 GM090483 (to E. D. R.). This work was also supported by Howard Hughes Medical Institute Undergraduate Science Education Grant 52005881 (to A. S. B.). Research support was also provided by the proteomics facility of the Nevada-INBRE Program, National Institutes of Health Grant P20 RR016464.
S. L. Lusetti, unpublished work.
- DSB
- DNA double-stranded break
- ssDNA
- single-stranded DNA
- dsDNA
- double-stranded DNA
- ATPγS
- adenosine 5′-O-(thiotriphosphate)
- DTT
- dithiothreitol
- AEBSF-HCl
- 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride.
REFERENCES
- 1.Cox M. M., Goodman M. F., Kreuzer K. N., Sherratt D. J., Sandler S. J., Marians K. J. (2000) Nature 404, 37–41 [DOI] [PubMed] [Google Scholar]
- 2.Kuzminov A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 8461–8468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michel B., Flores M. J., Viguera E., Grompone G., Seigneur M., Bidnenko V. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 8181–8188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuzminov A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 8241–8246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sargentini N. J., Smith K. C. (1988) J. Bacteriol. 170, 2392–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lloyd R. G., Picksley S. M., Prescott C. (1983) Mol. Gen. Genet. 190, 162–167 [DOI] [PubMed] [Google Scholar]
- 7.Sargentini N. J., Smith K. C. (1983) Radiat. Res. 93, 461–478 [PubMed] [Google Scholar]
- 8.Picksley S. M., Attfield P. V., Lloyd R. G. (1984) Mol. Gen. Genet. 195, 267–274 [DOI] [PubMed] [Google Scholar]
- 9.Rostas K., Morton S. J., Picksley S. M., Lloyd R. G. (1987) Nucleic Acids Res. 15, 5041–5049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finch P. W., Chambers P., Emmerson P. T. (1985) J. Bacteriol. 164, 653–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neher S. B., Villén J., Oakes E. C., Bakalarski C. E., Sauer R. T., Gygi S. P., Baker T. A. (2006) Mol. Cell 22, 193–204 [DOI] [PubMed] [Google Scholar]
- 12.Meddows T. R., Savory A. P., Grove J. I., Moore T., Lloyd R. G. (2005) Mol. Microbiol. 57, 97–110 [DOI] [PubMed] [Google Scholar]
- 13.Sargentini N. J., Smith K. C. (1986) Radiat. Res. 107, 58–72 [PubMed] [Google Scholar]
- 14.Rocha E. P., Cornet E., Michel B. (2005) PLoS Genet. 1, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang G., Maier R. J. (2008) Infect. Immun. 76, 153–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stohl E. A., Criss A. K., Seifert H. S. (2005) Mol. Microbiol. 58, 520–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kidane D., Sanchez H., Alonso J. C., Graumann P. L. (2004) Mol. Microbiol. 52, 1627–1639 [DOI] [PubMed] [Google Scholar]
- 18.Funayama T., Narumi I., Kikuchi M., Kitayama S., Watanabe H., Yamamoto K. (1999) Mutat. Res. 435, 151–161 [DOI] [PubMed] [Google Scholar]
- 19.Sanchez H., Alonso J. C. (2005) Nucleic Acids Res. 33, 2343–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grove J. I., Wood S. R., Briggs G. S., Oldham N. J., Lloyd R. G. (2009) DNA Rep. (Amst.) 8, 1434–1443 [DOI] [PubMed] [Google Scholar]
- 21.Sanchez H., Kidane D., Castillo Cozar M., Graumann P. L., Alonso J. C. (2006) J. Bacteriol. 188, 353–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirano T. (2005) Philos. Trans. R. Soc. Lond. B Biol. Sci. 360, 507–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ström L., Lindroos H. B., Shirahige K., Sjögren C. (2004) Mol. Cell 16, 1003–1015 [DOI] [PubMed] [Google Scholar]
- 24.Hirano T. (2002) Genes Dev. 16, 399–414 [DOI] [PubMed] [Google Scholar]
- 25.Jessberger R. (2002) Nat. Rev. Mol. Cell Biol. 3, 767–778 [DOI] [PubMed] [Google Scholar]
- 26.Williams R. S., Williams J. S., Tainer J. A. (2007) Biochem. Cell Biol. 85, 509–520 [DOI] [PubMed] [Google Scholar]
- 27.Connelly J. C., de Leau E. S., Leach D. R. (1999) Nucleic Acids Res. 27, 1039–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Connelly J. C., de Leau E. S., Leach D. R. (2003) DNA Rep. (Amst.) 2, 795–807 [DOI] [PubMed] [Google Scholar]
- 29.Kimura K., Rybenkov V. V., Crisona N. J., Hirano T., Cozzarelli N. R. (1999) Cell 98, 239–248 [DOI] [PubMed] [Google Scholar]
- 30.Cortés-Ledesma F., de Piccoli G., Haber J. E., Aragón L., Aguilera A. (2007) Cell Cycle 6, 914–918 [DOI] [PubMed] [Google Scholar]
- 31.Losada A., Hirano T. (2001) Curr. Biol. 11, 268–272 [DOI] [PubMed] [Google Scholar]
- 32.Ampatzidou E., Irmisch A., O'Connell M. J., Murray J. M. (2006) Mol. Cell. Biol. 26, 9387–9401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watrin E., Peters J. M. (2006) Exp. Cell Res. 312, 2687–2693 [DOI] [PubMed] [Google Scholar]
- 34.De Piccoli G., Cortes-Ledesma F., Ira G., Torres-Rosell J., Uhle S., Farmer S., Hwang J. Y., Machin F., Ceschia A., McAleenan A., Cordon-Preciado V., Clemente-Blanco A., Vilella-Mitjana F., Ullal P., Jarmuz A., Leitao B., Bressan D., Dotiwala F., Papusha A., Zhao X., Myung K., Haber J. E., Aguilera A., Aragón L. (2006) Nat. Cell Biol. 8, 1032–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sherratt D. J. (2003) Science 301, 780–785 [DOI] [PubMed] [Google Scholar]
- 36.Hiraga S. (2000) Annu. Rev. Genet. 34, 21–59 [DOI] [PubMed] [Google Scholar]
- 37.Mascarenhas J., Sanchez H., Tadesse S., Kidane D., Krisnamurthy M., Alonso J. C., Graumann P. L. (2006) BMC Mol. Biol. 7, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soppa J., Kobayashi K., Noirot-Gros M. F., Oesterhelt D., Ehrlich S. D., Dervyn E., Ogasawara N., Moriya S. (2002) Mol. Microbiol. 45, 59–71 [DOI] [PubMed] [Google Scholar]
- 39.Hirano M., Anderson D. E., Erickson H. P., Hirano T. (2001) EMBO J. 20, 3238–3250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hirano M., Hirano T. (2004) EMBO J. 23, 2664–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cox M. M., Battista J. R. (2005) Nat. Rev. Microbiol. 3, 882–892 [DOI] [PubMed] [Google Scholar]
- 42.Blasius M., Sommer S., Hübscher U. (2008) Crit. Rev. Biochem. Mol. Biol. 43, 221–238 [DOI] [PubMed] [Google Scholar]
- 43.Garger S. J., Griffith O. M., Grill L. K. (1983) Biochem. Biophys. Res. Commun. 117, 835–842 [DOI] [PubMed] [Google Scholar]
- 44.Pugh B. F., Schutte B. C., Cox M. M. (1989) J. Mol. Biol. 205, 487–492 [DOI] [PubMed] [Google Scholar]
- 45.Robu M. E., Inman R. B., Cox M. M. (2004) J. Biol. Chem. 279, 10973–10981 [DOI] [PubMed] [Google Scholar]
- 46.Shibata T., Cunningham R. P., Radding C. M. (1981) J. Biol. Chem. 256, 7557–7564 [PubMed] [Google Scholar]
- 47.Mertens N., Remaut E., Fiers W. (1995) Biotechnology 13, 175–179 [DOI] [PubMed] [Google Scholar]
- 48.Lusetti S. L., Wood E. A., Fleming C. D., Modica M. J., Korth J., Abbott L., Dwyer D. W., Roca A. I., Inman R. B., Cox M. M. (2003) J. Biol. Chem. 278, 16372–16380 [DOI] [PubMed] [Google Scholar]
- 49.Blasius M., Buob R., Shevelev I. V., Hubscher U. (2007) BMC Mol. Biol. 8, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nagashima K., Kubota Y., Shibata T., Sakaguchi C., Shinagawa H., Hishida T. (2006) J. Biol. Chem. 281, 30941–30946 [DOI] [PubMed] [Google Scholar]
- 51.Graumann P. L., Knust T. (2009) Chromosome Res. 17, 265–275 [DOI] [PubMed] [Google Scholar]
- 52.van der Linden E., Sanchez H., Kinoshita E., Kanaar R., Wyman C. (2009) Nucleic Acids Res. 37, 1580–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hirano T. (2006) Nat. Rev. Mol. Cell Biol. 7, 311–322 [DOI] [PubMed] [Google Scholar]
- 54.Kinoshita E., van der Linden E., Sanchez H., Wyman C. (2009) Chromosome Res. 17, 277–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arumugam P., Nishino T., Haering C. H., Gruber S., Nasmyth K. (2006) Curr. Biol. 16, 1998–2008 [DOI] [PubMed] [Google Scholar]
- 56.Tanaka M., Earl A. M., Howell H. A., Park M. J., Eisen J. A., Peterson S. N., Battista J. R. (2004) Genetics 168, 21–33 [DOI] [PMC free article] [PubMed] [Google Scholar]